Abstract
We previously demonstrated the susceptibility of pheasants to infection with influenza A viruses of 15 hemagglutinin (HA) subtypes: 13/23 viruses tested were isolated for ≥14 days, all in the presence of serum-neutralizing antibodies; one virus (H10) was shed for 45 days postinfection. Here we confirmed that 20% of pheasants shed low-pathogenic influenza viruses for prolonged periods. We aimed to determine why the antibody response did not clear the virus in the usual 3 to 10 days, because pheasants serve as a long-term source of influenza viruses in poultry markets. We found evidence of virus replication and histological changes in the large intestine, bursa of Fabricius, and cecal tonsil. The virus isolated 41 days postinfection was antigenically distinct from the parental H10 virus, with corresponding changes in the HA and neuraminidase. Ten amino acid differences were found between the parental H10 and the pheasant H10 virus; four were in potential antigenic sites of the HA molecule. Prolonged shedding of virus by pheasants results from a complex interplay between the diversity of virus variants and the host response. It is often argued that vaccination pressure is a mechanism that contributes to the generation of antigenic-drift variants in poultry. This study provided evidence that drift variants can occur naturally in pheasants after prolonged shedding of virus, thus strengthening our argument for the removal of pheasants from live-bird retail markets.
Live bird markets have been associated with avian influenza viruses since 1924 (22). By the early 1990s, live bird markets in the United States were recognized as the “missing link” in the epidemiology of avian influenza virus (20). Long associated with the emergence of highly pathogenic H5 and H7 influenza viruses, live bird markets are also a source of low-pathogenic avian influenza viruses, and they teem with a mix of poultry species such as chickens, pigeons, ducks, geese, quail, guinea fowl, chukar partridges, and pheasants (14, 16). Much is known about the relationship between influenza viruses and the major poultry species—ducks, geese, chickens, and pigeons—sold in the live markets. Less has been known until recently regarding the replication of influenza A viruses and the minor poultry species, such as chukar partridges and pheasants, which can serve as long-term sources of influenza viruses in live poultry markets.
We previously reported that pheasants supported the replication of influenza viruses of 15 different hemagglutinin (HA) subtypes. Moreover, experimentally inoculated pheasants shed virus for prolonged periods in the presence of high levels of serum-neutralizing antibodies (9). Thirteen of the 23 viruses previously tested were isolated for 14 days; one virus (H10) was shed for 45 days postinfection. In North America, pheasants, peafowl, geese, and chukar partridges account for 15% of poultry sold in live bird markets (16). In addition, between 2002 and 2003, nearly half a million pheasants were imported into Hong Kong to be sold in live bird markets (R. G. Webster, unpublished data). Prolonged shedding of virus, even in a small percentage of the pheasant population, has implications for the market system where these birds are kept for days to weeks, because pheasants can serve as a long-term source of influenza viruses in this setting. Therefore, it is important to understand how influenza virus could be shed from pheasants for prolonged periods.
The length of time that influenza A viruses can be shed depends on the subtype of the virus; the host's species, age, and immune status; and the presence of concurrent infections (6). However, the birds in our previous study produced high levels of serum-neutralizing antibodies to the virus regardless of the length of time the virus was shed (9). Antibodies produced against the HA are usually neutralizing and the primary immune mediator for protection in the host against the disease. In addition, a hemagglutination inhibition (HI) antibody titer of 1:40 is considered to be protective against infection with influenza A viruses, so we hypothesized that the virus must be replicating in an immunologically privileged site.
The fact that we detected prolonged shedding only from cloacal swabs of pheasants may signify virus replication in the lower intestine, kidneys, and/or the bursa of Fabricius. Several reports of the replicative ability of duck viruses in chickens indicated that the viruses preferentially replicate in kidney and digestive tract tissues (3). The nucleoprotein's presence in the kidney identified the kidney as an important site for replication of low-pathogenic avian influenza virus (21). The bursa has also been suggested as the primary site of influenza virus replication in birds. Virus has been isolated at a high rate from the bursae of both turkeys (90%) and ducks (70%) intravenously inoculated with influenza A viruses (3). In addition, high titers of a human-duck recombinant influenza virus were recovered from the bursae of ducks experimentally inoculated with the virus, while lower levels of virus were detected in the feces. The presence of high titers of virus in the bursa suggests that this lymphoid organ, not the intestinal tract, could be the primary site of virus replication (7). In pheasants, influenza virus may have established itself in an immunologically privileged site where immunologic mechanisms may have been less efficient at virus elimination, resulting in prolonged shedding.
Changes in the antigenic and pathogenic properties of influenza viruses are favored when a virus completes multiple replication cycles in newly introduced susceptible hosts. The high mutational rate of the influenza virus genome (105.5 per replication cycle for any single nucleotide) has favored its epidemiological success in nature, because it allows the virus to evolve quickly and escape the host immune response (23). Mutations in the antigenic determinants of the major surface glycoproteins, HA and neuraminidase (NA), cause antigenic drift among influenza viruses. In human influenza viruses, antigenic drift occurs continually at a rate of <1% per year (4). However, in wild bird populations, avian viruses are said to be in evolutionary stasis, because they have mutated infrequently over time (1). Antigenic variation in domestic poultry populations has yet to be described. The development of antigenic drift variants in pheasants could have serious consequences, because birds are the primary hosts of all influenza strains that have been introduced into mammals. In the current study, we aimed to determine (i) how the virus survived for prolonged periods in pheasants in the presence of high levels of serum-neutralizing antibodies and (ii) whether drift variants emerged as a result of the prolonged shedding of virus from pheasants. We isolated viruses from the large intestine, cecal tonsil, and bursa of Fabricius and observed histological changes in these organs. We also found that virus shed from pheasants for prolonged periods was antigenically distinct from the parent strain, and this was confirmed by sequence analysis of the HA glycoprotein.
MATERIALS AND METHODS
Viruses.
The viruses used in this study were sent to the repository of St. Jude Children's Research Hospital from World Health Organization laboratories, particularly the one at Hong Kong University. The A/Duck/Hong Kong/562/79 (H10N9) virus was the same virus stock that was used in our previous study. Stock viruses were grown in 10-day-old embryonated chicken eggs for 48 h at 35°C.
Animals and experimental infections.
Six-week-old Chinese ring-necked pheasants (Phasianus colchicus) (Ideal Poultry, Cameron, TX) were used for this study. Serum samples from each bird were collected and tested before inoculation to ensure that the birds were serologically negative (HI titer, <1:10) for avian influenza virus. To further characterize prolonged shedding of virus from pheasants, we used the virus that was shed for the longest period (45 days) in our previous study (A/Duck/Hong Kong/562/79 [H10N9]) (DK H10). A total of 40 pheasants were used in this study in order to determine the frequency of prolonged shedding from this avian species. Twenty-three pheasants were inoculated with 107 50% egg infectious doses (EID50) of DK H10 in a total volume of 1.0 ml of phosphate-buffered saline (PBS) intraocularly (0.2 ml), intranasally (0.4 ml), and intratracheally (0.4 ml). One hour after inoculation, two or three uninoculated pheasants were placed in the cage with the inoculated pheasants for a total of 17 contact pheasants. The birds were observed daily, and tracheal and cloacal swabs were obtained on days 3, 5, 7, and 10 postinoculation (p.i.) to test for the presence of virus. Swabs were also obtained at additional time points, every other day beginning on day 12 p.i., to determine the length of time the birds shed virus. To confirm the finality of virus shedding, pheasants were swabbed until the samples were negative three consecutive times. Three 10-day-old embryonated chicken eggs were inoculated with 0.1 ml of the sample medium for each swab collected and then incubated for 48 h at 35°C, after which time allantoic fluid from each egg was evaluated to detect influenza virus infection by the hemagglutination test using 0.5% chicken red blood cells (CRBCs). If at least one of the three eggs was positive by the hemagglutination test, the bird from which the sample was collected was considered positive for influenza virus infection. Serial 10-fold dilutions were made from 10−1 to 10−5 of the cloacal samples, and the infectivity of each sample was determined by calculating the EID50 by the Reed and Muench method (18). Virus titers of <101 were recorded as zero. All studies were performed in an animal facility approved for use by the United States Department of Agriculture and certified by the Association for the Assessment and Accreditation of Laboratory Animal Care.
Determination of the site of viral replication.
To determine the site of viral persistence in the pheasant, 40 pheasants were inoculated as described above with the same dose of DK H10 (107 EID50/ml) intranasally, intraocularly, and intratracheally. Every other day, beginning on day 5 p.i., one pheasant was sacrificed and underwent necropsy; however, one pheasant was sacrificed before the other birds were inoculated. It was used as a control. Tracheal and cloacal swabs were collected from each bird before euthanization. Tissue samples from the following organs were collected in 10% buffered formalin for histological examination: lungs, spleen, bursa, small intestine, pancreas, reproductive tissue (ovary or testis), large intestine, kidney, and cecal tonsil.
Formalin-fixed tissues were embedded in paraffin wax, sectioned (4 to 5 μm thick), and placed on glass slides. Tissues were examined histologically by using routine hematoxylin and eosin staining to detect lesions consistent with viral infection. A small section of each tissue listed was also collected in 1.5-ml tubes and homogenized in PBS containing antibiotics. The tissue homogenates were used for virus isolation and PCR analysis. Tissue homogenates were stored at −70°C. After being thawed, tissue samples were centrifuged for 5 min at 10,000 rpm. Tissue homogenates were titrated 10−1 to 10−5 in PBS containing antibiotics, and three 10-day-old embryonated chicken eggs were inoculated with 0.1 ml of each dilution of homogenate for each organ harvested. After incubation at 35°C for 48 h, allantoic fluid from each egg was tested for hemagglutination using 0.5% CRBCs. Virus titers in organs were calculated by the method of Reed and Muench and expressed as EID50 (log10/ml) (18). RNA was also extracted from tissue homogenates by using the RNeasy kit (QIAGEN, Inc., Valencia, CA). One-step reverse transcriptase PCR (RT-PCR) was used to amplify the M gene, because it is the most abundant virus gene segment. PCR products were visualized on a 1% agarose gel containing ethidium bromide.
Production of ferret antisera.
We obtained four adult male ferrets that were seronegative (HI titer, <1:10) for H10 influenza A virus and the currently circulating human H1N1 influenza A virus from the ferret-breeding program at St. Jude Children's Research Hospital (Memphis, TN). The two ferrets used for production of antisera to the pheasant H10 41-day isolate (Ph H10) had low HI antibody titers (1:20 and 1:40) against A/NY/55/04 (H3N2) before infection with the H10 isolate. Each of two ferrets used for production of antisera were inoculated intranasally with either 106 EID50 of DK H10 in 1.0 ml of PBS or 1.0 ml of a 1:10 dilution of the Ph H10 41-day isolate. Fourteen days after inoculation, we collected 1.0 ml of blood and tested the serum in an HI assay against the homologous virus to determine the levels of serum antibodies. Ferrets initially infected with the DK H10 virus received a booster dose (0.5 ml of a 1:10 dilution of the virus) in the footpad 23 days p.i. (dpi). Seven days postboost, 1.0 ml of blood was collected from the ferrets, and the serum was tested in an HI assay to determine serum antibody levels. Ferrets were bled by cardiac puncture either 12 days postboost (DK H10) or 21 dpi (Ph H10).
Serological analysis by HI and virus neutralization assays.
Postinfection ferret sera were treated with receptor-destroying enzyme and tested in an HI assay with 0.5% CRBCs as previously described (19). Viruses were diluted to contain 4 agglutinating units in sterile PBS.
Virus neutralization (VN) assays were performed in Madin-Darby canine kidney (MDCK) cells plated in 96-well tissue culture plates, as previously described (10). The 50% tissue culture infectious dose (TCID50) was determined for each virus. Briefly, 10-fold serial dilutions of the virus were made from 10−1 to 10−10 in 1× minimal essential medium (Invitrogen, CA) with 4% bovine serum albumin and TPCK (l-[tosylamido-2-phenyl] ethyl chloromethyl ketone)-treated trypsin (Worthington Biochemical Corporation, Lakewood, NJ) (1 μg/ml). Dilutions of the virus were added to the MDCK cells (4 wells for each dilution; 200 μl/well), and the cells were incubated for 48 h at 35°C. The contents of each well were tested for hemagglutination by incubating 50 μl of tissue culture supernatant with 50 μl of 0.5% CRBCs for 30 min. The TCID50 was calculated by the Reed and Muench method (18).
Postinfection ferret serum samples were treated with receptor-destroying enzyme and heat inactivated as mentioned above. Twofold serial dilutions of serum samples, beginning at 1:40, were made in 1× minimal essential medium containing 4% bovine serum albumin and TPCK-treated trypsin (Worthington Biochemical Corporation) (1 μg/ml). The homologous virus was standardized to contain 200 TCID50s, an equal volume of virus was added to each serum dilution, and the mixture was incubated for 60 min at 35°C. Confluent monolayers of MDCK cells plated in 96-well plates were washed three times with PBS, and 200 μl of the virus-antiserum mixture was added to each well (4 wells per mixture). After incubation at 35°C for 72 h, 50 μl of supernatant from each well was tested for HA activity using 0.5% CRBCs. Geometric mean titers were calculated for each serum sample.
Sequencing and analysis of sequence data.
Except where otherwise noted, sequencing materials were obtained using kits from QIAGEN, Inc. RNA was extracted from 200 μl of allantoic fluid using the RNeasy kit. The eight virus gene segments were amplified using the One-Step RT-PCR kit and influenza universal primers (8). PCR products were purified using the QIAquick PCR purification kit. When amplicons contained secondary bands, they were gel purified using the QIAquick gel extraction kit in accordance with the manufacturer's recommended protocol. The Hartwell Center for Bioinformatics and Biotechnology at St. Jude Children's Research Hospital determined the sequences using rhodamine or dRhodamine dye terminator cycle-sequencing ready reaction kits with the enzyme AmpliTaq DNA polymerase FS (Perkin-Elmer Applied Biosystems, Inc., Foster City, CA) and synthetic nucleotides.
Samples were analyzed on Applied Biosystems 3700 DNA analyzers. DNA sequences were analyzed using the Lasergene sequence analysis software package (DNASTAR, Madison, WI).
RESULTS
What is the frequency of prolonged shedding?
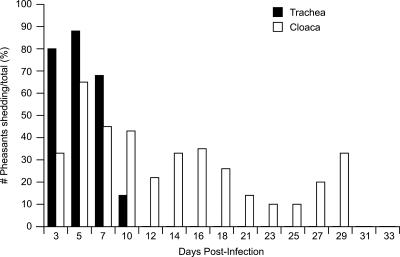
Previously we showed that low-pathogenic influenza viruses were detectable from cloacal swabs of experimentally inoculated pheasants for prolonged periods with no apparent clinical disease signs (9). Because we had such a small sample size in our first study and only one of five pheasants shed A/Duck/Hong Kong/562/79 (H10N9) for 45 dpi, we used a larger group of pheasants (n = 40) this time to determine the frequency of prolonged shedding in this species. None of the inoculated or contact birds showed clinical disease signs during the 14-day observation period. Virus was detected in tracheal and cloacal swabs of both contact and inoculated pheasants for 10 dpi, after which virus was detected solely from cloacal swabs (Fig. 1). By day 3 p.i., virus was detected in samples from 80% of the pheasants (all 23 inoculated pheasants and 7/17 contact pheasants). On day 5 p.i., 87.5% of the pheasants (12/17 contact birds and 100% of inoculated birds) were shedding virus. The percentage of pheasants shedding virus dropped to its lowest (9.5%) on day 23 p.i. Virus was detected in samples from 20% of the pheasants until day 27 p.i. (Fig. 1). All pheasants were negative for virus 31 dpi, and the finality of virus shedding was confirmed on day 33 p.i.
FIG. 1.
Percentage of pheasants shedding A/Duck/Hong Kong/562/79 (H10N9). The percentage of positive pheasants was the number of pheasants shedding virus out of the total number of birds in the study (n = 40). Virus shedding was examined until each pheasant tested negative for three consecutive samplings. One pheasant (pheasant 27) that was still shedding virus was found dead 27 dpi.
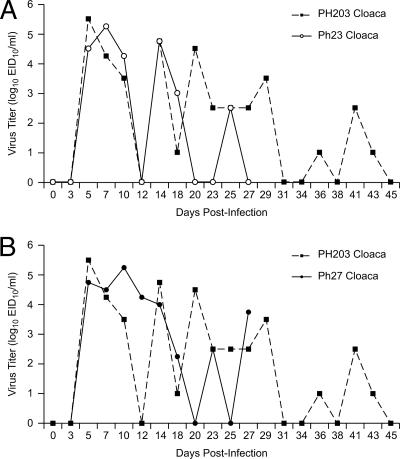
We determined virus titers from cloacal swabs taken at various times from two pheasants (pheasants 27 and 23) that shed virus for the longest time (27 and 29 dpi) and compared them to virus titers from cloacal swabs of the contact (pheasant 203) that shed virus for 45 dpi during our previous study. As shown in Fig. 2A and B, the erratic pattern of shedding mimicked that of pheasant 203, peaking on days 5 (pheasant 27; 104.75 EID50/ml) and 7 (pheasant 23; 105.25 EID50/ml) p.i. Titers dropped to undetectable levels by day 20 p.i. Virus titers peaked again on days 23 (pheasant 27) and 25 (pheasant 23) p.i. (102.5 EID50/ml). Virus titers dropped to undetectable levels again on days 25 (pheasant 27) and 27 (pheasant 23) p.i. The pheasants were swabbed until the virus was no longer detectable on three consecutive occasions. Its cage mates killed pheasant 27, which was still shedding virus on day 27; consequently, we could not determine how long the bird might have continued to shed virus had it lived.
FIG. 2.
Comparison of virus titers from cloacal swabs of pheasants (previous and current study) infected with A/Duck/Hong Kong/562/79 (H10N9). All virus titers of <101 EID50 were graphed as zero. (A) Comparison of virus titers for pheasant 203 and pheasant 23. (B) Comparison of virus titers for pheasant 203 and pheasant 27.
What is the site of influenza A virus replication in pheasants? (i) Virus isolation.
The environment in which the virus replicates will determine whether it is subjected to immune selection pressure. To determine the site of virus replication in the pheasant, 40 pheasants were inoculated with the same dose of DK H10 (107 EID50/ml) intranasally, intraocularly, and intratracheally. Necropsy was performed on six randomly selected pheasants between 5 dpi and 35 dpi (Table 1). Virus was isolated from tracheal and/or cloacal swabs collected from three of the six birds before euthanization. Virus was isolated from the large intestine (103.75 EID50/ml) and cecal tonsil (104.5 EID50/ml) of one bird by using the standard egg inoculation procedure; the sample was obtained from pheasant 50, which was euthanized 5 dpi. Virus was not detected in the lungs, kidney, spleen, bursa, pancreas, small intestine, or ovary of this bird. No virus was isolated from any of the organ homogenates from the other five pheasants that were sacrificed.
TABLE 1.
Virus isolation, RT-PCR, and histopathology results after inoculation of pheasants with A/Duck/Hong Kong/562/79 (H10N9)
| Pheasant no. | dpi | Resulta for the following tissue:
|
||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Swabs
|
Trachea/lung
|
Kidney
|
Spleen
|
Bursa
|
Pancreas
|
Small intestine
|
Large intestine
|
Cecal tonsil
|
Reproductive
|
|||||||||||||||||||||
| Trachea | Cloaca | VI | RT-PCR | HP | VI | RT-PCR | HP | VI | RT-PCR | HP | VI | RT-PCR | HP | VI | RT-PCR | HP | VI | RT-PCR | HP | VI | RT-PCR | HP | VI | RT-PCR | HP | VI | RT-PCR | HP | ||
| 50 | 5 | + | + | − | + | + | − | − | − | − | − | − | − | + | − | − | − | − | − | − | − | + | + | + | + | + | NS | − | + | − |
| 2 | 14 | + | − | − | − | − | − | − | − | − | − | − | − | − | + | − | − | − | − | − | − | − | − | − | − | − | NS | − | − | − |
| 9 | 14 | − | − | − | − | − | − | − | − | − | − | − | − | − | + | − | − | − | − | − | − | − | − | + | − | − | NS | − | − | NS |
| 21 | 18 | − | − | − | − | − | − | − | NS | − | − | − | − | − | + | − | − | − | − | − | − | − | − | + | − | − | NS | − | − | − |
| 27b | 27 | − | + | − | − | + | − | − | − | − | − | NT | − | − | NT | − | − | − | − | − | NT | − | − | NT | − | − | NT | − | − | − |
| 32 | 35 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | + | − | − | NS | − | − | − |
‖+, positive for virus; −, negative for virus. VI, virus isolation; HP, histopathology; NS, no sample available; NT, not tested (autolysis of tissue precluded a detailed histological examination).
Pheasant 27 was found dead 27 dpi.
(ii) RT-PCR.
Because virus titers may have been below the level of detection by culture in embryonated chicken eggs, RT-PCR was performed on RNA extracted from organ homogenates. The M gene was chosen for amplification, because it is the most abundant virus gene segment. The M gene segment was amplified from samples of the large intestine, cecal tonsil, ovary, lung, and bursa of pheasant 50, which was sacrificed 5 dpi (Table 1). RT-PCR did not yield M gene amplicons from organs harvested from any of the other pheasants sacrificed in this study.
(iii) Gross lesions.
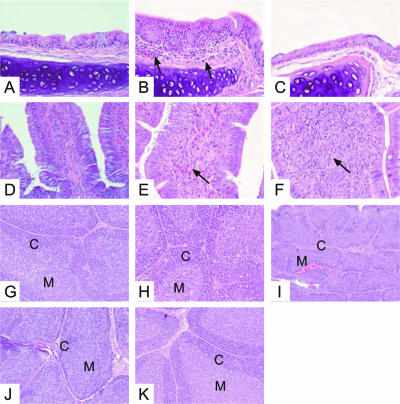
Routine hematoxylin and eosin staining was used to histologically examine the tissues for lesions consistent with viral infection. Gross lesions were not apparent in any tissues of the pheasants on which necropsy was performed. Autolysis precluded histologic examination of samples of the spleen, small intestine, large intestine, cecal tonsil, and bursa of pheasant 27, found dead 27 dpi. Virus-induced microscopic lesions, observed in the trachea, large intestine, and bursa, differed depending on the duration of infection. At day 5 p.i., infiltrates of lymphocytes, plasma cells, heterophils, and macrophages were observed in the submucosa of the trachea (Fig. 3B). The tracheal mucosa remained intact but was mildly hyperplastic. In the large intestine, we observed infiltrates of heterophils. Fewer lymphocytes, plasma cells, and macrophages were observed in the lamina propria, submucosa, and muscularis mucosae. Foci extended into the serosal surface and mesenteric fat (Fig. 3E). Tracheitis resolved as the time postinfection progressed (Fig. 3C), and the infiltrate in the large intestine evolved from a primarily heterophilic to a lymphoplasmacytic and histiocytic population that persisted through the latest necropsy time point, 35 dpi (Fig. 3F). At day 5 p.i., the architecture of the bursa was normal; however, increased numbers of degenerate cells consistent with apoptosis were observed in the cortex and medulla (Fig. 3H). Changes in the bursa, observed beginning on day 14 p.i. (Fig. 3I and J), consisted of depletion of small lymphocytes in the cortex, characterized by a thin, often discontinuous zone of cortical lymphocytes, resulting in collapse of the parenchyma and a decrease in the overall size of the bursa. By day 35 p.i., recovery of lymphoid tissue of the bursa was observed, and its histologic appearance was consistent with a normal resting state (Fig. 3K). Cecal tonsil samples were not available for histologic examination.
FIG. 3.
Histopathology of organs from pheasants. (A through C) Hematoxylin and eosin-stained tracheae (magnification, ×400). (A) Uninfected control. (B) At day 5 p.i., infiltrates of lymphocytes, plasma cells, macrophages, and a few heterophils can be observed in the submucosa (arrows), along with mild hyperplasia (arrowhead). (C) This inflammatory response gradually diminishes during the course of the infection and is completely resolved by day 35 p.i. (D through F) Hematoxylin and eosin-stained large intestines (magnification, ×400). (D) Uninfected control. (E) At day 5 p.i., the infiltrate in the large intestine is composed primarily of heterophils and involves the lamina propria and submucosa (arrow). (F) The inflammation in the large intestine persists at day 35 p.i. However, during the course of infection, the composition of the inflammatory response has changed to lymphocytes, plasma cells, and macrophages (arrow). (G through K) Hematoxylin and eosin-stained bursae (magnification, ×200). (G) Uninfected control. (H) Early in infection (day 5 p.i.), increased cell death is observed in the cortex (C) and medulla (M). (I and J) At days 14 and 18 p.i., respectively, lymphoid depletion is observed with collapse of the parenchyma. (K) By day 35 p.i., there is recovery of the lymphoid tissue.
Does long-term shedding of influenza virus from pheasants result in antigenic drift? (i) Antigenic analysis of virus isolated from pheasants at different time points.
Experimentally inoculated pheasants shed the DK H10 influenza virus in the presence of high titers of serum-neutralizing antibody (mean neutralizing antibody titer, 1:640). In our first study, sera collected 14 dpi from pheasants infected with the DK H10 virus reacted well with the parental H10 virus (1:40 to 1:80 with CRBCs and 1:160 to 1:640 with horse red blood cells) (9). Antisera produced in ferrets against the DK H10 and Ph H10 41-day viruses were used in an HI assay with the DK H10 parental strain and virus isolates from the pheasants with peak virus titers (14, 20, and 41 dpi) to determine whether the viruses isolated from pheasants at different times postinfection differed from the H10 parental strain. The Ph H10 41-day isolate seemed to be more immunogenic in ferrets than was the DK H10 parental strain; less of the Ph H10 41-day isolate was required to produce a high level of serum antibodies in ferrets (1:160 to 1:320). In addition, because the postinfection serum has a titer of 1:20, ferrets inoculated with the DK H10 virus had to receive a virus boost to produce the same level of serum antibodies as that produced by the Ph H10 41-day isolate.
The DK H10 and Ph H10 41-day isolate could not be differentiated from each other by using the sera produced against the H10 parental strain; however; a fourfold difference was observed between the Ph H10 20-day and 41-day isolates (Table 2). A marked difference in HI antibody titers was observed between the two viruses (DK H10 and Ph H10 41-day) when tested against the sera produced against the pheasant isolate. The DK H10 virus did not react (titer, <1:10) with the postinfection ferret sera produced against Ph H10 41-day isolates, which produced titers of 1:160 to 1:320 against this serum.
TABLE 2.
HI results for virus shed from pheasants for 45 dpi
| Virusa | HI titer for the following antiserum (1:10):
|
No. of amino acid differences in the HA | |
|---|---|---|---|
| F. 1115 (αDK/HK/562/79 [H10N9]) | F. 1209 (αPh203C 41D [H10N9]) | ||
| DK H10 parental | 160 | <10 | 0 |
| Ph203C 14d | 160 | 40 | 5 |
| Ph203C 20d | 80 | <10 | 7 |
| Ph203C 41d | 320 | 160-320 | 10 |
14d, 20d, and 41d, virus shed at days 14, 20, and 41 postinfection, respectively.
To verify the HI results, the DK H10 virus and the Ph H10 41-day isolate were tested in a VN assay against the same antiserum that was used in the HI assay. It is noteworthy that the Ph H10 41-day isolate grew poorly in MDCK cells. The TCID50 for this virus was 4 log units lower (103.4/ml) than that of the parental DK H10 isolate (107.2/ml). When the two viruses were tested against the ferret antiserum produced against the H10 parental strain, the VN titer of the DK H10 strain (1:4,305) was 2.4-fold higher than that of the Ph H10 41-day isolate (1:1,810). The DK H10 parental strain was not neutralized (titer, <1:80) with the antiserum produced against the Ph H10 41-day isolate, while the Ph H10 isolate had a VN titer of 1:190 against this antiserum. These results, along with the HI data, showed that the H10 virus isolated from the pheasant 41 dpi differed antigenically from the H10 parental virus, suggesting that antigenic variants were selected throughout the course of virus replication in this pheasant.
(ii) Sequence analysis of virus isolated from pheasants at different times.
Sequence analysis of viruses provides additional evidence of antigenic drift. To determine whether the sequences of gene segments in the original DK H10 virus differed from those of the Ph H10 41-day isolate, we sequenced and compared all eight gene segments of each virus. We did the same with gene segments from virus isolated from pheasants at various times during shedding, as well as the eight gene segments of a virus shed for a short term (A/Duck/Hong Kong/Y280/97 [H9N2]) and a virus isolated 7 dpi from a pheasant inoculated with the H9 virus. We observed no amino acid differences between the original H9 virus and the H9 virus isolated from the pheasant 7 dpi (data not shown). Short-term shedding of virus did not result in selection of genetically distinguishable variants.
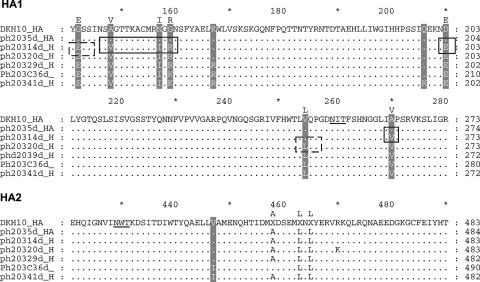
We observed 12 nucleotide differences between the HA sequence of the original DK H10 stock and the Ph H10 41-day isolate. Ten were nonsynonymous (sense); the remaining two were synonymous (silent). The nonsynonymous nucleotide changes produced 10 amino acid changes. Because the 3-dimensional structure of the H10 HA has not been determined, the location of amino acid changes between the DK H10 and Ph H10 41-day isolate were modeled on the A/HK/1/68 (H3N2) 3-dimensional structure. In the DK H10 virus, 9 of the 10 differences map to the globular head of the HA, and 1 occurs in the HA2 (V447I) based on alignment with H3 sequence data. Four of the amino acid changes map to antigenic sites of the HA molecule: N157I, G159R (K), Q206K, and D210E. The first two reside in the most obvious antibody-binding site, which seems to be the most immunogenic site of the HA. All four are close to the sialic acid binding site. A fifth (A148V) occurs near the receptor-binding site. None alter glycosylation sites on the HA molecule. In addition, the 10 amino acid changes occur successively during shedding from the pheasant. Five occur by day 14 p.i., two more by day 20 p.i., one more at day 29, and another at day 36 p.i. All 10 are present in the 41-day isolate (Fig. 4).
FIG. 4.
Alignment of HA amino acid sequences of pheasant H10 isolates with the corresponding sequences of the DK H10 virus. The top consensus sequence is that of the A/Duck/Hong Kong/562/79 (H10N9) parental virus. Successive sequences are the HA sequences of viruses isolated from the pheasant at various times postinfection (5 days [5d], 14d, 20d, 29d, 36d, and 41d). Underlined residues are potential glycosylation sites. Amino acid changes are highlighted. Boxes with solid lines indicate changes that occur at 14 dpi; boxes with dashed lines indicate changes that occur at 20 dpi.
The HA of the virus isolated from pheasant 23 (Ph23 H10) was also sequenced to determine whether the same amino acid changes occurred in the HA of the H10 virus shed on day 27 p.i. in our current study. Of the six nucleotide differences between the DK H10 virus and the Ph23 H10 isolate, five resulted in four amino acid differences: S143P, N157I, G159R (K), and G175E. Like the differences observed in the Ph H10 41-day isolate, these amino acid differences occurred successively; three were present by 16 dpi and the fourth appeared by 25 dpi, although the 16-dpi difference at position 159 changed again at 25 dpi (data not shown). Of the four amino acid differences found between the original DK H10 isolate and the Ph23 H10 isolate, two (N157I and G159R [K]) were similar to those found in the Ph H10 41-day isolate, and the other three were unique to the virus isolated from pheasant 23.
Differences were also found in the other gene segments of the Ph H10 41-day isolate, with the exception of the PB2 gene and M2. Three concurrent changes occurred in the NA (H157Y, E259K, and T375I) based on comparison with N2 sequence data, none within sites of potential interaction with antibodies (2). One of the amino acid changes (H157Y) was located near the catalytic site of the NA; none altered presumptive glycosylation sites. There was one amino acid change in PB1 (P50S), and there were two amino acid changes each in PA (E126G and G631S), NP (M105V and S377N), and M1 (H110Y and A155T). Three amino acid changes occurred in the NS2 protein (F30S, S51A, and Q118K). The amino acid changes in the internal genes may provide an additional growth advantage to this virus.
DISCUSSION
We previously reported prolonged shedding of low-pathogenic influenza A viruses from experimentally inoculated pheasants in the presence of high levels of neutralizing serum antibodies (9). Other avian species rarely shed influenza viruses more than 10 dpi; here we showed that 20% of pheasants shed low-pathogenic influenza A viruses for prolonged periods. We detected virus in the large intestine, the cecal tonsil, and the bursa of Fabricius, the primary lymphoid organ for B-cell development and differentiation. We also characterized the biological properties of the influenza viruses isolated from pheasants after prolonged shedding. The virus isolated from pheasants was antigenically distinct from the parental virus used as the inoculum, and corresponding amino acid changes were observed in the virus surface glycoproteins HA and NA. Several changes in the HA mapped to potential antigenic sites. As a result, prolonged shedding of influenza A virus from pheasants produced antigenic drift variants.
The survival of viruses during their interaction with a host is challenged by diverse cellular environments in different tissues, as well as by host defense mechanisms. Originally we suspected that influenza viruses were able to replicate in pheasants for prolonged periods because the virus was replicating in an immunologically privileged site and thus the virus was hidden from attack by the immune response. However, in this study, we found evidence that contradicted our original hypothesis. Although we observed no conclusive evidence for the site of prolonged virus replication in pheasants, we were able to narrow down potential sites. During the early stages of infection of pheasants, we observed some evidence for replication of influenza virus in the large intestine, cecal tonsil, and bursa of Fabricius; histologic changes were observed later. The fact that data from each time point were from a different pheasant may help to explain why we did not isolate virus at each time point: some pheasants were no longer shedding when they were randomly selected for harvesting of organs. The cecal tonsil of birds, a patch of tissue proximal to the cecum, is similar in function to the mammalian palatine tonsil. Made up of both T and B cells, the cecal tonsil produces antibodies, functions as a secondary lymphoid organ, and has a sentinel role. In poultry, the bursa is the primary lymphoid organ responsible for the creation and expansion of the antibody repertoire. As a peripheral lymphoid organ, it is the site in which uptake of antigenic material by anal sucking allows for the constant exchange of the contents of the cloaca and bursa. Continued virus replication in an organism requires a supply of susceptible cells and the ability of the virus to survive the host immune response (5). B cells in the bursa proliferate extensively within the organ's follicles until it involutes at the age of 15 to 24 weeks, just before the sexual maturity of the bird. The microenvironment of the bursa may serve as the ideal site for the continual replication of influenza A virus. Necropsy was performed on only 6 of 40 pheasants in this study; therefore, further studies are needed to conclusively demonstrate the site where influenza virus is maintained in pheasants.
The H10 virus isolated from the pheasant late in infection was antigenically distinct from the parental virus in the inoculum. Zou et al. have shown that a lower ratio of amino acid to nucleotide substitutions can be expected when a negative selection process is in effect, while the reverse is true for positive selection (27). In our study, the ratio of amino acid to nucleotide substitutions was 10:12 (83.3%); therefore, the HA of the DK H10 virus was under significant positive selection pressure, with the virus acquiring mutations over the course of prolonged replication in the pheasant. Each amino acid change in the HA may have induced conformational changes that altered the antibody-binding sites in such a way that the HA was no longer recognized by the antibody, allowing the continual replication of the escape mutants. The locations of the amino acid changes modeled on the H3 HA 3-dimensional structure are approximate; however, 4 of 10 amino acid changes in the Ph H10 41-day isolate HA gene mapped to regions close to the sialic acid binding site, and changes in these regions might alter the interaction of the virus with host cell receptors. Sequence analysis of H3 viruses indicated that differences in receptor specificity could be accounted for by a single amino acid substitution at position 226 in the receptor-binding pocket of the HA (19). Receptor specificity seems to be controlled by different amino acids in influenza viruses of different subtypes, because specificity in H1 isolates did not correlate with differences in amino acid residues at position 226. In addition to amino acid changes in antigenic regions of the HA, the Ph H10 41-day isolate did not grow as well in MDCK cells as the original DK H10 isolate. The Ph H10 41-day isolate was also more immunogenic in ferrets, in which it induced high levels of serum antibody titers after the initial inoculation with the virus. The ferrets inoculated with the DK H10 isolate had to receive a boost of virus to produce a similar antibody response. These data suggest that the amino acid changes that occurred during prolonged shedding from pheasants might have altered the receptor-binding specificity of the DK H10 virus. Alteration of the viruses' receptor-binding specificity could have important consequences, such as transmission of the virus to humans. To verify this observation, we need to conduct further studies, including studies to measure the growth of the virus in MDCK cells over time and to measure the binding affinity of the virus for specific glycans.
Some animals may have a restricted antibody response. In some cases, an ineffective host immune response such as tolerance, immunosuppression, or production of nonneutralizing antibodies may allow the virus to survive. Although the pheasant that shed virus for 45 dpi produced a high level of antibodies, sufficient to neutralize the virus, by 14 dpi (1:452), we found evidence of lymphocyte depletion in the bursae of pheasants over time that may have led to immunosuppression in the pheasant. Several studies have demonstrated the emergence of escape mutants in the presence of monoclonal anti-HA antibodies when the concentration of antibodies was low, resulting in partial neutralization of the virus (11, 12, 26). A subneutralizing antibody response confronting a high virus load would fail to clear the virus, and this failure would allow escape mutants to survive and establish persistent infection. In addition, it has been shown that a single amino acid substitution in HA1 can account for reduced antibody interactions (12). Alternatively, circulating antibodies may have had a low affinity for HA antigenic sites. Among the avian species, the immune system of the chicken has been studied most extensively. Chickens have limited germ line potential and, therefore, limited antibody diversity. It was determined that a single λ-like light-chain isotype makes up ≥95% of chicken immunoglobulin light chains and a single variable (Vλ) gene is used by at least 95% of chicken B cells (17). Moreover, most of the variation in the antibody repertoire of chickens is generated during antigen-independent replication of B cells in the bursa. Pheasant immunoglobulin allotypes have been shown to be similar to those of the chicken, although each pheasant haplotype was associated with fewer class II B genes than those of the domestic chicken (25). In the pheasant, the antibody repertoire could be more restricted, and/or antibodies that could bind to the H10 antigenic sites may not have had sufficient binding strength to completely neutralize the virus, which helps explain why the virus was not cleared after the induction of an antibody response in the pheasant.
The HA of human influenza A viruses undergoes antigenic drift that allows the virus to incrementally escape the host's adaptive immune response. Escape mutants have typically emerged during persistent infection, after prolonged passage of viruses in vitro, usually with highly selected fractions or low concentrations of antiserum, or in vivo, where vaccination resulted in subprotective immunity (11). In contrast, avian influenza viruses have retained a relatively stable antigenic structure, and antigenic drift has not been observed in avian influenza viruses isolated from wild bird populations. In addition, the development of drift variants in domestic poultry has not been described, owing in part to their short productive lives, and because vaccination against influenza viruses remained uncommon. Multilineage antigenic drift was observed in the Mexican lineage of avian influenza viruses after a widespread program of poultry vaccination was implemented in an attempt to eradicate the virus in Mexico (13). This study highlighted the effect of long-term vaccine use on the evolution of avian influenza viruses in domestic poultry. Additionally, neither a commercial oil emulsion vaccine nor standardized reverse genetics resulted in vaccine that provided sterilizing immunity in chickens (15, 24). Low levels of virus replication in the presence of subneutralizing doses of antibodies help drive antigenic drift, especially when the vaccine strain is heterologous to the challenge or infecting virus strain. Thus, vaccine pressure is one mechanism for production of drift variants in poultry. Here we report naturally occurring escape mutants arising after prolonged shedding from a poultry species infected with live virus. And we provide evidence that escape mutants can arise within one individual bird in the absence of vaccination pressure. The prolonged replication of influenza viruses in pheasants not only allows influenza viruses to be transmitted to other poultry in the marketplace but adds to the genetic diversity of influenza viruses in this setting. This study strengthens our argument for the removal of pheasants from live bird retail markets.
Acknowledgments
We thank John Franks, Patrick Seiler, Hui-Ling Yen, and Yolanda Griffin for excellent technical assistance. We thank Christoph Scholtissek for many helpful discussions; Margaret Carbaugh, senior scientific editor, for editorial assistance; and Carol Walsh for manuscript preparation.
This study was supported by U.S. Public Health Service grant AI95357 and by the American Lebanese Syrian Associated Charities (ALSAC).
Footnotes
Published ahead of print on 31 January 2007.
REFERENCES
- 1.Air, G. M., A. J. Gibbs, W. G. Laver, and R. G. Webster. 1990. Evolutionary changes in influenza B are not primarily governed by antibody selection. Proc. Natl. Acad. Sci. USA 87:3884-3888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Colman, P. M., J. N. Varghese, and W. G. Laver. 1983. Structure of the catalytic and antigenic sites in influenza virus neuraminidase. Nature 303:41-44. [DOI] [PubMed] [Google Scholar]
- 3.Condobery, P. K., and R. D. Slemons. 1992. Biological properties of waterfowl-origin type A influenza viruses in chickens. Avian Dis. 36:17-23. [PubMed] [Google Scholar]
- 4.Cox, N. J., and Y. Kawaoka. 1998. Orthomyxoviruses: influenza, p. 385-433. In L. Collier, A. Balows, and M. Sussman (ed.), Topley & Wilson's microbiology and microbial infections, 9th ed. Arnold, London, United Kingdom.
- 5.Domingo, E., E. Baranowski, C. M. Ruiz-Jarabo, A. M. Martin-Hernandez, J. C. Saiz, and C. Escarmis. 1998. Quasispecies structure and persistence of RNA viruses. Emerg. Infect. Dis. 4:521-527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Easterday, B. C., V. S. Hinshaw, and D. A. Halvorson. 1997. Influenza, p. 583-606. In B. W. Calneck et al. (ed.), Diseases of poultry, 10th ed. Iowa State University Press, Ames.
- 7.Hinshaw, V. S., R. G. Webster, C. W. Naeve, and B. R. Murphy. 1983. Altered tissue tropism of human-avian reassortant influenza viruses. Virology 128:260-263. [DOI] [PubMed] [Google Scholar]
- 8.Hoffmann, E., J. Stech, Y. Guan, R. G. Webster, and D. R. Perez. 2001. Universal primer set for the full-length amplification of all influenza A viruses. Arch. Virol. 146:2275-2289. [DOI] [PubMed] [Google Scholar]
- 9.Humberd, J., Y. Guan, and R. G. Webster. 2006. Comparison of the replication of influenza A viruses in Chinese ring-necked pheasants and chukar partridges. J. Virol. 80:2151-2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kida, H., L. E. Brown, and R. G. Webster. 1982. Biological activity of monoclonal antibodies to operationally defined antigenic regions on the hemagglutinin molecule of A/Seal/Massachusetts/1/80 (H7N7) influenza virus. Virology 122:38-47. [DOI] [PubMed] [Google Scholar]
- 11.Lambkin, R., L. McLain, S. E. Jones, S. L. Aldridge, and N. J. Dimmock. 1994. Neutralization escape mutants of type A influenza virus are readily selected by antisera from mice immunized with whole virus: a possible mechanism for antigenic drift. J. Gen. Virol. 75:3493-3502. [DOI] [PubMed] [Google Scholar]
- 12.Laver, W. G., G. M. Air, R. G. Webster, W. Gerhard, C. W. Ward, and T. A. Dopheide. 1980. The mechanism of antigenic drift in influenza virus: sequence changes in the haemagglutinin of variants selected with monoclonal hybridoma antibodies. Philos. Trans. R. Soc. Lond. B 288:313-326. [DOI] [PubMed] [Google Scholar]
- 13.Lee, C. W., D. A. Senne, and D. L. Suarez. 2004. Effect of vaccine use in the evolution of Mexican lineage H5N2 avian influenza virus. J. Virol. 78:8372-8381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu, M., S. He, D. Walker, N. Zhou, D. R. Perez, B. Mo, F. Li, X. Huang, R. G. Webster, and R. J. Webby. 2003. The influenza virus gene pool in a poultry market in South central china. Virology 305:267-275. [DOI] [PubMed] [Google Scholar]
- 15.Liu, M., J. M. Wood, T. Ellis, S. Krauss, P. Seiler, C. Johnson, E. Hoffmann, J. Humberd, D. Hulse, Y. Zhang, R. G. Webster, and D. R. Perez. 2003. Preparation of a standardized, efficacious agricultural H5N3 vaccine by reverse genetics. Virology 314:580-590. [DOI] [PubMed] [Google Scholar]
- 16.Panigrahy, B., D. A. Senne, and J. C. Pedersen. 2002. Avian influenza virus subtypes inside and outside the live bird markets, 1993-2000: a spatial and temporal relationship. Avian Dis. 46:298-307. [DOI] [PubMed] [Google Scholar]
- 17.Pink, J. R. 1986. Counting components of the chicken's B cell system. Immunol. Rev. 91:115-128. [DOI] [PubMed] [Google Scholar]
- 18.Reed, L. J., and H. A. Muench. 1938. A simple method for estimating fifty percent endpoints. Am. J. Hyg. 27:493-497. [Google Scholar]
- 19.Rogers, G. N., J. C. Paulson, R. S. Daniels, J. J. Skehel, I. A. Wilson, and D. C. Wiley. 1983. Single amino acid substitutions in influenza haemagglutinin change receptor binding specificity. Nature 304:76-78. [DOI] [PubMed] [Google Scholar]
- 20.Senne, D. A., J. E. Pearson, and B. Panigrahy. 1992. Live poultry markets: a missing link in the epidemiology of avian influenza, p. 50-58. In Proceedings of the Third International Symposium on Avian Influenza. U.S. Animal Health Association, Richmond, VA.
- 21.Slemons, R. D., and D. E. Swayne. 1990. Replication of a waterfowl-origin influenza virus in the kidney and intestine of chickens. Avian Dis. 34:277-284. [PubMed] [Google Scholar]
- 22.Stubbs, E. L. 1965. Fowl plague, 5th ed. Iowa State University Press, Ames.
- 23.Webster, R. G., and W. G. Laver. 1980. Determination of the number of nonoverlapping antigenic areas on Hong Kong (H3N2) influenza virus hemagglutinin with monoclonal antibodies and the selection of variants with potential epidemiological significance. Virology 104:139-148. [DOI] [PubMed] [Google Scholar]
- 24.Webster, R. G., R. J. Webby, E. Hoffmann, J. Rodenberg, M. Kumar, H. J. Chu, P. Seiler, S. Krauss, and T. Songserm. 2006. The immunogenicity and efficacy against H5N1 challenge of reverse genetics-derived H5N3 influenza vaccine in ducks and chickens. Virology 351:303-311. [DOI] [PubMed] [Google Scholar]
- 25.Wittzell, H., T. von Schantz, R. Zoorob, and C. Auffray. 1994. Molecular characterization of three MHC class II B haplotypes in the ring-necked pheasant. Immunogenetics 39:395-403. [DOI] [PubMed] [Google Scholar]
- 26.Yewdell, J. W., A. J. Caton, and W. Gerhard. 1986. Selection of influenza A virus adsorptive mutants by growth in the presence of a mixture of monoclonal antihemagglutinin antibodies. J. Virol. 57:623-628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zou, S., I. Prud'homme, and J. M. Weber. 1997. Evolution of the hemagglutinin gene of influenza B virus was driven by both positive and negative selection pressures. Virus Genes 14:181-185. [DOI] [PubMed] [Google Scholar]