Abstract
The effect of Epstein-Barr virus (EBV) SM protein on EBV gene expression was examined using a recombinant EBV strain with the SM gene deleted and DNA microarrays representing all known EBV coding regions. Induction of lytic EBV replication in the absence of SM led to expression of approximately 40% of EBV genes, but a block in expression of over 50% of EBV genes. Contrary to previous findings, several early genes were SM dependent, and lytic EBV DNA replication did not occur in the absence of SM. Notably, two genes essential for lytic EBV DNA replication, BSLF1 and BALF5, encoding EBV DNA primase and polymerase, respectively, were SM dependent. Lytic DNA replication was partially rescued by ectopic expression of EBV primase and polymerase, but virion production was not. Rescue of DNA replication only enhanced expression of a subset of late genes, consistent with a direct requirement for SM for late gene expression in addition to its contribution to DNA replication. Therefore, while SM is essential for most late gene expression, the proximate block to virion production by the EBV SM deletion strain is an inability to replicate linear DNA. The block to DNA replication combined with the direct effect of SM on late gene expression leads to a global deficiency of late gene expression. SM also inhibited BHRF1 expression during productive replication in comparison to that of cells induced into lytic replication in the absence of SM. Thus, SM plays a role in multiple steps of lytic cycle EBV gene expression and that it is transcript-specific in both activation and repression functions.
Each human herpesvirus expresses an RNA binding protein during the lytic phase of replication that regulates viral RNA accumulation by transcriptional or posttranscriptional mechanisms. This family of proteins is represented by SM in Epstein-Barr virus (EBV), ICP27 in herpes simplex virus (HSV), ORF4 in varicella-zoster virus (VZV), UL69 in human cytomegalovirus (hCMV), and ORF57 in Kaposi's sarcoma-associated herpesvirus (KSHV) (25). The corresponding protein in EBV, HSV, VZV, and KSHV has been shown to be essential for virus production (8, 15, 17, 48). Several of these proteins have multiple mechanisms of action in regulating both viral and cellular gene expression. VZV ORF4, HSV ICP27, and hCMV UL69 have been reported to affect transcription (23, 38, 57). ICP27, UL69, ORF57, and SM regulate viral mRNA export and stability (3, 16, 22, 29, 47, 49). ICP27 and SM also appear to affect host cell mRNA splicing (18, 29, 46, 50). However, the reason that these various proteins are essential for productive virus replication is not well understood.
A major function for this family of proteins is likely to be their role in facilitating export of intronless viral transcripts. Spliced transcripts can gain access to the major cellular pathway for export of mRNAs by the formation of protein complexes near the splice junction (for review, see reference 42). These exon junction complexes (EJCs) include several proteins which facilitate association with TAP, a central mediator of nuclear export. Intronless transcripts, unless they contain constitutive transport elements that directly bind cellular export proteins, are relatively inefficiently exported. ICP27, SM, and ORF57 are capable of forming complexes with Ref/Aly, a component of the EJC, although the nature of the association may vary among the different viral proteins (5, 6, 30). CMV UL69 forms complexes with UAP56, another EJC component, and ICP27 may also directly associate with TAP (5, 29). EBV SM, HSV ICP27, KSHV ORF57, and hCMV UL69 proteins also are capable of nucleocytoplasmic shuttling, as evinced by in vitro heterokaryon shuttling assays (3, 5, 7, 11, 28, 49, 51, 54). It has been proposed, therefore, that viral regulatory proteins such as EBV SM serve to compensate for inefficient export of unspliced viral transcripts. By binding to viral mRNAs and to cellular export proteins simultaneously, SM family members could provide access for unspliced transcripts, which comprise the majority of herpesviral lytic cycle RNAs, to cellular nuclear export pathways (27).
Several aspects of SM, ORF57, and ICP27 function remain to be explained by this model. The first is the apparent specificity of action of several of these proteins. In reporter assays, SM and ORF57 demonstrate target specificity, enhancing expression of both heterologous and viral transcripts with various efficiencies (26, 45, 47, 51). Although RNA binding in vivo and in vitro has been demonstrated, no stringent sequence-specific binding has been established for any of these proteins (21, 45). In one study, in which a yeast three-hybrid assay was used to demonstrate RNA-protein interactions, short RNAs corresponding to 17 specific HSV mRNAs of all kinetic classes were identified as binding ICP27 with greater affinity than other HSV mRNAs (53). These RNAs did not contain a specific ICP27-binding element in common, but based on their sequences, it was suggested that higher-order structures consisting of G-rich sequences may bind ICP27 with greater affinity. Additionally, in vivo binding of specific transcripts by SM did not correlate with the ability of SM to enhance their accumulation (45).
A second aspect of the function of these proteins is their effect on splicing. ICP27 acts as an inhibitor of host cell splicing (4, 18). ICP27 inhibits essential splicing cofactor activity by inhibiting their phosphorylation and may do so by functionally sequestering an SR protein kinase (50). SM also decreases expression of spliced reporter genes at the posttranscriptional level, although the mechanism of this effect has not been established (46, 47).
The gene specificity (or lack thereof) and multifunctional nature of this family of proteins make a priori prediction of their effects on viral and cellular gene expression difficult. Pinpointing the role of these proteins on the tightly regulated patterns of gene expression during lytic replication is therefore also challenging. A global analysis of ICP27 function has been performed by comparing the patterns of HSV gene expression by oligonucleotide array and revealed that ICP27 has positive and negative effects on both cellular and viral genes (55). An examination of several EBV genes in a recombinant EBV strain with the SM gene deleted indicated that expression of some unspliced EBV genes was not enhanced by SM, whereas expression of others was (2). One conclusion of this study was that SM is essential for expression of at least two late genes (coding for VCA and gp350) that would be required for encapsidation and infection. In the present study, a DNA array representing all identifiable EBV open reading frames (ORFs) was used to examine the global effects of SM on EBV gene expression and to identify those steps in the viral replicative cycle where SM may play an essential role.
MATERIALS AND METHODS
Cell lines, plasmids, and transfections.
SM-KO cells are HEK293 cells carrying an EBV bacmid with the SM gene deleted expressing green fluorescent protein (GFP) (15). SM-KO cells were maintained as monolayer cultures in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum (Gibco). SM-KO transfections were performed with TransIT293 reagent (Mirus) according to the manufacturer's protocol. All transfection mixtures were adjusted to contain equal amounts of DNA by addition of pCDNA3 vector plasmid (Invitrogen) as required. SM, BALF4, and BZLF1 (Z) expression vectors have been previously described (34, 47). EBV BALF5 and BSLF1 expression vectors were constructed by PCR amplification of the respective ORFs from B95-8 DNA and cloned into pCDNA3. B95-8 is a permissive lymphoblastoid cell line derived from EBV-infected marmoset B lymphocytes (33). Raji is an EBV-positive human Burkitt's lymphoma cell line (41). Both B95-8 and Raji cells were grown in RPMI 1640 containing 10% fetal bovine serum (Gibco).
Gardella gel analysis.
Cells were harvested 60 h after transfection, washed, resuspended in loading dye (15% Ficoll, 1× Tris-borate-EDTA, 200 μg/μl RNAse A), and 2 × 106 live cells were loaded into each well of a lysing gel containing 2 mg/ml of pronase E, 2% sodium dodecyl sulfate, and 0.8% agarose in Tris-borate-EDTA. Cells were overlaid with 40 μl of lysis buffer (1 mg/ml pronase E, 1% sodium dodecyl sulfate, 5% Ficoll). Electrophoresis was performed at 30 V at 4°C for 6.5 h, and the gel was further electrophoresed at 100 V for 17.5 h to separate the episomal and linear viral DNAs. The gel was then analyzed by Southern blotting with a radiolabeled BamHI W fragment of EBV DNA.
Virus and cell DNA preparation.
SM-KO cells were transfected with BALF4, BZLF1, BSLF1, BALF5, and SM expression vectors or pCDNA3 in various combinations as indicated in each experiment. At 4 days posttransfection, 12 × 106 SM-KO cells were centrifuged and washed, and total cell DNA was isolated using DNAzol (Molecular Research Center, Inc.) as per the manufacturer's protocol. EBV virion DNA was prepared by filtration of the cell culture supernatant followed by centrifugation to pellet virus. The viral pellet was digested with DNase I, followed by protease K digestion. Virion DNA was extracted with phenol and purified by passage over a Sephadex G-50 column.
Quantitative PCR.
Real-time PCR was performed using an ABI prism 7900 sequence detection system (Applied Biosystems). PCR was performed in triplicate and carried out in a total of 20 μl of PCR mixture containing equal amounts of template DNA, 10 μl of 2× Taqman Universal PCR master mix, and 1 μl of 20× PCR mix (Assays-by-Design). The following BamHI W-specific primers and probe were designed and used for measurement of EBV DNA: BamHI W forward primer (5′-CCCTGCTCCTCTCCAACCT-3′; nucleotides 22461 to 22479), BamHI W reverse primer (5′-GGCTGGCCTGGTGGAC-3′; nucleotides 28661 to 28676), and BamHI W Taqman probe (Fam-5′-CTCCACCCTAGACCCC-3′; nucleotides 19411 to 19426). Amplification of human β-actin was used as an internal normalization control. The primers used for BMRF1 real-time PCR were described previously (20).
RNA preparation and analysis.
RNA was isolated from cells at 0, 6, 12, 24, and 48 h posttransfection. Cells were washed and lysed in RNA-bee (Tel-Tech, Inc.) and isolated using QIAGEN RNeasy mini columns as per the manufacturer's protocol. For Northern blotting, 5 μg of RNA was electrophoresed in a denaturing formaldehyde-agarose gel, transferred to charged nylon membrane, and hybridized to probe labeled with 32P. Gene-specific probes were generated by gel purification of fragments excised from gene expression plasmids.
Immunofluorescence microscopy and immunoblotting.
SM-KO cells were plated 2 days prior to transfection on glass coverslips for immunofluorescence microscopy. Forty-eight hours after transfection, SM-KO cells were washed and fixed in ice-cold 100% methanol for 10 min, air dried, and stored at −20°C. Fixed cells were blocked in 20% goat serum and stained with anti-gp350 antibody (BMA17.3) (56) and Alexa Fluor 594-conjugated anti-mouse immunoglobulin G antibodies.
Immunoblotting was performed with monoclonal antibody to BHRF1 protein (5BΔ11) (37) and horseradish peroxidase-conjugated secondary antibody, followed by chemiluminescence detection (Pierce).
Virus passage.
Supernatant was harvested 4 days after transfection, cleared by centrifugation, and filtered through a 0.45-μm cellulose acetate membrane prior to incubation with an equal volume of Raji cells at a final concentration of 2.5 × 105 cells/ml. Cells were examined for GFP expression by microscopy 2 days after infection.
EBV microarray construction.
Microarrays containing DNA fragments representing all EBV translational ORFs were generated by the Functional Genomics Laboratory of the Hartwell Center for Bioinformatics and Biotechnology at St. Jude Children's Research Hospital. EBV DNA fragments containing the 3′ 280 to 860 bp of each ORF of the EBV genome (B95-8 isolate) (1) were amplified from viral genomic DNA or cDNA by PCR and then cloned into the plasmid vector pCR2.1-TOPO (Invitrogen, Carlsbad, CA). DNAs from the EBNA-2 and EBNA-3A, -3B, and -3C ORFs of the type 2 EBV isolate AG876 were also included on the array, as the genes encoding these proteins differ significantly in DNA sequence from their counterparts in the genomes of type 1 EBV isolates (e.g., B95-8) (9, 10, 39). DNA fragments representing ORFs LF1, LF2, and LF3 (36), present within the 12.5-kbp region deleted from the B95-8 EBV genome, were amplified from EBV genomic DNA isolated from either the Akata (LF1 and LF2) or AG876 (LF3) Burkitt's lymphoma cell lines. A list of the EBV DNA fragments present on the microarray and their genomic coordinates is available (see the supplemental material). For each DNA clone, four separate PCRs were performed to amplify the EBV DNA inserts, which were then purified using the QIAGEN Qiaquick PCR purification kit according to the manufacturer's instructions. For quality control, an aliquot of DNA from each PCR was subjected to agarose gel electrophoresis, and its nucleotide sequence was verified. For printing of arrays, purified PCR products were resuspended in 20 μl of 3× SSC (1× SSC is 15 mM sodium citrate plus 150 mM NaCl) and spotted a total of 32 times per clone onto poly-l-lysine-coated glass slides using an Omnigrid 100 (Genomic Solutions, Huntington, United Kingdom) with 16 CMP4 pins (Telechem International, Sunnyvale, CA). After printing, slides were processed and blocked according to the protocol for postprocessing of arrays at http:\\derisilab.ucsf.edu/.
Microarray analysis of EBV gene expression.
RNA to be subjected to microarray analysis was purified as described above. Microarray-based detection of EBV gene expression, including RNA quality control, labeling with Cy3 and Cy5 dye, hybridization, washing, scanning, and data analysis were performed by the Functional Genomics Laboratory at St. Jude Children's Research Hospital. Briefly, total cellular RNA (5 μg per sample) was amplified with T7 RNA polymerase, followed by a second round of amplification with 2 μg of product from the first amplification, using the Ambion (Austin, TX) Message Amp II aRNA Amplification v.0506 kit according to the manufacturer's protocol. Dye labeling was performed with 5 μg amplified RNA using the method for “reverse transcription and aminoallyl labeling of RNA” (M004.10) at http://www.hartwellcenter.org/bio_services/fungen/cDNA.php#PROTOCOLS. Hybridization and washing procedures were performed according to array hybridization protocol M005.7 (http://www.hartwellcenter.org/bio_services/fungen/cDNA.php#PROTOCOLS).
Microarray data analysis.
The hybridized slides were scanned using an Axon (Axon Corp., Union City, CA) 4000B dual-channel scanner to generate a multi-TIFF image, and the images were analyzed by using Axon GenePix 6.0 image analysis software. For each spot that was flagged as meeting the qualitative spot criterion by the image software and having a signal-to-noise ratio of greater than 1.5 in at least one channel, the ratio was calculated from the background subtracted median signal of the two channels. To remove the intensity-specific bias, the global lowess normalization was then performed within Spotfire DecisionSite for Microarray Analysis (version 8.2.1; Spotfire, Somerville, MA). The normalized ratio was further scaled by the ratio of the β-actin probe on the array. The results of the probe replicates were combined by taking the mean of the log ratios. Any probe that had less than 25% replicates passing the quality control steps was excluded. The final ratios representing the relative gene expression changes were used to construct a hierarchical clustering map within Spotfire using the unweighted average clustering method and the correlation similarity measure.
RESULTS
EBV gene expression during lytic replication induced in the presence and absence of SM.
293 cells infected with a recombinant SM deletion strain (SM-KO) were induced to permit lytic replication by transfection of a BZLF1 expression vector (Z). Parallel transfections were performed with Z and an SM expression vector (SM) or with vector (C) alone. RNA was isolated from all sets of cells 24 and 48 h after transfection. Four different array comparisons were performed: between RNA from cells transfected with (i) C versus Z at 24 h, (ii) C versus Z at 48 h, (iii) Z versus Z plus SM at 24 h, and (iv) Z versus Z plus SM at 48 h. Each RNA sample was dye labeled, and pairs of RNA samples were hybridized to EBV microarrays as described in Materials and Methods. The results are represented as a cluster analysis in Fig. 1A. Several findings regarding SM effects of EBV gene expression were immediately apparent.
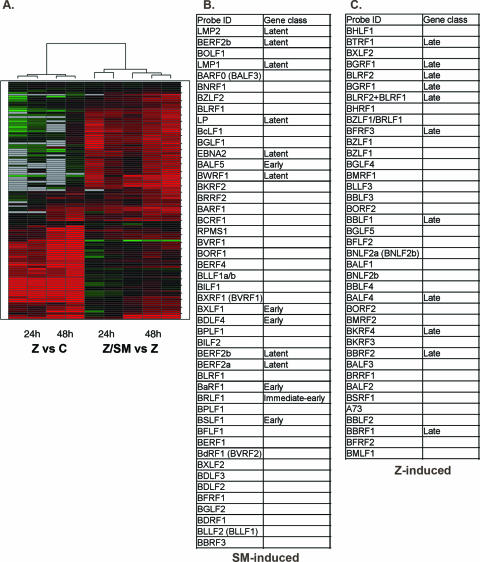
FIG. 1.
Cluster analysis of EBV gene expression after induction of lytic replicative cycle in the presence and absence of SM. (A) Results of simultaneous hybridization to arrays using RNAs from SM-KO cells transfected with different expression plasmids are shown graphically. RNAs were harvested from cells at 0, 6, 12, 24, 48, and 60 h posttransfection. Two RNAs were hybridized to each array to compare global EBV gene expression in the presence or absence of a particular EBV gene. The transfected DNAs representing vector (C), Z, or Z plus SM (Z/SM) and the time of harvest of each RNA are shown below the map. The effect of Z expression on EBV gene expression compared to that of empty vector is therefore represented by the left side of the cluster map. Similarly, the effect of SM plus Z compared to that of Z alone is shown on the right. Two or three independent transfections were performed for each time point. Genes whose expression is increased are in red, those not detectably changed are in black, and those whose expression is decreased are in green. The changes (fold) and other array data for each gene are provided (see Fig. S1 in the supplemental material). (B) SM-dependent genes. Genes whose expression was significantly increased by transfection of SM plus Z compared to transfection of Z alone and whose expression was not significantly increased by Z compared to vector are listed. The gene class of each transcript where clearly established is also listed. All genes not listed as latent or early were either late transcripts or transcripts whose kinetic class has not been formally demonstrated. ORFs in parentheses represent genes that cannot be distinguished by the probe. (C) SM-independent genes. Genes whose expression was significantly increased by transfection of Z alone and whose expression was not significantly increased by transfection of SM plus Z are listed. All genes not listed as late were early lytic transcripts or transcripts whose kinetic class has not been formally demonstrated.
First, expression of approximately 50% of the ORFs was SM dependent. Approximately 30% (in the top right quadrant of the cluster map) were not induced by transfection of Z alone to any significant degree at either 24 or 48 h (see Z versus C, Fig. 1). However, in the presence of SM, expression of these genes was induced at both 24 and 48 h. This group of genes is shown in Fig. 1B. Of the SM-dependent genes, the majority (33/47) consist of late lytic cycle genes. Interestingly, the latency-associated EBNA2, EBNA-LP, EBNA3B, EBNA3C, LMP1, and LMP2 transcripts were also upregulated, consistent with the published finding that latent gene expression increases during lytic replication (58). In addition, five early lytic cycle genes were in the group of SM-dependent genes. These include genes coding for two essential components of the DNA replication complex, BSLF1 and BALF5, the EBV primase and DNA polymerase, respectively. Also, BDLF4, the conserved homologue of ORF31 in murine gammaherpesvirus 68 (MHV-68), which has been shown to be essential for MHV-68 lytic replication (24), was induced by SM. These findings raised the possibility that SM expression might be required for EBV DNA replication (see below).
Second, it was clear from the analysis that approximately 40% of the genes were upregulated by Z alone (Fig. 1C). As might be expected, these included the majority of the early genes. Somewhat surprisingly, at least 12 of the Z-induced genes were late genes, consistent with previous findings that Z expression can activate late promoters in the absence of DNA replication (12, 52). Examination of the heat map also revealed that some Z-induced genes displayed a temporal pattern of expression, with 12 genes being maximally induced by 24 h, whereas the remainder showed a further induction by Z at 48 h. Of the Z-induced genes, although most were not further induced by SM, 11 showed increased expression by 48 h in the presence of SM. This group, seen in the bottom right quadrant of the heat map, which included both late and early genes, can therefore be classified as SM responsive but not SM dependent, whereas the remainder of the Z-induced genes were completely SM independent. No latency-associated genes were significantly induced by Z alone at either time point.
Finally, several genes appeared to be down-regulated by SM compared to their expression in the presence of Z alone. Most of these were not consistently or strongly decreased by SM. However, BHRF1, the antiapoptotic bcl-2 homolog, was significantly less well expressed in the presence of SM (approximately 16% of the level expressed in the presence of Z alone), suggesting that SM may play a negative role in regulating the expression of BHRF1 during lytic EBV replication.
An intrinsic limitation of such an array, or an analysis with a quantitative PCR array, is that several sets of EBV lytic genes are coterminal and thus overlap at the 3′ portion of the transcripts. While probes can be designed that are unique for the 5′ transcript, it is not possible to design probes specific for the 3′ transcripts. Thus, if signals from all three probe sets are increased relative to their comparators, it is only possible to state that the most 5′ gene is induced. However, if the genes do not covary, it is possible to draw conclusions regarding the induction of individual genes. Of seven sets of lytic cycle genes known to have coterminal 3′ ends, three did not covary. For example, BFLF1, a late gene, was SM dependent, whereas BFLF2, an early gene, was SM independent. In two cases, two of the genes covaried, whereas the third behaved differently: BDLF4 and BGLF1 were apparently SM dependent, but BGLF2 was SM independent; and LMP1 was upregulated by SM, but BNLF2a and BNLF2b were not. In the remaining two sets, the genes covaried, so it was only possible to draw conclusions regarding the most 5′ gene. These were the BDLF1-3 transcripts and the BGLF4, BGLF5, and BBLF1 transcripts.
SM is required for lytic EBV DNA replication.
The SM dependence of two genes (BALF5 and BSLF1) known to be required for linear EBV DNA replication suggested that lytic cycle DNA replication might be strictly SM dependent. In order to compare production of linear EBV DNA molecules in the presence and absence of SM, Gardella gel analysis was performed on SM-KO cells transfected with either empty vector (C), Z, or Z plus SM. Forty-eight hours after transfection, live cells were loaded into wells of an in situ lysing gel and subjected to electrophoresis to separate linear and episomal EBV DNA molecules followed by Southern blotting to detect EBV DNA (Fig. 2). As expected, transfection with vector alone led to no production of linear EBV DNA. Transfection with Z also led to no significant increase in the amount of linear EBV DNA. In contrast, Z-plus-SM-transfected cells exhibited robust production of linear EBV DNA, comparable to that induced by tetradecanoyl phorbol acetate (TPA) treatment of EBV-infected B95-8 B cells. Thus, lytic EBV DNA replication is critically dependent on SM.
FIG. 2.
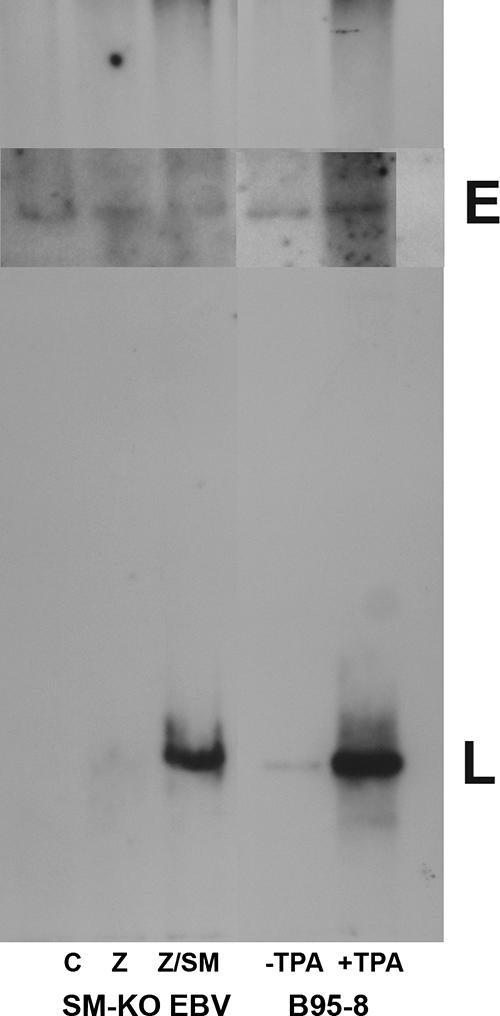
Lytic DNA replication is SM dependent. SM-KO EBV-infected 293 cells were transfected with either vector plasmid (C), Z, or Z plus SM (Z/SM) and analyzed by Gardella gel electrophoresis 48 h posttransfection. EBV DNA was detected by Southern blotting with radiolabeled BamHI W fragment of EBV DNA. Untreated B95-8 cells (−TPA) and B95-8 cells induced to replicate lytically by treatment with TPA (+TPA) were used as controls. L, linear genomes. In this experiment, longer exposure was required to visualize the episomal genome bands (inset E).
EBV primase and DNA polymerase partially rescue lytic EBV DNA replication.
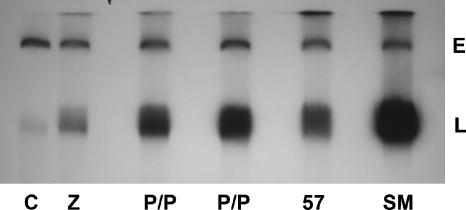
Since it is known that linear EBV DNA replication in vitro requires the viral primase (BSLF1) and DNA polymerase (BALF5), and expression of both was SM dependent, it was possible that the defect in EBV DNA replication in SM-KO EBV could be rescued by these genes. Alternatively, it was possible that additional gene products or SM itself would be required to rescue DNA replication. In order to test this hypothesis, Gardella gel analysis was performed on SM-KO cells transfected with Z plus either empty vector, BSLF1 plus BALF5, or SM. As shown in Fig. 3, BSLF1 and BALF5 increased replication over that achieved with Z alone, whereas SM increased replication further. Thus, although BSLF1 and BALF5 were capable of rescuing DNA replication, the level of DNA replication did not reach that achieved in cells transfected with SM. These data indicated that the defect in DNA replication, while at least partially due to a lack of DNA polymerase and primase expression, was also due to other effects of SM, potentially on other, nonessential genes involved in DNA replication. We have previously shown that KSHV ORF57, a homolog of EBV SM, cannot rescue virus production of SM-KO EBV. It was therefore of interest to determine whether ORF57 could rescue DNA replication. While ORF57 did enhance DNA replication slightly, it was less effective than BSLF1/BALF5, indicating that one reason for its inability to functionally substitute for EBV SM is an inability to efficiently enhance EBV DNA replication.
FIG. 3.
Lytic DNA replication is partially rescued by the EBV DNA primase and polymerase. SM-KO cells were transfected with empty vector (C), Z alone (Z), or Z plus either DNA primase and polymerase (P/P), KSHV ORF57 (56), or SM. Live cells were then analyzed by Gardella gel electrophoresis. The amounts of BALF5 and BSLF1 plasmid used for transfection were varied (lane 3, 0.2 μg of each plasmid; lane 4, 0.5 μg of each plasmid). The locations of episomal EBV DNA (E) and linear EBV DNA (L) within the gel are shown to the right.
The SM-associated defect in virus production cannot be rescued by enhanced lytic DNA replication.
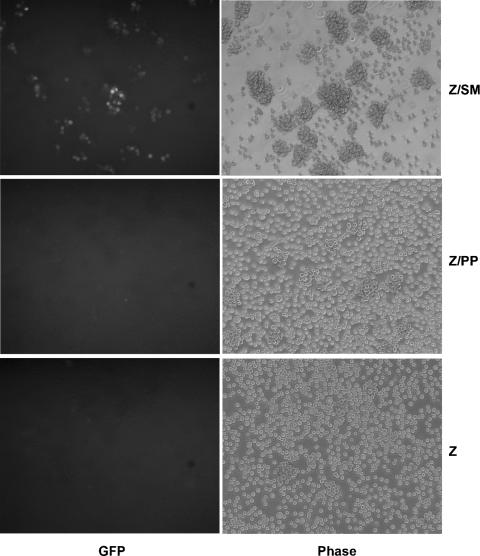
Since late gene expression is associated with DNA replication, it was possible that most or all SM late gene dependence could be indirectly due to the block in DNA replication in the absence of SM. Thus, it was possible that EBV virus production could be at least partially rescued by DNA replication alone. In order to test this possibility, we measured infectious virus production by SM-KO-infected cells rescued by transfection of BALF5 and BSLF1. SM-KO cells were transfected with Z plus either empty vector, BALF5 and BSLF1, or SM. Cell culture supernatants were harvested 72 h after transfection, filtered, and used to infect Raji cells. Forty-eight hours after infection, Raji cells were inspected by fluorescence microscopy to detect infected cells, which would be green due to GFP expression from the recombinant EBV. As expected, Raji cells exposed to supernatant from vector- or Z-transfected SM-KO cells expressed no detectable GFP, whereas infection with supernatant from Z-plus-SM-transfected cells yielded numerous green Raji cells in each field (Fig. 4). However, supernatant from BSLF1- and BALF5-transfected SM-KO cells did not yield detectable GFP expression in target Raji cells (Fig. 4). These data demonstrated that DNA replication alone is not sufficient to rescue the defects in gene expression in SM deletion recombinants that prevent productive EBV replication.
FIG. 4.
Infectious virus production cannot be rescued by DNA replication. SM-KO cells were transfected with Z and BALF4 plus either SM (Z/SM), BSLF1 plus BALF5 (Z/PP), or empty vector (Z/C). Cell culture supernatants were harvested 72 h posttransfection, filtered, and used to infect Raji cells, which were examined by fluorescence microscopy 48 h after infection. Cells visualized with UV light are shown to the left, and phase-contrast images of the same fields are shown to the right. The plasmids used to transfect the cells used as the source of each supernatant are shown to the right of each panel. GFP-positive cells are clearly evident in Raji cells infected with supernatant from Z-plus-SM-transfected cells (top left).
SM is required for production of extracellular virion particles.
Although it was likely that no EBV particles were produced by BALF5/BSLF1-transfected cells due to a lack of essential structural proteins, it was nevertheless possible that defective EBV particles were produced from the DNA replicated in the presence of BALF5/BSLF1 that were not detected by the Raji infection assay. In order to exclude this possibility, extracellular viral DNA was prepared and analyzed. Supernatant from SM-KO cells transfected with Z plus either empty vector, BALF5 and BSLF1, or SM was sequentially digested with DNase and protease K, followed by DNA purification. DNA preparations were then analyzed by quantitative PCR. As shown in Fig. 5, extracellular EBV DNA was detectable in supernatants from Z-plus-SM-transfected cells but not from cells transfected with either Z plus vector or Z plus BALF5/BSLF1. These data demonstrate that replicated EBV DNA is not processed into extracellular virions in the absence of SM, indicating that late gene expression dependent on SM is required for production of infectious virus.
FIG. 5.
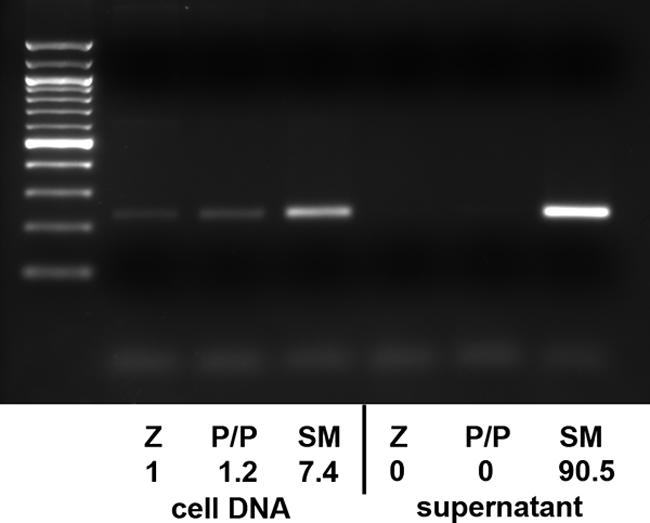
Detection of extracellular EBV virion DNA. SM-KO cells were transfected with Z alone (Z), Z plus BSLF1/BALF5 (P/P), or Z plus SM (SM). DNA was prepared from both cells and extracellular medium 72 h after transfection. Virion DNA was prepared from supernatant by DNase treatment followed by protease digestion prior to DNA isolation. PCR was performed with primers corresponding to the BamHI W fragment of EBV DNA, and products were analyzed by gel electrophoresis and staining with ethidium bromide. A 100-bp molecular size standard ladder is shown to the left. The same amount of DNA used for this analysis was also quantitated by quantitative PCR using a dye-labeled BamHI W probe. The relative amounts of EBV DNA as determined by the quantitative PCR are shown below each lane.
Essential late gene expression is dependent on SM.
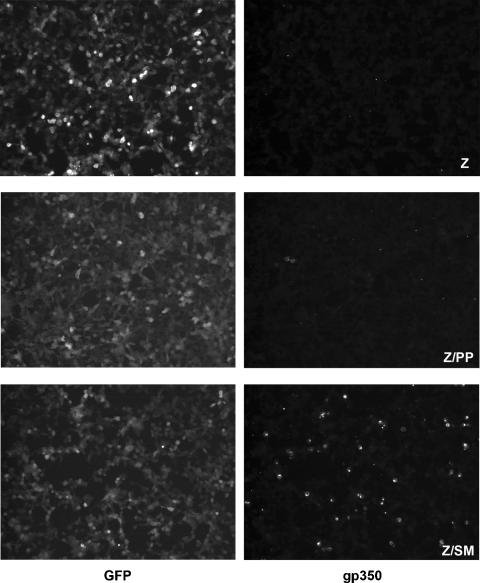
Despite the inability of DNA replication to rescue virion production, it was still possible that some late gene expression could be rescued by DNA replication alone in the absence of SM. Therefore, we examined the expression of a late gene whose product is essential for infectious virus production, the gp350 membrane antigen for which antibodies are readily available and which was identified as SM dependent in our array experiments and in a previous study (2). To determine whether gp350 was expressed when DNA replication occurred but in the absence of SM, we examined by immunofluorescence microscopy SM-KO cells transfected with either Z plus C, Z plus BALF5/BSLF1, or Z plus SM. Z-plus-SM-transfected cells demonstrated staining with gp350 antibodies in approximately 10% of cells, whereas none of the Z-plus-C or Z-plus-BALF5/BSLF1-transfected cells were positive (Fig. 6). These data demonstrate that efficient gp350 expression is indeed SM dependent and cannot be rescued by DNA replication alone.
FIG. 6.
Effect of SM on late gene expression. SM-KO cells were transfected with Z plus empty vector (Z), Z plus BALF5/BSLF1 (Z/PP), or Z plus SM (Z/SM). Cells were fixed and stained with anti-gp350 monoclonal antibody and Alexa Fluor-labeled secondary antibody prior to fluorescence microscopy (shown in the right panels and labeled gp350). All SM-KO cells are GFP positive, and corresponding GFP images are shown in the left panels (GFP). Expression of gp350 was only detected in the Z-plus-SM-transfected cells.
Late gene expression is dependent on both DNA replication and SM.
Since it was clear that gp350 expression was SM and not DNA replication dependent, in order to determine whether other late lytic cycle genes are similarly SM dependent, levels of gene expression between Z/BALF5/BSLF1-transfected cells and Z/SM-transfected cells were compared. The array comparisons were between RNAs from Z versus Z/BALF5/BSLF1 transfections and Z/BALF5/BSLF1 versus Z/SM transfections. Such an experiment is intrinsically limited by the fact that DNA replication is more robust when SM-KO virus is rescued with SM than with BALF5/BSLF1 (see above). Nevertheless, the analysis revealed that there is a subset of genes whose expression was not significantly increased by BALF5/BSLF1 alone but which was induced by SM (Fig. 7). Conversely, expression of approximately 12 late genes was significantly induced by BALF5/BSLF1 transfection alone. These data indicate that while there is an independent enhancing effect of DNA replication on late gene expression, efficient accumulation of the majority of late gene transcripts is SM dependent.
FIG. 7.
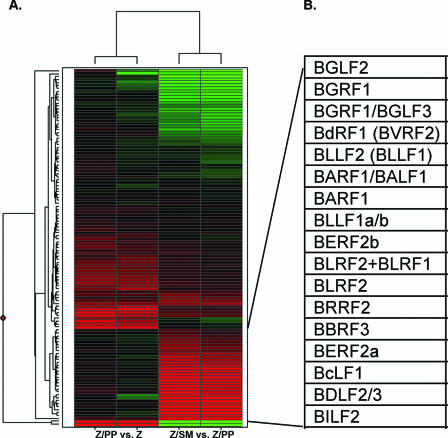
Comparison of levels of EBV gene expression after induction of lytic DNA replication in the presence and absence of SM. (A) Results of simultaneous hybridization to EBV arrays using RNAs from SM-KO cells transfected with different expression plasmids are shown graphically. Two RNAs were hybridized to each array to compare global EBV gene expression in the presence or absence of specific EBV genes. One set of arrays compared RNAs from Z-transfected SM-KO cells (Z) to RNAs from Z-plus-BALF5 and -BSLF1-transfected SMKO cells (Z/PP). The second set of arrays compared RNAs from Z-plus-SM-transfected cells (Z/SM) to RNAs from Z-plus-BALF5- and -BSLF1-transfected cells (Z/PP). The genes with increased expression are in red, those that did not detectably change are in black, and those with decreased expression are in green. The changes (fold) and other array data for each gene are provided (see Fig. S2 in the supplemental material). (B) Genes highly induced by Z/SM over those induced by Z and primase/polymerase. The genes comprising this group of SM-dependent and replication-independent (bottom right of the cluster map) genes are listed.
SM may affect expression of genes which are not strictly SM dependent.
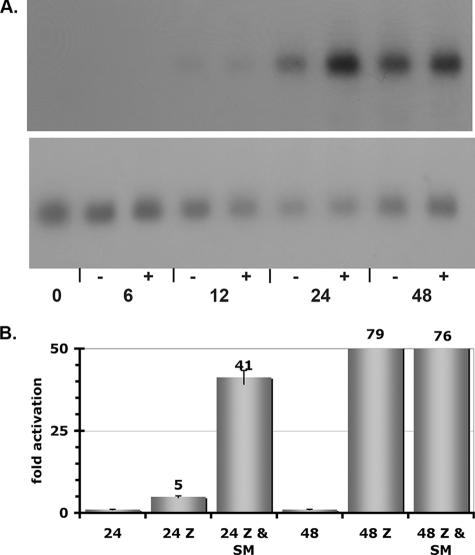
The gene encoding the EBV DNA processivity factor, BMRF1, has been used in several studies of SM function and behaves as a highly responsive SM target gene in transfection experiments (35). It was therefore somewhat surprising that in the context of EBV replication, BMRF1 was only slightly additionally induced by SM (over Z alone) in the array measurements. It was possible, however, that SM might have an effect on the kinetics of BMRF1 expression during EBV replication that was not evident in the array analyses. In order to assess this possibility, SM-KO cells were transfected with either Z or Z plus SM and RNA was harvested at various time points after transfection and analyzed by Northern blotting (Fig. 8). Z/SM-transfected cells expressed more BMRF1 at 24 h after transfection than Z-transfected cells, but this difference was negligible by 48 h when levels of BMRF1 accumulation were comparable in both sets of cells. To confirm this result, the experiment was repeated and quantitative real-time PCR was performed on RNA harvested at 24 and 48 h after transfection. The results, shown in Fig. 8B, confirm that SM-transfected cells express more BMRF1 RNA at the earlier time point. This experiment indicates that SM effects may include modulation of the temporal course of gene expression during lytic replication, which may also affect genes that are not absolutely SM dependent.
FIG. 8.
Effect of SM on the kinetics of BMRF1 expression. (A) SM-KO cells were transfected with either Z alone (−) or Z plus SM (+), and samples were harvested at the indicated time in hours posttransfection. RNA was isolated and analyzed by Northern blotting with BMRF1 probe (top panel). The blot was stripped and reprobed with GAPDH (glyceraldehyde-3-phosphate dehydrogenase) probe (bottom panel). (B) The ratio of BMRF1 RNA in the presence and absence of SM was determined by quantitative real-time PCR. Activation (fold) by Z or Z and SM over vector is shown, and the time of harvest is shown on the x axis.
SM may negatively regulate BHRF1 expression during lytic replication.
Although many genes were upregulated by SM, only one gene, BHRF1, was strongly down-regulated by SM during lytic replication, as revealed by our microarray analysis. Since the transcriptional and posttranscriptional regulation of BHRF1 is complex (37, 40), we used immunoblotting to ask whether SM affected BHRF1 protein levels during replication. As in previous experiments, SM-KO 293 cells were transfected with either C, Z, or Z plus SM, and protein lysates were prepared at several time points after transfection. Lysates were also made from B95-8 cells treated with TPA to induce EBV replication as a positive control and reference for the level of BHRF1 expression achieved in this standard model of EBV lytic infection. At each time point where BHRF1 expression was detectable, SM-transfected cells expressed less BHRF1 protein than cells transfected with Z alone, indicating that SM has a negative effect on BHRF1 expression (Fig. 9). In B95-8 cells treated with TPA, the levels of BHRF1 rose rapidly but stabilized after 24 h, whereas the levels continued to rise in SM-KO cells transfected with Z in the absence of SM, suggesting that SM may act to limit the levels of BHRF1 induced by Z during the course of lytic replication.
FIG. 9.
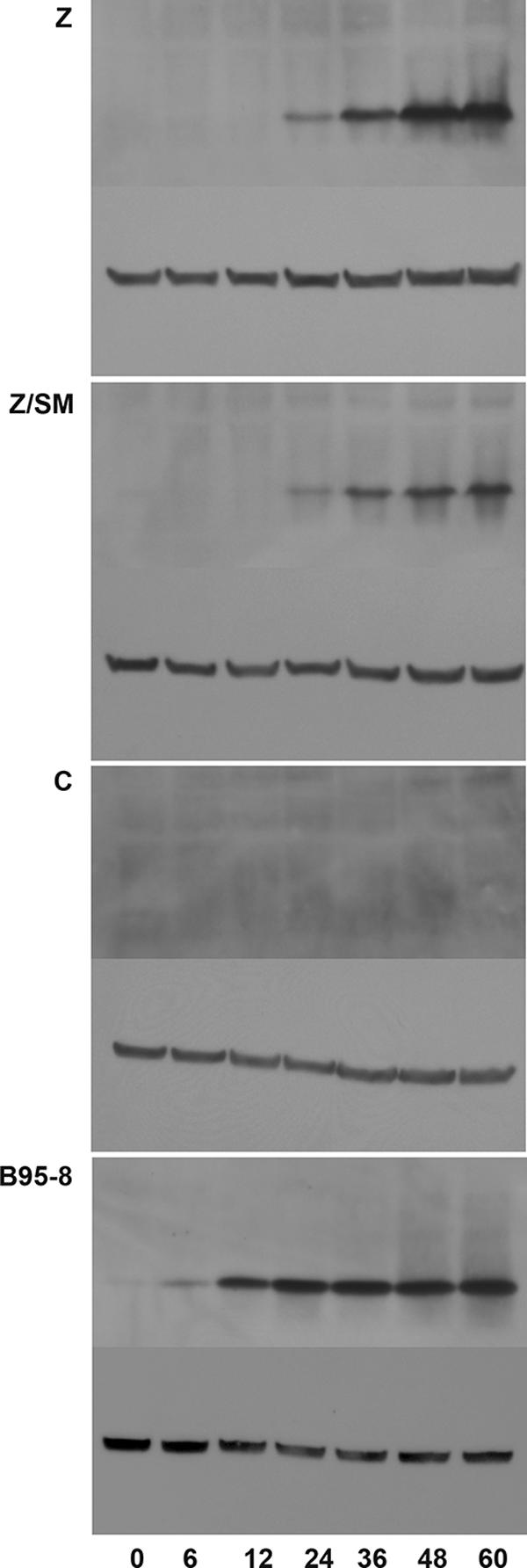
Effect of SM on BHRF1 expression. SM-KO cells were transfected with either empty vector (C), Z, or Z plus SM (Z/SM) and harvested at 0, 6, 12, 24, 48, and 60 h posttransfection (times shown at bottom). Protein lysates were analyzed by immunoblotting with anti-BHRF1 antibody. Each blot was stripped and reprobed with antiactin antibody as a loading control (shown below each panel). A time course of BHRF1 expression in B95-8 cells induced to replicate EBV by treatment with TPA is also shown for comparison (B95-8).
DISCUSSION
EBV SM protein is known to be essential for production of infectious EBV. However, the role that it plays in regulating EBV gene expression to allow productive replication remains to be fully defined. Establishing the functions of SM in lytic replication is complicated by the fact that although SM is an RNA-binding protein with a posttranscriptional mechanism of action, relatively little is known of its RNA target specificity. Previous work has shown that SM, while acting on a number of reporter genes and EBV lytic cycle genes, does exhibit gene specificity (2, 26, 45, 51). For example, SM enhances accumulation of chloramphenicol acetyltransferase but not β-galactosidase mRNA (26). However, no specific RNA sequence motif has been identified that is required for SM binding. Possible explanations for SM specificity include underlying differences in the stability or exportability of target mRNAs, with SM preferentially enhancing the accumulation of unstable or poorly exported intronless mRNAs. Alternatively, unidentified RNA secondary structures may exist which act as determinants of SM binding.
In contrast to previously published findings, we demonstrate that lytic DNA replication and production of linear EBV genomes do not occur in the absence of SM. Two early genes essential for EBV DNA replication, coding for EBV primase and polymerase, were shown to be SM dependent. In this respect, SM is similar to ICP27, as an ICP27 mutant of HSV (Δ27 HSV) also fails to replicate viral DNA (31). The DNA replication defect in Δ27 HSV has been attributed to its failure to efficiently express six of the genes required for HSV lytic replication (23). Interestingly, expression of one essential gene, coding for ICP8, does not seem to require ICP27. The situation therefore parallels that in EBV; although the SM-dependent replication complex genes are fewer (polymerase and primase), there seems to be a selective dependence of the essential DNA replication genes on SM. Why only a subset of the genes that act coordinately to carry out DNA replication is regulated by SM is not obvious, but it is clear that SM may act as a key element of the temporal regulation of DNA replication.
By transfection of the primase and polymerase genes, a partial but significant rescue of DNA replication in the SM-KO virus could be accomplished (Fig. 3). The level of DNA replication was approximately 20 to 25% of that achieved with transfection of SM. Previous studies have demonstrated replication of EBV oriLyt using cosmids that expressed six EBV genes: BALF5 (DNA polymerase), BMRF1 (polymerase processivity factor), BALF2 (single-stranded DNA-binding protein), BSLF1 (primase), BBLF4 (helicase), and BBLF2/3 (helicase-primase-associated protein) (14). Although initially it was thought that SM was directly involved in replication, it was subsequently demonstrated that the role of SM was in fact indirect, enhancing expression of the proteins involved in the actual DNA replication (13). Our data suggest that the role of SM in vivo is similar to its role in replication assays in vitro. However, it is also clear that SM enhances DNA replication by mechanisms in addition to enhancing expression of these six minimally essential genes, since full rescue of DNA replication requires SM. It is likely that other genes induced by SM, such as those coding for thymidine kinase and the small unit of ribonucleotide reductase, which, while not absolutely essential, contribute to efficient replication in vivo.
Nevertheless, the level of DNA replication achieved by expression of primase and polymerase should have been sufficient to allow virus production, albeit at a reduced level, if the block to DNA replication were the primary reason for the lack of late gene expression observed in the absence of SM. However, transfection of polymerase and primase genes was unable to rescue infectious virus production, under conditions where even production of 1% of wild-type levels of virus should have been detectable (as GFP expression within infected Raji cells). Furthermore, noninfectious virus was also not produced, indicating that there are additional genes whose expression is SM dependent and not merely a consequence of the block to DNA replication in the absence of SM. Thus, it is clear that while DNA replication enhances late gene expression, possibly by effects on genome copy number as well as on transcription, SM is required for full productive late gene expression.
Rescue of DNA replication by transfected polymerase and primase allowed us to examine whether late genes were activated solely by DNA replication. These experiments clearly demonstrated that expression of a number of late lytic genes is stimulated by DNA replication alone in the absence of SM. In essence, these can be considered true γ2 genes of EBV in that their expression was enhanced by DNA replication per se. While some genes could be clearly shown to increase with DNA replication (seen in the middle left portion of the cluster map in Fig. 7), they were further stimulated by SM expression. Conversely, there was also a group of late genes whose expression was not stimulated by DNA replication but was enhanced by SM (shown in the bottom third of the map in Fig. 7 and listed in Fig. 7B). Such a stimulation of γ gene expression can also be distinguished from effects on viral DNA replication in the case of HSV ICP27 (19, 43, 44). A previous study had shown that SM-KO EBV did not synthesize gp350 or VCA, both of which would be essential for infectious virus production (2). In addition to the genes coding for these proteins, the current study has identified several other structural genes which are SM dependent, demonstrating that SM is directly required for efficient expression of multiple late genes.
It should be noted that there are some inherent limitations to the experiments described here. First, the temporal regulation of Z and SM likely does not represent that occurring during EBV reactivation in vivo (modeled in vitro here), in that both genes were expressed by transfection. In addition, these studies were performed in 293 cells, which may not accurately represent EBV replication in B cells or naturally infected epithelial cells. In addition, while this analysis has permitted the identification of essential roles of SM in DNA replication, virus production, and late gene expression, there are likely to be more subtle aspects of SM regulation of the replicative cycle as demonstrated by the kinetics of BMRF1 expression, where SM led to an earlier accumulation of BMRF1 mRNA.
The negative regulation of BHRF1 by SM was unexpected and intriguing. It is tempting to speculate that an antiapoptotic effect of BHRF1 expression is useful during an initial period of EBV DNA synthesis and protein production but is counterproductive at later times prior to cell lysis. A parallel again exists in HSV, where expression of certain early genes continues to increase unchecked in Δ27 mutants, suggesting that ICP27 also plays a viral gene-suppressive role. Certain early herpesvirus genes may be repressed by genome replication (31, 32). Whether effects on DNA replication or direct effects on BHRF1 transcription or processing are involved in the regulation of BHRF1 by SM remains to be determined.
The large number of transcripts whose accumulation is enhanced by SM raises the question of how SM specificity might be determined. There are clearly many transcripts that are strongly activated by Z and do not require SM for high-level expression. One possible explanation is that SM binding is nonspecific but only has effects on transcripts that are inherently poorly expressed. According to such a model, individual EBV transcripts vary in their exportability or stability, with SM enhancing the accumulation of poorly expressed transcripts. Conversely, SM would have relatively little effect on the levels of transcripts that are constitutively well expressed. Such differences in the ability of intronless mRNAs to serve as export substrates clearly exist and may be attributable to the presence of constitutive transport elements in some transcripts that are bound by cellular RNA-binding proteins. Alternative explanations that are not mutually exclusive also may apply. Although SM has been shown to exert posttranscriptional effects, transcriptional effects have not been ruled out. Nuclear run-on analyses have only been performed with transfected cell nuclei, where the effect of SM on a limited number of promoters has been examined (35, 47). Thus, it remains possible that SM, like its homologs in HSV and CMV, may have transcriptional effects on specific EBV promoters. The genes identified here provide a starting point to compare SM-responsive and SM-independent transcripts on the basis of these parameters.
In summary, the data presented above demonstrate that there is a clear demarcation of gene expression in the absence of SM. Lytic DNA replication is essentially curtailed without SM due to its effect on some early genes. While DNA replication does stimulate some late gene expression, SM is also required for late gene expression independent of its effects on DNA replication. Therefore, due to indirect (by permitting DNA replication) and direct effects on late transcripts, few late EBV lytic cycle genes are expressed in the absence of SM. SM also has kinetic effects on gene expression, leading to earlier expression of some genes and repressing the expression of one gene with antiapoptotic function. These multiple effects on the pattern of lytic cycle EBV gene expression explain the complete absence of infectious virus production in the absence of SM.
Supplementary Material
Acknowledgments
This work was supported by U.S. Public Health Service grants CA073544 and CA056639 to J.T.S. and CA081133 and CA119905 to S.S. and Cancer Center Support grant CA21765 (to St. Jude Children's Research Hospital) from the National Cancer Institute and the American Lebanese Syrian Associated Charities (ALSAC).
We thank Elliott Kieff for the gift of cloned EBV DNA fragments used for array construction, E. B. Daniel Henson for technical assistance, Deanna Naeve for design and production of the EBV DNA microarray, Granger Ridout for the processing of RNA for microarray analysis, and Dirk Dittmer for assistance with quantitative PCR.
Footnotes
Published ahead of print on 7 February 2007.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Baer, R., A. T. Bankier, and M. D. Biggin. 1984. DNA sequence and expression of the B95-8 Epstein-Barr virus genome. Nature 310:207-211. [DOI] [PubMed] [Google Scholar]
- 2.Batisse, J., E. Manet, J. Middeldorp, A. Sergeant, and H. Gruffat. 2005. Epstein-Barr virus mRNA export factor EB2 is essential for intranuclear capsid assembly and production of gp350. J. Virol. 79:14102-14111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bello, L. J., A. J. Davison, M. A. Glenn, A. Whitehouse, N. Rethmeier, T. F. Schulz, and J. Barklie Clements. 1999. The human herpesvirus-8 ORF 57 gene and its properties. J. Gen. Virol. 80:3207-3215. [DOI] [PubMed] [Google Scholar]
- 4.Bryant, H. E., S. E. Wadd, A. I. Lamond, S. J. Silverstein, and J. B. Clements. 2001. Herpes simplex virus IE63 (ICP27) protein interacts with spliceosome-associated protein 145 and inhibits splicing prior to the first catalytic step. J. Virol. 75:4376-4385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen, I.-H. B., L. Li, L. Silva, and R. M. Sandri-Goldin. 2005. ICP27 recruits Aly/REF but not TAP/NXF1 to herpes simplex virus type 1 transcription sites although TAP/NXF1 is required for ICP27 export. J. Virol. 79:3949-3961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen, I.-H. B., K. S. Sciabica, and R. M. Sandri-Goldin. 2002. ICP27 interacts with the RNA export factor Aly/REF to direct herpes simplex virus type 1 intronless mRNAs to the TAP export pathway. J. Virol. 76:12877-12889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, L., G. Liao, M. Fujimuro, O. J. Semmes, and S. D. Hayward. 2001. Properties of two EBV Mta nuclear export signal sequences. Virology 288:119-128. [DOI] [PubMed] [Google Scholar]
- 8.Cohen, J. I., T. Krogmann, J. P. Ross, L. Pesnicak, and E. A. Prikhod'ko. 2005. Varicella-zoster virus ORF4 latency-associated protein is important for establishment of latency. J. Virol. 79:6969-6975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dambaugh, T., K. Hennessy, L. Chamnankit, and E. Kieff. 1984. U2 region of Epstein-Barr virus DNA may encode Epstein-Barr nuclear antigen 2. Proc. Natl. Acad. Sci. USA 81:7632-7636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dolan, A., C. Addison, D. Gatherer, A. J. Davison, and D. J. McGeoch. 2006. The genome of Epstein-Barr virus type 2 strain AG876. Virology 350:164-170. [DOI] [PubMed] [Google Scholar]
- 11.Farjot, G., M. Buisson, M. Duc Dodon, L. Gazzolo, A. Sergeant, and I. Mikaelian. 2000. Epstein-Barr virus EB2 protein exports unspliced RNA via a Crm-1-independent pathway. J. Virol. 74:6068-6076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Feederle, R., M. Kost, M. Baumann, A. Janz, E. Drouet, W. Hammerschmidt, and H. J. Delecluse. 2000. The Epstein-Barr virus lytic program is controlled by the co-operative functions of two transactivators. EMBO J. 19:3080-3089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fixman, E. D., G. S. Hayward, and S. D. Hayward. 1995. Replication of Epstein-Barr virus oriLyt: lack of a dedicated virally encoded origin-binding protein and dependence on Zta in cotransfection assays. J. Virol. 69:2998-3006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fixman, E. D., G. S. Hayward, and S. D. Hayward. 1992. trans-acting requirements for replication of Epstein-Barr virus ori-Lyt. J. Virol. 66:5030-5039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gruffat, H., J. Batisse, D. Pich, B. Neuhierl, E. Manet, W. Hammerschmidt, and A. Sergeant. 2002. Epstein-Barr virus mRNA export factor EB2 is essential for production of infectious virus. J. Virol. 76:9635-9644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gupta, A. K., V. Ruvolo, C. Patterson, and S. Swaminathan. 2000. The human herpesvirus 8 homolog of Epstein-Barr virus SM protein (KS-SM) is a posttranscriptional activator of gene expression. J. Virol. 74:1038-1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Han, Z., and S. Swaminathan. 2006. Kaposi's sarcoma-associated herpesvirus lytic gene ORF57 is essential for infectious virion production. J. Virol. 80:5251-5260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hardy, W. R., and R. M. Sandri-Goldin. 1994. Herpes simplex virus inhibits host cell splicing, and regulatory protein ICP27 is required for this effect. J. Virol. 68:7790-7799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hibbard, M. K., and R. M. Sandri-Goldin. 1995. Arginine-rich regions succeeding the nuclear localization region of the herpes simplex virus type 1 regulatory protein ICP27 are required for efficient nuclear localization and late gene expression. J. Virol. 69:4656-4667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hilscher, C., W. Vahrson, and D. P. Dittmer. 2005. Faster quantitative real-time PCR protocols may lose sensitivity and show increased variability. Nucleic Acids Res. 33:e182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hiriart, E., L. Bardouillet, E. Manet, H. Gruffat, F. Penin, R. Montserret, G. Farjot, and A. Sergeant. 2003. A region of the Epstein-Barr virus (EBV) mRNA export factor EB2 containing an arginine-rich motif mediates direct binding to RNA. J. Biol. Chem. 278:37790-37798. [DOI] [PubMed] [Google Scholar]
- 22.Hiriart, E., G. Farjot, H. Gruffat, M. V. Nguyen, A. Sergeant, and E. Manet. 2003. A novel nuclear export signal and a REF interaction domain both promote mRNA export by the Epstein-Barr virus EB2 protein. J. Biol. Chem. 278:335-342. [DOI] [PubMed] [Google Scholar]
- 23.Jean, S., K. M. LeVan, B. Song, M. Levine, and D. M. Knipe. 2001. Herpes simplex virus 1 ICP27 is required for transcription of two viral late (gamma 2) genes in infected cells. Virology 283:273-284. [DOI] [PubMed] [Google Scholar]
- 24.Jia, Q., T.-T. Wu, H.-I. Liao, V. Chernishof, and R. Sun. 2004. Murine gammaherpesvirus 68 open reading frame 31 is required for viral replication. J. Virol. 78:6610-6620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Karlin, S., E. S. Mocarski, and G. A. Schachtel. 1994. Molecular evolution of herpesviruses: genomic and protein sequence comparisons. J. Virol. 68:1886-1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kenney, S., J. Kamine, E. Holley-Guthrie, E. C. Mar, J. C. Lin, D. Markovitz, and J. Pagano. 1989. The Epstein-Barr virus immediate-early gene product, BMLF1, acts in trans by a posttranscriptional mechanism which is reporter gene dependent. J. Virol. 63:3870-3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Koffa, M. D., J. B. Clements, E. Izaurralde, S. Wadd, S. A. Wilson, I. W. Mattaj, and S. Kuersten. 2001. Herpes simplex virus ICP27 protein provides viral mRNAs with access to the cellular mRNA export pathway. EMBO J. 20:5769-5778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lischka, P., O. Rosorius, E. Trommer, and T. Stamminger. 2001. A novel transferable nuclear export signal mediates CRM1-independent nucleocytoplasmic shuttling of the human cytomegalovirus transactivator protein pUL69. EMBO J. 20:7271-7283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lischka, P., Z. Toth, M. Thomas, R. Mueller, and T. Stamminger. 2006. The UL69 transactivator protein of human cytomegalovirus interacts with DEXD/H-box RNA helicase UAP56 to promote cytoplasmic accumulation of unspliced RNA. Mol. Cell. Biol. 26:1631-1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Malik, P., D. J. Blackbourn, and J. B. Clements. 2004. The evolutionarily conserved Kaposi's sarcoma-associated herpesvirus ORF57 protein interacts with REF protein and acts as an RNA export factor. J. Biol. Chem. 279:33001-33011. [DOI] [PubMed] [Google Scholar]
- 31.McCarthy, A. M., L. McMahan, and P. A. Schaffer. 1989. Herpes simplex virus type 1 ICP27 deletion mutants exhibit altered patterns of transcription and are DNA deficient. J. Virol. 63:18-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McMahan, L., and P. A. Schaffer. 1990. The repressing and enhancing functions of the herpes simplex virus regulatory protein ICP27 map to C-terminal regions and are required to modulate viral gene expression very early in infection. J. Virol. 64:3471-3485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miller, G., and M. Lipman. 1973. Release of infectious Epstein-Barr virus by transformed marmoset leukocytes. Proc. Natl. Acad. Sci. USA 70:190-194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Neuhierl, B., R. Feederle, W. Hammerschmidt, and H. J. Delecluse. 2002. Glycoprotein gp110 of Epstein-Barr virus determines viral tropism and efficiency of infection. Proc. Natl. Acad. Sci. USA 99:15036-15041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nicewonger, J., G. Suck, D. Bloch, and S. Swaminathan. 2004. Epstein-Barr virus (EBV) SM protein induces and recruits cellular Sp110b to stabilize mRNAs and enhance EBV lytic gene expression. J. Virol. 78:9412-9422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Parker, B. D., A. Bankier, S. Satchwell, B. Barrell, and P. J. Farrell. 1990. Sequence and transcription of Raji Epstein-Barr virus DNA spanning the B95-8 deletion region. Virology 179:339-346. [DOI] [PubMed] [Google Scholar]
- 37.Pearson, G. R., J. Luka, L. Petti, J. Sample, M. Birkenbach, D. Braun, and E. Kieff. 1987. Identification of an Epstein-Barr virus early gene encoding a second component of the restricted early antigen complex. Virology 160:151-161. [DOI] [PubMed] [Google Scholar]
- 38.Perera, L. P., S. Kaushal, P. R. Kinchington, J. D. Mosca, G. S. Hayward, and S. E. Straus. 1994. Varicella-zoster virus open reading frame 4 encodes a transcriptional activator that is functionally distinct from that of herpes simplex virus homolog ICP27. J. Virol. 68:2468-2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Petti, L., J. Sample, F. Wang, and E. Kieff. 1988. A fifth Epstein-Barr virus nuclear protein (EBNA3C) is expressed in latently infected growth-transformed lymphocytes. J. Virol. 62:1330-1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pfitzner, A. J., E. C. Tsai, J. L. Strominger, and S. H. Speck. 1987. Isolation and characterization of cDNA clones corresponding to transcripts from the BamHI H and F regions of the Epstein-Barr virus genome. J. Virol. 61:2902-2909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pulvertaft, J. V. 1965. A study of malignant tumours in Nigeria by short-term tissue culture. J. Clin. Pathol. 18:261-273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reed, R., and H. Cheng. 2005. TREX, SR proteins and export of mRNA. Curr. Opin. Cell Biol. 17:269-273. [DOI] [PubMed] [Google Scholar]
- 43.Rice, S. A., and D. M. Knipe. 1990. Genetic evidence for two distinct transactivation functions of the herpes simplex virus α protein ICP27. J. Virol. 64:1704-1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rice, S. A., and V. Lam. 1994. Amino acid substitution mutations in the herpes simplex virus ICP27 protein define an essential gene regulation function. J. Virol. 68:823-833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ruvolo, V., A. K. Gupta, and S. Swaminathan. 2001. Epstein-Barr virus SM protein interacts with mRNA in vivo and mediates a gene-specific increase in cytoplasmic mRNA. J. Virol. 75:6033-6041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ruvolo, V., L. Sun, K. Howard, S. Sung, H.-J. Delecluse, W. Hammerschmidt, and S. Swaminathan. 2004. Functional analysis of Epstein-Barr virus SM protein: identification of amino acids essential for structure, transactivation, splicing inhibition, and virion production. J. Virol. 78:340-352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ruvolo, V., E. Wang, S. Boyle, and S. Swaminathan. 1998. The Epstein-Barr virus nuclear protein SM is both a post-transcriptional inhibitor and activator of gene expression. Proc. Natl. Acad. Sci. USA 95:8852-8857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sacks, W. R., C. C. Greene, D. P. Aschman, and P. A. Schaffer. 1985. Herpes simplex virus type 1 ICP27 is an essential regulatory protein. J. Virol. 55:796-805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sandri-Goldin, R. M. 1998. ICP27 mediates HSV RNA export by shuttling through a leucine-rich nuclear export signal and binding viral intronless RNAs through an RGG motif. Genes Dev. 12:868-879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sciabica, K. S., Q. J. Dai, and R. M. Sandri-Goldin. 2003. ICP27 interacts with SRPK1 to mediate HSV splicing inhibition by altering SR protein phosphorylation. EMBO J. 22:1608-1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Semmes, O. J., L. Chen, R. T. Sarisky, Z. Gao, L. Zhong, and S. D. Hayward. 1998. Mta has properties of an RNA export protein and increases cytoplasmic accumulation of Epstein-Barr virus replication gene mRNA. J. Virol. 72:9526-9534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Serio, T. R., J. L. Kolman, and G. Miller. 1997. Late gene expression from the Epstein-Barr virus BcLF1 and BFRF3 promoters does not require DNA replication in cis. J. Virol. 71:8726-8734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sokolowski, M., J. E. Scott, R. P. Heaney, A. H. Patel, and J. B. Clements. 2003. Identification of herpes simplex virus RNAs that interact specifically with regulatory protein ICP27 in vivo. J. Biol. Chem. 278:33540-33549. [DOI] [PubMed] [Google Scholar]
- 54.Soliman, T. M., R. M. Sandri-Goldin, and S. J. Silverstein. 1997. Shuttling of the herpes simplex virus type 1 regulatory protein ICP27 between the nucleus and cytoplasm mediates the expression of late proteins. J. Virol. 71:9188-9197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stingley, S. W., J. J. Garcia Ramirez, S. A. Aguilar, K. Simmen, R. M. Sandri-Goldin, P. Ghazal, and E. K. Wagner. 2000. Global analysis of herpes simplex virus type 1 transcription using an oligonucleotide-based DNA microarray. J. Virol. 74:9916-9927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tanner, J., Y. Whang, J. Sample, A. Sears, and E. Kieff. 1988. Soluble gp350/220 and deletion mutant glycoproteins block Epstein-Barr virus adsorption to lymphocytes. J. Virol. 62:4452-4464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Winkler, M., S. A. Rice, and T. Stamminger. 1994. UL69 of human cytomegalovirus, an open reading frame with homology to ICP27 of herpes simplex virus, encodes a transactivator of gene expression. J. Virol. 68:3943-3954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yuan, J., E. Cahir-McFarland, B. Zhao, and E. Kieff. 2006. Virus and cell RNAs expressed during Epstein-Barr virus replication. J. Virol. 80:2548-2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.