Abstract
The membrane-proximal external region (MPER) of human immunodeficiency virus type 1 (HIV-1) gp41 bears the epitopes of two broadly neutralizing antibodies (Abs), 2F5 and 4E10, making it a target for vaccine design. A third Ab, Fab Z13, had previously been mapped to an epitope that overlaps those of 2F5 and 4E10 but only weakly neutralizes a limited set of primary isolates. Here, libraries of Fab Z13 variants displayed on phage were engineered and affinity selected against an MPER peptide and recombinant gp41. A high-affinity variant, designated Z13e1, was isolated and found to be ∼100-fold improved over the parental Fab not only in binding affinity for the MPER antigens but also in neutralization potency against sensitive HIV-1. Alanine scanning of MPER residues 664 to 680 revealed that N671 and D674 are crucial for peptide recognition as well as for the neutralization of HIV-1 by Z13e1. Ab competition studies and truncation of MPER peptides indicate that Z13e1 binds with high affinity to an epitope between and overlapping with those of 2F5 and 4E10, with the minimal peptide epitope WASLWNWFDITN. Still, Z13e1 remained about an order of magnitude less potent than 4E10 against several isolates of pseudotyped HIV-1. The sum of our molecular analyses with Z13e1 suggests that the segment on the MPER of gp41 between the 2F5 and 4E10 epitopes is exposed on the functional envelope trimer but that access to the specific Z13e1 epitope within this segment is limited. Thus, the ability of MPER-bearing immunogens to elicit potent HIV-1-neutralizing Abs may depend in part on recapitulating the particular constraints that the functional envelope trimer imposes on the segment of the MPER to which Z13e1 binds.
A crucial aim in human immunodeficiency virus (HIV) type 1 (HIV-1) vaccine design is to elicit neutralizing antibodies (Abs) against primary isolates of HIV-1 (9, 25, 29). Although high titers of HIV-1-specific Abs are readily elicited through immunization or during natural infection, neutralizing Abs are typically poorly represented (15, 26, 41, 53). HIV-1-neutralizing Abs bind to the envelope glycoproteins, gp120 and gp41 (Env), but are unique in their ability to recognize functional and non-covalently associated heterotrimers of these two glycoproteins (18, 36, 43, 54, 56). Unfortunately, the functional Env trimer is labile such that immunogens based on it, and HIV-1 virions themselves, contain monomeric or irrelevant (nonfunctional) forms of gp120 and gp41 that tend to elicit mostly nonneutralizing Abs (3, 17, 19, 23, 31, 34, 38). One sought-after solution is to engineer homogeneous immunogens that mimic the functional envelope trimer or key epitopes thereof. Since the structural details of the functional envelope trimer are lacking, monoclonal Abs (MAbs) with HIV-1-neutralizing activity have been important tools for characterizing candidate envelope mimetic vaccines.
Two human MAbs, 2F5 and 4E10, neutralize a broad range of HIV-1 primary isolates and target neighboring epitopes on the membrane-proximal external region (MPER) of gp41 (4, 32, 48, 64). Due in part to the remarkable neutralizing activities of 2F5 and 4E10, the MPER of gp41 has become an important target for HIV-1 vaccine design. However, eliciting broadly neutralizing Abs against the MPER by use of designed mimetics has proven to be very challenging. For example, peptides corresponding to the core epitope of 2F5, as coupled to a protein carrier or genetically grafted into various protein scaffolds, have generally failed to elicit neutralizing Ab against primary HIV-1 (2, 28, 60). These difficulties may be due to steric blocks by the membrane and the quaternary structure of the envelope trimer that limit the access of many elicited Abs to the native epitopes on the MPER. Alternatively, success may be limited by conformational differences between the peptide epitope and the corresponding site on the native MPER. Immunological tolerance-suppression of neutralizing Abs due autoantigen reactivity has also been proposed to explain the poor immunogenicity of the 2F5 and 4E10 epitopes (21, 60). The prevalence of 2F5- and 4E10-like Abs in the sera of HIV-1 infected individuals is indeed extremely low, indicating that these particular epitopes are poorly immunogenic during natural infection (references 14 and 57; also G. F. Shaw, Bibollet-Ruche, J. Decker, H. Li, P. Goepfert, M. Peeters, S. Allen, E. Hunter, J. Robinson, and P. Kwong, presented at the 13th Conference on Retroviruses and Opportunistic Infections, Colorado Conference Center, Denver, CO, 2006). However, it is not known whether other Abs directed to the MPER would neutralize as potently or be as difficult to elicit as 2F5 or 4E10.
Antigen engineering has been viewed as a way to boost the relative immunogenicity of the MPER of gp41 and to elicit 2F5- and 4E10-like Abs. However, designing successful mimics of the MPER is not straightforward, especially considering that many details of native Env structure remain unknown. Available data indicate that the core epitope of 2F5 on the MPER appears to be in an extended conformation with a type I β-turn about the three-residue motif DKW, which is deeply buried in the combining site of 2F5 (33). In contrast, the 4E10 peptide epitope has a largely helical fold that terminates at the conserved residues that have the most contact with the Ab (i.e., “WF” of the NWFDIT motif) (10). Cryo-electron microscopy tomography has recently been used to model HIV-1 envelope glycoprotein trimers; however, there is a discrepancy over whether the gp41 MPERs are separated (as in a tripod), with each MPER consisting of a membrane-associated “foot” and a short “leg” that connects the foot to the bulk of the trimer (59), or whether the MPERs pack against one another to form a single stalk at the base of the trimer (58). Additional gaps in the structural information about the MPER include the degree of mobility of the MPER, how it folds to connect with the transmembrane domain and the rest of the envelope trimer, and the way in which the 2F5 and 4E10 epitopes connect to one another.
A third human MAb, Fab Z13, appears to bind to a portion of the MPER between the 2F5 and 4E10 epitopes. This Ab was selected from a Fab phage display library that was prepared from the bone marrow RNA of an asymptomatic individual with strong and broadly neutralizing serum Ab titers (30, 37, 52, 64). It was found to have a continuous core epitope that ostensibly coincides with that of 4E10, but Z13 recognition of a peptide corresponding to the MPER can be blocked by both 4E10 and 2F5 (64). The neutralization potency of Fab Z13 is less than that observed with immunoglobulin Gs (IgGs) 2F5 and 4E10 by use of a peripheral blood mononuclear cell-based assay, and its activity depends on a somewhat variable residue (position 674) in the MPER for which 4E10 activity is apparently unaffected (64). Because the epitope of Z13 at least partially overlaps with those of both 2F5 and 4E10, an analysis of the Z13 epitope should shed light on the exposure of all three epitopes on the MPER (64).
Since Fab Z13 is less potent than 2F5 and 4E10 against HIV-1, we attempted to improve its affinity for gp41, possibly also improving its neutralization potency. Random mutations were introduced into the complementarity-determining region (CDR) L3 of Fab Z13 in a phage display library (1), and high-affinity variants of Z13 were selected. We finely mapped the specificity and neutralizing activity of a representative high-affinity variant, Z13e1, in both Fab and IgG formats. We found that Z13e1 binds extremely well to peptides and recombinant gp41 that bear MPER sequences overlapping the 2F5 and 4E10 epitopes but that Z13e1 has only moderate neutralization potency. These results suggest that the segment between the 2F5 and 4E10 epitopes on the MPER of native gp41 must be exposed to neutralizing Ab but that the binding of Z13e1 to this segment on the Env trimer is limited. These findings and IgG Z13e1 should be useful in further clarifying the structural details of the MPER of gp41 and in designing and prescreening vaccine candidates.
MATERIALS AND METHODS
Materials.
Butoxycarbonyl amino acids and MBHA (p-methylbenzhydrylamine) resin were obtained from Peptides International (Louisville, KY). All solvents (high-performance liquid chromatography [HPLC]-grade N,N-dimethylformamide, dichloromethane, and acetonitrile) of high purity were purchased from Fisher. Trifluoroacetic acid was obtained from Halocarbon Products (River Edge, NJ). HF was purchased from Matheson Gas (Cucamonga, CA). The following reagents were obtained from the National Institutes of Health AIDS Research and Reference Reagent Program: pNL4-3.Luc.R-E- (13) (contributed by N. Landau) and U87.CD4 (CCR5+ or CXCR4+) cells (5) (contributed by H. Deng and D. Littman). Env complementation plasmids JR-FL and JR-CSF were previously described (49, 62), and the MN-encoding plasmid was kindly provided by John Moore and Simon Beddows. The plasmids encoding JR2 Env and MPER Ala substitutions were described previously (61). Recombinant gp41JR-FL (amino acids [aa] 535 to 681) was produced as a C-terminal fusion to the maltose-binding protein (MBP) in Escherichia coli and purified on an amylose column (63). HIV Ig was provided by John Mascola (VRC, Bethesda, MD). 2F5 (32), 4E10 (7), and 2G12 (51) IgGs were generously provided by Hermann Katinger, Gabriela Stiegler, and Renate Kunert. Soluble CD4 and D49 IgG (murine) (17) were obtained from the IAVI NAC.
Z13 Fab mutagenesis library construction.
Two saturation mutagenesis libraries were prepared in the phage display format targeting five residues each in the CDR L3, as described previously (1). The “N” and “C” libraries randomize residues Q90 to W94 and D93 to T97, respectively. To serve as the template for PCR mutagenesis, the pComb3H plasmid encoding the wild-type (wt) Z13 Fab (1, 64) was mutated to have a stop codon (UAA) in the (kappa) light chain gene using the following primers: Z13L3STOPFOR (CTGTCAGCAGCGTAGCGACTAACCCCGGACGTTCGGCCAAG) and Z13L3STOPREV (CTTGGCCGAACGTCCGGGGTTAGTCGCTACGCTGCTGACAG). The initial mutagenesis PCRs (PCR 1) were performed using the above-described template and the primers Z13L3_N_LIBFOR [GAGGATTTTGCAGTTTATTACTGTCAG(NNK)5CCCCGGACGTTCGGCCAAG] plus HG33 [CCTTATTAGCGTTTGCCATC] and Z13L3_C_LIBFOR [GCAGTTTATTACTGTCAGCAGCGTAGC(NNK)5TTCGGCCAAGGGACCAAGG] plus HG33, respectively. The remaining DNA segments to be spliced onto the 5′ end of the mutagenesis DNA fragments were prepared using the primers RSC-F (GAGGAGGAGGAGGAGGAGGCGGGGCCCAGGCGGCCGAGCTC) plus Z13L3_N_LIBREV (CTGACAGTAATAAACTGCAAAATCCT) and RSC-F plus Z13L3_C_LIBREV (GCTACGCTGCTGACAGTAATAAACTGC), respectively (PCR 2). The products of PCRs 1 and 2 were then linked using splicing overlap extension PCR (22) to generate the mutagenesis library cassette. Taq DNA polymerase (Promega) was used for all PCRs, and the product DNAs were gel extracted (QIAquick kit) prior to and following the final SfiI 15-fold overdigestion. The SfiI-digested cassette was subcloned into pComb3X (Carlos Barbas, TSRI) that was similarly cleaved, as described previously (1). Electroporation of XL1-Blue cells yielded 2.3 ×107 and 4.6 × 106 transformants, and a restriction digest of 10 randomly picked clones yielded 10 and 8 correct inserts, for the N and C libraries, respectively.
Affinity selection of Z13 Fab phage display libraries.
Ninety-six-well polystyrene plates (Corning) were coated overnight at 4°C with 50 μl phosphate-buffered saline (PBS) containing either 100 ng M41xt (gp41JRFL fused to MBP [63]) or 1 μg streptavidin. After blocking with 4% nonfat dry milk (NFDM) in PBS, the wells coated with streptavidin were treated with 50 μl PBS containing the biotinylated peptide SLWNWFDITNWLWRRK(biotin)-NH2 (2 μg/ml) and incubated for 15 min at room temperature (RT). The wells were washed, ∼1011 input phage of the N and C libraries in 50 μl PBS containing 1% bovine serum albumin (BSA) were added, and the plate was incubated for 2 h at 37°C. The wells were washed 10 times with PBS containing 0.2% NFDM and 0.05% Tween 20 over a period of 180 min, and the remaining phage were first eluted using a 10-min incubation with 0.1 ml of 0.1 M glycine-HCl (pH 2.2) and then pH neutralized by the addition of 6 μl of 2 M Tris-HCl. The surviving phage were used to infect 2 ml fresh XL1-Blue cells (optical density [OD], 0.5), and the cultures were shaken for 2 h at 250 rpm and 37°C. VCSM13 helper phage were added and the cultures shaken overnight at 37°C before the phage were subjected to the subsequent round of affinity selection according to previously described methods (1). Individual clones were picked from rounds 4, 5, and 6, the eluted phage were used to infect TOP10F′ cells (Invitrogen), and plasmid was prepared for DNA sequencing (TSRI Center for Nucleic Acids Research). In parallel, the same clones were used to prepare crude Fab supernatant for testing by enzyme-linked immunosorbent assay (ELISA), using crude Fab Z13 wt as a control.
Construction and purification of IgG1 Z13e1.
The DNAs encoding the heavy chain (Fd fragment) and light chain of Z13e1 were each excised from the pComb3X phagemid vector (1) as XhoI-SpeI and SacI-XbaI fragments, respectively, and subcloned sequentially into a mammalian IgG expression vector which contains compatible restriction sites, pIgG (8). The pIgG-Z13e1 DNA was verified by DNA sequencing and used with the FuGENE6 transfection reagent (Roche, Indianapolis, IN) to transfect Chinese hamster ovary (CHO-K1) cells. Stable transfected clones were amplified under selection using methionine sulfoximine (Sigma, St. Louis, MO). The clone with the highest production of IgG1 Z13e1, as determined using ELISA with cell culture supernatant, was chosen for scale-up in the CellCube System (Corning, Inc., Corning, NY) and purified by affinity chromatography with protein A (Pharmacia, Uppsala, Sweden). Purified IgG1 Z13e1 was concentrated and dialyzed against PBS. Ab purity and concentration were determined using sodium dodecyl sulfate-polyacrylamide gel electrophoresis and the absorbance at 280 nm.
Fab Z13e1 preparation by pepsin treatment of IgG.
Antigen-binding fragment Z13e1 was obtained by pepsin digestion of IgG Z13e1 followed by reduction of interchain disulfide bridges. Pepsin (Sigma) was resuspended in 0.1 M sodium citrate (pH 3.5) at a concentration of 0.5 mg/ml. The pepsin solution was added to IgG Z13e1 (4.2 mg/ml) to give a final ratio of 4% (wt/wt) pepsin, and the reaction mixture was incubated at 37°C for 3 h. The digestion reaction was stopped with a 1/10 volume of 1 M Tris (pH 10). The interchain disulfide bridges of the F(ab′)2 fragments were reduced by the addition of 20 mM cysteine followed by further incubation at 37°C for 4 h and then acetylation with 20 mM iodoacetamide at 37°C for 1 h.
Fab Z13e1 was purified using sequential affinity and size exclusion chromatography. First, the digested sample was loaded onto a HiTrap Protein A HP column (GE Healthcare) equilibrated in 20 mM sodium phosphate (pH 7.0). The nonbound material was then loaded onto a HiTrap Protein G HP column (GE Healthcare), also equilibrated in 20 mM sodium phosphate (pH 7.0). The Fab was eluted using 0.1 M citric acid, pH 3.0, and immediately neutralized with a 1/10 volume of 1 M Tris (pH 10). The Fab fragment was separated from the unreduced F(ab′)2 by use of a Superdex 200 HR16-60 column (GE Healthcare) equilibrated in PBS (pH 7.0) and then concentrated by ultrafiltration.
Peptide synthesis.
The peptides were synthesized manually using solid-phase peptide methodology on a C-terminal amide yielding MBHA resin with in situ neutralization cycles for butoxycarbonyl solid-phase peptide synthesis (46) as reported previously (6). Solubilizing tails were introduced on the C-terminal end of the peptide to allow for easier synthesis of multiple compounds. Following chain assembly, the peptides were cleaved from the resin with HF and 10% anisole for 1 h at 0°C. The peptides were purified by HPLC. Preparative reversed-phase HPLC was performed on a Waters 4000 HPLC system using Vydac C18 columns (10 mm, 5.0 by 25 cm) and a Gilson UV detector. Linear gradients of acetonitrile in water-0.1% trifluoroacetic acid were used to elute bound peptides. Peptides were characterized by electrospray ionization mass spectrometry on an API-III triple quadrupole mass spectrometer (Sciex, Thornhill, Ontario, Canada). Peptide masses were calculated from the experimental mass/charge (m/z) ratios from all of the observed protonation states of a peptide by using MacSpec software (Sciex). All observed peptide masses agreed with the calculated average masses to within 0.5 Da.
CD.
For circular dichroism (CD) spectroscopy, an Aviv model 203-02 spectropolarimeter was used, with cells of 0.1 cm in length, a wavelength step of 0.5 nm, and a bandwidth of 1.0 nm. One to three scans were reported. The exact peptide concentrations were determined by UV measurements at 280 nm on a Gilson UV detector, model 116.
ELISAs. (i) In-solution peptide competition.
Fifty percent inhibitory concentrations (IC50s) were determined by competitive ELISA using fixed concentrations of the biotinylated epitope peptide, SLWNWFDITNWLWRRK(biotin)-NH2 (20 nM) and IgG Z13e1 (0.3 nM), which was previously shown to generate a nonsaturating signal, and various concentrations of gp41 competitor peptides. Briefly, microwells were coated overnight at 4°C with 50 μl PBS containing neutravidin (4 μg/ml; Pierce). Wells were washed four times with PBS containing 0.05% Tween 20 and then blocked with 4% NFDM in PBS for 1 h at 37°C. Meanwhile, different concentrations of the competing peptide (in a threefold dilution series starting at 10 μM) were mixed with the biotinylated peptide (20 nM) in PBS containing 0.4% NFDM-0.02% Tween and immediately (within 1 min) combined with IgG Z13e1 (0.3 nM). The mixture was incubated at 37°C for 2 h in a 96-well round-bottom plate separately from the ELISA plate being blocked. After the blocked plate was washed, the mixture of Z13e1, biotinylated peptide, and competing peptide was added to the wells. After 20 min at RT, the wells were washed five times, and a 1:1,000 dilution of goat anti-human IgG F(ab′)2-horseradish peroxidase conjugate (Pierce) was added. Following incubation at RT for 45 min, the wells were washed five times and developed by adding 50 μl of tetramethylbenzidine (TMB) solution (Pierce) according to the manufacturer's instructions. Wells containing TMB solution were stopped after ∼10 to 20 min before the maximal signals reached an optical density of ∼1.8, by adding 50 μl of H2SO4 (0.2 M), and the optical density at 450 nm was read on a microplate reader (Molecular Devices). The concentration of competitor peptide corresponding to a half-maximal signal (IC50) was determined by interpolation of the resulting binding curve. Each peptide competitor was tested in duplicate in at least two separate experiments.
(ii) Ab competition ELISA.
Microwells were coated overnight at 4°C with 0.2 μg M41xt in 50 μl of PBS. The wells were washed twice and blocked with 4% NFDM in PBS for 1 h at 37°C. Twenty-five microliters of competitor Ab diluted in PBS containing 0.4% NFDM and 0.02% Tween 20 was added to the wells immediately followed by an equal volume of biotinylated Ab (final concentrations of biotinylated Abs were as follows: for Z13e1, 8 pg/ml; for 2F5, 40 pg/ml; for 4E10, 40 pg/ml). Following a 2-h incubation at 37°C, the wells were washed five times, and a streptavidin-horseradish peroxidase conjugate (1:1,000; Jackson ImmunoResearch) was added to the wells and incubated for 1 h. The wells were washed five times and then developed with TMB solution as described above.
(iii) CL ELISAs.
Protocol A was as follows (20). Microwells were coated with 1.5 μg cardiolipin (CL; Sigma) in 50 μl 100% ethanol and left to evaporate overnight at 4°C. Other antigens (4 μg/ml in PBS) were coated on wells overnight at 4°C without evaporation. The wells were washed twice with PBS and blocked for 1 h at RT with 10% adult bovine serum (ABS) in PBS. The wells were washed twice with PBS, and Ab diluted in 10% ABS-PBS was added to the wells for 1 h at RT. Following two washes with PBS, goat anti-human IgG-alkaline phosphatase conjugate (1:1,000 in 10% ABS-PBS) was added to the wells and incubated for 1 h at RT. The wells were washed thrice with PBS, the plate was developed with substrate at 37°C, and the OD at 405 nm was read on a plate reader. Protocol B was as follows (reference 21 and B. Haynes, personal communication). Microwells were coated with CL at 1.35 μg/well (4E10, Z13e1) or 10 μg/well (2F5) in 50 μl 100% ethanol and allowed to evaporate for 45 min at 55°C. Other antigens were coated at 4 μg/ml in PBS without evaporation. The wells were washed once with PBS and blocked with 3% BSA in PBS for 2 h at RT. The wells were washed twice with PBS, and Ab in dilution buffer (3% BSA, 2% normal goat serum, 0.05% Tween 20 in PBS) was added to the wells for 1 h at RT. Following four washes with 0.05% Tween in PBS, goat anti-human IgG-alkaline phosphatase conjugate (1:1,000 in dilution buffer) was added to the wells and incubated for 1 h at RT. The wells were washed four times with 0.05% Tween in PBS, the plate was developed at RT, and the OD at 405 nm was read on a plate reader.
BN-PAGE.
A modified Blue Native (BN)-PAGE protocol (44, 47) was used as recently described (31). Pseudotyped HIV-1 virions were mixed with monovalent Fab for 5 min. The concentration of Fab used was reported as that in the final sample at the time of loading. To liberate Env, pseudotyped HIV-1 virions were incubated in an equal volume of solubilization buffer (0.12% Triton X-100 in 1 mM EDTA-1.5 M aminocaproic acid) and 1 μl of a protease inhibitor cocktail (Sigma). An equal volume of 2× sample buffer containing 100 mM MOPS (morpholinepropanesulfonic acid), 100 mM Tris-HCl (pH 7.7), 40% glycerol, and 0.1% Coomassie blue was then added. Samples were loaded onto a 4 to 12% Bis-Tris NuPAGE gel (Invitrogen). Samples were electrophoresed at 4°C for 3 h at 100 V with 50 mM MOPS-50 mM Tris, pH 7.7, containing 0.002% Coomassie blue as the cathode buffer and the same buffer without Coomassie blue as the anode buffer. The gel was then Western blotted onto polyvinylidene difluoride. Excess Coomassie blue dye was removed after blotting by washing with 30% methanol-10% acetic acid then 100% methanol. The blot was then transferred to blocking buffer (4% NFDM in PBS) for 30 min and probed using 1 μg/ml each of MAbs 2G12, b12, and 447-52D (anti-gp120 cocktail). Goat anti-human Fc-alkaline phosphatase conjugate was used to detect the MAbs at 1:3,000 (Jackson).
HIV-1 neutralization assays.
Neutralization assays were performed in the following two formats: (i) HIV-1MN, Fab-versus-IgG, and Ala scan-pseudotyped neutralization assays, and (ii) Monogram neutralization assay.
(i) HIV-1MN, Fab-versus-IgG, and Ala scan-pseudotyped neutralization assays.
Pseudotyped HIV-1 mutants (MN, JR-FL, JRCSF, JR2, and JR2 Ala MPER mutants), competent for a single round of infection, were generated by cotransfection of 293T cells with the luciferase reporter plasmid pNL4-3.Luc.R-E- and Env complementation, as described previously (61). The pseudotyped virus was assayed for neutralization using U87.CD4.CCR5 cells as target cells (or U87.CD4.CXCR4 cells for MN virus) (5). Different concentrations of Ab were added (1:1 by volume) to HIV-1, and the mixture was incubated for 1 h at 37°C prior to transferring (1:1 by volume) to the target cells. The assay was developed using luciferase reagent (Promega) following a 48- to 72-h incubation at 37°C, and the luminescence in relative light units was measured using an Orion microplate luminometer (Berthold Detection Systems). The extent of virus neutralization was determined as a percentage reduction of viral infectivity against that of an Ab-free control. All experiments were performed in triplicate and repeated at least twice with similar results.
(ii) Monogram neutralization assay.
Pseudotyped viruses capable of a single round of infection were produced were produced by cotransfecting HEK293 cells with plasmids encoding Env libraries plus an HIV subgenomic vector that contains a firefly luciferase indicator gene (4, 41). Recombinant pseudotyped viruses were harvested 48 h posttransfection and incubated for 1 h at 37°C with serial fourfold dilutions of Ab. U87 cells that express CD4 plus the CCR5 and CXCR4 coreceptors were inoculated with virus-Ab dilutions. Virus infectivity was determined 72 h postinoculation by measuring the amount of luciferase activity expressed in infected cells. Recombinant viruses pseudotyped with Env proteins from amphotropic murine leukemia virus were used to control for nonspecific neutralization. Neutralizing activity is reported as the concentration or dilution of each MAb or plasma required to confer inhibition of infection (IC50) as follows: {percent inhibition = [1 − (luciferase + Ab/luciferase − Ab)] × 100}.
RESULTS
Identification of Z13 affinity-enhanced Fab Z13e1.
Our previous experiments with Fab Z13 showed that it bound to gp41 with lower affinity than 2F5 and 4E10, although a precise quantitative comparison was not made due to the difference in valency between Fab Z13 and IgGs 2F5 and 4E10; Fab Z13 also neutralized HIV-1 with limited potency (64). To improve the affinity of Fab Z13 against the MPER of gp41 via affinity maturation techniques, we chose to use the phage display format. CDR L3 is generally near the center of an Ab-combining site and is a hot spot for somatic hypermutation that makes it a reasonable choice for mutagenesis to affinity mature an Ab in vitro (1, 11). Thus, we engineered two separate mutagenesis libraries of Fab Z13 targeting five residues each in CDR L3. For each library, splicing overlap extension PCR and Ab-specific primers containing NNK codons were used to generate diversity in five consecutive positions in L3 such that any amino acid is possible at each of the five positions, producing a theoretical (amino acid) diversity of 205 or 3.2 × 106 per library. The N library spans the five N-terminal L3 residues Q90 to W94, and the C library includes five residues slightly shifted toward the C-terminal end of L3, D93 to T97, such that there are two overlapping residues between the two libraries. The libraries were affinity selected in parallel against a translational fusion protein of recombinant gp41JRFL (aa 535 to 681) fused to the C terminus of the MBP, named M41xt, as well as against a biotinylated peptide [SLWNWFDITNWLWRRK(biotin)-NH2] that was captured on immobilized streptavidin.
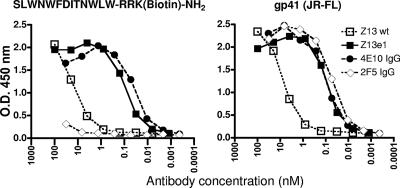
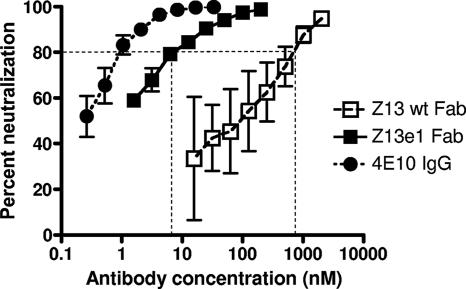
After several rounds of affinity selection, Fab Z13 variants were picked from rounds 4, 5, and 6, and initial ELISAs using crude Fab indicated that a diverse panel of high-affinity Fabs had been selected (data not shown). Following DNA sequence analysis, several unique Fabs were overexpressed, purified, and used in ELISAs, which indicated that several variants had half-maximal binding titers ∼10- to 100-fold greater than that of the parental Fab Z13 (Table 1). Although no clear overall consensus was found among the CDR L3 sequences selected, similarities exist among several clones. Several Fab variants had comparable binding affinities, but Z13e1 was selected for further study, as it was the most frequently selected sequence and exhibited affinities in various ELISA formats slightly higher than those of other clones. Moreover, in an initial experiment using a single-round infectivity assay, Fab Z13e1 neutralized pseudotyped HIV-1MN and HIV-1JR-FL with the highest potency (Table 1). A careful side-by-side comparison of the parental Fab Z13 and Fab Z13e1 against the selecting antigens using a direct ELISA, and using a single-round infectivity assay against HIV-1MN, shows a 2-order-of-magnitude enhancement both in binding affinity and in neutralization potency at the IC80 for the MN strain (Fig. 1 and 2).
TABLE 1.
Half-maximal binding affinities and neutralization potencies of Fab Z13e1 and other affinity-enhanced variants of Fab Z13
| Ab | CDR L3 sequence | Half-maximal binding titers (nM)a
|
Neutralization IC50s (nM)b
|
||
|---|---|---|---|---|---|
| Rec. gp41 | Bio. peptide | HIV-1MN | HIV-1JR-FL | ||
| Z13 (wt) | QQRSDWPRT | 8.6 | 10 | 100 | >2,000 |
| Z13e1 | .A.LLL.Q. | 0.07 | 0.09 | <2.0 | 190 |
| Z13e4 | .SLMSM.Q. | 0.10 | 0.13 | 3.0 | 320 |
| Z13e6 | .SLLAL... | 0.13 | 0.10 | 3.6 | 240 |
| Z13e10 | .SLLVL... | 0.13 | 0.10 | 11 | 340 |
| Z13e12 | ....LHDG. | 0.10 | ND | ND | ND |
| Z13e15 | NDc | 0.40 | 0.44 | 11 | 600 |
| Z13e17 | .TGSAL... | 0.19 | ND | ND | ND |
| Z13A1 | ......Q.. | 2.0 | 37 | ND | ND |
| IgG 4E10 (positive control MAb) | 0.07 | 0.03 | 1.0 | 33 | |
Titers of Fabs against recombinant [Rec.] gp41JRFL [M41xt] and biotinylated [Bio.] peptide [69-3; SLWNWFDITNWLWRRK(biotin)-NH2] were determined in an ELISA.
Neutralization assay using single-round infectious HIV-1 pseudotyped with pNL4-3Luc and U87.CD4.CXCR4 (MN) or CCR5 (JR-FL) cells.
ND, not determined.
FIG. 1.
Apparent binding affinities of parental Fab Z13 (wt) and affinity-enhanced Fab Z13e1, IgG 2F5, and IgG 4E10 against a biotinylated peptide [SLWNWFDITNWLWRRK(biotin)-NH2] that is captured on immobilized streptavidin, as well as against immobilized M41xt, a recombinant gp41JRFL [aa 535 to 681] fused to MBP, by use of a direct ELISA.
FIG. 2.
Neutralization of pseudotyped HIV-1MN by Fab Z13, Fab Z13e1, and 4E10 IgG in U87.CD4.CXCR4 target cells in a single-cycle infectivity assay. The ∼100-fold improvement in the neutralization potency of Fab Z13e1 relative to that of Fab Z13 is indicated at the level of IC80.
Determination of the peptide epitope of Fab Z13e1.
In a prior study, we finely mapped the peptide epitope of IgG 4E10 (6). We used a similar approach here to determine the fine specificity of Z13e1. We found that whereas Z13e1 binds poorly to the 4E10-optimized peptide 94-1 (IC50, >10,000 nM), it binds with high affinity (IC50, 15 nM) to an N-terminally extended peptide, 179-4 (LLELDKWASLWNWFDITNWLWYIKKKK-NH2), the affinity for which is comparable to that of 4E10 for the 94-1 peptide (IC50, 20 nM). This affinity is a ∼35-fold improvement over that observed with the previously described Z13 peptide epitope, designated KGND (IC50, 390 nM; Table 2) (64). In addition, whereas 4E10 binds poorly to the peptide 178-3 (WASLWNWFDITNKKKK-NH2), which has six fewer C-terminal MPER residues than peptides 179-4 and 94-1, Z13e1 binds with near-maximal affinity to 178-3 (IC50, 30 nM; Table 2). The above-described results suggest that the core of the optimized Z13e1 peptide epitope (roughly WASLWNWFDITN) is N terminal to that of 4E10 (NWFDITNWLWYIK [6]) but C terminal to that of 2F5 (ELLELDKWASLWN [2]).
TABLE 2.
Amino acid sequences and Z13e1 and 4E10 binding data for a series of truncated MPER peptides
| Peptide | Sequencea | IC50 (nM)b
|
|
|---|---|---|---|
| 4E10 | Z13e1 | ||
| 94-1 | NWFDITNWLWYIKKKK | 15 | >10,000 |
| 132 | SLWNWFDITNWLWYIKKKK | 25 | 110 |
| 160 | DKWASLWNWFDITNWLWYIKKKK | 90 | 15 |
| 179-4 | LLELDKWASLWNWFDITNWLWYIKKKK | 240 | 15 |
| 178-4 | DKWASLWNWFDITNKKKK | >10,000 | 45 |
| 178-3 | WASLWNWFDITNKKKK | >10,000 | 30 |
| 178-2 | SLWNWFDITNKKKK | >10,000 | 550 |
| KGND | KGWNWFDITNWGK-OH | >10,000 | 390 |
Boldface indicates residues that correspond to gp41 MPER.
The concentration of peptide needed to inhibit by 50% the maximal binding signal that is generated by the MAb and the biotinylated peptide, SLWNWFDITNWLWRRK(biotin)-NH2, in the absence of any competitor peptide, as determined using an ELISA.
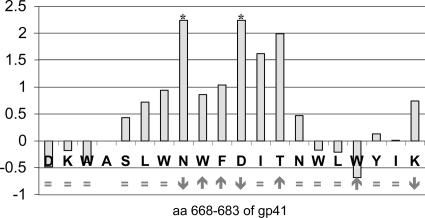
A series of Ala mutant peptides that scan the MPER residues 664 to 683 were used in the above-described competition ELISA format. Replacement of either N671 or D674 with Ala knocks out Z13e1 recognition (Fig. 3). Several other alanine replacements, including W670, W672, and F673 (∼10-fold), I675 (∼40-fold), and T676 (∼100-fold), diminish Z13e1 binding by 10-fold or more. Other alanine substitutions had moderate to no effect on Z13e1 affinity (e.g., D674A, K665A, W666A, S668A, L669A, N677A, W678A, L679A, Y681A, I682A, and K683A), whereas W680A enhanced Z13e1 binding by severalfold. Analysis of peptide secondary structure by CD spectroscopy showed that the parental peptides and many of the Ala analogs were helical in solution with essentially indistinguishable CD spectra. However, substitutions N671A, D674A, and K683A disrupted structure due to probable aggregation and, interestingly, all of these variant peptides had diminished affinity for Z13e1. Alanine replacements W672A, F673A, T676A, and W680A enhanced the observed helical content relative to the parental peptide, but no clear correlation between enhanced helicity and Z13e1 binding is apparent (Fig. 3).
FIG. 3.
The relative Z13e1 binding affinities and helical contents for a series of alanine-substituted MPER peptides. Each bar represents the log of the ratio of the IC50 of the Ala mutant to the IC50 of the isomorphous reference peptide. Higher bars indicate poorer relative binding for each Ala mutant at the indicated residue position (e.g., a value of 1 means the Ala substitution causes a 10-fold drop in affinity of Z13e1 for the Ala mutant peptide). The effect of the Ala replacement on the CD spectra is shown below each bar as follows: no change, =; loss of helical content due to probable aggregation, ↓; or enhanced helical content, ↑. The asterisks indicate a minimum value corresponding to the highest concentration of Ala mutant peptide tested in the competition assay (10 mM) for which an IC50 was not reached. The reference peptide for Ala W672A, F673A, I675A, T676A, and W680A mutants was SLWNWFDITNWLWKKKK; the reference peptide DKWASLWNWFDITNWLWYIKKKK was used for D664A, K665A, and W666A mutants; and reference peptide SLWNWFDITNWLWYIKKKK was used for the other Ala mutants.
Z13e1 competition with 2F5 and 4E10.
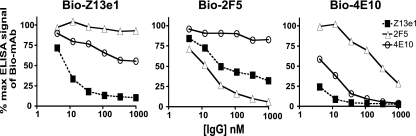
Next, we tested the ability of IgGs Z13e1, 2F5, and 4E10 to compete with each other for binding to M41xt, the MBP-gp41JR-FL ectodomain fusion protein, by use of a competition ELISA format. Biotinylated IgGs were prepared and added to microwells that were coated with M41xt and pretreated with different concentrations of (nonbiotinylated) Z13e1, 2F5, and 4E10 as competitor MAbs. As expected, all three Abs could compete very effectively with their biotinylated counterparts for M41xt recognition, although Z13e1 was even more effective than 4E10 in inhibiting biotinylated 4E10 (Fig. 4). In fact, Z13e1 was able to efficiently block both biotinylated 4E10 and biotinylated 2F5, but 2F5 and 4E10 were less effective inhibitors of biotinylated Z13e1 recognition of M41xt (Fig. 4). These results suggest that Z13e1 binds to recombinant HIV-1 gp41 with very high affinity to an epitope that impinges on the other two, but perhaps with greater overlap with the 4E10 epitope.
FIG. 4.
Competition ELISA using IgGs Z13e1, 2F5, and 4E10 against their biotinylated (Bio) counterparts for recognition of immobilized M41xt (MBP fused to gp41JRFL ectodomain; aa 535 to 681). Biotinylated IgG Z13e1 (left panel), biotinylated IgG 4E10 (middle panel), and biotinylated IgG 2F5 (right panel) were added to wells that were coated with immobilized M41xt and pretreated with various concentrations of competing MAb. Each data point represents the mean of duplicate samples.
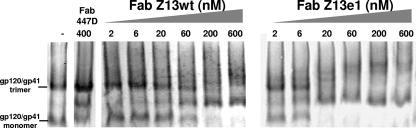
Z13e1 binds to trimeric, virion-associated envelope glycoprotein.
HIV-1 envelope glycoprotein that is gently liberated from the virion surface with a mild detergent runs as a mixture of trimeric and monomeric species of gp120-gp41 heterodimers by use of BN-PAGE (31). In this system, the gel mobility of each species can be “shifted” using envelope-specific Abs (31). We used Fabs Z13 (wt) and Z13e1 against pseudotyped HIV-1JR-FL at graded concentrations and found that both Fabs were able to shift the bands corresponding to both the monomer and the trimer species (Fig. 5). As might be expected, Fab Z13e1 was able to completely shift both envelope glycoprotein species at a much lower concentration than parental Fab Z13 (wt) (1 μg/ml versus 10 to 30 μg/ml). In contrast, the 447-52D control Ab against gp120 (V3) clearly binds to monomers, but trimer binding was negligible, in accordance with its modest neutralization potency against HIV-1JR-FL (4, 12).
FIG. 5.
Abilities of Fabs Z13 (wt) and Z13e1 to bind to monomeric and trimeric forms of virion-liberated HIV-1JRFL envelope glycoproteins (gp120-gp41), as determined using a BN-PAGE band shift assay. The leftmost lane and the one next to it show gp120/gp41 trimer and monomer bands in the absence of any added MAb and in the presence of 20 μg/ml Fab 447-52D (against V3 on gp120), respectively.
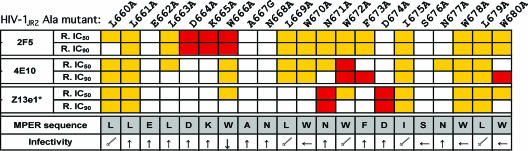
Neutralization of HIV-1 with Z13e1. (i) Neutralization of HIV-1JR2 MPER alanine mutants.
We recently reported on the effects of alanine substitutions in the MPER on the ability of 2F5 and 4E10 to neutralize the pseudotyped virus, HIV-1JR2, by use of a single-round infectivity assay (61). These HIV-1JR2 MPER Ala mutants were all found to be equally sensitive to IgG b12 and the fusion-inhibiting peptide T20 (61). Using the same Ala mutant viruses, we found that N671A and D674A mutants are uniquely resistant to Z13e1 but remain sensitive to the other two MAbs, whereas L660A, L661A, I675A, W678A, and L679A mutants are all hypersensitive to neutralization by Z13e1, 2F5, and 4E10. The remaining MPER Ala mutants are about as sensitive as parental HIV-1JR2 to Z13e1 (Fig. 6). Interestingly, W666A and W672A mutants, which were completely resistant to neutralization by 2F5 and 4E10, respectively, are more sensitive than the parental virus to Z13e1. In general, similar to our previous findings with 2F5 and 4E10, only a very small number of residues (i.e., two) within the linear epitope of the MPER are crucial in order for Z13e1 to neutralize HIV-1JR2 (Fig. 6), even though many alanine substitutions diminish Z13e1 binding to MPER peptides (Fig. 3).
FIG. 6.
Relative neutralization sensitivities of MPER Ala mutants of pseudotyped HIV-1JR2 to Z13e1, 4E10, and 2F5 in the single-round infectivity assay using U87.CD4.CCR5 target cells. Each HIV-1JR2 MPER Ala mutant bears a single Ala substitution in the MPER of gp41 (L660 to W680; primary sequence is shown). The 2F5 and 4E10 data are taken from a previous study (61), and the Z13e1 data are from the present study (*). R. IC50 and R. IC90 refer to the ratios IC50 for mutant/IC50 for parental JR2 and IC90 for mutant/IC90 for parental JR2, respectively, so that mutant viruses are scored as >3 (neutralization resistant; red boxes), between 1/3 and 1 (neutralization sensitivity unchanged; white boxes), and less than 1/3 (neutralization hypersensitive; yellow boxes). The results shown represent the averages of three independent experiments performed in triplicate. The infectivity data are taken from a previous study: Ala mutant viruses are scored as having infectivities that are either similar to (↑) or ∼30% (←), <30% (↙), or <10% (↓) than that observed for parental HIV-1JR2 infectivity (61).
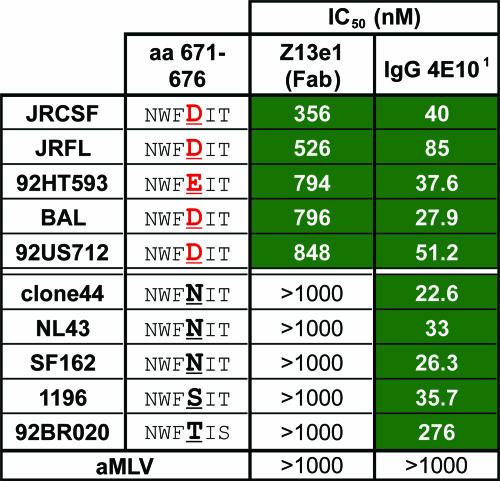
(ii) Fab Z13e1 in the Monogram assay.
Fab Z13e1 was also assayed in an independent laboratory against a small panel of 10 clade B primary isolates by use of a single-cycle infectivity assay (4). Five out of 10 clade B primary isolates were neutralized by Fab Z13e1 (IC50, <1,000 nM) in this format, which, on average, was about 14-fold less potent at the IC50 level than with 4E10 IgG against the same isolates on a molar scale (Fig. 7). We note that the sensitive isolates, but not the resistant ones, bore either an Asp or a Glu residue at position 674, which was also found to be critical in our Ala scan analyses (Fig. 6). Asp is the most commonly observed residue at position 674 in the Los Alamos database (591 out of 979 sequences; http://www.hiv.lanl.gov/content/index [accessed 4 July 2006]).
FIG. 7.
Neutralization of various pseudotyped clade B primary isolates of HIV-1 by Fab Z13e1 and 4E10 IgG by use of a single-cycle infectivity assay (Monogram). 1, The IC50 data for IgG 4E10 were taken from a previous study performed using the same assay under identical conditions (4); the IC50s for Fab Z13e1 are new data. The IC50s that are less than 1,000 nM are shaded green for emphasis. The residues at position 674 are in bold and underlined, as this position is crucial for Z13e1 recognition (Fig. 3 and 6), and the residues D674 and E674, which are permissive for Z13e1 neutralization, are in red for emphasis. An amphotropic murine leukemia virus (aMLV) envelope-pseudotyped virus was used as a control to rule out nonspecific neutralization.
(iii) Neutralization potencies of IgG and Fab versions of Z13e1 and 4E10.
A side-by-side comparison of Fab and whole IgGs of Z13e1 and 4E10 against pseudotyped HIV-1MN in the single-round infectivity assay showed a very modest (two- to threefold) improvement in potency in going from the Fab to the IgG for both Z13e1 and 4E10 (Table 3). Against the primary isolates HIV-1JR-FL and HIV-1JR-CSF, the differences in neutralization potencies between the Fabs and IgGs of Z13e1 and 4E10 were even less apparent (i.e., 0.6- to 1.9-fold changes) (Table 3). Thus, for Z13e1 and 4E10, although there are some differences in neutralization potencies with the Fab and IgG formats, the effects appear to be subtle. We note that the neutralization potencies observed in these assays with Z13e1 (Fab and IgG) were typically ∼7- to 10-fold lower than those with 4E10 Fab and IgG.
TABLE 3.
Neutralization by Z13e1 and 4E10 (Fab and IgG) of pseudotyped HIV-1 by use of a single-round infectivity assay with U87.CD4.CXCR4 (HIV-1MN) or U87.CD4.CCR5 (HIV-1JR-FL and HIV-1JR-CSF) target cells
| Ab or comparison | Concn (nM) or neutralization potency (n-fold) for indicated straina:
|
|||||
|---|---|---|---|---|---|---|
| HIV-1MN
|
HIV-1JR-FL
|
HIV-1JR-CSF
|
||||
| IC50 | IC90 | IC50 | IC80 | IC50 | IC90 | |
| Z13e1 Fab | 3.8 | 145 | 220 | 1,650 | 550 | 3,200 |
| Z13e1 IgG | 1.8 | 47 | 285 | 2,600 | 600 | 4,000 |
| Z13e1 Fab to Z13e1 IgG | 2.1 | 3.1 | 0.8 | 0.6 | 0.9 | 0.8 |
| 4E10 Fab | 1.4 | 20.5 | 35 | 140 | 120 | 650 |
| 4E10 IgG | 0.8 | 7.5 | 45.5 | 235 | 63 | 400 |
| 4E10 Fab to 4E10 IgG | 1.8 | 2.7 | 0.8 | 0.6 | 1.9 | 1.6 |
Values in lightface are the Ab concentrations required to diminish virus infectivity by 50, 80, or 90%. For HIV-1MN and HIV-1JR-FL, each value represents the mean of two independent experiments performed in triplicate. For HIV-1JR-CSF, each value represents the mean of triplicate samples (single experiment). Values in boldface are Fab-to-IgG neutralization potency changes (n-fold).
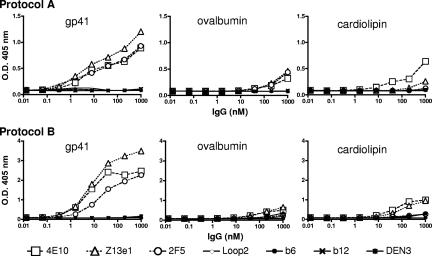
Z13e1 exhibits weak nonspecific binding to various antigens in an ELISA format.
A recent report has suggested that 4E10 and to a lesser extent 2F5 have some reactivity against CL under certain defined ELISA conditions (21). Determining the specific CL cross-reactivity of an Ab is complicated by the difficulty in differentiating true CL reactivity and nonspecific or artifactual binding interactions that lead to false-positive binding (20). Nevertheless, we wanted to evaluate the CL reactivities of 2F5 and 4E10 in the same assay, with Z13e1 IgG now included for comparison. At the same time, we performed a standardized ELISA that has been used in a multicentered study for determining CL reactivity, though we note that this assay too has shortcomings, since the CL is not presented in a stable lipid bilayer (20). In both protocols, all three anti-MPER MAbs showed weak cross-reactivity with CL, but only 4E10 bound to CL better than to ovalbumin in one of the assay formats (Fig. 8). Significantly, all three MAbs bound to gp41 with a titer ∼100-fold higher than that to either CL or ovalbumin. The primary specificity of the three MPER MAbs is therefore to gp41, with very weak cross-reactivity to these other antigens in conditions of limiting detergent.
FIG. 8.
The anti-MPER MAbs Z13e1, 4E10, and 2F5 may react weakly and somewhat nonspecifically with unrelated antigens in an ELISA in which detergent is limiting. IgG b6 against the CD4 binding site of gp120 (42) was used as a negative control. Two protocols were used, including that of Harris et al. (20) (top panels) and that of Haynes et al. (21) (bottom panels).
DISCUSSION
In this study, we identified a high-affinity variant of Fab Z13 through targeted random mutagenesis of CDR L3 and affinity selection against recombinant gp41 and an MPER peptide. The enhanced variant Z13e1 binds to the selecting antigens with affinity ∼100-fold greater than that of parental Fab, which is respectable and not atypical for in vitro affinity maturation experiments (40, 45, 55). In theory, it should be possible to further optimize Z13e1 affinity, although further improvements in neutralization potency may require mutation of other Z13 CDRs and more technically challenging selections using specific antigens that present the MPER of gp41 as in a native trimeric and/or membrane context. There may also be a limitation in neutralization potency by Z13 variants due to steric restrictions to the angle of approach of the Fab arm to its MPER epitope or due to other factors. Nevertheless, the apparent affinity of Z13e1 to optimized MPER peptides (IC50, ∼10 nM) is within the range of binding affinities typically observed for MAbs generated in vivo, such as 2F5 and 4E10 (2, 6). We note that whereas other variants besides Z13e1 were selected, Z13e1 was largely representative of these, though perhaps it had slightly higher affinity. Thus, no strong-binding clones that were nonneutralizing were found, and conversely, no low-affinity clones that had strong neutralizing activity were found. Hence, for the clones we tested at least, improved affinity to MPER antigens was predictive of improved neutralization potency, although Z13e1 neutralizes the sensitive HIV-1MN strain over an order of magnitude more potently than it neutralizes the MPER-sequence-matched primary virus, HIV-1JR-FL.
The mapping studies all suggest strongly that Z13e1 binds to an epitope between those of 2F5 and 4E10. In the peptide truncation analysis, the shortest peptide epitope for Z13e1 for which Ab affinity was near optimum was the 12-mer motif (indicated in boldface) within the peptide 178-3 (WASLWNWFDITNKKKK; IC50, ∼30 nM). The first six and the last seven residues of this 12-mer motif have been implicated in 2F5 and 4E10 recognition, respectively (2, 6, 10, 33). In addition, Z13e1 is clearly able to block the binding of both 2F5 and 4E10 to recombinant gp41. The Ala substitutions N671A and D674A both knock out Z13e1 peptide binding and neutralization, suggesting that the Z13e1 epitope may overlap more with that of 4E10, the crucial residues of which are W672 and F673 (10, 61). Since 4E10 recognizes a largely helical epitope, it has been suggested that this helical region bears distinct “neutralizing” and “nonneutralizing” faces, since access to the MPER would presumably be restricted to an exposed face on the functional trimer (6, 10). If the MPER of gp41 were strictly helical throughout the Z13e1 epitope, that would place the residues most critical to Z13e1 recognition, N671 and D674, on the putative “nonneutralizing” face of the helix, away from W672, the residue that is most buried in the 4E10 paratope (Fig. 9). Thus, an explanation for the relatively modest neutralization potency of Z13e1 may be that its helical epitope impinges on the nonneutralizing face of the MPER, which is occluded by neighboring MPERs. However, a nonhelical epitope for Z13e1 cannot be ruled out. Indeed, the 4E10 core epitope loses a-helical character toward its N-terminal residues (6, 10), and the 2F5-bound peptide adopts an extended conformation (33), suggesting either that there is significant flexibility in the MPER of gp41 or that a distinct structural transition occurs in the segment between the 2F5 and 4E10 epitopes (60). In either case, the high-affinity epitope conformation(s) that is available to Z13e1 on recombinant gp41 and MPER peptides does not appear to be persistently present on the cognate epitope found on the native trimer (as evidenced by the low neutralization potency of Z13e1). A structure of Z13e1 in complex with a peptide that overlaps with those used in the structure determinations of 2F5 and 4E10 (10, 33) should help to explain the limited ability of Z13e1 to bind to the native Env trimer and refine the existing structural models of the MPER (59, 60).
FIG. 9.
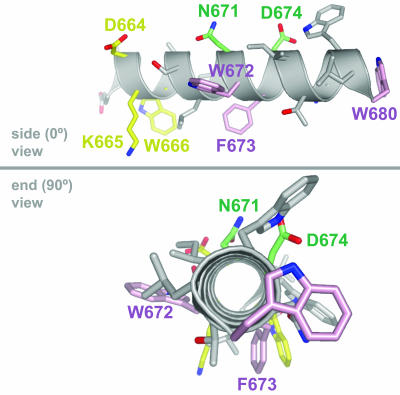
Hypothetical model of the MPER of HIV-1 gp41 as an ideal alpha helix, in side (0°) and end-on (90°) views, places 4E10 and Z13e1 crucial binding residues on opposite faces of the helix. The MPER residues that are crucial in the neutralizing activity of 2F5, Z13e1, and 4E10 are shown in yellow, green, and pink, respectively. The model of the MPER sequence DKWASLWNWFDITNWLW was created using the INSIGHT II package (Accelrys, Inc., San Diego, CA) and the PyMOL program (The PyMOL Molecular Graphics System; DeLano Scientific, San Carlos, CA) (16).
It is perhaps not surprising that Fab Z13e1 was able to bind both monomeric and trimeric Env species by use of the BN-PAGE gel shift assay because Z13e1 recognizes a continuous epitope. Given the established relationship between neutralization and the ability of Ab to bind to trimeric envelope glycoprotein (18, 31, 36, 38, 43, 54, 56), the neutralizing activity of Fab Z13e1 most likely derives from its ability to bind to the trimeric envelope, even though monomer binding affinity is strong and is improved to an extent similar to that seen for trimer binding affinity.
The Fab and IgG formats of Z13e1 appeared to be roughly equivalent in neutralization potency on a molar scale, differing by only threefold or less. Such a result may reflect the inability of two Fab arms of the IgG to bind simultaneously to a functional Env trimer(s), although other steric factors affecting the accessibility to receptor-bound envelope trimers may also influence neutralization by IgG (27). We also found that neutralization of HIV-1JR-FL by 4E10 was not greatly improved in going from the Fab to the IgG format. This result is in apparent conflict with a previously published result in which IgG 4E10 was found to be severalfold more potent than Fab against HIV-1JR-FL (33). The reason for this discrepancy is not clear, although a different assay format and target cells were used in the previous study.
As with an earlier study involving 2F5 and 4E10 (21), we found that Z13e1 bound weakly and somewhat nonspecifically to different antigens in an ELISA setup in which detergent was limiting, although its reactivity was typically less than that of 4E10 (nonspecific binding with 2F5 was the least of the three). It is presently unclear what this low-level and nonspecific binding means in terms of vaccine design involving the MPER of gp41. However, if there were tolerance mechanisms that would limit the elicitation of neutralizing anti-MPER Abs, they would less likely be a result of Ab cross-reactivity with a specific host antigen than be attributable to a more general phenomenon associated with their specificity for a membrane-proximal epitope. A casual inspection of the primary sequences of the heavy- and light-chain CDRs of Z13e1 reveals a relatively high content of hydrophobic relative to charged residues in the heavy-chain CDRs (16 hydrophobic versus 4 charged residues), particularly in CDRs H1 and H3 (64). The subtle “stickiness” of Z13e1 may relate to these hydrophobic CDRs, which could facilitate recognition of a hydrophobic epitope very close to a membrane. Nevertheless, Z13e1 and the other anti-MPER MAbs bind with very high (low-nanomolar) affinity to their MPER peptide epitopes. A more detailed analysis of the interaction of 2F5 and 4E10 with CL and other lipid antigens is in progress (Erin Scherer, MBZ, DRB). Isolating and studying more Abs against the MPER of gp41 will also help to determine if such Abs are subject to a strict requirement for the type of weak and nonspecific reactivity to unrelated antigens that we observed with the existing three MPER MAbs.
The concentration of neutralizing Ab required to protect against challenge with HIV has been suggested to be on the order of 100-fold greater than that required to completely neutralize the virus in a classical peripheral blood mononuclear cell-based assay (35). Furthermore, neutralizing Ab alone is likely to have limited control over an established HIV-1 infection (39). In fact, the potential of 2F5 and 4E10 to contribute to protection against HIV-1 has been questioned due to a clinical study in which a passively administered cocktail of 2G12, 2F5, and 4E10 showed a limited ability to control viral rebound in several HIV-1-infected patients (50). Since the potency of Z13 was so readily improved in this study, a similar approach might enable improvements to the potencies of 2F5 or 4E10, although the affinities of 2F5 and 4E10 for gp41 peptides are already quite high (Kds in the low nanomolar range), so improvements to these MAbs may be more challenging. In order to determine the in vitro neutralization titers that will have to be elicited by immunogens based on the MPER, it will be important to establish the individual doses of 2F5 and 4E10 required for protection in vivo.
Using HIV-2 chimeric viruses that bear HIV-1 MPER sequences, MPER-specific neutralizing Abs have recently been detected in the sera of certain HIV-1 infected individuals; these Abs do not recognize the 2F5 or 4E10 epitopes (G. F. Shaw, Bibollet-Ruche, J. Decker, H. Li, P. Goepfert, M. Peeters, S. Allen, E. Hunter, J. Robinson, and P. Kwong, presented at the 13th Conference on Retroviruses and Opportunistic Infections, Colorado Conference Center, Denver, CO, 2006). Perhaps these Abs, like Z13e1, recognize epitopes between those of 2F5 and 4E10. If Abs are more easily elicited to the segment between the 2F5 and 4E10 epitopes than to these two epitopes themselves, it will be important to determine their neutralization potencies. We speculate that the segment of the MPER to which Z13e1 binds may elicit Abs that are more potent than Z13e1. Such Abs would have to bind with affinities greater than that of Z13e1 to the functional Env trimer, regardless of their affinities for MPER peptides or recombinant gp41 (60). MPER immunogens designed to elicit potently neutralizing Abs will need to reproduce the structural features that limit access to the Z13e1 epitope but preserve access to the 2F5 and 4E10 epitopes, as found on the functional Env trimer.
An HIV-1 envelope mimetic against which in vitro affinity maturation of a neutralizing MAb leads to diminished neutralization potency is probably a poor vaccine candidate. This concept was appreciated in a prior study in which the anti-gp120 neutralizing Ab, b12, was affinity optimized against monomeric gp120, eventually leading to decreased neutralization potency (24). In the present study, affinity maturation of Z13 against recombinant gp41 and an MPER peptide did translate to improved neutralization potency, but the potency of Z13e1 is still less than those of 2F5 and 4E10, which bind to these antigens in vitro with affinities comparable to that of Z13e1. The observed modest neutralization potency of IgG Z13e1, despite its very strong binding affinity for MPER peptides and recombinant gp41, strongly suggests that there are limitations in exposure of the Z13e1 epitope, and possibly others like it, on the native envelope trimer. Firmer conclusions as to whether these limitations are due to steric occlusion of a static conformation of the MPER or to short-lived access to a transient epitope on a flexible MPER await crystallographic studies with a Z13e1-peptide complex. Z13e1 Fab may also be useful as a probe in cryo-electron microscopy studies to determine whether the MPERs within the native Env trimer are strictly separated as in a “tripod,” are bundled together as in a single “trunk,” or perhaps participate in a more dynamic equilibrium between these two states. Finely optimizing the presentation of the “neutralizing face” and native conformation of the MPER may be crucial in order to elicit Abs with neutralization potencies equal to or exceeding those of 2F5 and 4E10.
Acknowledgments
We acknowledge Christos Petropoulos and Monogram for performing neutralization assays. We thank R. Pantophlet for supplying reagents, D. Tehrani and M. Elliott for technical assistance, E. Scherer for useful discussions, B. Haynes for sharing details of the CL-binding assay protocol, and L. Hangartner for assistance with figures.
We acknowledge support from the following: an AmFAR fellowship and fellowship F06-SRI-260 of the University AIDS Research Program at the University of California (F.M.B.); the Neutralizing Antibody Consortium of the International AIDS Vaccine Initiative; NIH AI 058725 (M.B.Z.), AI 33292 (D.R.B.), GM-46192 (I.A.W.), and RO1 AI 58763; Bill and Melinda Gates Foundation Vaccine Immune Monitoring Consortium grant 38619; and the AIDS and Infectious Disease Science Center of the Torrey Pines Institute for Molecular Studies (J.M.B.), P01 GM048870 (P.E.D.).
Footnotes
Published ahead of print on 7 February 2007.
REFERENCES
- 1.Barbas, C. F., III, D. R. Burton, J. K. Scott, and G. J. Silverman. 2001. Phage display: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 2.Barbato, G., E. Bianchi, P. Ingallinella, W. H. Hurni, M. D. Miller, G. Ciliberto, R. Cortese, R. Bazzo, J. W. Shiver, and A. Pessi. 2003. Structural analysis of the epitope of the anti-HIV antibody 2F5 sheds light into its mechanism of neutralization and HIV fusion. J. Mol. Biol. 330:1101-1115. [DOI] [PubMed] [Google Scholar]
- 3.Binley, J. M., H. J. Ditzel, C. F. Barbas III, N. Sullivan, J. Sodroski, P. W. Parren, and D. R. Burton. 1996. Human antibody responses to HIV type 1 glycoprotein 41 cloned in phage display libraries suggest three major epitopes are recognized and give evidence for conserved antibody motifs in antigen binding. AIDS Res. Hum. Retrovir. 12:911-924. [DOI] [PubMed] [Google Scholar]
- 4.Binley, J. M., T. Wrin, B. Korber, M. B. Zwick, M. Wang, C. Chappey, G. Stiegler, R. Kunert, S. Zolla-Pazner, H. Katinger, C. J. Petropoulos, and D. R. Burton. 2004. Comprehensive cross-clade neutralization analysis of a panel of anti-human immunodeficiency virus type 1 monoclonal antibodies. J. Virol. 78:13232-13252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bjorndal, A., H. Deng, M. Jansson, J. R. Fiore, C. Colognesi, A. Karlsson, J. Albert, G. Scarlatti, D. R. Littman, and E. M. Fenyo. 1997. Coreceptor usage of primary human immunodeficiency virus type 1 isolates varies according to biological phenotype. J. Virol. 71:7478-7487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brunel, F. M., M. B. Zwick, R. M. Cardoso, J. D. Nelson, I. A. Wilson, D. R. Burton, and P. E. Dawson. 2006. Structure-function analysis of the epitope for 4E10, a broadly neutralizing human immunodeficiency virus type 1 antibody. J. Virol. 80:1680-1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buchacher, A., R. Predl, K. Strutzenberger, W. Steinfellner, A. Trkola, M. Purtscher, G. Gruber, C. Tauer, F. Steindl, A. Jungbauer, et al. 1994. Generation of human monoclonal antibodies against HIV-1 proteins; electrofusion and Epstein-Barr virus transformation for peripheral blood lymphocyte immortalization. AIDS Res. Hum. Retrovir. 10:359-369. [DOI] [PubMed] [Google Scholar]
- 8.Burton, D. R., J. Pyati, R. Koduri, S. J. Sharp, G. B. Thornton, P. W. Parren, L. S. Sawyer, R. M. Hendry, N. Dunlop, P. L. Nara, et al. 1994. Efficient neutralization of primary isolates of HIV-1 by a recombinant human monoclonal antibody. Science 266:1024-1027. [DOI] [PubMed] [Google Scholar]
- 9.Burton, D. R., R. L. Stanfield, and I. A. Wilson. 2005. Antibody vs. HIV in a clash of evolutionary titans. Proc. Natl. Acad. Sci. USA 102:14943-14948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cardoso, R. M., M. B. Zwick, R. L. Stanfield, R. Kunert, J. M. Binley, H. Katinger, D. R. Burton, and I. A. Wilson. 2005. Broadly neutralizing anti-HIV antibody 4E10 recognizes a helical conformation of a highly conserved fusion-associated motif in gp41. Immunity 22:163-173. [DOI] [PubMed] [Google Scholar]
- 11.Chowdhury, P. S., and I. Pastan. 1999. Improving antibody affinity by mimicking somatic hypermutation in vitro. Nat. Biotechnol. 17:568-572. [DOI] [PubMed] [Google Scholar]
- 12.Conley, A. J., M. K. Gorny, J. A. Kessler II, L. J. Boots, M. Ossorio-Castro, S. Koenig, D. W. Lineberger, E. A. Emini, C. Williams, and S. Zolla-Pazner. 1994. Neutralization of primary human immunodeficiency virus type 1 isolates by the broadly reactive anti-V3 monoclonal antibody, 447-52D. J. Virol. 68:6994-7000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Connor, R. I., B. K. Chen, S. Choe, and N. R. Landau. 1995. Vpr is required for efficient replication of human immunodeficiency virus type-1 in mononuclear phagocytes. Virology 206:935-944. [DOI] [PubMed] [Google Scholar]
- 14.Crooks, E. T., P. L. Moore, D. Richman, J. Robinson, J. A. Crooks, M. Franti, N. Schulke, and J. M. Binley. 2005. Characterizing anti-HIV monoclonal antibodies and immune sera by defining the mechanism of neutralization. Hum. Antib. 14:101-113. [PMC free article] [PubMed] [Google Scholar]
- 15.Deeks, S. G., B. Schweighardt, T. Wrin, J. Galovich, R. Hoh, E. Sinclair, P. Hunt, J. M. McCune, J. N. Martin, C. J. Petropoulos, and F. M. Hecht. 2006. Neutralizing antibody responses against autologous and heterologous viruses in acute versus chronic human immunodeficiency virus (HIV) infection: evidence for a constraint on the ability of HIV to completely evade neutralizing antibody responses. J. Virol. 80:6155-6164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.DeLano, W. L. 2002. Unraveling hot spots in binding interfaces: progress and challenges. Curr. Opin. Struct. Biol. 12:14-20. [DOI] [PubMed] [Google Scholar]
- 17.Earl, P. L., C. C. Broder, R. W. Doms, and B. Moss. 1997. Epitope map of human immunodeficiency virus type 1 gp41 derived from 47 monoclonal antibodies produced by immunization with oligomeric envelope protein. J. Virol. 71:2674-2684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fouts, T. R., J. M. Binley, A. Trkola, J. E. Robinson, and J. P. Moore. 1997. Neutralization of the human immunodeficiency virus type 1 primary isolate JR-FL by human monoclonal antibodies correlates with antibody binding to the oligomeric form of the envelope glycoprotein complex. J. Virol. 71:2779-2785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gorny, M. K., and S. Zolla-Pazner. 2000. Recognition by human monoclonal antibodies of free and complexed peptides representing the prefusogenic and fusogenic forms of human immunodeficiency virus type 1 gp41. J. Virol. 74:6186-6192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harris, E. N., S. Pierangeli, and D. Birch. 1994. Anticardiolipin wet workshop report. Fifth International Symposium on Antiphospholipid Antibodies. Am. J. Clin. Pathol. 101:616-624. [DOI] [PubMed] [Google Scholar]
- 21.Haynes, B. F., J. Fleming, W. E. St Clair, H. Katinger, G. Stiegler, R. Kunert, J. Robinson, R. M. Scearce, K. Plonk, H. F. Staats, T. L. Ortel, H. X. Liao, and M. S. Alam. 2005. Cardiolipin polyspecific autoreactivity in two broadly neutralizing HIV-1 antibodies. Science 308:1906-1908. [DOI] [PubMed] [Google Scholar]
- 22.Horton, R. M., H. D. Hunt, S. N. Ho, J. K. Pullen, and L. R. Pease. 1989. Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension. Gene 77:61-68. [DOI] [PubMed] [Google Scholar]
- 23.Jiang, S., K. Lin, and M. Lu. 1998. A conformation-specific monoclonal antibody reacting with fusion-active gp41 from the human immunodeficiency virus type 1 envelope glycoprotein. J. Virol. 72:10213-10217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kessler, J. A., II, P. M. McKenna, E. A. Emini, C. P. Chan, M. D. Patel, S. K. Gupta, G. E. Mark III, C. F. Barbas III, D. R. Burton, and A. J. Conley. 1997. Recombinant human monoclonal antibody IgG1b12 neutralizes diverse human immunodeficiency virus type 1 primary isolates. AIDS Res. Hum. Retrovir. 13:575-582. [DOI] [PubMed] [Google Scholar]
- 25.Klausner, R. D., A. S. Fauci, L. Corey, G. J. Nabel, H. Gayle, S. Berkley, B. F. Haynes, D. Baltimore, C. Collins, R. G. Douglas, J. Esparza, D. P. Francis, N. K. Ganguly, J. L. Gerberding, M. I. Johnston, M. D. Kazatchkine, A. J. McMichael, M. W. Makgoba, G. Pantaleo, P. Piot, Y. Shao, E. Tramont, H. Varmus, and J. N. Wasserheit. 2003. Medicine. The need for a global HIV vaccine enterprise. Science 300:2036-2039. [DOI] [PubMed] [Google Scholar]
- 26.Kostrikis, L. G., Y. Cao, H. Ngai, J. P. Moore, and D. D. Ho. 1996. Quantitative analysis of serum neutralization of human immunodeficiency virus type 1 from subtypes A, B, C, D, E, F, and I: lack of direct correlation between neutralization serotypes and genetic subtypes and evidence for prevalent serum-dependent infectivity enhancement. J. Virol. 70:445-458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Labrijn, A. F., P. Poignard, A. Raja, M. B. Zwick, K. Delgado, M. Franti, J. Binley, V. Vivona, C. Grundner, C. C. Huang, M. Venturi, C. J. Petropoulos, T. Wrin, D. S. Dimitrov, J. Robinson, P. D. Kwong, R. T. Wyatt, J. Sodroski, and D. R. Burton. 2003. Access of antibody molecules to the conserved coreceptor binding site on glycoprotein gp120 is sterically restricted on primary human immunodeficiency virus type 1. J. Virol. 77:10557-10565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liang, X., S. Munshi, J. Shendure, G. Mark III, M. E. Davies, D. C. Freed, D. C. Montefiori, and J. W. Shiver. 1999. Epitope insertion into variable loops of HIV-1 gp120 as a potential means to improve immunogenicity of viral envelope protein. Vaccine 17:2862-2872. [DOI] [PubMed] [Google Scholar]
- 29.Mascola, J. R. 2003. Defining the protective antibody response for HIV-1. Curr. Mol. Med. 3:209-216. [DOI] [PubMed] [Google Scholar]
- 30.Moore, J. P., Y. Cao, J. Leu, L. Qin, B. Korber, and D. D. Ho. 1996. Inter- and intraclade neutralization of human immunodeficiency virus type 1: genetic clades do not correspond to neutralization serotypes but partially correspond to gp120 antigenic serotypes. J. Virol. 70:427-444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moore, P. L., E. T. Crooks, L. Porter, P. Zhu, C. S. Cayanan, H. Grise, P. Corcoran, M. B. Zwick, M. Franti, L. Morris, K. H. Roux, D. R. Burton, and J. M. Binley. 2006. Nature of nonfunctional envelope proteins on the surface of human immunodeficiency virus type 1. J. Virol. 80:2515-2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Muster, T., F. Steindl, M. Purtscher, A. Trkola, A. Klima, G. Himmler, F. Ruker, and H. Katinger. 1993. A conserved neutralizing epitope on gp41 of human immunodeficiency virus type 1. J. Virol. 67:6642-6647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ofek, G., M. Tang, A. Sambor, H. Katinger, J. R. Mascola, R. Wyatt, and P. D. Kwong. 2004. Structure and mechanistic analysis of the anti-HIV-1 antibody 2F5 in complex with its epitope. J. Virol. 78:10724-10737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pantophlet, R., and D. R. Burton. 2006. GP120: target for neutralizing HIV-1 antibodies. Annu. Rev. Immunol. 24:739-769. [DOI] [PubMed] [Google Scholar]
- 35.Parren, P. W., P. A. Marx, A. J. Hessell, A. Luckay, J. Harouse, C. Cheng-Mayer, J. P. Moore, and D. R. Burton. 2001. Antibody protects macaques against vaginal challenge with a pathogenic R5 simian/human immunodeficiency virus at serum levels giving complete neutralization in vitro. J. Virol. 75:8340-8347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Parren, P. W., I. Mondor, D. Naniche, H. J. Ditzel, P. J. Klasse, D. R. Burton, and Q. J. Sattentau. 1998. Neutralization of human immunodeficiency virus type 1 by antibody to gp120 is determined primarily by occupancy of sites on the virion irrespective of epitope specificity. J. Virol. 72:3512-3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Parren, P. W., M. Wang, A. Trkola, J. M. Binley, M. Purtscher, H. Katinger, J. P. Moore, and D. R. Burton. 1998. Antibody neutralization-resistant primary isolates of human immunodeficiency virus type 1. J. Virol. 72:10270-10274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Poignard, P., M. Moulard, E. Golez, V. Vivona, M. Franti, S. Venturini, M. Wang, P. W. Parren, and D. R. Burton. 2003. Heterogeneity of envelope molecules expressed on primary human immunodeficiency virus type 1 particles as probed by the binding of neutralizing and nonneutralizing antibodies. J. Virol. 77:353-365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Poignard, P., R. Sabbe, G. R. Picchio, M. Wang, R. J. Gulizia, H. Katinger, P. W. Parren, D. E. Mosier, and D. R. Burton. 1999. Neutralizing antibodies have limited effects on the control of established HIV-1 infection in vivo. Immunity 10:431-438. [DOI] [PubMed] [Google Scholar]
- 40.Rajpal, A., N. Beyaz, L. Haber, G. Cappuccilli, H. Yee, R. R. Bhatt, T. Takeuchi, R. A. Lerner, and R. Crea. 2005. A general method for greatly improving the affinity of antibodies by using combinatorial libraries. Proc. Natl. Acad. Sci. USA 102:8466-8471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Richman, D. D., T. Wrin, S. J. Little, and C. J. Petropoulos. 2003. Rapid evolution of the neutralizing antibody response to HIV type 1 infection. Proc. Natl. Acad. Sci. USA 100:4144-4149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Roben, P., J. P. Moore, M. Thali, J. Sodroski, C. F. Barbas III, and D. R. Burton. 1994. Recognition properties of a panel of human recombinant Fab fragments to the CD4 binding site of gp120 that show differing abilities to neutralize human immunodeficiency virus type 1. J. Virol. 68:4821-4828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sattentau, Q. J., and J. P. Moore. 1995. Human immunodeficiency virus type 1 neutralization is determined by epitope exposure on the gp120 oligomer. J. Exp. Med. 182:185-196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schagger, H., W. A. Cramer, and G. von Jagow. 1994. Analysis of molecular masses and oligomeric states of protein complexes by blue native electrophoresis and isolation of membrane protein complexes by two-dimensional native electrophoresis. Anal. Biochem. 217:220-230. [DOI] [PubMed] [Google Scholar]
- 45.Schier, R., A. McCall, G. P. Adams, K. W. Marshall, H. Merritt, M. Yim, R. S. Crawford, L. M. Weiner, C. Marks, and J. D. Marks. 1996. Isolation of picomolar affinity anti-c-erbB-2 single-chain Fv by molecular evolution of the complementarity determining regions in the center of the antibody binding site. J. Mol. Biol. 263:551-567. [DOI] [PubMed] [Google Scholar]
- 46.Schnolzer, M., P. Alewood, A. Jones, D. Alewood, and S. B. Kent. 1992. In situ neutralization in Boc-chemistry solid phase peptide synthesis. Rapid, high yield assembly of difficult sequences. Int. J. Pept. Protein Res. 40:180-193. [DOI] [PubMed] [Google Scholar]
- 47.Schulke, N., M. S. Vesanen, R. W. Sanders, P. Zhu, M. Lu, D. J. Anselma, A. R. Villa, P. W. Parren, J. M. Binley, K. H. Roux, P. J. Maddon, J. P. Moore, and W. C. Olson. 2002. Oligomeric and conformational properties of a proteolytically mature, disulfide-stabilized human immunodeficiency virus type 1 gp140 envelope glycoprotein. J. Virol. 76:7760-7776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stiegler, G., R. Kunert, M. Purtscher, S. Wolbank, R. Voglauer, F. Steindl, and H. Katinger. 2001. A potent cross-clade neutralizing human monoclonal antibody against a novel epitope on gp41 of human immunodeficiency virus type 1. AIDS Res. Hum. Retrovir. 17:1757-1765. [DOI] [PubMed] [Google Scholar]
- 49.Sullivan, N., Y. Sun, Q. Sattentau, M. Thali, D. Wu, G. Denisova, J. Gershoni, J. Robinson, J. Moore, and J. Sodroski. 1998. CD4-induced conformational changes in the human immunodeficiency virus type 1 gp120 glycoprotein: consequences for virus entry and neutralization. J. Virol. 72:4694-4703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Trkola, A., H. Kuster, P. Rusert, B. Joos, M. Fischer, C. Leemann, A. Manrique, M. Huber, M. Rehr, A. Oxenius, R. Weber, G. Stiegler, B. Vcelar, H. Katinger, L. Aceto, and H. F. Gunthard. 2005. Delay of HIV-1 rebound after cessation of antiretroviral therapy through passive transfer of human neutralizing antibodies. Nat. Med. 11:615-622. [DOI] [PubMed] [Google Scholar]
- 51.Trkola, A., M. Purtscher, T. Muster, C. Ballaun, A. Buchacher, N. Sullivan, K. Srinivasan, J. Sodroski, J. P. Moore, and H. Katinger. 1996. Human monoclonal antibody 2G12 defines a distinctive neutralization epitope on the gp120 glycoprotein of human immunodeficiency virus type 1. J. Virol. 70:1100-1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vujcic, L. K., and G. V. Quinnan, Jr. 1995. Preparation and characterization of human HIV type 1 neutralizing reference sera. AIDS Res. Hum. Retrovir. 11:783-787. [DOI] [PubMed] [Google Scholar]
- 53.Wei, X., J. M. Decker, S. Wang, H. Hui, J. C. Kappes, X. Wu, J. F. Salazar-Gonzalez, M. G. Salazar, J. M. Kilby, M. S. Saag, N. L. Komarova, M. A. Nowak, B. H. Hahn, P. D. Kwong, and G. M. Shaw. 2003. Antibody neutralization and escape by HIV-1. Nature 422:307-312. [DOI] [PubMed] [Google Scholar]
- 54.Wyatt, R., and J. Sodroski. 1998. The HIV-1 envelope glycoproteins: fusogens, antigens, and immunogens. Science 280:1884-1888. [DOI] [PubMed] [Google Scholar]
- 55.Yang, W. P., K. Green, S. Pinz-Sweeney, A. T. Briones, D. R. Burton, and C. F. Barbas III. 1995. CDR walking mutagenesis for the affinity maturation of a potent human anti-HIV-1 antibody into the picomolar range. J. Mol. Biol. 254:392-403. [DOI] [PubMed] [Google Scholar]
- 56.Yang, X., S. Kurteva, S. Lee, and J. Sodroski. 2005. Stoichiometry of antibody neutralization of human immunodeficiency virus type 1. J. Virol. 79:3500-3508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yuste, E., H. B. Sanford, J. Carmody, J. Bixby, S. Little, M. B. Zwick, T. Greenough, D. R. Burton, D. D. Richman, R. C. Desrosiers, and W. E. Johnson. 2006. Simian immunodeficiency virus engrafted with human immunodeficiency virus type 1 (HIV-1)-specific epitopes: replication, neutralization, and survey of HIV-1-positive plasma. J. Virol. 80:3030-3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zanetti, G., J. A. Briggs, K. Grunewald, Q. J. Sattentau, and S. D. Fuller. 2006. Cryo-electron tomographic structure of an immunodeficiency virus envelope complex in situ. PLoS Pathogens 2:e83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhu, P., J. Liu, J. Bess, Jr., E. Chertova, J. D. Lifson, H. Grise, G. A. Ofek, K. A. Taylor, and K. H. Roux. 2006. Distribution and three-dimensional structure of AIDS virus envelope spikes. Nature 441:847-852. [DOI] [PubMed] [Google Scholar]
- 60.Zwick, M. B. 2005. The membrane-proximal external region of HIV-1 gp41: a vaccine target worth exploring. AIDS 19:1725-1737. [DOI] [PubMed] [Google Scholar]
- 61.Zwick, M. B., R. Jensen, S. Church, M. Wang, G. Stiegler, R. Kunert, H. Katinger, and D. R. Burton. 2005. Anti-human immunodeficiency virus type 1 (HIV-1) antibodies 2F5 and 4E10 require surprisingly few crucial residues in the membrane-proximal external region of glycoprotein gp41 to neutralize HIV-1. J. Virol. 79:1252-1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zwick, M. B., R. Kelleher, R. Jensen, A. F. Labrijn, M. Wang, G. V. Quinnan, Jr., P. W. Parren, and D. R. Burton. 2003. A novel human antibody against human immunodeficiency virus type 1 gp120 is V1, V2, and V3 loop dependent and helps delimit the epitope of the broadly neutralizing antibody immunoglobulin G1 b12. J. Virol. 77:6965-6978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zwick, M. B., H. K. Komori, R. L. Stanfield, S. Church, M. Wang, P. W. Parren, R. Kunert, H. Katinger, I. A. Wilson, and D. R. Burton. 2004. The long third complementarity-determining region of the heavy chain is important in the activity of the broadly neutralizing anti-human immunodeficiency virus type 1 antibody 2F5. J. Virol. 78:3155-3161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zwick, M. B., A. F. Labrijn, M. Wang, C. Spenlehauer, E. O. Saphire, J. M. Binley, J. P. Moore, G. Stiegler, H. Katinger, D. R. Burton, and P. W. Parren. 2001. Broadly neutralizing antibodies targeted to the membrane-proximal external region of human immunodeficiency virus type 1 glycoprotein gp41. J. Virol. 75:10892-10905. [DOI] [PMC free article] [PubMed] [Google Scholar]