Abstract
The nucleocapsid (N) protein of infectious bronchitis virus (IBV) localizes to the cytoplasm and nucleolus and contains an eight-amino-acid nucleolar retention motif. In this study, a leucine-rich nuclear export signal (NES) (291-LQLDGLHL-298) present in the C-terminal region of the IBV N protein was analyzed by using alanine substitution and deletion mutagenesis to investigate the relative contributions that leucine residues make to nuclear export and where these residues are located on the structure of the IBV N protein. The analysis indicated that Leu296 and Leu298 are required for efficient nuclear export of the protein. Structural information indicated that both of these amino acids are available for interaction with protein complexes involved in this process. However, export of N protein from the nucleus/nucleolus was not inhibited by leptomycin B treatment, indicating that N protein nuclear export is independent of the CRM1-mediated export pathway.
An emerging paradigm is that positive-strand RNA viruses, whose primary site of replication is the cytoplasm, may have a nuclear/nucleolar component to their life cycle (14). This was recently illustrated by the finding that up to 20% of RNA-dependent RNA polymerase activity was found within the nuclei of cells infected with several different examples of flaviviruses (41). In addition, many diverse positive-strand RNA virus capsid proteins localize to the nucleus and/or a dynamic subnuclear structure called the nucleolus in either virus-infected cells or cells overexpressing the capsid protein (12, 13, 34, 43).
On first inspection, these observations are unusual given that the site of replication and virus assembly is the cytoplasm. If the subcellular localization of the capsid protein (or a portion of it) is in the nucleus, then the protein is not available for RNA synthesis, encapsidation, and assembly and therefore progeny virus production might be less efficient. Why capsid proteins localize to the nucleus is unknown and may be a case of molecular mimicry in which the proteins contain amino acid motifs which are similar to nuclear localization signals (NLSs) (34). However, if such proteins do target the nucleus as part of a virus replication strategy then they will contain appropriate targeting signals (34). These will include not only NLSs and nucleolar localization signals (NoLSs; also referred to as nucleolar accumulation or retention signals) but perhaps more importantly nuclear export signals (NESs). Whichever of the former two nuclear import possibilities may be operating, the capsid protein will require trafficking to the cytoplasm.
The coronavirus nucleocapsid (N) protein is no exception and can localize to both the cytoplasm and nucleus/nucleolus (4, 6, 15, 25, 26, 45). However, in the case of severe acute respiratory syndrome coronavirus there are differing reports of subcellular localization in both virus-infected cells and cells expressing the N protein (21, 31, 38, 40, 46). Likewise, several different N proteins of arterivirus (which, together with the coronaviruses, forms part of the nidoviruses) have been shown to localize to the nucleus/nucleolus (32, 39). Mutagenesis studies of the arterivirus porcine reproductive and respiratory syndrome virus N protein NLS revealed that nuclear localization of the protein is a fundamental part of the virus life cycle (20).
The avian infectious bronchitis virus (IBV) coronavirus N protein is a phosphoprotein (3, 18) which binds viral RNAs with high affinity (3, 37). The subcellular localization of the N protein within a cell can be either cytoplasmic or both nucleolar and cytoplasmic. Deletion and substitution mutagenesis delineated an NoLS (71-WRRQARFK-78) in the N-terminal region of the protein that was necessary and sufficient to direct nucleolar localization of the N protein (30). Bioinformatic analysis and deletion mutagenesis indicated a NES (291-LQLDGLHL-298) in the C-terminal region of the protein and that individual leucine residues may be important for localization (30). Thus, the IBV N protein contained signals to signal both nucleolar localization and subsequent export from the nucleus.
A leucine-rich NES is one of the most common and well-characterized NESs and in general is represented by an approximately 11-amino-acid leucine-rich signal, typified by the LxxxLxxLxL motif. In this case, a number of hydrophobic amino acids can substitute for L and the spacer regions (x) can vary in number (22, 27). The soluble receptor for this motif was shown to be exportin 1 (CRM1), and transport of proteins dependent on this motif is inhibited by leptomycin B (LMB). Although the IBV N protein does not precisely resemble this generic signal, variations of this motif have been demonstrated in both cellular and viral proteins (19, 22, 27). While the majority of the active leucine-rich motifs discovered operate via the CRM1-mediated pathway, there are some examples that use other, as yet unknown, export mechanisms.
In the case of CRM1-dependent leucine-rich motifs (so-called “Rev-type” export signals), both the specific leucine and its structural context can contribute to the efficiency of nuclear export (10). The same is also true of cellular proteins, whose trafficking out of the nucleus is mediated by binding to cellular mRNAs (e.g., the KNS NES found in hnRNP K) (23) and which are CRM1 independent (10). In this study, we investigated the relative contribution of each leucine residue in the leucine-rich motif to nuclear export in both a mammalian and an avian cell line and whether export of the IBV N protein is dependent on CRM1.
MATERIALS AND METHODS
Cell culture.
Vero (monkey-derived kidney epithelial) and DF1 (avian embryo fibroblast) cells were grown at 37°C with 5% CO2 in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum and penicillin-streptomycin (5). Thirty-millimeter tissue culture dishes were seeded with 2 × 105 cells 24 h prior to transfection.
Plasmid construction.
The N gene from the Beaudette strain (accession number AAA46214) of IBV served as the template for PCR of region and subregion constructs. All of the amino acid numbers used in the nomenclature of constructs relate to this 409-amino-acid protein. Production of NES deletion constructs pEGFP-IBVNΔ291-298 and pEGFP-IBVNΔ268-275 was carried out as described previously (30). Overlap PCR mutagenesis was used to produce the NES alanine substitution mutant proteins. The outer set of primers comprised a 5′ EcoRI restriction site and a 3′ BamHI restriction site, i.e., Jae1forward (GAATTCATGGCAATGGCAAGCGGTAAAGCAGCTGGA) and Jae1reverse (GGATCCTCAAAGTTCATTCTCTCCTAGATGC), as used in previous studies (30, 46). Internal primers for the first round of overlap mutagenesis PCR were as follows: L291A forward (GAAGTAGAGTGACACCCAAAGCCCAACTAGATGGGCTTCACTT) and L291A reverse (AAGTGAAGCCCATCTAGTTGGGCTTTGGGTGTCACTCTACTTC), L293A forward (GAGTGACACCCAAACTTCAAGCCGATGGGCTTCACTTGAGA) and L293A reverse (TCTCAAGTGAAGCCCATCGGCTTGAAGTTTGGGTGTCACTC), L296A forward (CCCAAACTTCAACTAGATGGGGCCCACTTGAGATTTGAATTTACTAC) and L296A reverse (GTAGTAAATTCAAATCTCAAGTGGGCCCCATCTAGTTGAAGTTTGGG), and L298A forward (CTTCAACTAGATGGGCTTCACGCCAGATTTGAATTTACTACTGTGGT) and L298A reverse (ACCACAGTAGTAAATTCAAATCTGGCGTGAAGCCCATCTAGTTGAAG). PCR products were purified and subcloned into the pCR2.1 TOPO vector (Invitrogen, United Kingdom). DNA was purified by alkaline lysis and digested with EcoRI and BamHI before being ligated into pEGFP-C2 (Clontech) with T4 DNA ligase (Invitrogen, United Kingdom) in accordance with the manufacturer's instructions. Double-alanine substitution mutant pEGFP-IBVN296L→A,298L→A was produced through sequential alanine substitution processes.
Live-cell imaging.
Vero and DF1 cells were transfected with 1 μg DNA to 5 μg polyethylenimine. DNA and polyethylenimine were mixed in a total volume of 200 μl serum-free Dulbecco's modified Eagle's medium and incubated at 20°C for 30 min. Transfection mix was added dropwise to cells and incubated at 37°C with 5% CO2 for 24 h. Live-cell imaging was performed with a Nikon Eclipse TS100 microscope with the filter B-2A and excitation at 450 to 490 nm for enhanced cyan fluorescent protein (ECFP) and enhanced green fluorescent protein (EGFP). Fluorescence and bright-field images were captured with a Nikon Digital Sight DS-L1.
Confocal imaging.
Confocal sections of fixed samples were captured on an LSM510 META microscope (Carl Zeiss Ltd., Germany) equipped with 40× and 63× numerical aperture 1.4 oil immersion lenses. Pinholes were set to allow optical sections of 1 μm to be acquired. EGFP was excited with the 488-nm argon laser line running at 2%, and emission was collected through an LP505 filter. All fluorescence was measured in the linear range as the detector is a photomultiplier, and the range indicator was used to ensure that no saturated pixels were obtained on image capture. Images were averaged eight times.
Relative fluorescence analysis.
In order to analyze the relative fluorescence intensity of each fluorescent fusion protein in the cytoplasm, nucleus, or nucleolus, 25 live-cell images of Vero and DF1 cells expressing the given construct were collected. ImageJ software (NIH) was used to convert the images to grey scale, and the bright-field images were used to demarcate each cellular compartment. These boundaries were applied to the grey-scale image, and the fluorescence intensity was measured, averaged for the area of the cellular compartment. All measurements were standardized with regard to background fluorescence. Graphs showing analytical data show the mean intensities over the 25 cells sampled and the standard error within the group.
FRAP analysis.
Fluorescence recovery after photobleaching (FRAP) experiments were performed with live Vero cells transfected with the appropriate cDNA. FRAP was performed with an LSM510 META confocal microscope with a 63× 1.4 numerical aperture oil immersion lens and a heated stage set to 37°C. Pinhole was set to allow 1-μm optical sections to be acquired. Cells were maintained in CO2-independent medium (GIBCO) for the duration of the live-cell imaging experiments. In order to compare nuclear export trafficking dynamics, selective photobleaching of EGFP within the cytoplasmic region of interest was achieved with a 488-nm argon laser running at 100% power. EGFP was excited with the 488-nm argon laser running at 1% power in all pre- and postbleaching images so as to minimize any background bleaching during filming. Images were acquired every 500 ms for a total time of 180 s. Bleach time and bleach area varied because of cell-to-cell variation. Optimal conditions for bleaching and imaging were established with the appropriate controls. All images were taken within the linear range with the range indicator palette of the LSM510 software (Carl Zeiss Ltd.). Relative fluorescence was determined with the region-of-interest tool of the LSM510 software (Carl Zeiss Ltd.).
LMB treatment.
Cells were transfected with either pEGFP-IBVN or pECFP-IBVNR3. At 18 h posttransfection, cells were washed with 1 ml phosphate-buffered saline and either treated with 2.5 ng/ml LMB diluted with medium or left in unsupplemented cell culture medium for a further 6 h. Localization patterns of EGFP-IBVN and ECFP-IBVNR3 were examined by live-cell imaging at this time. LMB activity was shown with a previously described luciferase assay (44) carried out in triplicate.
RESULTS
Comparison of the trafficking of N protein and NES deletion mutant proteins in Vero and DF1 cells.
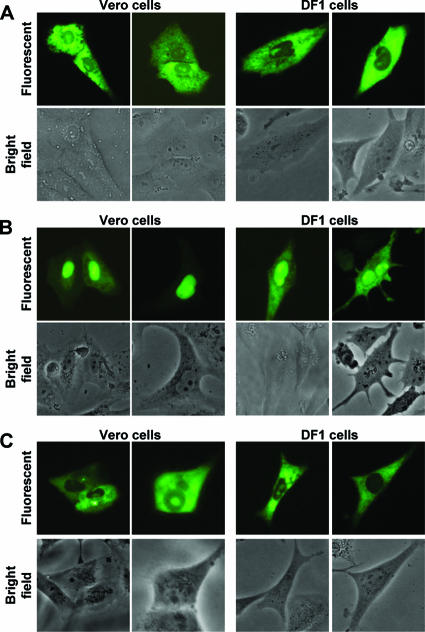
In this study, the IBV N protein NES was characterized in two cell types (where appropriate), a continuous African green monkey kidney epithelial cell line (Vero) which supports the replication of IBV (Beaudette-US strain) (1) and an immortalized chicken embryo fibroblast cell line (DF1). This was done to investigate potential differences between the trafficking profiles of the IBV N protein in the two cell lines as described for the IBV 3b nonstructural protein (28). To determine whether differential subcellular localization of N protein occurred between the two cell types, Vero and DF1 cells were transfected with pEGFP-IBVN (30, 46), which leads to the expression of IBV N protein fused C terminally to EGFP, and visualized 24 h posttransfection by live-cell imaging. Two examples of the subcellular localization of EGFP-IBVN are shown in both Vero and DF1 cells (Fig. 1A) cells; both the fluorescence and corresponding bright-field images are presented. The distribution of EGFP-IBVN in Vero and DF1 cells corresponded to the previously described localization in Vero cells; localization was either to the cytoplasm or to the cytoplasm and the nucleolus (6, 15, 30). This indicated that there was no difference in the subcellular localization of N protein between the two cell lines.
FIG. 1.
Live-cell imaging of the subcellular localization of (A) EGFP-IBVN, (B) EGFP-IBVNdel291-298, and (C) EGFP-IBVNdel268-275 proteins in Vero and DF1 cells (indicated) at 24 h posttransfection. Two examples are shown for each cell type. Both fluorescence and corresponding bright-field images of the same cells are shown.
To investigate whether the NES motif was active in the two cell types, the amino acid sequence 291-LQLDGLHLR-298 or a control eight-amino-acid hydrophobic region (268-VTAMLNLV-275) was deleted in the context of the EGFP-IBV N protein (pEGFP-IBVNΔ291-298 and pEGFP-IBVNΔ268-275, respectively) (30). Vero or DF1 cells were transfected with these plasmids, and the subcellular localization of the proteins was visualized by live-cell imaging. In Vero cells, EGFP-IBVNΔ291-298 localized predominately to the nucleolus and nucleus (as has been described previously (30) and a similar pattern was observed in DF1 cells (Fig. 1B) but with some signal in the cytoplasm. However, in both cell types EGFP-IBVNΔ291-298 localized to the nucleus, which was not observed in cells expressing EGFP-IBVN (Fig. 1A). In both Vero and DF1 cells which expressed EGFP-IBVNΔ268-275 (Fig. 1C), no nuclear localization was observed (as has been described previously for Vero cells (30) and the distribution of the fluorescent fusion protein was similar to that of wild-type N protein fused to EGFP—either cytoplasmic or cytoplasmic and nucleolar (Fig. 1A)—thus confirming that the 291-LQLDGLHLR-298 motif could act as a potential NES and was not dependent on any one cell type.
Contribution of leucine residues to nuclear export.
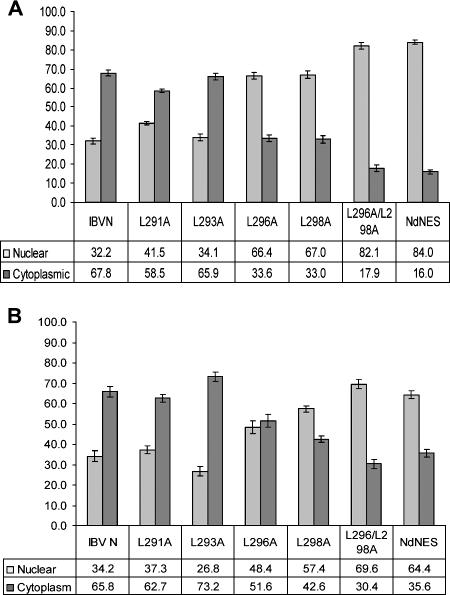
To identify which of the leucine residues present within the NES were functionally important in the context of the full-length protein, a systematic alanine substitution mutagenesis analysis was undertaken by previously described methodologies (30), producing plasmids pEGFP-IBVN291L→A, pEGFP-IBVN293L→A, pEGFP-IBVN296L→A, and pEGFP-IBVN298L→A. Expression of proteins from these plasmids would result in the translation of fluorescent fusion proteins in which Leu291, Leu293, Leu296, and Leu298 had been sequentially replaced with alanine. Vero and DF1 cells were transfected with these constructs and observed by live-cell imaging and confocal microscopy 24 h posttransfection. The ratio of protein between the nucleus and the cytoplasm (N/C) was measured by ImageJ analysis on 25 live cells for each construct (Fig. 2) and compared to the ratio of EGFP-IBVN between the nucleus and the cytoplasm by live-cell imaging. The data indicated that EGFP-IBVN291L→A and EGFP-IBVN293L→A localized predominantly to the cytoplasm or to the cytoplasm and the nucleolus (Fig. 3 and 4; Vero and DF1 cells, respectively), which is similar to EGFP-IBVN (Fig. 1A). The average respective N/C ratios in both Vero and DF1 cells indicated no significant difference between EGFP-IBVN (Vero cells = N32/C68; DF1 cells = N34/C66) and EGFP-IBVN291L→A (Vero cells = N41/C59; DF1 cells = N37/C63) or EGFP-IBVN293L→A (Vero cells = N34/C66; DF1 cells = N27/C73). Likewise, Vero and DF1 cell data were not significantly different. However, both the EGFP-IBVN296L→A and EGFP-IBVN298L→A proteins localized to the cytoplasm, nucleus, and nucleolus with some variability in the distribution of protein among these compartments (Fig. 3 and 4, Vero and DF1 cells, respectively). The average respective N/C ratios in both Vero and DF1 cells indicated a significant difference between the distribution of EGFP-IBVN (Vero cells = N32/C68; DF1 cells = N34/C66) and that of EGFP-IBVN296L→A (Vero cells = N66/C34; DF1 cells = N48/C52) or EGFP-IBVN298L→A (Vero cells = N67/C33; DF1 cells = N57/C43). The data indicated that nuclear accumulation of EGFP-IBVN296L→A and EGFP-IBVN298L→A was greater in Vero cells than in DF1 cells. However, because both EGFP-IBVN296L→A and EGFP-IBVN298L→A accumulated in the nucleus in both cell types, this indicated that these amino acids were involved in nuclear export.
FIG. 2.
Relative fluorescence in the nucleus and cytoplasm of (A) Vero cells and (B) DF1 cells expressing EGFP-IBVN (designated IBV N), EGFP-IBVN291L→A (designated L291A), EGFP-IBVN293L→A (designated L293A), EGFP-IBVN296L→A (designated L296A), EGFP-IBVN298L→A (designated L298A), EGFP-IBVN296L→A,298L→A (designated L296/298A), and EGFP-IBVNΔ291-298 (designated NdNES).
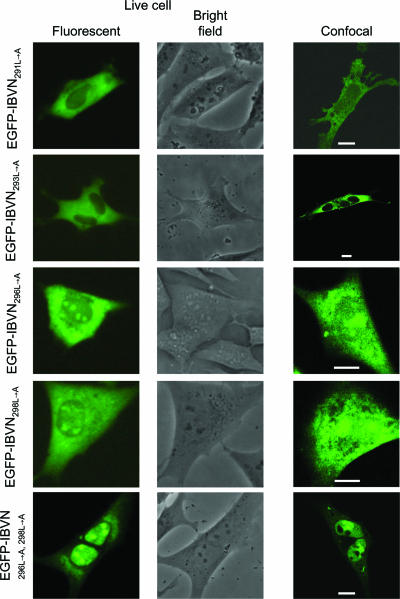
FIG. 3.
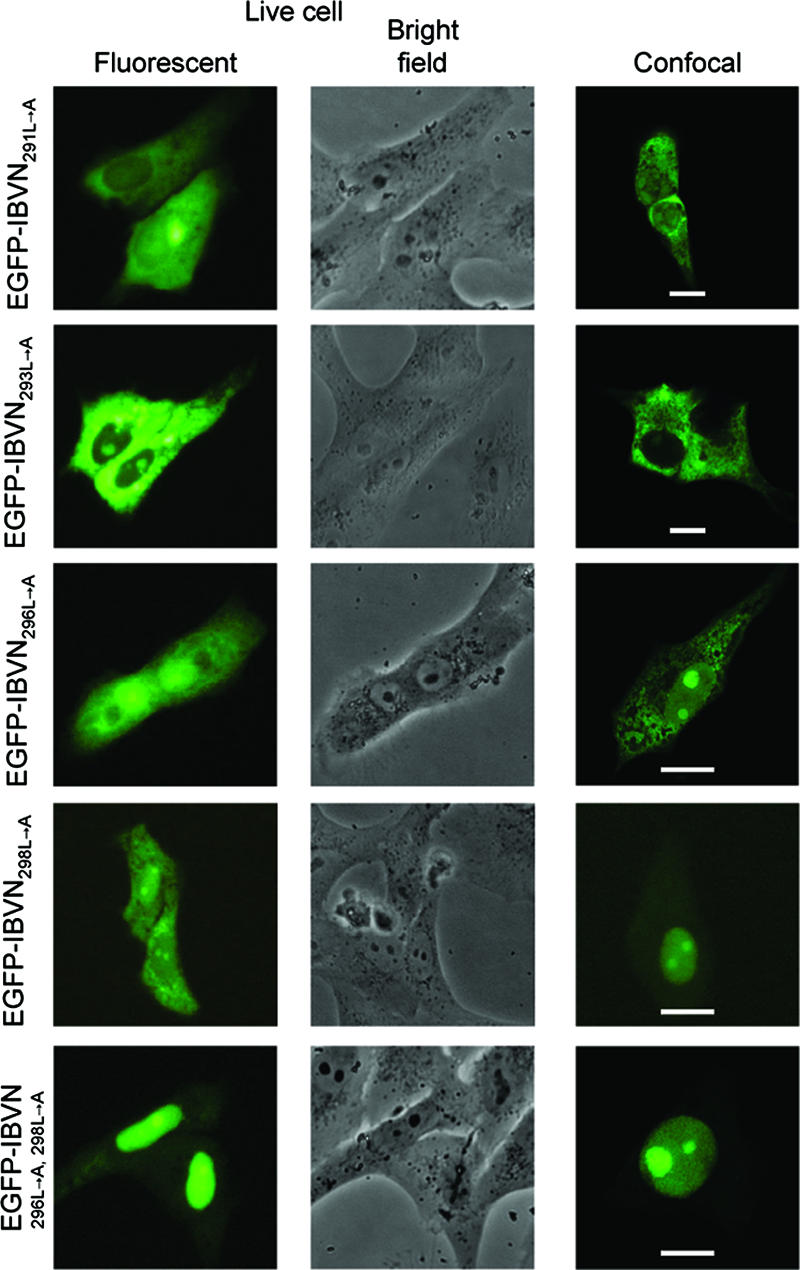
Live-cell and confocal imaging of the subcellular localization of EGFP-IBVN291L→A, EGFP-IBVN293L→A, EGFP-IBVN296L→A, EGFP-IBVN298L→A, and EGFP-IBVN296L→A,298L→A proteins expressed in Vero cells 24 h posttransfection. Both fluorescence and corresponding bright-field images of the same live cells are shown. Fluorescence confocal microscope images are also shown, and the scale bars indicate 10 μm.
FIG. 4.
Live-cell and confocal imaging of the subcellular localization of EGFP-IBVN291L→A, EGFP-IBVN293L→A, EGFP-IBVN296L→A, EGFP-IBVN298L→A, and EGFP-IBVN296L→A,296L→A proteins expressed in DF1 cells 24 h posttransfection. Both fluorescence and corresponding bright-field images of the same live cells are shown. Fluorescence confocal microscope images are also shown, and the scale bars indicate 10 μm.
Given that alanine substitutions at positions Leu296 and Leu298 resulted in increased nuclear localization of the fluorescent fusion protein, substitution mutagenesis was used to create plasmid pEGFP-IBVN296L→A,298L→A, whose expression would lead to a fluorescent fusion protein in which both Leu296 and Leu298 had been replaced with alanine (protein EGFP-IBVN296L→A,298L→A). Expression of this protein in both Vero and DF1 cells led to cytoplasmic, nuclear, and nucleolar localization (Fig. 3 and 4, respectively) with nuclear-to-cytoplasm ratios of N82/C18 and N70/C30, respectively. These data are significantly different from the distribution of EGFP-IBVN (Vero cells = N32/C68; DF1 cells = N34/C66) and indicated increased nuclear accumulation of the EGFP-IBVN296L→A,298L→A fusion protein. This ratio is not significantly different from the nuclear-to-cytoplasm ratio for EGFP-IBVNΔ291-298 (Vero cells = N84/C16; DF1 cells = N64/C35) but significantly higher than those for EGFP-IBVN296L→A and EGFP-IBVN298L→A. Therefore, the data indicated that both Leu296 and Leu298 were necessary for NES function and therefore efficiency of nuclear export of the EGFP-IBVN.
LQLDGLHL is the only NES operating in the N protein.
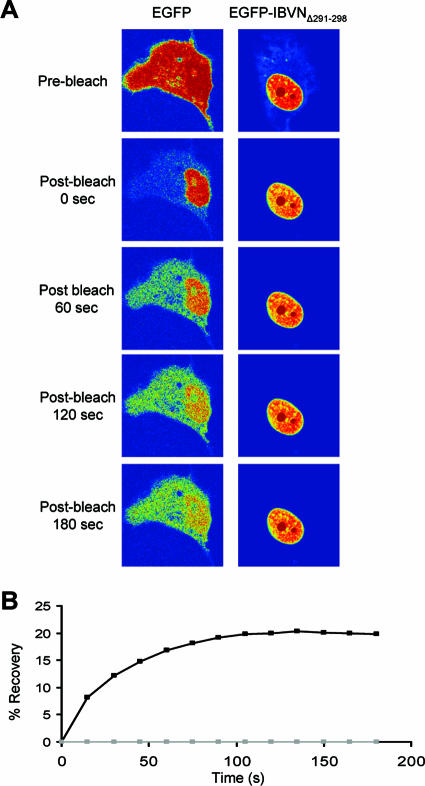
To investigate whether the NES signal located between amino acids 291 and 298 was the only functional NES, the expression construct pEGFP-IBVNΔ291-298 was used in a FRAP analysis. EGFP-IBVNΔ291-298 has been shown to localize predominantly to the nucleus/nucleolus (Fig. 1B). If this protein contained an alternative active NES, then the distribution would also be cytoplasmic. To investigate this, Vero cells were transfected with pEGFP-IBVNΔ291-298 and the subcellular localization of EGFP IBVNΔ291-298 was compared to that of EGFP by FRAP 24 h posttransfection. FRAP on the cytoplasm showed that EGFP reached equilibrium between the cytoplasm and the nucleus after 138 s whereas the EGFP-IBVNΔ291-298 protein remained nuclear/nucleolar throughout the time course (Fig. 5 shows a representative time course image [A] and a graph summarizing five data sets [B]). These data indicated that amino acids 291 to 298 mediated nuclear export and that there was no other NES motif that could replace this lost motif.
FIG. 5.
(A) FRAP analysis of EGFP-IBVNΔ291-298 protein and EGFP in which the cytoplasm has been photobleached at 24 h posttransfection. Images were sampled postbleaching three times a second for 180 s. EGFP-IBVNΔ291-298 protein and EGFP were false colored to show their concentration by using the rainbow feature of the Zeiss LSM imaging software, where red is high intensity and blue is low intensity. The experiment was repeated five times, and a representative image from each cell series is shown. (B) Average FRAP data within the cytoplasm are presented graphically; note that the error bars have been removed for clarity.
The nuclear export of N protein is not CRM1 mediated.
To determine if the IBV N protein was exported via the CRM1-mediated pathway, the subcellular localization of the EGFP-IBVN was compared between Vero cells left untreated or treated with 2.5 ng/ml LMB. To confirm that the LMB treatment was active in these experiments, we used an RNA export assay based on the export of human immunodeficiency virus type 1 (HIV-1) REV, which uses the CRM1 pathway (44). A luciferase gene was placed within an inefficiently spliced intron that contained the REV response element (creating plasmid pLucsalrre). The pre-mRNA is normally retained in the nucleus, and the exported, spliced mRNA lacks the luciferase gene. Export is facilitated through coexpressing HIV-1 Rev protein under the control of a cytomegalovirus promoter (plasmid pCMV-Rev), which ensures that the pre-mRNA is actively exported through the CRM1 pathway and luciferase activity would therefore be detected. The presence of LMB would inhibit REV-dependent mRNA export, confirming that LMB treatment of cells was effectively blocking the CRM1-dependent export pathway. Therefore, in order to assess the activity of LMB, Vero cells were transfected with pLucsalrre with or without LMB and also in the absence or presence of pCMV-Rev (Fig. 6A). The data indicated that, in the absence of pCMV-Rev, there was no significant difference in luciferase activity between LMB-treated and untreated cells, and this level was taken as background activity. In contrast, when the pCMV-Rev protein was present, the level of luciferase was 90% less in LMB-treated cells compared to that in untreated cells, indicating that LMB inhibited CRM1-mediated export.
FIG. 6.
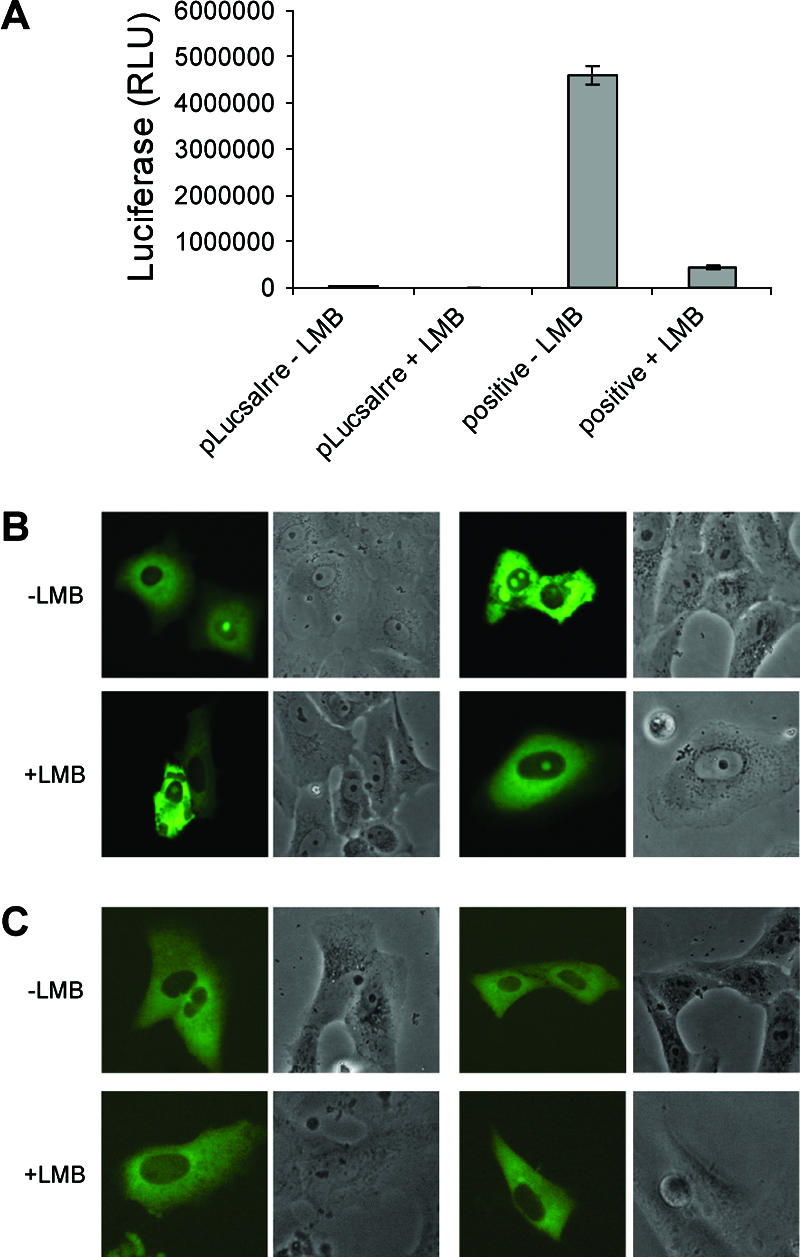
(A). Histogram showing the activity of LMB in relation to its ability to inhibit CRM1-mediated export of HIV-1 Rev protein. The first two columns are the negative controls, where the luciferase reporter RNA construct is expressed in the absence of Rev protein, whereas in the two columns on the right Rev is expressed in combination with the luciferase reporter RNA construct. The presence and absence of LMB are designated by plus and minus signs, respectively. RLU, relative light units. Live-cell fluorescence and corresponding bright-field images of Vero cells expressing either (B) EGFP-IBVN or (C) ECFP-IBVNR3 protein. Two representative cells are shown for each protein in the presence or absence of LMB (indicated as plus or minus LMB).
To determine whether the export of N protein was CRM1 dependent, Vero cells were transfected with pEGFP-IBVN, treated with LMB where appropriate, and visualized by live-cell imaging 24 h posttransfection. The data demonstrated that EGFP-IBVN in both the presence and absence of LMB (Fig. 6B) localized to both the cytoplasm and the nucleolus. Two examples of cells expressing EGFP-IBVN are shown together with corresponding bright-field images for each treatment. Had trafficking been CRM1 dependent, then the protein would have accumulated in the nucleus. To confirm this observation, we made use of a plasmid which expressed the C-terminal region of IBV N protein fused C terminal of ECFP, which contains the NES (30), whose expression product (ECFP-IBVNR3) localizes to the cytoplasm (30). This fusion protein is below the size exclusion of the nuclear pore complex and therefore, without any trafficking signals, would be predicted to localize to the cytoplasm and nucleus (similar to EGFP and ECFP). No difference was observed in Vero cells expressing ECFP-IBVNR3 in the presence or absence of LMB (Fig. 6C). Two examples of cells expressing ECFP-IBVNR3 are shown together with corresponding bright-field images for each treatment. Taken together, the data suggest that the trafficking of IBV N protein from the nucleus to the cytoplasm is CRM1 independent.
DISCUSSION
NESs in positive-strand RNA virus capsid proteins are not well characterized, nor are the pathways which determine their export from the nucleus/nucleolus. In this study, we have made use of the trafficking of IBV N protein from the cytoplasm to the nucleolus to the cytoplasm to investigate this type of signal. Substitution mutagenesis indicated that, in the previously identified NES (30), only two leucines contributed to the activity of this motif, Leu296 and Leu298. This is in agreement with a number of leucine-rich NESs that have been characterized in which one or two leucine (or other hydrophobic) residues determine the phenotype of nuclear export, such as with the morbillivirus nucleoprotein (35) and Rous sarcoma virus Gag protein (36). Although a number of viral proteins contain multiple functional NESs, such as the bovine herpesvirus type 1 UL47 protein (42), both live-cell imaging and dynamic tracking suggested that only one NES (containing Leu296 and Leu298) was functional in the trafficking of IBV N protein from the nucleus to the cytoplasm.
In many cases, the structural context of these leucine residues is important, especially in binding to the partner protein complexes involved in export, and two defined characteristics of an active NES are accessibility and flexibility (19, 29). The solid-state structure of the C-terminal region of the IBV N protein has been determined with the Gray strain (accession number AAB24054) (17), which has 93.6% identity with the N protein from the Beaudette-US strain (used in this study). With this structural information, the positions of Leu291, Leu296, and Leu298 were mapped (Fig. 7A). This was not possible for Leu293, as this is replaced with a proline in the Gray strain. However, the above substitution mutagenesis revealed that this leucine had no role in nuclear export of the N protein. Leu291, -296, and -298 were available for interaction within this structure when the protein was present as a monomer. However, Leu296 and Leu298 are masked when the C-terminal region is present as a dimer (Fig. 7B). Thus, different conformations of N protein may determine its subcellular localization by exposing different trafficking signals, such as the NoLS located in the N-terminal region (30) and the NES.
FIG. 7.
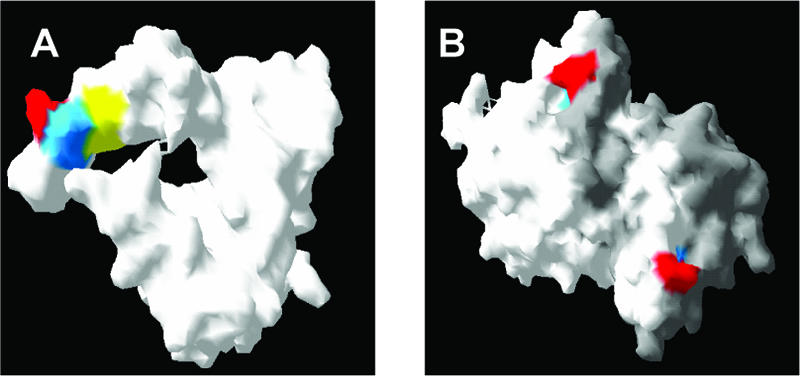
Molecular surface of a monomer (A) and dimer (B) of the C-terminal region of the Gray strain IBV N protein in which Leu291 is shown in red, Leu296 is shown in blue, and Leu298 is shown in yellow. Individual monomers are shown in white and gray for the purpose of differentiation.
The data indicated that the IBV N protein used a CRM1-independent nuclear export pathway. Leucine- and hydrophobic-residue-rich export motifs can operate via a non-CRM1-mediated pathway, for example, the nucleoprotein of the morbillivirus canine distemper virus (35). In contrast, LMB treatment of cells infected with the related arterivirus equine arteritis virus resulted in the accumulation of N protein predominately in the nucleus/nucleolus, whereas the subcellular localization of this protein in cells not treated with LMB is cytoplasmic and nucleolar (39). Likewise, prediction algorithms were used to identify a motif resembling an NES that has been identified in the PRRSV N protein (33). Treatment of cells with LMB expressing the PRRSV N protein fused to EGFP resulted in all cells exhibiting the phenotype of cytoplasmic and nucleolar localization of this protein, rather than in approximately 50 to 70% of the cells not treated with LMB (34).
The mechanism by which N protein is trafficked from the nucleus to the cytoplasm is unknown, and several pathways alternative to those mediated by CRM1 are possible. For example, the herpesvirus ORF57 protein localizes to the nucleolus/nucleus and shuttles to the cytoplasm (2, 9) and, as part of its function, is trafficked out of the nucleus via an interaction with cellular mRNA export factor TAP (Tip-associated protein) (44). The Epstein-Barr virus mRNA export factor EB2 traffics from the nucleus to the cytoplasm in a non-CRM1-dependent manner (8) and complexes with the cellular export factors REF and TAP (11). In some cases, both CRM1-dependent and -independent signals are used, as is the case with the nuclear-cytoplasmic shuttling activity of the African swine fever virus p37 protein (7). With the influenza A virus NS2 protein, an NES is crucial for nuclear export but through direct interaction with Ran-GTP rather than CRM1 (24).
Two amino acids were defined in this study that together were essential for the trafficking of IBV N protein to the cytoplasm. Similar to that proposed for the nuclear/nucleolar trafficking signal of nidovirus N proteins (20, 30), and for the export of the influenza A virus NS2 protein (16), mutation of the NES in the IBV N protein and subsequent disruption of cellular trafficking may be an attractive target for the generation of attenuated recombinant vaccines.
Acknowledgments
This work was supported by BBSRC Committee studentship BBSSP200310434 to J.A.H. The confocal facility in the Astbury Centre for Structural Molecular Biology was funded by the Wellcome Trust.
We thank members of the coronavirus laboratory for critically reviewing the manuscript and suggesting appropriate experiments, Jae-Hwan You in our laboratory for providing the pEGFP-IBVN clone, and Adrian Whitehouse for help setting up the LMB activity assay.
Footnotes
Published ahead of print on 3 January 2007.
REFERENCES
- 1.Alonso-Caplen, F. V., Y. Matsuoka, G. E. Wilcox, and R. W. Compans. 1984. Replication and morphogenesis of avian coronavirus in Vero cells and their inhibition by monensin. Virus Res. 1:153-167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boyne, J. R., and A. Whitehouse. 2006. Nucleolar trafficking is essential for nuclear export of intronless herpesvirus mRNA. Proc. Natl. Acad. Sci. USA 103:15190-15195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen, H., A. Gill, B. K. Dove, S. R. Emmett, F. C. Kemp, M. A. Ritchie, M. Dee, and J. A. Hiscox. 2005. Mass spectroscopic characterization of the coronavirus infectious bronchitis virus nucleoprotein and elucidation of the role of phosphorylation in RNA binding using surface plasmon resonance. J. Virol. 79:1164-1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen, H., T. Wurm, P. Britton, G. Brooks, and J. A. Hiscox. 2002. Interaction of the coronavirus nucleoprotein with nucleolar antigens and the host cell. J. Virol. 76:5233-5250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dove, B. K., G. Brooks, K. A. Bicknell, T. Wurm, and J. A. Hiscox. 2006. Cell cycle perturbations induced by infection with the coronavirus infectious bronchitis virus and their effect on virus replication. J. Virol. 80:4147-4156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dove, B. K., J.-H. You, M. L. Reed, S. R. Emmett, G. Brooks, and J. A. Hiscox. 2006. Changes in nucleolar architecture and protein profile during coronavirus infection. Cell. Microbiol. 8:1147-1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eulálio, A., I. Nunes-Correia, A. L. Carvalho, C. Faro, V. Citovsky, J. Salas, M. L. Salas, S. Simoes, and M. C. de Lima. 2006. Nuclear export of African swine fever virus p37 protein occurs through two distinct pathways and is mediated by three independent signals. J. Virol. 80:1393-1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Farjot, G., M. Buisson, M. Duc Dodon, L. Gazzolo, A. Sergeant, and I. Mikaelian. 2000. Epstein-Barr virus EB2 protein exports unspliced RNA via a Crm-1-independent pathway. J. Virol. 74:6068-6076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goodwin, D. J., K. T. Hall, A. J. Stevenson, A. F. Markham, and A. Whitehouse. 1999. The open reading frame 57 gene product of herpesvirus saimiri shuttles between the nucleus and cytoplasm and is involved in viral RNA nuclear export. J. Virol. 73:10519-10524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Henderson, B. R., and A. Eleftheriou. 2000. A comparison of the activity, sequence specificity, and CRM1-dependence of different nuclear export signals. Exp. Cell Res. 256:213-224. [DOI] [PubMed] [Google Scholar]
- 11.Hiriart, E., G. Farjot, H. Gruffat, M. V. Nguyen, A. Sergeant, and E. Manet. 2003. A novel nuclear export signal and a REF interaction domain both promote mRNA export by the Epstein-Barr virus EB2 protein. J. Biol. Chem. 278:335-342. [DOI] [PubMed] [Google Scholar]
- 12.Hiscox, J. A. 2002. Brief review: the nucleolus—a gateway to viral infection? Arch. Virol. 147:1077-1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hiscox, J. A. 2003. The interaction of animal cytoplasmic RNA viruses with the nucleus to facilitate replication. Virus Res. 95:13-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hiscox, J. A. 2007. RNA viruses: hijacking the dynamic nucleolus. Nat. Rev. Microbiol. 5:119-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hiscox, J. A., T. Wurm, L. Wilson, D. Cavanagh, P. Britton, and G. Brooks. 2001. The coronavirus infectious bronchitis virus nucleoprotein localizes to the nucleolus. J. Virol. 75:506-512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Iwatsuki-Horimoto, K., T. Horimoto, Y. Fujii, and Y. Kawaoka. 2004. Generation of influenza A virus NS2 (NEP) mutants with an altered nuclear export signal sequence. J. Virol. 78:10149-10155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jayaram, H., H. Fan, B. R. Bowman, A. Ooi, J. Jayaram, E. W. Collisson, J. Lescar, and B. V. Prasad. 2006. X-ray structures of the N- and C-terminal domains of a coronavirus nucleocapsid protein: implications for nucleocapsid formation. J. Virol. 80:6612-6620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jayaram, J., S. Youn, and E. W. Collisson. 2005. The virion N protein of infectious bronchitis virus is more phosphorylated than the N protein from infected cell lysates. Virology 339:127-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.la Cour, T., L. Kiemer, A. Molgaard, R. Gupta, K. Skriver, and S. Brunak. 2004. Analysis and prediction of leucine-rich nuclear export signals. Protein Eng. Design Selection 17:527-536. [DOI] [PubMed] [Google Scholar]
- 20.Lee, C., D. Hodgins, J. G. Calvert, S. K. Welch, R. Jolie, and D. Yoo. 2006. Mutations within the nuclear localization signal of the porcine reproductive and respiratory syndrome virus nucleocapsid protein attenuate virus replication. Virology 346:238-250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li, F. Q., H. Xiao, J. P. Tam, and D. X. Liu. 2005. Sumoylation of the nucleocapsid protein of severe acute respiratory syndrome coronavirus. FEBS Lett. 579:2387-2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Macara, I. G. 2001. Transport into and out of the nucleus. Microbiol. Mol. Biol. Rev. 65:570-594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Michael, W. M., P. S. Eder, and G. Dreyfuss. 1997. The K nuclear shuttling domain: a novel signal for nuclear import and nuclear export in the hnRNP K protein. EMBO J. 16:3587-3598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Neumann, G., M. T. Hughes, and Y. Kawaoka. 2000. Influenza A virus NS2 protein mediates vRNP nuclear export through NES-independent interaction with hCRM1. EMBO J. 19:6751-6758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ning, Q., S. Lakatoo, M. F. Liu, W. M. Yang, Z. M. Wang, M. J. Phillips, and G. A. Levy. 2003. Induction of prothrombinase fgl2 by the nucleocapsid protein of virulent mouse hepatitis virus is dependent on host hepatic nuclear factor-4 alpha. J. Biol. Chem. 278:15541-15549. [DOI] [PubMed] [Google Scholar]
- 26.Ning, Q., M. F. Liu, P. Kongkham, M. M. C. Lai, P. A. Marsden, J. Tseng, B. Pereira, M. Belyavskyi, J. Leibowitz, M. J. Phillips, and G. Levy. 1999. The nucleocapsid protein of murine hepatitis virus type 3 induces transcription of the novel fgl2 prothrombinase gene. J. Biol. Chem. 274:9930-9936. [DOI] [PubMed] [Google Scholar]
- 27.Ossareh-Nazari, B., C. Gwizdek, and C. Dargemont. 2001. Protein export from the nucleus. Traffic 2:684-689. [DOI] [PubMed] [Google Scholar]
- 28.Pendleton, A. R., and C. E. Machamer. 2006. Differential localization and turnover of infectious bronchitis virus 3b protein in mammalian versus avian cells. Virology 345:337-345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Petosa, C., G. Schoehn, P. Askjaer, U. Bauer, M. Moulin, U. Steuerwald, M. Soler-Lopez, F. Baudin, I. W. Mattaj, and C. W. Muller. 2004. Architecture of CRM1/exportin1 suggests how cooperativity is achieved during formation of a nuclear export complex. Mol. Cell 16:761-775. [DOI] [PubMed] [Google Scholar]
- 30.Reed, M. L., B. K. Dove, R. M. Jackson, R. Collins, G. Brooks, and J. A. Hiscox. 2006. Delineation and modelling of a nucleolar retention signal in the coronavirus nucleocapsid protein. Traffic 7:833-848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rowland, R. R., V. Chauhan, Y. Fang, A. Pekosz, M. Kerrigan, and M. D. Burton. 2005. Intracellular localization of the severe acute respiratory syndrome coronavirus nucleocapsid protein: absence of nucleolar accumulation during infection and after expression as a recombinant protein in Vero cells. J. Virol. 79:11507-11512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rowland, R. R., R. Kerwin, C. Kuckleburg, A. Sperlich, and D. A. Benfield. 1999. The localisation of porcine reproductive and respiratory syndrome virus nucleocapsid protein to the nucleolus of infected cells and identification of a potential nucleolar localization signal sequence. Virus Res. 64:1-12. [DOI] [PubMed] [Google Scholar]
- 33.Rowland, R. R. R., P. Schneider, Y. Fang, S. Wootton, D. Yoo, and D. A. Benfield. 2003. Peptide domains involved in the localization of the porcine reproductive and respiratory syndrome virus nucleocapsid protein to the nucleolus. Virology 316:135-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rowland, R. R. R., and D. Yoo. 2003. Nucleolar-cytoplasmic shuttling of PRRSV nucleocapsid protein: a simple case of molecular mimicry or the complex regulation by nuclear import, nucleolar localization and nuclear export signal sequences. Virus Res. 95:23-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sato, H., M. Masuda, R. Miura, M. Yoneda, and C. Kai. 2006. Morbillivirus nucleoprotein possesses a novel nuclear localization signal and a CRM1-independent nuclear export signal. Virology 352:121-130. [DOI] [PubMed] [Google Scholar]
- 36.Scheifele, L. Z., E. P. Ryan, and L. J. Parent. 2005. Detailed mapping of the nuclear export signal in the Rous sarcoma virus Gag protein. J. Virol. 79:8732-8741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Spencer, K. A., and J. A. Hiscox. 2006. Characterisation of the RNA binding properties of the coronavirus infectious bronchitis virus nucleocapsid protein amino-terminal region. FEBS Lett. 580:5993-5998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Surjit, M., R. Kumar, R. N. Mishra, M. K. Reddy, V. T. Chow, and S. K. Lal. 2005. The severe acute respiratory syndrome coronavirus nucleocapsid protein is phosphorylated and localizes in the cytoplasm by 14-3-3-mediated translocation. J. Virol. 79:11476-11486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tijms, M. A., Y. van der Meer, and E. J. Snijder. 2002. Nuclear localization of non-structural protein 1 and nucleocapsid protein of equine arteritis virus. J. Gen. Virol. 83:795-800. [DOI] [PubMed] [Google Scholar]
- 40.Timani, K. A., Q. Liao, L. Ye, Y. Zeng, J. Liu, Y. Zheng, X. Yang, K. Lingbao, J. Gao, and Y. Zhu. 2005. Nuclear/nucleolar localization properties of C-terminal nucleocapsid protein of SARS coronavirus. Virus Res. 114:23-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Uchil, P. D., A. V. Kumar, and V. Satchidanandam. 2006. Nuclear localization of flavivirus RNA synthesis in infected cells. J. Virol. 80:5451-5464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Verhagen, J., M. Donnelly, and G. Elliott. 2006. Characterization of a novel transferable CRM-1-independent nuclear export signal in a herpesvirus tegument protein that shuttles between the nucleus and cytoplasm. J. Virol. 80:10021-10035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Weidman, M. K., R. Sharma, S. Raychaudhuri, P. Kundu, W. Tsai, and A. Dasgupta. 2003. The interaction of cytoplasmic RNA viruses with the nucleus. Virus Res. 95:75-85. [DOI] [PubMed] [Google Scholar]
- 44.Williams, B. J., J. R. Boyne, D. J. Goodwin, L. Roaden, G. M. Hautbergue, S. A. Wilson, and A. Whitehouse. 2005. The prototype gamma-2 herpesvirus nucleocytoplasmic shuttling protein, ORF 57, transports viral RNA through the cellular mRNA export pathway. Biochem. J. 387:295-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wurm, T., H. Chen, P. Britton, G. Brooks, and J. A. Hiscox. 2001. Localization to the nucleolus is a common feature of coronavirus nucleoproteins and the protein may disrupt host cell division. J. Virol. 75:9345-9356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.You, J.-H., B. K. Dove, L. Enjuanes, M. L. DeDiego, E. Alvarez, G. Howell, P. Heinen, M. Zambon, and J. A. Hiscox. 2005. Sub-cellular localisation of the severe acute respiratory syndrome coronavirus nucleocapsid protein. J. Gen. Virol. 86:3303-3310. [DOI] [PubMed] [Google Scholar]