Abstract
The family Avsunviroidae comprises four viroid species with the ability to form hammerhead ribozymes that mediate self-cleavage of the multimeric plus and minus strands resulting from replication in the chloroplast through a symmetric rolling-circle mechanism. Research on these RNAs is restricted by their host range, which is limited to the plants wherein they were initially identified and some closely related species. Here we report cleavage and ligation in transplastomic Chlamydomonas reinhardtii expressing plus- and minus-strand dimeric transcripts of representative members of the family Avsunviroidae. Despite the absence of viroid RNA-RNA transcription, the C. reinhardtii-based system can be used to address intriguing questions about viroid RNA processing and, in particular, about the cellular factors involved in cleavage and ligation.
Species of the Avsunviroidae family, Avocado sunblotch viroid (ASBVd) (19), Peach latent mosaic viroid (PLMVd) (18), Chrysanthemum chlorotic mottle viroid (CChMVd) (23), and Eggplant latent viroid (ELVd) (12), comprise a unique class of plant pathogens because ASBVd and PLMVd, and most likely CChMVd and ELVd, replicate in the chloroplast (13). Like nuclear viroids (family Pospiviroidae), they are formed of small, circular RNAs (247 to 401 nucleotides) not encoding any protein but able to autonomously infect their hosts (11, 15, 28). Although ASBVd, PLMVd, CChMVd, and ELVd do not contain the central conserved region characteristic of the family Pospiviroidae, they can form hammerhead ribozymes that mediate self-cleavage of multimeric plus and minus strands resulting from symmetric rolling-circle replication (3, 8). The four members of the family Avsunviroidae have a restricted host range: they infect only the plants wherein they were discovered and related species (12, 13), a feature limiting the study of these intriguing RNAs. Although no viroid infects Arabidopsis thaliana, nuclear transformation of this model plant with dimeric cDNA constructs of members of the family Pospiviroidae has revealed its potential in investigation of viroid-host interactions (7). Here, we report that transplastomic Chlamydomonas reinhardtii expressing dimeric viroid transcripts can be used to explore viroid-host interactions in the family Avsunviroidae.
Transformation of C. reinhardtii chloroplasts with dimeric viroid cDNAs.
Transplastomic C. reinhardtii cell lines expressing different plus-strand viroid transcripts were obtained. These transcripts, which resemble replicative intermediates and are highly infectious when inoculated mechanically into their hosts (5), were from the genera Avsunviroid (ASBVd), Pelamoviroid (CChMVd), and Elaviroid (ELVd) of the family Avsunviroidae and from Citrus exocortis viroid (CEVd) (15) of the family Pospiviroidae. A cell line expressing a dimeric minus-strand ASBVd transcript was also included.
Dimeric viroid cDNAs were inserted between the XbaI and EcoRI sites of the chloroplast transformation vector pCrc+157 (27), replacing the β-glucuronidase open reading frame. The recombinant plasmids, in which the insertions were preceded by the promoter, the 5′ untranslated region, and the beginning of the open reading frame of the rbcL gene (from −70 to +157) and followed by the 3′ untranslated region of the psaB gene from C. reinhardtii (Fig. 1), were particle bombarded (1) into atpB-deficient C. reinhardtii strain CC-373 (ac-uc-2-21). Photoautotrophic transformants were selected, and for each construct, ten lines were grown in high-salt minimal medium at 32°C, with 12/12 h of day/night and bubbling with 2% CO2-enriched air (27). Slot and Southern blot hybridizations of DNA preparations with 32P-labeled viroid-specific riboprobes detected the expected sequences and facilitated choosing one highly homoplasmic line per construct.
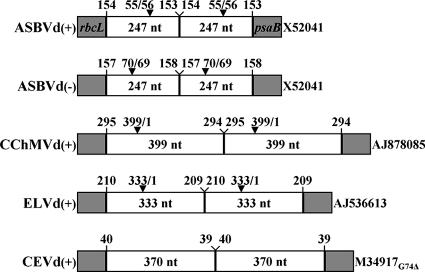
FIG. 1.
Bar diagram representing the viroid transcripts expressed in the chloroplasts of transgenic C. reinhardtii. The viroid RNAs and their polarities [(+), plus-strand; (−), minus-strand] are indicated on the left, and the database accession numbers of the specific sequence variants are indicated on the right (the subscript in the CEVd sequence refers to a deletion with respect to database entry). Gray bars represent the C. reinhardtii rbcL 5′ and psaB 3′ terminal regions (not to scale), and open bars represent viroid monomeric units (to scale). Arrowheads indicate the viroid ribozyme self-cleavage sites. The positions at which the viroid sequences start and end in the transcript, the size of each monomeric-viroid unit, and hammerhead self-cleavage sites are also indicated. nt, nucleotides.
ASBVd, CChMVd, and ELVd dimeric RNAs, but not CEVd dimeric RNA, are processed in C. reinhardtii chloroplasts.
To confirm the expression of viroid transcripts, the selected transplastomic lines were analyzed by Northern blot hybridization. Cells from saturated cultures (50 ml) were sedimented, resuspended in buffer (0.1 M Tris-HCl [pH 9.0], 0.1 M NaCl, 10 mM EDTA, and 0.1 M 2-mercaptoethanol), and extracted with phenol-chloroform and sodium dodecyl sulfate. The aqueous phase was adjusted to 35% ethanol, and the RNAs were fractionated on 0.2 g of nonionic cellulose (CF11; Whatman) (8). The eluted RNAs were separated by polyacrylamide gel electrophoresis (PAGE) in 5% gels in Tris-borate-EDTA buffer containing 8 M urea, blotted onto nylon membranes, and hybridized at 70°C in the presence of 50% formamide with strand-specific 32P-labeled riboprobes (8).
Transcripts from members of the family Avsunviroidae were expressed in transplastomic C. reinhardtii and correctly processed to monomeric linear (ml) and circular (mc) RNAs, as revealed by comigration with the forms from ASBVd-infected avocado (Persea americana Mill.), CChMVd-infected chrysanthemum (Dendranthema grandiflora Tzvelez), and ELVd-infected eggplant (Solanum melongena L.) (Fig. 2A to D). The plus-strand dimeric CEVd transcript was expressed but remained unprocessed, according to the lack of signals comigrating with the mc and ml RNAs from CEVd-infected gynura (Gynura aurantiaca DC.) (Fig. 2E).
FIG. 2.
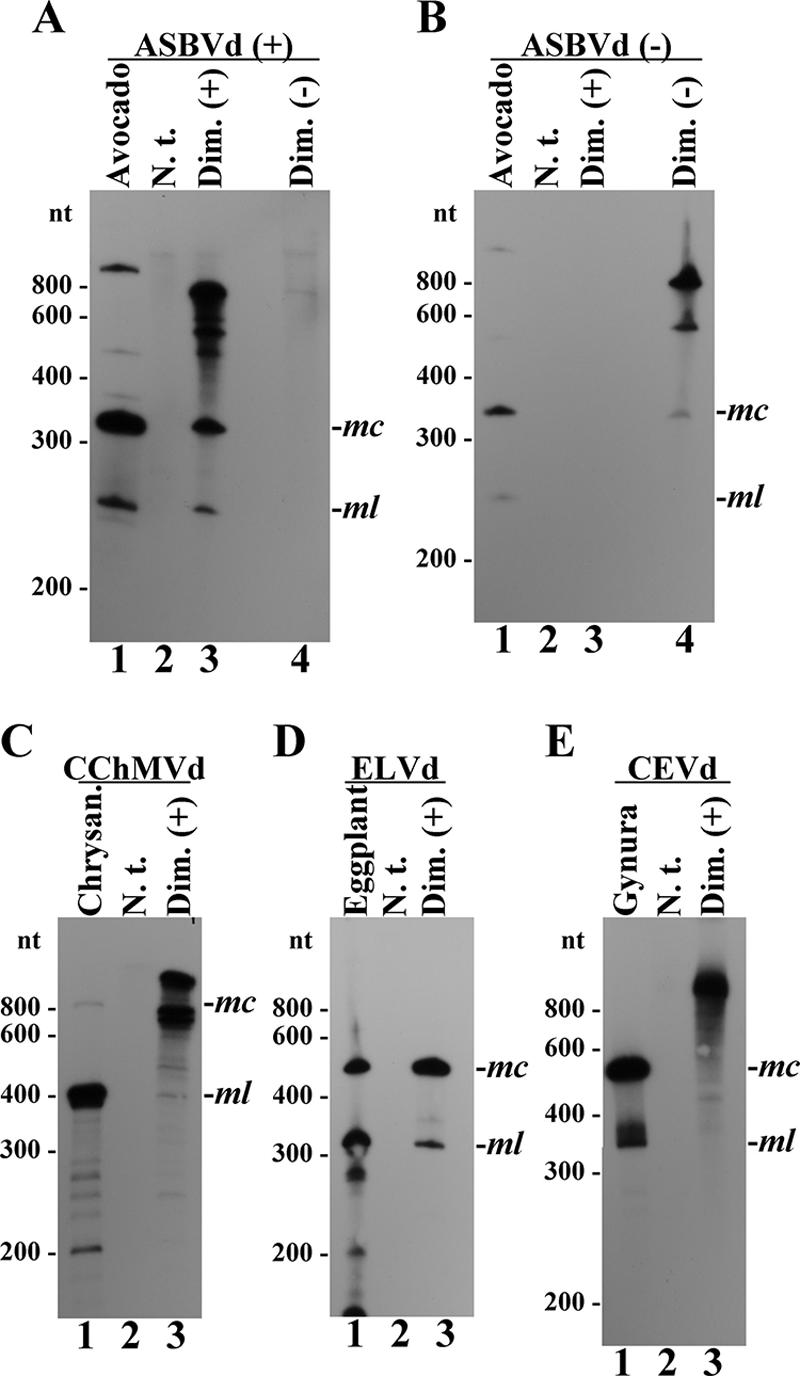
Northern blot hybridization analysis of transplastomic C. reinhardtii expressing dimeric viroid RNAs. Viroid RNA preparations were fractionated by denaturing PAGE and hybridized with riboprobes to detect ASBVd (+) (A), ASBVd (−) (B), CChMVd (+) (C), ELVd (+) (D), and CEVd (+) (E). Lanes 1, controls of ASBVd-infected avocado (A and B), CChMVd-infected chrysanthemum (C), ELVd-infected eggplant (D), and CEVd-infected gynura (E). Lanes 2, controls of nontransformed C. reinhardtii (A to E). Lanes 3, transplastomic C. reinhardtii expressing dimeric ASBVd (+) (A and B), CChMVd (+) (C), ELVd (+) (D), and CEVd (+) (E). Lanes 4, transplastomic C. reinhardtii expressing dimeric ASBVd (−) (A and B). The positions of linear RNA markers, with their sizes in nucleotides, are indicated on the left of each panel, and the positions of the monomeric circular (mc) and linear (ml) forms of the different viroids are indicated on the right of each panel. (+), plus-strand transcript; (−), minus-strand transcript; N. t., nontransformed C. reinhardtii Dim. (+), C. reinhardtii expressing dimeric viroid plus-strand transcripts; Dim. (−), C. reinhardtii expressing dimeric viroid minus-strand transcripts.
To corroborate these results, the RNA preparations from C. reinhardtii cell lines expressing dimeric plus-strand RNAs from ASBVd, CChMVd, and ELVd were separated by double PAGE (23) in nondenaturing 5% gels in Tris-acetate-EDTA, with the segment of this gel containing the mc and ml forms being applied on top of a second denaturing 5% gel (in 0.25× Tris-borate-EDTA and 8 M urea). Under these conditions, the mc viroid RNAs comigrating with their ml counterparts in the first gel are retarded in the second. Northern blot hybridization of the RNAs from the second gel showed that bands attributed to mc and ml viroid RNAs (Fig. 2) comigrated again with the mc and ml forms from their infected plants (Fig. 3). However, because of the higher resolution of double PAGE, the intense bands that migrated below the dimeric plus-strand CChMVd transcript in single-denaturing PAGE (Fig. 2C, lane 3) should be generated by partial self-cleavage products of the dimeric plus-strand CChMVd transcript, because the band corresponding to the mc CChMVd form in double PAGE was visible only after prolonged exposure (Fig. 3B, lane 3). Therefore, transplastomic C. reinhardtii expressing dimeric viroid RNAs replicating in the chloroplasts of their host plants were cleaved and ligated, but the control viroid replicating in the nuclei was not.
FIG. 3.
Northern blot hybridization analysis of transplastomic C. reinhardtii expressing different dimeric viroid RNAs after separation by double PAGE. Viroid RNA preparations were separated by two consecutive PAGE steps, first under nondenaturing conditions, with the segment of this gel containing the mc and ml viroid forms being cut and applied on top of a second denaturing gel. The RNAs from the second gel were blotted and hybridized with riboprobes to detect ASBVd (+) (A), CChMVd (+) (B), and ELVd (+) (C). Lanes 1, controls of ASBVd-infected avocado (A), CChMVd-infected chrysanthemum (B), and ELVd-infected eggplant (C). Lanes 2, controls of nontransformed C. reinhardtii (A to C). Lanes 3, transplastomic C. reinhardtii expressing dimeric ASBVd (+) transcripts (A), CChMVd (+) transcripts (B), and ELVd (+) transcripts (C). The positions of the monomeric circular (mc) and linear (ml) forms of the different viroids are indicated on the left of each panel. The bands with faster mobility in panel C, lane 1, most likely result from degradation products of the ml form generated during purification. (+), plus-strand transcript; (−), minus-strand transcript; N. t., nontransformed C. reinhardtii; Dim. (+), C. reinhardtii expressing dimeric viroid plus-strand transcripts.
Processing efficiency in C. reinhardtii chloroplasts differs among members of the family Avsunviroidae.
Examination of the hybridizations revealed that processing efficiency was dependent on the particular viroid. Transplastomic C. reinhardtii expressing the dimeric plus-strand ASBVd transcript showed a prominent species with the slow mobility expected for the primary transcript, two faster-migrating species corresponding to mc and ml ASBVd RNAs, and others presumably generated by partial self-cleavage of the primary transcript (Fig. 2A, lane 3). Because ml ASBVd RNA results from double self-cleavage of the primary transcript mediated by a double-hammerhead structure (16), self-cleavage at either of the two single sites should generate RNAs with mobilities intermediate between those of the primary transcript and the mc form and smaller fragments (Fig. 1). Circularization of the ml ASBVd RNA yields the mc form that in C. reinhardtii, as in ASBVd-infected avocado, accumulates more than its ml counterpart (Fig. 2A, lanes 1 and 3). A similar situation was observed with the C. reinhardtii cell line expressing the dimeric minus-strand ASBVd transcript, although processing appeared less efficient, with minor amounts of mc and ml minus-strand ASBVd RNAs being detected (Fig. 2B, lane 4), the latter after prolonged exposure (data not shown).
The dimeric plus-strand CChMVd transcript expressed in C. reinhardtii was processed to its ml and mc forms with even lower efficiency. The intense bands observed below that generated by the primary transcript indicated that the latter self-cleaved efficiently at either of its two self-cleavage sites but only to a limited extent at both sites simultaneously, judging from the weak intensity of the band produced by the ml form (Fig. 2C and 3B, lanes 3). This result was unexpected because the plus-strand CChMVd single-hammerhead structure is stable (23). Indeed, when the same construct was transcribed in vitro under control of the T7 promoter, each hammerhead individually and both concurrently self-cleaved efficiently (data not shown). Possible explanations for this dissimilar behavior in vivo and in vitro are outlined below. The CChMVd mc form from C. reinhardtii was visible only after prolonged exposure (data not shown), paralleling the situation of CChMVd-infected chrysanthemum, in which this form accumulates less than its ml counterpart (Fig. 2C and 3B, lanes 1) (23).
In contrast, self-cleavage of dimeric plus-strand ELVd RNA in C. reinhardtii was efficient at both self-cleavage sites: only two prominent bands corresponding to the processing products of the primary transcript, mc and ml ELVd RNAs, were observed (Fig. 2D, lane 3). This efficient self-cleavage could result from either the high catalytic activity of the plus-strand ELVd hammerhead during transcription (12) or from recognition by a cellular factor, particularly an RNA chaperone. The ratio of ELVd mc to ml forms in C. reinhardtii was higher than in ELVd-infected eggplant, indicating efficient circularization (Fig. 2D, compare lanes 1 and 3).
C. reinhardtii chloroplasts expressing dimeric ASBVd plus and minus strands do not support RNA-RNA transcription.
To determine whether C. reinhardtii chloroplasts support viroid RNA-RNA transcription in addition to correct processing, the presence of viroid cRNA in C. reinhardtii expressing dimeric ASBVd plus- and minus-strand transcripts was investigated. Northern blot hybridizations showed no signal when an ASBVd plus-strand-specific probe was used to examine the C. reinhardtii cell line expressing dimeric ASBVd minus-strand transcripts (Fig. 2A, lane 4) and when an ASBVd minus-strand-specific probe was used to examine the cell line expressing dimeric ASBVd plus-strand transcripts (Fig. 2B, lane 3). Faint bands in the upper parts of these lanes are most likely nonspecific. C. reinhardtii chloroplasts are therefore incompetent for initiating and/or elongating viroid RNAs on a cRNA template, implying that viroid replication could not be completed. Although this may be regarded as a limitation, it presents some advantages (see below).
Potential of transplastomic C. reinhardtii for studying in vivo RNA processing in the family Avsunviroidae.
The unicellular green alga C. reinhardtii is a model organism with a sequenced genome and a panoply of tools that include transformation of the nuclear, chloroplast, and mitochondrial genomes (17). Despite progress in dissecting the replication of chloroplastic viroids, many aspects remain poorly understood, particularly the host factors involved. This has prompted us to explore the potential for expressing dimeric viroid RNAs in transformed C. reinhardtii chloroplasts. Viroids replicate in their hosts through an RNA-RNA rolling-circle mechanism (3). In the symmetric pathway followed by members of the family Avsunviroidae (8), mc plus- and minus-strand RNAs serve as templates for synthesis of their complementary oligomeric RNAs, which are processed to the mc RNAs. Therefore, replication comprises three steps: transcription, cleavage, and ligation. Regarding the first step, available data indicate that a nuclear-encoded chloroplast RNA polymerase (NEP) transcribes ASBVd and PLMVd RNAs, starting at defined regions in each polarity strand (10, 24; M. E. Rodio, S. Delgado, A. De Stradis, M. D. Gómez, R. Flores, and F. Di Serio, unpublished data). Expression of dimeric ASBVd plus- or minus-stranded RNAs in C. reinhardtii chloroplasts failed to incite accumulation of their complementary strands, showing that ASBVd, and possibly the other members of its family, are not transcribed. This failure could result from the apparent lack of any NEP-like activity in C. reinhardtii (2).
Cleavage in the family Avusnviroidae is mediated by hammerhead structures embedded in both polarity strands (14, 19, 26), with available evidence indicating that these ribozymes operate in vitro and in vivo (8, 10, 23, 24). However, critical aspects of hammerhead ribozymes are unknown, as illustrated by the recent finding of interactions between peripheral regions of natural hammerheads that increase their self-cleavage at the low-magnesium concentrations existing in vivo (9, 20). Expression of dimeric ASBVd plus- and minus-strand RNAs and dimeric CChMVd plus- and ELVd plus-strand RNAs in C. reinhardtii chloroplasts showed proper self-cleavage to their ml forms. Whereas this is consistent with previous in vitro results (14, 19, 26), the extent of the reaction was dependent on the viroid RNA. Moreover, no relationship could be established between the extent of the reaction and self-cleavage mediated by either single- or double-hammerhead structures, indicating the involvement of auxiliary factors from the alga. Because RNA folding occurs during transcription, some viroid RNAs (i.e., CChMVd) could be trapped in conformations catalytically inactive for self-cleavage or display low affinity for cellular RNA chaperones that presumably assist self-cleavage in vivo. In this context, a protein binding ASBVd in vivo behaves as an RNA chaperone that stimulates in vitro ASBVd self-cleavage (6). Alternatively, some of the self-cleavage products could lack elements of a secondary structure that stabilizes transcripts in C. reinhardtii chloroplasts (27) and be degraded.
Ligation has been proposed to occur autocatalytically in PLMVd, giving rise in vivo to a 2′,5′-phosphodiester bond (4). However, the mc plus-strand RNA isolated from ASBVd-infected tissue does not appear to have such an atypical bond (D. Molina-Serrano, R. Flores, and J. A. Daròs, unpublished data). Therefore, the involvement of a host RNA ligase producing a 3′,5′-phosphodiester bond (21), which might also result from the hammerhead ribozyme catalyzing not only cleavage but also ligation (perhaps assisted by host proteins) (25), must also be considered. Because ligation of viroid ml to mc RNAs occurs in transformed C. reinhardtii chloroplasts and because the ratio between the two forms is similar to that existing in planta, the transplastomic system could serve to identify cellular factors mediating this reaction in vivo. Moreover, since there is no viroid RNA-RNA transcription in transgenic C. reinhardtii, the effects of mutations in the viroid primary transcript specifically affecting cleavage and/or ligation can be evaluated.
In summary, we believe that the experimental system presented here can facilitate the study of RNA motifs and cellular factors involved in viroid replication in a chloroplastic physiological context. Moreover, expressing foreign genes in the chloroplast is an environmentally-friendly alternative to nuclear transformation (22), and RNAs of the family Avsunviroidae, the only ones able to enter and replicate in this organelle, could served as vectors for this purpose.
Acknowledgments
This work was supported by grants AGL2004-06311-C02-01 (to J.-A.D.), BFU2005-06808/BMC (to R.F.) and BMC2003-03209 (to M.S.) from Ministerio de Educación y Ciencia (MEC), and by ACOMP06/141 from Generalitat Valenciana (GV). D.M. and L.S. received predoctoral fellowships from MEC and GV, respectively.
Footnotes
Published ahead of print on 7 February 2007.
REFERENCES
- 1.Blowers, A. D., L. Bogorad, K. B. Shark, and J. C. Sanford. 1989. Studies on Chlamydomonas chloroplast transformation: foreign DNA can be stably maintained in the chromosome. Plant Cell 1:123-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bohne, A. V., V. Irihimovitch, A. Weihe, and D. B. Stern. 2006. Chlamydomonas reinhardtii encodes a single sigma70-like factor which likely functions in chloroplast transcription. Curr. Genet. 49:333-340. [DOI] [PubMed] [Google Scholar]
- 3.Branch, A. D., and H. D. Robertson. 1984. A replication cycle for viroids and other small infectious RNAs. Science 223:450-455. [DOI] [PubMed] [Google Scholar]
- 4.Côte, F., D. Lévesque, and J. P. Perreault. 2001. Natural 2′,5′-phosphodiester bonds found at the ligation sites of peach latent mosaic viroid. J. Virol. 75:19-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cress, D. E., M. C. Kiefer, and R. A. Owens. 1983. Construction of infectious potato spindle tuber viroid cDNA clones. Nucleic Acids Res. 11:6821-6835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Daròs, J. A., and R. Flores. 2002. A chloroplast protein binds a viroid RNA in vivo and facilitates its hammerhead-mediated self-cleavage. EMBO J. 21:749-759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Daròs, J. A., and R. Flores. 2004. Arabidopsis thaliana has the enzymatic machinery for replicating representative viroid species of the family Pospiviroidae. Proc. Natl. Acad. Sci. USA 101:6792-6797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Daròs, J. A., J. F. Marcos, C. Hernández, and R. Flores. 1994. Replication of avocado sunblotch viroid: evidence for a symmetric pathway with two rolling circles and hammerhead ribozyme processing. Proc. Natl. Acad. Sci. USA 91:12813-12817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De la Peña, M., S. Gago, and R. Flores. 2003. Peripheral regions of natural hammerhead ribozymes greatly increase their self-cleavage activity. EMBO J. 22:5561-5570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Delgado, S., A. E. Martínez de Alba, C. Hernández, and R. Flores. 2005. A short double-stranded RNA motif of Peach latent mosaic viroid contains the initiation and the self-cleavage sites of both polarity strands. J. Virol. 79:12934-12943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Diener, T. O. 2003. Discovering viroids—a personal perspective. Nat. Rev. Microbiol. 1:75-80. [DOI] [PubMed] [Google Scholar]
- 12.Fadda, Z., J. A. Daròs, C. Fagoaga, R. Flores, and N. Duran-Vila. 2003. Eggplant latent viroid, the candidate type species for a new genus within the family Avsunviroidae (hammerhead viroids). J. Virol. 77:6528-6532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Flores, R., J. A. Daròs, and C. Hernández. 2000. The Avsunviroidae family: viroids containing hammerhead ribozymes. Adv. Virus Res. 55:271-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Flores, R., C. Hernández, M. De la Peña, A. Vera, and J. A. Daròs. 2001. Hammerhead ribozyme structure and function in plant RNA replication. Methods Enzymol. 341:540-552. [DOI] [PubMed] [Google Scholar]
- 15.Flores, R., C. Hernández, A. E. Martínez de Alba, J. A. Daròs, and F. Di Serio. 2005. Viroids and viroid-host interactions. Annu. Rev. Phytopathol. 43:117-139. [DOI] [PubMed] [Google Scholar]
- 16.Forster, A. C., C. Davies, C. C. Sheldon, A. C. Jeffries, and R. H. Symons. 1988. Self-cleaving viroid and newt RNAs may only be active as dimers. Nature 334:265-267. [DOI] [PubMed] [Google Scholar]
- 17.Harris, E. H. 2001. Chlamydomonas as a model organism. Annu. Rev. Plant Physiol. Plant Mol. Biol. 52:363-406. [DOI] [PubMed] [Google Scholar]
- 18.Hernández, C., and R. Flores. 1992. Plus and minus RNAs of peach latent mosaic viroid self-cleave in vitro via hammerhead structures. Proc. Natl. Acad. Sci. USA 89:3711-3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hutchins, C. J., P. D. Rathjen, A. C. Forster, and R. H. Symons. 1986. Self-cleavage of plus and minus RNA transcripts of avocado sunblotch viroid. Nucleic Acids Res. 14:3627-3640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Khvorova, A., A. Lescoute, E. Westhof, and S. D. Jayasena. 2003. Sequence elements outside the hammerhead ribozyme catalytic core enable intracellular activity. Nat. Struct. Biol. 10:708-712. [DOI] [PubMed] [Google Scholar]
- 21.Kikuchi, Y., K. Tyc, W. Filipowicz, H. L. Sänger, and H. J. Gross. 1982. Circularization of linear viroid RNA via 2′-phosphomonoester,3′,5′-phosphodiester bonds by a novel type of RNA ligase from wheat-germ and Chlamydomonas. Nucleic Acids Res. 10:7521-7529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maliga, P. 2004. Plastid transformation in higher plants. Annu. Rev. Plant Biol. 55:289-313. [DOI] [PubMed] [Google Scholar]
- 23.Navarro, B., and R. Flores. 1997. Chrysanthemum chlorotic mottle viroid: unusual structural properties of a subgroup of self-cleaving viroids with hammerhead ribozymes. Proc. Natl. Acad. Sci. USA 94:11262-11267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Navarro, J. A., and R. Flores. 2000. Characterization of the initiation sites of both polarity strands of a viroid RNA reveals a motif conserved in sequence and structure. EMBO J. 19:2662-2670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nelson, J. A., I. Shepotinovskaya, and O. C. Uhlenbeck. 2005. Hammerheads derived from sTRSV show enhanced cleavage and ligation rate constants. Biochemistry 44:14577-14585. [DOI] [PubMed] [Google Scholar]
- 26.Prody, G. A., J. T. Bakos, J. M. Buzayan, I. R. Schneider, and G. Bruening. 1986. Autolytic processing of dimeric plant-virus satellite RNA. Science 231:1577-1580. [DOI] [PubMed] [Google Scholar]
- 27.Suay, L., M. L. Salvador, E. Abesha, and U. Klein. 2005. Specific roles of 5′ RNA secondary structures in stabilizing transcripts in chloroplasts. Nucleic Acids Res. 33:4754-4761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tabler, M., and M. Tsagris. 2004. Viroids: petite RNA pathogens with distinguished talents. Trends Plant Sci. 9:339-348. [DOI] [PubMed] [Google Scholar]