Abstract
Human transcriptional coactivator p75/lens epithelium-derived growth factor (LEDGF) binds human immunodeficiency virus type 1 (HIV-1) integrase (IN). We studied the effects of LEDGF on the assembly and activity of HIV-1 synaptic complexes, which, upon association with a target, mediate concerted integration of viral DNA substrates in vitro. We found that while augmenting single-ended viral DNA integration into target DNA, the host factor was able to either stimulate or abrogate concerted integration in a concentration-dependent manner. LEDGF modestly stimulated (two- to threefold) concerted integration at low molar ratios to IN (<1). The modest stimulation was independent of solution conditions and several different viral DNA substrates. In solution, concerted integration was inhibited if the molar ratios of LEDGF to IN were >1, apparently due to the disruption of IN-IN interactions essential for the formation of active synaptic complexes prior to their association with a circular target. The isolated IN binding domain of LEDGF was sufficient to stimulate and inhibit concerted integration, as observed with full-length protein, albeit at lower efficiencies. Our data show that LEDGF differentially affects IN-DNA complexes mediating single-ended viral DNA integration and synaptic complexes mediating concerted integration. Synaptic complexes associated with target, termed strand transfer complexes, are resistant to disruption by high concentrations of LEDGF. The results suggest that LEDGF may influence HIV-1 integration in vivo.
Retroviruses are unique in that they insert DNA replicas of their genomes into host cell chromosomal DNA. Following reverse transcription of the viral RNA genome into linear DNA, preintegration complexes (PIC) that contain the viral integrase (IN) and several other viral and cellular proteins are formed (2). The PIC traverses the nuclear envelope and associates with chromosomes, where IN inserts the viral genome. Isolated cytoplasmic PIC can insert the viral DNA ends in a concerted fashion into exogenously supplied target DNA substrates in vitro (3, 5, 6, 22, 33, 50).
Major advances have occurred in identifying target sites and understanding the basis for target site selectivity exhibited by human immunodeficiency virus type 1 (HIV-1), murine leukemia virus (MLV), and avian sarcoma-leukemia virus in human chromosomes (25, 34, 36, 37, 51). HIV-1 predominantly integrates within genes, having a preference for those that are actively transcribing. MLV prefers integration into or near transcription start sites, while avian sarcoma-leukemia virus exhibits a slight preference for genes and no preference for transcription start sites. These results suggest that HIV-1 and MLV may interact with chromatin-associated factors and/or transcriptional cofactors that direct the PIC to transcription units in the case of HIV-1 and to or near transcriptional start sites in the case of MLV (16). Simian immunodeficiency virus selectivity for integration into human chromosomes is similar to that of HIV-1 (14). Retrovirus site selection has implications for human gene therapy (46).
Several cellular proteins were observed to cofractionate with the PIC; their proposed roles range from trafficking and transport of the PIC into the nucleus to structural maintenance and selection of host target sites (40, 45). A human transcriptional coactivator, lens epithelium-derived growth factor (LEDGF)/p75 (17), was identified as a high-affinity interactor with ectopically expressed HIV-1 IN in human cells (9). LEDGF was later shown to associate with divergent feline immunodeficiency virus, but not MLV IN (30). Functional HIV-1 and feline immunodeficiency virus PICs could be immunoprecipitated with anti-LEDGF antibody (30). LEDGF specifically interacts with the catalytic core domain (CCD) of HIV-1 IN at a stoichiometric ratio of 1 (7, 10, 31). The HIV-1 IN binding domain (IBD) in LEDGF maps to a highly conserved region within the C-terminal region of the protein (7, 8, 10). Depletion of LEDGF by RNA interference (RNAi) results in a loss of stable association between IN and chromatin, suggesting that LEDGF tethers IN to cellular DNA (15, 30, 32, 44). Furthermore, a HIV-1 IN mutant (Q168A) deficient for binding to LEDGF failed to associate with chromosomes (15).
RNAi-mediated depletion of LEDGF appears to have little effect on HIV-1 replication (30, 42), although other LEDGF depletion studies suggested a two- to fourfold reduction in HIV-1 replication, with the block occurring at the integration step (43). In another LEDGF depletion study, an average 53% reduced infection and a 74% decrease in integration was observed in HIV-1-infected macrophages, suggesting a modest involvement of LEDGF in integration (52). Further intensified RNAi and dominant-negative protein approaches suggested that LEDGF is an essential HIV-1 integration cofactor (29). Lastly, genomic analyses of HIV-1-infected cells depleted of LEDGF compared to nondepleted cells indicated that the protein is involved in the selection of chromosomal integration sites (13). These studies suggest that LEDGF is important for HIV-1 integration, although its precise functions remain to be established.
The in vitro biochemical dissection of retroviral integration provides important mechanistic insights into the integration process in vivo. Synaptic complexes consisting of purified IN and viral DNA substrates can efficiently insert two viral DNA ends in a concerted fashion into circular target DNA (12, 47-49) in a process here termed full-site integration (Fig. 1). The insertion of one DNA substrate end into target DNA produces a circular half-site (CHS) integration product. IN in detergent-lysed HIV-1 virions (19), as well as recombinant HIV-1 IN, catalyzes these reactions (20, 23, 26, 38, 39). Although LEDGF has been demonstrated to stimulate gross strand transfer activity of HIV-1 IN in vitro (8-10, 41), the effect of the cofactor on full-site integration has not been addressed. In this report, we show that the concentration of LEDGF modulates synaptic complexes, but not strand transfer complexes (STC) (Fig. 1), that produce full-site products. Modest stimulation of full-site integration (two- to threefold) is observed at molar ratios of LEDGF to IN of <1, while at ratios of >1, LEDGF disrupts IN-IN interactions necessary for the assembly of synaptic complexes in solution.
FIG. 1.
Assembly of HIV-1 synaptic complexes and production of DNA integration products. A linear donor containing U3 and U5 ends was assembled with IN at 14°C to form a synaptic complex (top). Circular target DNA was added to allow strand transfer activity at 37°C. The STC produces the full-site integration product. Other products produced by different IN-DNA complexes include CHS and donor-donor products equal to twice the length of the original donor and other Y-type structures. The arrows indicate the different orders of addition of LEDGF. At the bottom, different substrates having U5-U3 LTR ends or only U5 LTR ends used in this study are shown.
MATERIALS AND METHODS
Production and labeling of DNA substrates.
The linear 4.1-kbp and 0.48-kbp DNA substrates containing HIV-1 wild-type (wt) U3 and U5 long terminal repeat (LTR) attachment ends were previously described (38, 39). NdeI digestion of the plasmids produced linear substrates with 3′-OH recessed ends containing a 5′-TA overhang (Fig. 1). To obtain a 3.4-kbp single-ended 3′-OH recessed U5 substrate, the NdeI-linearized and -dephosphorylated 4.1-kbp plasmid was digested with SacI. The DNA donor substrates were 5′ end labeled on the noncatalytic strand with [γ-32]ATP and T4 polynucleotide kinase. The specific activities were ∼1,500 to 2,000 cpm (Cerenkov) per ng of double-end-labeled donor substrates and ∼1,000 cpm for single-end-labeled substrates. The fragments were isolated from agarose gels and extracted using the QIAquick Gel Extraction Kit (QIAGEN, Valencia, CA) or electroelution. Supercoiled pBSK2Δ-Zeo (2.7 kbp), kindly provided by Karen Moreau (35), was used as target DNA.
A HIV-1 linear 4.7-kbp DNA containing wt U3 and U5 LTR blunt ends (11) was also used for strand transfer. ScaI digestion of the plasmid produced a substrate containing HIV-1 natural DNA blunt ends (11). To obtain single-ended U5 and U3 blunt-end substrates, the ScaI-linearized and -dephosphorylated plasmid was further digested with NcoI to yield a 1.6-kbp U5 substrate (Fig. 1) and a 2.4-kbp U3 substrate. Preprocessed natural DNA fragments containing 5′-AC overhangs were constructed by ligation of 93-bp double-stranded oligonucleotides containing HIV-1 U5 LTR sequences to a pUC19-derived fragment. The ligated DNA fragments were purified by agarose gel electrophoresis and end labeled. The synthetic 5′-AC fragment was 1.5 kbp in length, and the nonspecific end was blunt ended (Fig. 1).
Integration assay.
The HIV-1 integration assays with all of the DNA substrates (Fig. 1) were performed as described previously (38). Briefly, HIV-1 IN was assembled with the donor substrate in the presence of 20 mM HEPES (pH 7.0), 5 mM dithiothreitol, 10 mM MgCl2, 25 μM ZnCl2, 50 mM NaCl, and 10% polyethylene glycol (6,000 Da) at 14°C for 15 min. The NaCl concentration was varied between 50 mM and 125 mM as indicated. The IN and donor concentrations are described for each experiment. The standard reaction volumes were 200 μl. The reactions were initiated by addition of 500 ng of supercoiled target DNA, and the mixtures were incubated for 2 h at 37°C. The above-mentioned reactions were stopped by the addition of EDTA, sodium dodecyl sulfate (SDS), and proteinase K to final concentrations of 25 mM, 0.5%, and 1 mg/ml, respectively. Equivalent amounts of labeled integration products (∼10,000 to 20,000 cpm) were subjected to 0.8% or 1.5% agarose gel electrophoresis in Tris-borate-EDTA (TBE) buffer for 10 to 16 h at 100 V, depending upon the size of the donor. Note that the full-site and CHS products migrated at different positions on the two different agarose gels. The gels were dried and exposed to a phosphorimager screen. Full-site and CHS products were quantified using a Storm 860 system (Amersham Biosciences). To evaluate the differential effects of LEDGF upon the assembly and catalytic properties of synaptic complexes and STC, LEDGF was either added to assembled synaptic complexes at 14°C or bound with HIV-1 IN at 14°C prior to the addition of the donor (Fig. 1). In other sets of experiments, the target was mixed with assembled synaptic complexes prior to the addition of LEDGF. Lastly, preformed STC were allowed to form for 2 h at 37°C and were then challenged with LEDGF prior to analysis on native 0.7% agarose gels.
Identification and evaluation of STC with native agarose gels.
The synaptic complexes were assembled as described above. After the addition of target, STC were allowed to form at 37°C for 2 h. The reactions were stopped by adding 20 mM EDTA, and the products were subjected to electrophoresis on a 0.7% native agarose gel in TBE buffer without 1 M urea (27). Two-dimensional gel electrophoresis of complexes separated on agarose gels was performed as described previously (27).
Purification of recombinant proteins.
IN from the pNY clone was expressed in Escherichia coli BL21(DE3) cells and purified to near homogeneity as described previously (38). Concentrations of HIV-1 IN were determined using Rous sarcoma virus (RSV) IN as a standard (12). LEDGF was purified as previously described (9, 10), and its concentration was determined using the molar extinction coefficient value of 16,500 M−1 cm−1, determined from its amino acid sequence. Isolated IBDs of LEDGF (residues 347 to 471) and full-length mutant LEDGF (D336N) were prepared as described previously (10).
Analysis of integration products.
The integration products obtained with HIV-1 IN and the 4.1-kb donor substrate were previously analyzed by EcoRI digestion and DNA sequencing of the viral DNA-host junctions (38). The full-site integration (9.5 kbp) products obtained with the single-ended U5 donor (3.4 kb) were also digested with EcoRI and yielded the correct 2.9-kb U5-target-U5 product (38). The 1.6-kbp DNA substrate containing blunt-ended U5 produced the predicted 5.9-kbp full-site product that, upon digestion by either HpaI, MluI, or Bsu36I, produced the predicted full-site DNA fragments (data not shown). The viral DNA-host junctions of the full-site integration products derived from coupled U5-U5 and U5-U3 substrates were sequenced. The U5-U5 full-site products were isolated from a 0.8% agarose gel and purified by electroelution. The integration products were amplified by PCR using U5 LTR sequence as primers (forward and reverse, 5′ GTCAGTGTGGAAAATCTCTAGC 3′) using PfuUltra Hotstart PCR Master Mix (Stratagene). The PCR cycling parameters included initial denaturation at 95°C for 2 min, followed by 30 cycles at 95°C for 1 min, 56°C for 1 min, and 72°C for 3.5 min. Final extension was for 10 min at 72°C. PCR products (∼2.7 kb) were purified from agarose gels with a Qiaquick Gel extraction kit and cloned in Zero Blunt PCR vector (Invitrogen). Recombinant clones were sequenced using custom primers (KKPBlunt244, 5′ GGTGACGCGTTAGAATACTCAAGC 3′, and ccd-R, 5′ GCCCCGGCGTGTCAATAATATC 3′) to analyze the LTR-target junction and host site duplications. The U5-U3 full-site products were isolated from the gel and digested with BglII and BamHI. The 3.1-kb products were gel purified, self-ligated, and used to transform E. coli DH5α cells. Recombinants were sequenced using the custom primers 6176U5BglII (5′ CTGACCTGCCAGATCTGCTAACTAGGGAACCCAC 3′) and 6176U3BamHI (5′ CGCCCGCCACTAGTGGATCCAAAGAATTCTATC 3′).
RESULTS
LEDGF, in a concentration-dependent manner, differentially interacts with synaptic complexes and other IN-DNA complexes mediating CHS integration.
Full-site integration by HIV-1 synaptic complexes in vitro allows functional analysis of putative cellular cofactors (Fig. 1). The production of full-site integration products by HIV-1 synaptic complexes is dependent upon the concentrations of HIV-1 IN in the reaction mixtures, the lengths and concentrations of the DNA substrates, the use of preprocessed or blunt-ended viral substrates, and the solution conditions (19, 20, 26, 27, 38, 39). We explored these parameters to investigate the effects of LEDGF on HIV-1 integration in vitro.
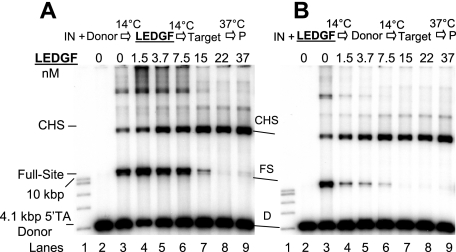
We first used a 4.1-kbp DNA substrate containing preprocessed 3′-OH recessed U3 and U5 ends with 5′-TA overhangs to examine the effects of LEDGF on the assembly and catalytic activity of HIV-1 synaptic complexes (Fig. 1). HIV-1 IN (5 nM) was assembled onto the 4.1-kbp donor (0.2 nM) prior to the addition of increasing concentrations of LEDGF into the assembly mixture, followed by analysis of strand transfer products (Fig. 2A). LEDGF at 1.5 and 3.7 nM stimulated full-site integration activity approximately 2.0- and 1.3-fold, respectively (Fig. 2A, lanes 4 and 5), in comparison to IN without LEDGF (lane 3). A minor donor-donor product that migrated slightly faster than the full-site product (38) was also affected by LEDGF (Fig. 2A). Higher concentrations of LEDGF (>7.5 nM) in the assembly mixture eliminated full-site integration activity, presumably by preventing assembly of the synaptic complex (Fig. 2A, lanes 7 to 9). In contrast, the production of CHS products was increased at all concentrations of LEDGF using the preprocessed 3′-OH substrate (Fig. 2A, lanes 3 to 9). Under the conditions of this assay, the CHS products were enhanced ∼5-fold (Fig. 2A, lane 9). Several other integration products migrating above the CHS products were inhibited, stimulated, or unaffected at various LEDGF concentrations. The data suggest that LEDGF, in a concentration-dependent manner, differentially interacts with synaptic complexes capable of full-site activity and with nucleoprotein complexes that possess CHS activity.
FIG. 2.
LEDGF differentially affects full-site and CHS integration. (A) Synaptic complexes were assembled with IN (5 nM) and the 4.1-kbp donor (0.2 nM) possessing U3 and U5 ends with 5′-TA overhangs. LEDGF was added at the indicated final concentrations, and the mixture was incubated for an additional 15 min at 14°C. Strand transfer was initiated by the addition of target and incubation for 2 h at 37°C. All lanes of Fig. 2A are identical to 2B except for the order of addition shown at the top. P, products. The DNA products and input donor are indicated on the left. Lanes 1 and 2 contained a 1-kbp DNA ladder (Promega) and donor only, respectively. Lane 3 contained IN only; it incorporated ∼13% and 4% of the donor into full-site (FS) and CHS products, respectively. The full-site products in lanes 4 to 6 in panel A were ∼30%, 16%, and 12%, respectively. The CHS products in lane 9 were ∼22%. (B) Conditions identical to those for panel A were employed, except that IN and LEDGF were preincubated together prior to the addition of the donor (top). The CHS products in lane 9 were 15%. The experiments in panels A and B were performed simultaneously.
The order of addition affects the assembly of the HIV-1 synaptic complex.
Binding of LEDGF to IN prior to addition of the donor with 5′-TA overhangs at 14°C essentially prevented the accumulation of the full-site product (Fig. 2B, lanes 4 to 9) in comparison to assembly in the absence of LEDGF (Fig. 2B, lane 3). These results suggest that binding of LEDGF to IN prevents the assembly of synaptic complexes, presumably by steric hindrance or conformational alterations in IN. In contrast, CHS products were enhanced at all LEDGF concentrations. The CHS activity was enhanced ∼4-fold at 37 nM LEDGF (Fig. 2B, lane 9).
Effects of DNA substrate and solution conditions on the properties of LEDGF.
A 3.4-kbp single-ended U5 donor (0.3 nM) containing preprocessed 5′-TA ends (Fig. 1) was assembled with HIV-1 IN (5 nM). Addition of LEDGF to assembled synaptic complexes slightly stimulated the production of full-site products at low LEDGF concentrations, while at higher LEDGF concentrations (10 nM), full-site integration was again disrupted (data not shown). On the other hand, the CHS products were reproducibly enhanced by LEDGF.
The interactions between LEDGF and IN within synaptic complexes are not overly sensitive to concentrations of components and solution conditions. A 0.48-kbp donor (2.5 nM) containing preprocessed HIV-1 U3 and U5 attachment sites with 5′-TA overhangs was assembled with HIV-1 IN (16 nM) on ice in the presence of 12% dimethyl sulfoxide and 100 mM NaCl (39). After assembly, various concentrations of LEDGF were added, followed by target DNA and incubation at 37°C (data not shown). The results suggest that even under significantly altered assay conditions, LEDGF interactions with assembled synaptic complexes were similar to those previously observed (Fig. 2A). In particular, the disruption of full-site integration occurred only at stoichiometric ratios of LEDGF to IN of >1.
Interactions of LEDGF with synaptic complexes containing DNA with natural blunt or 3′-OH recessed termini.
Processing of the natural HIV-1 DNA blunt ends by IN channels the reaction to concerted or full-site integration (26). Synaptic complexes were assembled using a 1.6-kbp donor (0.52 nM) containing a single U5 blunt end and IN (10 nM) at 14°C in 125 mM NaCl (Fig. 3). LEDGF at 2.5 nM and 5 nM stimulated production of the full-site integration products ∼2-fold (Fig. 3A, lanes 4 and 5) in comparison to its absence (Fig. 3A, lanes 3 and 10). Titration experiments established that the optimum stimulation (∼2-fold) of full-site integration required ∼3 nM LEDGF at 100 mM NaCl in the assembly mixtures (Table 1 and data not shown). Restriction analysis of the full-site products using the 1.6-kbp blunt-ended donor and DNA sequence analysis of the virus-host junctions verified that the full-site integration reaction had occurred (26). Sequence analysis of the donor-target junctions verified that the HIV-1 5-bp host site duplications were present with a fidelity of 76% for U5-U5 products (47 plasmid clones) and 88% for U5-U3 products (9 plasmid clones). The other recombinants possessed small deletions at the target site.
FIG. 3.
LEDGF stimulates HIV-1 synaptic complexes containing a donor with a natural U5 blunt end. (A) Synaptic complexes were assembled with a 1.6-kbp substrate containing a single U5 blunt end (0.52 nM) and IN (10 nM) in 125 mM NaCl at 14°C. LEDGF was added at the indicated concentrations (top), and the mixture was incubated for an additional 15 min. Strand transfer was initiated by the addition of a target and incubation for 2 h at 37°C. The DNA products are indicated on the left. Lane 1 contained Promega 1-kbp markers, and lane 2 contained donor only. Two controls with IN only (lanes 3 and 10) incorporated 5% and 1% of the donor into full-site and CHS products, respectively. The percentages of the donor incorporated into full-site products at 2.5 and 5 nM LEDGF (lanes 4 and 5) were 11% and 8%, respectively. Three similar independent experiments averaged ∼2.3-fold increase of full-site products at these LEDGF concentrations. A 3.2-kbp donor-donor product twice the size of the input donor was also produced (Fig. 1). The donor-donor products in lanes 4 and 5 were 6% and 2%, respectively. (B) Conditions identical to those for panel A were employed, except that IN and LEDGF were preincubated together prior to the addition of the donor. The experiments in panels A and B were performed simultaneously.
TABLE 1.
Summary of LEDGF stimulation of HIV-1 IN-mediated full-site integration
| Donor type | Stimulationa (n-fold) | P valueb | Molar ratioc (LEDGF-IN) |
|---|---|---|---|
| Preprocessed donors | |||
| 4.1-kbp 5′-TA (U5-U3) | 1.33 ± 0.15 | <0.001 | 0.6 |
| 1.5-kbp 5′-AC (U5) | 1.26 ± 0.07 | <0.03 | 0.6 |
| Blunt-ended donor | |||
| 1.6-kbp DNA 5′-AC (U5) | 2.35 ± 0.32 | <0.001 | 1.0 |
LEDGF stimulated HIV-1 full-site integration with different preprocessed 3′-OH recessed and blunt-ended substrates.
P values for stimulation by LEDGF were determined using and analysis of variance one-factor test.
The LEDGF concentration was determined as a monomer, while IN was a tetramer.
Higher concentrations of LEDGF prevented assembly of the synaptic complexes in the assembly buffer (Fig. 3A, lanes 6 to 9) with little observable effect on CHS production. Prior binding of LEDGF to IN before the addition of the 1.6-kbp blunt-ended donor almost completely prevented assembly of synaptic complexes, even at 5 nM LEDGF (Fig. 3B). Similar results were obtained using both order-of-addition schemes with LEDGF if a blunt-ended donor containing U3 LTR sequences was used (data not shown). The results suggest that LEDGF at low nM concentrations enhances the assembly of synaptic complexes with blunt ends and subsequent full-site integration activity. However, at higher concentrations, LEDGF efficiently prevented further assembly of synaptic complexes with the natural blunt-ended substrate.
The processing of DNA ends by IN in HIV-1 PIC occurs soon after reverse transcription and prior to transport of the PIC across the nuclear envelope (33). We tested HIV-1 synaptic complexes containing a 1.5-kbp donor possessing 3′-OH recessed ends with a natural 5′-AC overhang for interactions with LEDGF. The donor (0.42 nM) was assembled with IN (10 nM) at 14°C in 125 mM NaCl (Fig. 4). Titration of LEDGF demonstrated that there was slight stimulation (1.3-fold) of full-site integration activity (Fig. 4A, lanes 4 to 6), while full-site activity started to decrease at greater than 10 nM LEDGF in the assembly buffer (Fig. 4A, lanes 7 to 10, and B). Higher concentrations of LEDGF are required to inhibit full-site activity with a 3′-OH recessed ended (Fig. 4B) than with blunt-ended donors (Fig. 3). The results indicate that low concentrations of LEDGF modestly stimulated full-site integration, while higher concentrations of LEDGF prevented assembly of the synaptic complexes (see Fig. 10 for a model).
FIG. 4.
High concentrations of LEDGF disrupt synaptic complex formation in a quantitative manner in to IN-DNA complexes mediating CHS integration. (A) Synaptic complexes were assembled with a 1.5-kbp DNA substrate (0.52 nM) containing a single U5 end with a 5′-AC overhang and IN (10 nM) in 125 mM NaCl at 14°C. Lane 1 contained Promega 1-kbp markers, and lane 2 contained donor only. The amounts of the DNA fragment in lane 2 migrating below the 3-kbp marker in lane 1 varied between experiments (Fig. 3 and Fig. 6) and were apparently due to nonspecific interactions of the donor overhangs upon electrophoresis. LEDGF was added at the indicated final concentrations (top), and the mixture was incubated for an additional 15 min. Strand transfer was initiated by adding a target and incubation for 2 h at 37°C. The products are indicated on the left. (B) Graphic representation of the percentages of the donor incorporated into full-site, CHS, and 3.0-kbp donor-donor products.
FIG. 10.
A model for LEDGF interactions with the HIV-1 synaptic complex and STC. From left to right, HIV-1 IN is assembled with a viral DNA donor to form a synaptic complex. For simplicity, the two DNA donors have only one LTR end displayed. IN is represented as a dimer, while LEDGF is a monomer. At a low molar ratio of LEDGF to an IN tetramer in a synaptic complex, LEDGF modestly enhances the production of synaptic complexes that produce full-site integration products. For host site selection in vivo, a specific low stoichiometry between LEDGF and IN is implied (29). At a high molar ratio of LEDGF to IN in solution, LEDGF prevents the assembly of synaptic complexes. The STC is stable upon challenge by a high LEDGF concentration. The association and dissociation constants for LEDGF binding to IN within the synaptic complex in vitro are unknown but appear to be in the low nM range.
To further investigate the effect of LEDGF on full-site integration activity using the 1.6-kbp U5 blunt-ended donor, we executed another order-of-addition experiment in which LEDGF (50 nM) (Fig. 3) was or was not added at different times after initiation of strand transfer at 37°C. Addition of LEDGF at 30-, 45-, and 60-min time points after initiation of strand transfer had a minimal effect on the total amount of full-site product produced in 2 h at 37°C in comparison to the appropriate controls without LEDGF (data not shown). Addition of LEDGF at 0 and 15 min had a significant effect, similar to preincubation with LEDGF (Fig. 3). The results suggest that although LEDGF prevents assembly of the synaptic complex, the STC (Fig. 1) formed later in the reaction is apparently resistant to disruption by LEDGF (see Fig. 8).
FIG. 8.
STC are resistant to challenge by LEDGF. (A) Synaptic complexes were assembled with a 1.6-kbp substrate containing a single U5 blunt end (0.52 nM) and IN (15 nM) at 14°C. Samples were incubated at 37°C for 2 h after a target was added to enable the formation of STC. The samples were treated with 30 mM EDTA. In one set of samples, 50 nM LEDGF was added (+), and the samples were further incubated at 14°C for the indicated times. As controls in another set, there was no LEDGF added (−). The samples were subjected to electrophoresis on a native 0.7% agarose gel for 14 h. The STC and CHS are marked on the left. (B) A portion of the above-mentioned samples were deproteinized and subjected to electrophoresis on 0.7% agarose. The full-site (FS), CHS, and donor-donor products are marked on the right. (C) An aliquot from untreated STC (lane 4) was subjected to two-dimensional gel analysis as described in the legend to Fig. 7A. (D) An aliquot from LEDGF-treated sample (lane 5) was also subjected to two-dimensional gel analysis.
A subset of the donor-donor integration products respond to LEDGF in the same way as full-site products resulting from synaptic complexes.
Presumably, juxtaposition of two donor molecules is required prior to the presentation of target to produce the full-site product (Fig. 1). Assembly of RSV and avian myeloblastosis virus IN synaptic complexes occurs with two DNA donors containing 3′-OH recessed ends at 14°C without target present (1). Intriguingly, the production of HIV-1 3.2-kbp donor-donor products from two 1.6-kbp donors containing blunt ends (Fig. 3A, lanes 3 to 5; Fig. 5) and of 3-kbp donor-donor products from two 1.5-kbp donors with 5′-AC overhangs (Fig. 4 and 6) paralleled the production of full-site products, because each was affected by LEDGF. Approximately 60% of the complexes responsible for producing the donor-donor products with the 1.5-kbp 5′-AC donor were eliminated by LEDGF (Fig. 4B and Fig. 6), and they were completely eliminated by it with the 1.6-kbp DNA donor with blunt ends (Fig. 3 and Fig. 5) prior to strand transfer. Restriction and DNA sequence analyses of a 9.4-kbp donor-donor product from two 4.7-kbp substrates with blunt ends suggested that end joining occurs preferentially near the LTR ends (11). Similar results were obtained with restriction analysis of the 3.2-kbp donor-donor products obtained with the 1.6-kbp donor (data not shown). In contrast, the Y-type donor-donor products (Fig. 1) (11, 21, 39) migrating between 3 kbp and 4 kbp were not attenuated by LEDGF (Fig. 4 to 6). The modest stimulation and inhibition of the specific-size donor-donor integration products parallels that of the full-site products, suggesting that LEDGF modulates interactions between IN subunits bound at two LTR ends, but not with CHS and Y-type products containing one LTR end.
FIG. 5.
LEDGF-IBD affects full-site integration similarly to full-length LEDGF. Synaptic complexes were assembled with a 1.6-kbp substrate containing a single U5 blunt-ended donor (0.52 nM) and IN (10 nM) in 50 mM NaCl at 14°C. IBD at the indicated final concentrations (top) was added, and the mixture was incubated for an additional 15 min. Strand transfer was initiated by adding a target and incubation for 2 h at 37°C. Lanes 1 and 2 contained Promega 1-kbp markers and donor, respectively. Two controls with IN only (lanes 3 and 10) incorporated 16%, 7%, and 7% of the donor into full-site, CHS, and 3.2-kb donor-donor products, respectively. At 10 nM IBD (lane 5), IN incorporated 21%, 9%, and 9% of the donor, respectively, into these same products.
FIG. 6.
LEDGF is more effective than LEDGF-IBD for disrupting synaptic complexes. (A) IN (10 nM) and IBD at the indicated concentrations were incubated together for 15 min in 125 mM NaCl at 14°C. Synaptic complexes were formed with the addition of the 1.5-kbp 5′-AC donor (0.52 nM). Strand transfer was initiated by adding a target and incubation for 2 h at 37°C. Lane 1 contained Promega 1-kbp markers, and lane 2 contained donor only. Two controls with IN only (lanes 3 and 10) incorporated 8%, 8%, and 8% of the donor into full-site, CHS, and 3.0-kbp donor-donor products, respectively. At 75 nM IBD (lane 9), the same products were 4%, 10%, and 3%, respectively. The products are indicated on the left. (B) The assembly and assay conditions were the same as for panel A, except that LEDGF was used. IN incorporated 8%, 11%, and 8% of the donor into full-site, CHS, and 3.0-kbp donor-donor products, respectively (lane 3).
Interactions of isolated LEDGF-IBD with synaptic complexes are similar to those of full-length LEDGF.
LEDGF is a modular protein that interacts with HIV-1 IN via a small alpha-helical domain, IBD (residues 347 to 429), located within its C-terminal region (8, 10, 44). In addition to the IBD, the full-length protein contains other structural and functional elements, such as an N-terminal Pro-Trp-Trp-Pro domain, a nuclear localization signal, and a pair of putative AT hooks (8, 31, 41, 44). The IBD of LEDGF specifically interacts with the CCD and likely the N-terminal domain of HIV-1 IN (4, 7, 10, 31). Unlike full-length protein, IBD does not bind DNA in vitro (41). The IBD was essential but not sufficient to stimulate HIV-1 IN activity in uncoupled strand transfer assays (8).
Synaptic complexes were assembled with a 1.6-kbp DNA (0.42 nM) containing a single U5 blunt end and IN (10 nM) at 50 mM NaCl prior to the addition of a LEDGF fragment containing the IBD (residues 347 to 471) and further incubation at 14°C (Fig. 5). The NaCl concentration was lowered to 50 mM NaCl instead of 125 mM NaCl (Fig. 3 and 4) to help mediate more efficient 3′-OH processing of the blunt-ended substrate (26). Slight stimulation was observed for full-site integration and the specific 3.2-kbp donor-donor products up to 10 nM IBD. Thereafter, the full-site and the 3.2-kbp donor-donor integration reactions were essentially inhibited, while the CHS integration was unaffected. This result indicates that local allosteric and/or steric effects of the IN-LEDGF-IBD interactions are sufficient to prevent assembly of the synaptic complex.
Synaptic complexes containing a 5′-AC overhang display increased resistance to the inhibitory effects of LEDGF and LEDGF-IBD.
We wanted to determine if LEDGF and IBD binding to IN prior to assembly of the synaptic complex with the 5′-AC overhang donor was as effective as LEDGF binding to IN using the 4.1-kbp donor with a 5′TA overhang (Fig. 2B) or the 1.6-kbp blunt-ended donor (Fig. 3B). Interestingly, there was no significant inhibition of any strand transfer products by IBD at low concentrations when it was bound to IN prior to the addition of a preprocessed 1.5-kbp 5′-AC overhang donor (Fig. 6A, lanes 4 to 6). Approximately 50% inhibition of full-site products was observed only at higher IBD concentrations (Fig. 6A, lanes 8 and 9). Likewise, preincubation of LEDGF with IN prior to addition of the 5′-AC donor resulted in no inhibition, and only at higher concentrations of LEDGF was inhibition observed (Fig. 6B, lanes 8 to 10). The results suggest that (i) catalytic activities of HIV-1 IN are not affected by engagement of the LEDGF-binding site on the IN CCD per se; (ii) the association of IN with the 5′-AC donor end in synaptic complexes is higher than that observed with either the 5′-TA donor end (Fig. 2) or the natural blunt-ended DNA (Fig. 3), thus resulting in more efficient assembly and/or increased stability of the synaptic complexes; and (iii) LEDGF is more effective than IBD in preventing assembly of the synaptic complexes at higher nM concentrations (Fig. 6).
LEDGF does not disrupt the STC.
The time-dependent formation of the STC (Fig. 1) occurs through different stages that result in a stable IN-DNA complex containing the full-site integration product (27). The STC is stable upon electrophoresis in a 0.8% agarose gel with 1 M urea and is resistant to treatment with either 0.6 M NaCl, 0.1% NP-40, or 10 μg/ml heparin (27). We determined that the STC could be identified on 0.7% native agarose gels (Fig. 7A) and that it is resistant to disruption by LEDGF (Fig. 8). To produce the STC, IN (15 nM) was assembled with the 1.6-kbp U5 blunt-ended donor at 14°C for 15 min, followed by the addition of target and further incubation for 2 h at 37°C. Aliquots of the sample were subjected to 0.7% native agarose gel electrophoresis at 4°C (Fig. 7A, lane 3). Following separation of the complexes on the native gel (Fig. 7A, lane 3), the native gel was treated with SDS and was subjected to two-dimensional gel electrophoresis (Fig. 7A, right). The STC and CHS on the native gel gave rise to the predicted strand transfer products, as previously demonstrated by two-dimensional gel analysis (27). From phosphorimager analysis, the STC (Fig. 7A, lane 3) contained only full-site product with minor amounts of CHS and free donor (Fig. 7A, right). The two slower-migrating bands (Fig. 7A, lane 3) each contained ∼80% full-site product and other minor products, all of which were apparently looped together by IN in a nonspecific fashion (11, 21). The smeared products above the STC (Fig. 7A, lane 3) contained mostly full-site and 3.2-kbp donor-donor/Y-type products (Fig. 7A, right). The unknown higher-molecular-weight product probably represents integration of another donor into the full-site product or other DNA structures. A sample was also deproteinized and analyzed on 0.7% agarose to quantify the integration products (Fig. 7A, gel positioned below the two-dimensional gel). The percentages of the donor incorporated into full-site, CHS, and 3.2-kbp donor-donor/Y-type products were 19%, 6%, and 9%, respectively. Lastly, nearly equivalent quantities of STC containing the full-site product were evident on a native 0.7% agarose gel in comparison to the same sample on a gel containing 1 M urea, as determined by phosphorimager analysis (Fig. 7B). The data demonstrate that STC can be identified on native agarose gels even in the presence of nonspecific DNA binding by IN.
FIG. 7.
Formation of STC and their analysis by native agarose and two-dimensional gel electrophoresis. (A) STC were formed for 2 h at 37°C and treated with 20 mM EDTA, and the samples were subjected to electrophoresis on native 0.7% agarose gels in TBE buffer at 4°C for 16.5 h. STC were formed with IN at 15 nM (lane 3). STC and CHS are labeled on the left. A 1-kbp ladder and control DNA without IN were in lanes 1 and 2, respectively. The dots next to the native gel indicate that the slower-migrating bands contained full-site and other products, as well as free donor. The arrow indicates the direction of electrophoresis. (Right) An aliquot of the sample shown in lane 3 was subjected to electrophoresis on another native 0.7% agarose gel. After SDS treatment, the gel was run in the second dimension, as indicated at the top with an arrow. The 0.7% agarose gels for the native and deproteinized samples are positioned on the left and bottom of the two-dimensional gel, respectively. The full-site (FS), CHS, and 3.2-kbp donor-donor/Y-type structures and free donor are indicated on the deproteinized gel located at the bottom. The unknown larger-size product probably represents another integration event into full-site and/or CHS products. (B) STC were formed and subjected to 0.7% agarose gel electrophoresis without (−) and with (+) 1 M urea in the gel.
From our earlier results, we conclude that LEDGF can stimulate the assembly process at low nM concentrations, while at high concentrations, it prevents assembly of the synaptic complex. In preliminary studies, addition of 50 nM LEDGF to the assembly mixture (Fig. 3, lane 9) was found to essentially inhibit all full-site integration activity with the U5 blunt-ended donor, as well as detection of the STC in 0.7% native agarose gels (data not shown). Next, we determined that preformed STC are resistant to disruption by LEDGF. STC were formed with IN (15 nM) for 2 h at 37°C, followed by the addition of EDTA to prevent further catalysis. These samples were then subjected to challenge by LEDGF. In one set of samples, 50 nM LEDGF was added and incubated for different times at 14°C (Fig. 8A, lanes 5, 7, and 9). In the parallel set, samples were maintained at 14°C in the absence of LEDGF (Fig. 8A, lanes 4, 6, and 8). The samples were run on a native 0.7% agarose gel without (Fig. 8A) or with (Fig. 8B) deproteinization. From phosphorimager data, the STC and the two slower-migrating bands (Fig. 8A, lanes 5, 7, and 9) that contained full-site products (Fig. 7A, right panel) were essentially stable (∼85%) on native gels upon LEDGF challenge in comparison to their control lanes without LEDGF (Fig. 8A, lanes 4, 6, and 8). LEDGF also bound to the DNA in the STC, CHS, 3.2-kbp donor-donor, and free donor, because all these bands on the native agarose gel were shifted slightly upward (Fig. 8, lanes 5, 7, and 9). In all of the samples, the percentages of the donor incorporated into full-site and 3.2-kbp donor-donor products were essentially equivalent (30% and 15%, respectively) (Fig. 8B, lanes 3 to 9). Two-dimensional studies of the 15-min samples not treated (Fig. 8C) or treated (Fig. 8D) with LEDGF on the native gel (Fig. 8A, lanes 4 and 5, respectively) demonstrated that LEDGF did not modify the amounts of full-site, donor-donor, and other products in comparison to the untreated control, as defined by phosphorimager analysis. Two-dimensional analysis of the 30-min samples produced similar results (data not shown). The results suggest that LEDGF does not significantly disrupt the STC.
LEDGF does not modulate RSV IN-mediated full-site integration.
LEDGF binding appears to be specific to lentiviral IN (4, 30). Concordantly, a titration of LEDGF did not specifically affect recombinant RSV IN activities in vitro. There was no stimulation of CHS or full-site integration (Fig. 9). These results indicate that a direct interaction between LEDGF and HIV-1 IN is essential for the observed modest stimulation and disruption of IN-IN interactions necessary for full-site integration.
FIG. 9.
Titration of LEDGF with RSV IN. Recombinant RSV IN (5 nM) was assembled with a 3.4-kbp donor (0.21 nM) that contained a single wt U3 LTR end for 30 min on ice. LEDGF was added at the final concentration (top) for an additional 10 min. Supercoiled target was added, and the samples were incubated at 37°C for 20 min. The assay conditions were previously described (48), with the final NaCl concentration at 0.2 M. Lanes 1 and 10 were without LEDGF. The percentages of the donor incorporated into FS and CHS products in these two lanes averaged 31% and 3%, respectively. With increasing concentrations of LEDGF (lanes 2 to 9), the percentages of the donor incorporated into FS and CHS products averaged 33% and 2.6%, respectively. Part of the sample in lane 9 was lost.
DISCUSSION
LEDGF differentially modulates synaptic complexes capable of full-site integration and IN-DNA complexes producing CHS products in vitro.
LEDGF binds tightly and specifically to HIV-1 IN, primarily recognizing the CCD of the viral enzyme in the absence of viral DNA (7, 10, 32). Reconstituted synaptic complexes (26, 38, 39, 47, 48) proceed through a series of nucleoprotein complexes throughout the pathway to full-site integration (27). Through the monitoring of specific integration products, we found that the strand transfer activities of synaptic complexes are modulated, depending upon the structure and the concentration of LEDGF. In one order-of-addition scheme, IN was bound to viral DNA at 14°C prior to the addition of LEDGF. At low molar ratios of LEDGF to IN (<1), there was a modest stimulation of full-site product formation, presumably due to a facilitation of the assembly process that results in an ∼2- to 3-fold increase in the number of active synaptic complexes formed. At higher molar ratios (>1), LEDGF effected a concentration dependent attenuation of full-site integration. When present at high concentrations in solution, LEDGF disrupted IN-IN interactions necessary for assembly of synaptic complexes, resulting in a quantitative conversion to CHS products. LEDGF challenge experiments showed that the preformed STC is resistant to disruption (Fig. 10).
Correlations with integration studies in vivo.
In vivo knockdown studies have produced conflicting results regarding the role of LEDGF in HIV-1 replication. The depletion of LEDGF by RNAi appears to have little effect on virus replication, possibly because an estimated ∼2.5 × 104 copies of LEDGF persist per cell in these depletion experiments (28, 42). Estimation of the number of LEDGF copies in control HeLa-P4 cells is ∼4 × 105. In contrast, another LEDGF depletion study suggests a two- to fourfold reduction of HIV-1 replication, with the defect occurring at the integration step (43). Inhibition of HIV-1 integration (74%) in macrophages depleted of LEDGF (70 to 80%) suggests a modest involvement of LEDGF in integration (52). Minute levels of chromatin-associated LEDGF appear sufficient for HIV-1 integration (29). Recent knockout studies of LEDGF in mouse embryo fibroblasts (LEDGF−/−) infected by HIV-1 resulted in ∼5- to 10-fold reductions in virus titer, which, in a preliminary experiment, was found to directly correlate with a 5-fold reduction in PIC activity in vitro compared to results obtained with virus-infected control cells (N. Vandegraaff and A. Engelman, personal communication). The results suggest that the concentration and location of LEDGF in cells, in an undefined manner, modulates HIV-1 integration in vivo.
Our control reactions without LEDGF may represent PIC in cells devoid of LEDGF or salt-stripped PIC (42). The addition of LEDGF to synaptic complexes at low molar ratios of LEDGF to IN (<1) stimulated (two- to threefold) the assembly of synaptic complexes (Fig. 2 to 6) that mediated full-site integration (Table 1). The effects observed with LEDGF at low nM concentrations in our studies corroborate the above-mentioned knockout and knockdown studies of LEDGF in vivo. Possibly, this modest stimulation of full-site integration by LEDGF is due to its ability to promote the tethering of synaptic complexes to target DNA (15, 29, 30, 32, 44) and/or increase the binding affinity of HIV-1 IN to DNA (4). Importantly, the assembly of synaptic complexes (Fig. 2 to 6) proceeds normally in the presence of low concentrations of LEDGF, suggesting that, as in HIV-1 PIC which contain LEDGF (30), the juxtaposition of the viral ends is not hindered by this protein. The actual numbers of molecules of LEDGF in wt and depleted PIC in vivo are unknown. Integration activity in salt-stripped PICs is stimulated ∼40%, ∼6%, and ∼32% by the addition of LEDGF, IBD, and a LEDGF mutant (D366N), respectively, relative to the starting PIC activity (42). The stimulation of full-site integration observed at low concentrations of LEDGF correlates with these PIC reconstitution studies, implying a functional role for LEDGF in retrovirus integration.
Additional compelling evidence for the involvement of LEDGF in HIV-1 integration comes from knockdown studies of LEDGF that demonstrate a redistribution of integration target sites upon depletion of LEDGF in the infected cells (13). Although statistically significant, the redistribution of integration sites was not absolute, leading to ∼5 to 20% reduction of integration in transcription units, depending on the cell type, and ∼25% reduction of integration events into genes responsive to LEDGF (13). The partial redistribution of integration sites from transcription units to CG regions may suggest that overall integration is not always tightly controlled by LEDGF or by other cellular factors (13). These data suggest that IN is possibly involved in target site selection in cells that may proceed, in some cases, independently of cellular proteins (13, 34, 37). Synaptic complexes are fully functional in the absence of LEGDF (Fig. 2 to 8) (27, 38, 39), possibly supporting the observation that LEDGF has a modest involvement in HIV-1 integration in macrophages (52) and the partial redistribution of integration sites (13). Because LEDGF at low concentrations promotes legitimate full-site integration, it would be of interest to determine, under appropriate assay conditions, if LEDGF affects target site selection in vitro, thus extending the above-mentioned in vivo studies. It will also be interesting to determine if the fidelity of the HIV-1 5-bp host site duplications observed in vitro, which occur at ∼75% (18, 26, 38, 39), is significantly improved at low nM concentrations of LEDGF with several of the donor substrates used in this study. This will further show that low concentrations of LEDGF impart stability to the synaptic complex and, by analogy, to the PIC.
What are the concentration and structure requirements for LEDGF-mediated disruption of IN-IN interactions necessary for assembly of synaptic complexes and maintenance of STC?
The PIC traverses the cytoplasm and is transported into the nucleus, suggesting that it possesses structural stability. Synaptic complexes formed in vitro with avian IN are stable (1, 48), particularly when the HIV-1 synaptic complex is associated with a target, forming the STC (27). The assembly and stability of these structures is due to yet-unidentified IN-IN interactions that are required for these two macromolecular structures.
We performed order-of-addition experiments with LEDGF (Fig. 1) to investigate how it affects the assembly of the synaptic complex. Preincubation of LEDGF with HIV-1 IN prior to the addition of a donor possessing a 5′-TA overhang completely prevented the formation of synaptic complexes, while IN-DNA complexes producing CHS products were significantly stimulated with increasing concentrations of LEDGF (Fig. 2B). This steady increase in CHS products occurred in the near-complete absence of any synaptic complexes (Fig. 2B, lanes 4 to 9), suggesting LEDGF assists the binding of IN to single DNA ends (4). These results stand in contrast to the analogous situation in which the donor possesses a natural 5′-AC overhang (Fig. 6B). The synaptic complexes with the 5′-AC overhang donor induced stronger IN-IN interactions required for assembly compared to the synaptic complexes with a 5′-TA overhang (Fig. 2B). A recent report showed that the 5′ C is involved in a hydrogen bond with the Gln-148 of IN, stabilizing the conformation of the flexible loop adjacent to the active site of IN (24). Our results suggest that the configuration of IN associated with the natural 3′-OH recessed ends may be optimized for interacting with LEDGF.
In a different order-of-addition scheme, IN was assembled with viral DNA at 14°C prior to the addition of LEDGF. The tendency of LEDGF to more readily prevent the assembly of synaptic complexes using the blunt-ended 5′-AC donor substrate (Fig. 3) than using the 5′-AC overhang substrate (Fig. 4) further suggests that the IN-IN interactions associated with the latter substrate contribute to the stable structure of the IN-DNA complex found in the cytoplasmic PIC containing 3′-OH recessed ends. Titration experiments showed that LEDGF up to 100 nM does not inhibit 3′-OH processing of native blunt-ended donor by IN at 10 nM (data not shown). Upon assembly of IN with the 5′-AC overhang substrate prior to the addition of LEDGF, facilitation of assembly of synaptic complexes proceeds at molar ratios of LEDGF to IN of <1 (Fig. 4). In contrast with our experimental conditions, in which synaptic complexes were in equilibrium with high concentrations of LEDGF in solution (LEDGF/IN ratio, >1), PICs are exposed to the restrictions imposed by the complex cellular environment, where only minute amounts of LEDGF located on chromatin may interact with the PIC (29).
What are the structural requirements for LEDGF for both the modest stimulation of full-site products and assembly of the synaptic complex? The IBD of LEDGF specifically interacts with the CCD of IN (7, 10), but this specific interface is likely to represent only a portion of the total contacts between LEDGF and IN (32). Slight stimulation of full-site activity was observed with the IBD, while disruption of this activity was similar to LEDGF but less effective (Fig. 5 and 6). Even though the IBD fragment and LEDGF containing the D366N mutation do not interact with IN in pull-down experiments (10), a high concentration of full-length LEDGF containing this mutation appears to behave similarly to wt LEDGF with the native blunt-ended and 5′-AC overhang substrates (data not shown). However, unlike LEDGF, it does not modestly stimulate full-site integration, as reported earlier for PIC (42). Disruption of full-site integration by isolated IBD, full-length wt LEDGF, and its mutant D366N occurred essentially at the same molar ratio of each respective protein to IN. These results suggest that the interactions of LEDGF with IN in the context of synaptic complexes is extensive in disrupting subunit interactions necessary for assembly. However, preformed STC were essentially resistant to LEDGF at 50 nM (Fig. 8), which at that concentration completely prevented full-site integration when added before assembly of the synaptic complex (Fig. 2 to 6). It is unknown whether the resistance of STC to challenge by LEDGF is due to a lack of interaction with the CCD or other sites on IN subunits within the STC.
Acknowledgments
This work was supported by National Institutes of Allergy and Infectious Diseases grant AI31334 to D.P.G.
We thank Peter Cherepanov for his generous supply of wt and mutant LEDGF proteins and plasmids and for his discussions and comments on the manuscript. We thank Jacob Zahm for his comments and several experiments and Sibes Bera for producing the U5 and U3 LTR donor substrates that contained the 5′-AC overhang. We also thank Nickolas Vandergraaff and Alan Engelman for sharing results prior to publication.
Footnotes
Published ahead of print on 31 January 2007.
REFERENCES
- 1.Bera, S., A. C. Vora, R. Chiu, T. Heyduk, and D. P. Grandgenett. 2005. Synaptic complex formation of two retrovirus DNA attachment sites by integrase: a fluorescence energy transfer study. Biochemistry 44:15106-15114. [DOI] [PubMed] [Google Scholar]
- 2.Brown, P. O. 1997. Integration, p. 161-203. In J. M. Coffin, S. Hughes, and H. Varmus (ed.), Retroviruses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 3.Brown, P. O., B. Bowerman, H. E. Varmus, and J. M. Bishop. 1987. Correct integration of retroviral DNA in vitro. Cell 49:347-356. [DOI] [PubMed] [Google Scholar]
- 4.Busschots, K., J. Vercammen, S. Emiliani, R. Benarous, Y. Engelborghs, F. Christ, and Z. Debyser. 2005. The interaction of LEDGF/p75 with integrase is lentivirus-specific and promotes DNA binding. J. Biol. Chem. 280:17841-17847. [DOI] [PubMed] [Google Scholar]
- 5.Chen, H., and A. Engelman. 2001. Asymmetric processing of human immunodeficiency virus type 1 cDNA in vivo: implications for functional end coupling during the chemical steps of DNA transposition. Mol. Cell. Biol. 21:6758-6767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen, H., S. Q. Wei, and A. Engelman. 1999. Multiple integrase functions are required to form the native structure of the human immunodeficiency virus type I intasome. J. Biol. Chem. 274:17358-17364. [DOI] [PubMed] [Google Scholar]
- 7.Cherepanov, P., A. L. Ambrosio, S. Rahman, T. Ellenberger, and A. Engelman. 2005. Structural basis for the recognition between HIV-1 integrase and transcriptional coactivator p75. Proc. Natl. Acad. Sci. USA 102:17308-17313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cherepanov, P., E. Devroe, P. A. Silver, and A. Engelman. 2004. Identification of an evolutionarily conserved domain in human lens epithelium-derived growth factor/transcriptional co-activator p75 (LEDGF/p75) that binds HIV-1 integrase. J. Biol. Chem. 279:48883-48892. [DOI] [PubMed] [Google Scholar]
- 9.Cherepanov, P., G. Maertens, P. Proost, B. Devreese, J. Van Beeumen, Y. Engelborghs, E. De Clercq, and Z. Debyser. 2003. HIV-1 integrase forms stable tetramers and associates with LEDGF/p75 protein in human cells. J. Biol. Chem. 278:372-381. [DOI] [PubMed] [Google Scholar]
- 10.Cherepanov, P., Z. Y. Sun, S. Rahman, G. Maertens, G. Wagner, and A. Engelman. 2005. Solution structure of the HIV-1 integrase-binding domain in LEDGF/p75. Nat. Struct. Mol. Biol. 12:526-532. [DOI] [PubMed] [Google Scholar]
- 11.Cherepanov, P., D. Surratt, J. Toelen, W. Pluymers, J. Griffith, E. De Clercq, and Z. Debyser. 1999. Activity of recombinant HIV-1 integrase on mini-HIV DNA. Nucleic Acids Res. 27:2202-2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chiu, R., and D. P. Grandgenett. 2003. Molecular and genetic determinants of Rous sarcoma virus integrase for concerted DNA integration. J. Virol. 77:6482-6492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ciuffi, A., M. Llano, E. Poeschla, C. Hoffmann, J. Leipzig, P. Shinn, J. R. Ecker, and F. Bushman. 2005. A role for LEDGF/p75 in targeting HIV DNA integration. Nat. Med. 11:1287-1289. [DOI] [PubMed] [Google Scholar]
- 14.Crise, B., Y. Li, C. Yuan, D. R. Morcock, D. Whitby, D. J. Munroe, L. O. Arthur, and X. Wu. 2005. Simian immunodeficiency virus integration preference is similar to that of human immunodeficiency virus type 1. J. Virol. 79:12199-12204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Emiliani, S., A. Mousnier, K. Busschots, M. Maroun, B. Van Maele, D. Tempe, L. Vandekerckhove, F. Moisant, L. Ben-Slama, M. Witvrouw, F. Christ, J. C. Rain, C. Dargemont, Z. Debyser, and R. Benarous. 2005. Integrase mutants defective for interaction with LEDGF/p75 are impaired in chromosome tethering and HIV-1 replication. J. Biol. Chem. 280:25517-25523. [DOI] [PubMed] [Google Scholar]
- 16.Engelman, A. 2005. The ups and downs of gene expression and retroviral DNA integration. Proc. Natl. Acad. Sci. USA 102:1275-1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ge, H., Y. Si, and R. G. Roeder. 1998. Isolation of cDNAs encoding novel transcription coactivators p52 and p75 reveals an alternate regulatory mechanism of transcriptional activation. EMBO J. 17:6723-6729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goodarzi, G., R. Chiu, K. Brackmann, K. Kohn, Y. Pommier, and D. P. Grandgenett. 1997. Host site selection for concerted integration by human immunodeficiency virus type-1 virions in vitro. Virology 231:210-217. [DOI] [PubMed] [Google Scholar]
- 19.Goodarzi, G., G. J. Im, K. Brackmann, and D. Grandgenett. 1995. Concerted integration of retrovirus-like DNA by human immunodeficiency virus type 1 integrase. J. Virol. 69:6090-6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goodarzi, G., M. Pursley, P. Felock, M. Witmer, D. Hazuda, K. Brackmann, and D. Grandgenett. 1999. Efficiency and fidelity of full-site integration reactions using recombinant simian immunodeficiency virus integrase. J. Virol. 73:8104-8111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grandgenett, D. P., R. B. Inman, A. C. Vora, and M. L. Fitzgerald. 1993. Comparison of DNA binding and integration half-site selection by avian myeloblastosis virus integrase. J. Virol. 67:2628-2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hazuda, D. J., P. Felock, M. Witmer, A. Wolfe, K. Stillmock, J. A. Grobler, A. Espeseth, L. Gabryelski, W. Schleif, C. Blau, and M. D. Miller. 2000. Inhibitors of strand transfer that prevent integration and inhibit HIV-1 replication in cells. Science 287:646-650. [DOI] [PubMed] [Google Scholar]
- 23.Hindmarsh, P., T. Ridky, R. Reeves, M. Andrake, A. M. Skalka, and J. Leis. 1999. HMG protein family members stimulate human immunodeficiency virus type 1 and avian sarcoma virus concerted DNA integration in vitro. J. Virol. 73:2994-3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johnson, A. A., W. Santos, G. C. Pais, C. Marchand, R. Amin, T. R. Burke, Jr., G. Verdine, and Y. Pommier. 2006. HIV-1 integration requires the viral DNA end (5′-C) interaction with glutamine 148 of the integrase flexible loop. J. Biol. Chem. 281:461-467. [DOI] [PubMed] [Google Scholar]
- 25.Lewinski, M. K., and F. D. Bushman. 2005. Retroviral DNA integration—mechanism and consequences. Adv. Genet. 55:147-181. [DOI] [PubMed] [Google Scholar]
- 26.Li, M., and R. Craigie. 2005. Processing of viral DNA ends channels the HIV-1 integration reaction to concerted integration. J. Biol. Chem. 280:29334-29339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li, M., M. Mizuuchi, T. R. Burke, Jr., and R. Craigie. 2006. Retroviral DNA integration: reaction pathway and critical intermediates. EMBO J. 25:1295-1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Llano, M., S. Delgado, M. Vanegas, and E. M. Poeschla. 2004. Lens epithelium-derived growth factor/p75 prevents proteasomal degradation of HIV-1 integrase. J. Biol. Chem. 279:55570-55577. [DOI] [PubMed] [Google Scholar]
- 29.Llano, M., D. T. Saenz, A. Meehan, P. Wongthida, M. Peretz, W. H. Walker, W. Teo, and E. M. Poeschla. 2006. An essential role for LEDGF/p75 in HIV integration. Science 314:461-464. [DOI] [PubMed] [Google Scholar]
- 30.Llano, M., M. Vanegas, O. Fregoso, D. Saenz, S. Chung, M. Peretz, and E. M. Poeschla. 2004. LEDGF/p75 determines cellular trafficking of diverse lentiviral but not murine oncoretroviral integrase proteins and is a component of functional lentiviral preintegration complexes. J. Virol. 78:9524-9537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maertens, G., P. Cherepanov, Z. Debyser, Y. Engelborghs, and A. Engelman. 2004. Identification and characterization of a functional nuclear localization signal in the HIV-1 integrase interactor LEDGF/p75. J. Biol. Chem. 279:33421-33429. [DOI] [PubMed] [Google Scholar]
- 32.Maertens, G., P. Cherepanov, W. Pluymers, K. Busschots, E. De Clercq, Z. Debyser, and Y. Engelborghs. 2003. LEDGF/p75 is essential for nuclear and chromosomal targeting of HIV-1 integrase in human cells. J. Biol. Chem. 278:33528-33539. [DOI] [PubMed] [Google Scholar]
- 33.Miller, M. D., C. M. Farnet, and F. D. Bushman. 1997. Human immunodeficiency virus type 1 preintegration complexes: studies of organization and composition. J. Virol. 71:5382-5390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mitchell, R. S., B. F. Beitzel, A. R. Schroder, P. Shinn, H. Chen, C. C. Berry, J. R. Ecker, and F. D. Bushman. 2004. Retroviral DNA integration: ASLV, HIV, and MLV show distinct target site preferences. PLoS Biol. 2:e234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moreau, K., C. Faure, G. Verdier, and C. Ronfort. 2002. Analysis of conserved and non-conserved amino acids critical for ALSV (Avian leukemia and sarcoma viruses) integrase functions in vitro. Arch. Virol. 147:1761-1778. [DOI] [PubMed] [Google Scholar]
- 36.Narezkina, A., K. D. Taganov, S. Litwin, R. Stoyanova, J. Hayashi, C. Seeger, A. M. Skalka, and R. A. Katz. 2004. Genome-wide analyses of avian sarcoma virus integration sites. J. Virol. 78:11656-11663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schroder, A. R., P. Shinn, H. Chen, C. Berry, J. R. Ecker, and F. Bushman. 2002. HIV-1 integration in the human genome favors active genes and local hotspots. Cell 110:521-529. [DOI] [PubMed] [Google Scholar]
- 38.Sinha, S., and D. Grandgenett. 2005. Recombinant HIV-1 integrase exhibits a capacity for full-site integration in vitro that is comparable to that of purified preintegration complexes from virus-infected cells. J. Virol. 79:8208-8216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sinha, S., M. H. Pursley, and D. P. Grandgenett. 2002. Efficient concerted integration by recombinant human immunodeficiency virus type 1 integrase without cellular or viral cofactors. J. Virol. 76:3105-3113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Turlure, F., E. Devroe, P. A. Silver, and A. Engelman. 2004. Human cell proteins and human immunodeficiency virus DNA integration. Front. Biosci. 9:3187-3208. [DOI] [PubMed] [Google Scholar]
- 41.Turlure, F., G. Maertens, S. Rahman, P. Cherepanov, and A. Engelman. 2006. A tripartite DNA-binding element, comprised of the nuclear localization signal and two AT-hook motifs, mediates the association of LEDGF/p75 with chromatin in vivo. Nucleic Acids Res. 34:1663-1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vandegraaff, N., E. Devroe, F. Turlure, P. A. Silver, and A. Engelman. 2006. Biochemical and genetic analyses of integrase-interacting proteins lens epithelium-derived growth factor (LEDGF)/p75 and hepatoma-derived growth factor related protein 2 (HRP2) in preintegration complex function and HIV-1 replication. Virology 80:415-426. [DOI] [PubMed] [Google Scholar]
- 43.Vandekerckhove, L., F. Christ, B. Van Maele, J. De Rijck, R. Gijsbers, C. Van den Haute, M. Witvrouw, and Z. Debyser. 2006. Transient and stable knockdown of the integrase cofactor LEDGF/p75 reveals its role in the replication cycle of human immunodeficiency virus. J. Virol. 80:1886-1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vanegas, M., M. Llano, S. Delgado, D. Thompson, M. Peretz, and E. Poeschla. 2005. Identification of the LEDGF/p75 HIV-1 integrase-interaction domain and NLS reveals NLS-independent chromatin tethering. J. Cell Sci. 118:1733-1743. [DOI] [PubMed] [Google Scholar]
- 45.Van Maele, B., and Z. Debyser. 2005. HIV-1 integration: an interplay between HIV-1 integrase, cellular and viral proteins. AIDS Rev. 7:26-43. [PubMed] [Google Scholar]
- 46.Verma, I. M., and M. D. Weitzman. 2005. Gene therapy: twenty-first century medicine. Annu. Rev. Biochem. 74:711-738. [DOI] [PubMed] [Google Scholar]
- 47.Vora, A., S. Bera, and D. Grandgenett. 2004. Structural organization of avian retrovirus integrase in assembled intasomes mediating full-site integration. J. Biol. Chem. 279:18670-18678. [DOI] [PubMed] [Google Scholar]
- 48.Vora, A., and D. P. Grandgenett. 2001. DNase protection analysis of retrovirus integrase at the viral DNA ends for full-site integration in vitro. J. Virol. 75:3556-3567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vora, A. C., and D. P. Grandgenett. 1995. Assembly and catalytic properties of retrovirus integrase-DNA complexes capable of efficiently performing concerted integration. J. Virol. 69:7483-7488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wei, S. Q., K. Mizuuchi, and R. Craigie. 1997. A large nucleoprotein assembly at the ends of the viral DNA mediates retroviral DNA integration. EMBO J. 16:7511-7520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wu, X., Y. Li, B. Crise, and S. M. Burgess. 2003. Transcription start regions in the human genome are favored targets for MLV integration. Science 300:1749-1751. [DOI] [PubMed] [Google Scholar]
- 52.Zielske, S. P., and M. Stevenson. 2006. Modest but reproducible inhibition of human immunodeficiency virus type 1 infection in macrophages following LEDGFp75 silencing. J. Virol. 80:7275-7280. [DOI] [PMC free article] [PubMed] [Google Scholar]