Abstract
A direct involvement of the PreS domain of the hepatitis B virus (HBV) large envelope protein, and in particular amino acid residues 21 to 47, in virus attachment to hepatocytes has been suggested by many previous studies. Several PreS-interacting proteins have been identified. However, they share few common sequence motifs, and a bona fide cellular receptor for HBV remains elusive. In this study, we aimed to identify PreS-interacting motifs and to search for novel HBV-interacting proteins and the long-sought receptor. PreS fusion proteins were used as baits to screen a phage display library of random peptides. A group of PreS-binding peptides were obtained. These peptides could bind to amino acids 21 to 47 of PreS1 and shared a linear motif (W1T2X3W4W5) sufficient for binding specifically to PreS and viral particles. Several human proteins with such a motif were identified through BLAST search. Analysis of their biochemical and structural properties suggested that lipoprotein lipase (LPL), a key enzyme in lipoprotein metabolism, might interact with PreS and HBV particles. The interaction of HBV with LPL was demonstrated by in vitro binding, virus capture, and cell attachment assays. These findings suggest that LPL may play a role in the initiation of HBV infection. Identification of peptides and protein ligands corresponding to LPL that bind to the HBV envelope will offer new therapeutic strategies against HBV infection.
Attachment of virions to human hepatocyte membrane via the interaction of the viral envelope protein with a specific cell surface receptor is considered the initial step of hepatitis B virus (HBV) infection. Besides host-derived phospholipids, the envelope of an HBV virion contains virus-derived small (SHBs), middle (MHBs), and large (LHBs) surface proteins translated from distinct initiation codons while sharing a common carboxyl domain (i.e., SHBs). Consequently, LHBs contains an extra N-terminal PreS domain, which is further divided into the amino-PreS1 and carboxyl-PreS2 domains. MHBs contains the PreS2 domain but lacks the PreS1 domain. LHBs is associated mainly with infectious virions but can also be found in smaller amounts on rod-shaped subviral particles. On the other hand, the spherical subviral particles are composed mainly of SHBs. LHBs plays a pivotal role in the infection and budding stages of the HBV life cycle. LHBs proteins in the virion envelope exhibit mixed topologies, with their PreS domains protruding either inwardly or outwardly (32, 35). For successful viral attachment to hepatocytes, it is essential that the PreS domain of LHBs is on the virion surface.
The molecular mechanism of the attachment of HBV to hepatocytes remains unclear. Nevertheless, a generally accepted view is of the role of the PreS domain and in particular residues 21 to 47 of PreS1 (PreS121-47) in mediating HBV attachment to a putative viral receptor(s) on hepatocytes. Since Neurath et al. demonstrated the attachment of HBV to HepG2 cells mediated by PreS121-47 (29), several HBV binding proteins have been identified. Interleukin-6 was proposed to contain the recognition site(s) for PreS121-47 (30). A 31-kDa protein found in HepG2 cell lysate was shown to bind to PreS121-47 (13). More recently, human squamous cell carcinoma antigen 1, a membrane protein, was identified in HepG2 cells and shown to interact with PreS121-47 (14). In addition, immunoglobulin A receptor (31, 34), asialoglycoprotein receptor (43), transferrin receptor (18), apolipoprotein H (28, 42), polymerized serum albumin (12, 22), annexin V (19), fibronectin (3), and an 50-kDa serum glycoprotein (4) have also been found to interact with HBV, though not via PreS121-47. These proteins have no obvious sequence motif in common. Conclusive biological data have not yet been presented to validate any candidates as truly involved in HBV infection. Efforts to establish a cell line susceptible to HBV infection were usually not convincingly successful, since exogenous expression of a candidate receptor could not confer the susceptibility for virus infection. It has therefore been postulated that successful HBV infection, involving cell attachment, endocytosis, membrane fusion, and postfusion steps, may require multiple cellular cofactors (11). On the other hand, with the help of the rapid progresses in screening technology and bioinformatics, novel receptor or coreceptor candidates will continue to be discovered.
Undoubtedly, the study of virus-cell interaction will help the development of effective antiviral drugs (10). HBV attachment to hepatocytes is a potential target for antiviral intervention. Molecules or ligands specifically binding to HBV will likely interfere with viral attachment and hence reduce or block infection. Ideally, these molecules may mimic the structural features of the putative HBV receptor. Among various technologies for ligand discovery, phage display has evolved into one of the main approaches. Filamentous phage display libraries of random peptides have proven to be an excellent source of diverse ligands for many proteins as well as a rich source of information regarding structure-function relationship of proteins and protein-protein interactions (5, 39).
In this study, we used recombinant PreS fusion proteins as baits to screen a phage display library of random peptides, with an aim to identify a common PreS-binding motif(s) as a therapeutic candidate. Furthermore, this approach will lead to the discovery of novel HBV-interacting proteins and possibly the long-sought receptor. Several PreS-binding peptides were obtained, and three representative peptides bound predominantly to PreS121-47. These peptides contain a consensus motif that alone could bind to PreS specifically and capture HBV particles. Several human proteins containing such a motif were found through a BLAST search. Analysis of these proteins suggested that the lipoprotein lipase (LPL) represents a novel HBV binding protein interacting with the PreS region.
MATERIALS AND METHODS
Phage display library.
The gene for the pVIII protein of phage M13 was amplified by PCR with VCSM13 (Stratagene, La Jolla, CA) as the template and inserted into pCANTAB5E (Amersham Pharmacia, Uppsala, Sweden), replacing the original gene III. The new vector, designated pFuse8, was used as parental vector for pVIII-based phage display. For construction of the library, an antisense oligonucleotide (5′-ATTGCGGATCCACACC(MNN)12GCCTGCTGCCATTGCTGGCT-3′, where N is A, T, C, or G and M is A or C) was synthesized. A complementary primer (5′-CAGCAATGGCAGCAGGC-3′) was annealed to the 3′ end of the long oligonucleotide, followed by filling in with Klenow polymerase to generate double-stranded DNA, which was digested with BamHI and ligated into pFuse8 precut with SfiI and BamHI. Transformation was performed with XL1-Blue F′ competent cells (Stratagene) by electroporation. The phage library was established by coinfection of the transformed cells with helper phage VCSM13 (multiplicity of infection, 10:1) as previously described (48). To assess the randomness, a phagemid pool from the library was prepared for DNA sequencing with a T7 DNA sequencing kit (Amersham Pharmacia) according to the manufacturer's instruction.
PreS fusion proteins.
pThioHis-PreS and pTXB1-PreS were constructed for the production of thioredoxin-fused PreS (Thio-PreS) and chitin binding domain-fused PreS (CBD-PreS) in Escherichia coli, respectively, as described previously (14). Thio-PreS proteins were purified with the ProBond nickel-chelating Sepharose resin (Invitrogen, Carlsbad, CA) and desalted with Sephadex G25 (Amersham Pharmacia). Chitin resin was used for the preparation of CBD-PreS-coupled beads according to the manufacturer's instruction (New England Biolabs, Ipswich, MA). Protein concentrations were determined with the Bradford assay (New England Biolabs). For in vitro binding assays, the full-length and truncated PreSs were PCR amplified and inserted in pMAL-C2x (New England Biolabs) in frame with the maltose binding protein (MBP).
Library screening.
Five rounds of affinity screening were performed. In the first two rounds and the last round, microwells (Nunc, Roskilde, Denmark) were coated with Thio-PreS (1 μg/well) overnight in 0.1 M bicarbonate (pH 9.5) at 4°C and saturated with 10% nonfat milk in phosphate-buffered saline (PBS) for 2 h at room temperature, and then 1011 phage in 100 μl PBS were applied to the Thio-PreS-coated wells and incubated for 1 h at room temperature. CBD-PreS-coupled chitin resins were used in the third and the fourth rounds of screening. Briefly, 1011 phage were mixed with 30 μl of the CBD-PreS-coupled beads and incubated on a rotating wheel for 10 min at room temperature. After washing four times with PBS containing 0.05% Tween 20, the bound phage were eluted with glycine-HCl, pH 2.0. The acidic elution was quickly neutralized with 2 M Tris base solution. Eluted phages were propagated and precipitated with 0.1 volume of 20% polyethylene glycol 8000-3 M NaCl for the next round of panning.
GST pull-down assays.
Glutathione S-transferase (GST), GST-peptide, and GST fused to the carboxyl domain of LPL (LPLc) were produced in E. coli and purified with glutathione-Sepharose beads (Amersham Pharmacia) according to the manufacturer's instruction. In binding assays, 20 μl of beads fully loaded with GST-peptide or GST (in comparable amounts as verified by sodium dodecyl sulfate-polyacrylamide gel electrophoresis [SDS-PAGE]) were mixed with 10 μg of MBP-PreS in 400 μl PBS containing 0.05% Tween 20. After incubation on a rotating wheel for 1 h at room temperature, beads were washed thoroughly with PBS containing 0.05% Tween 20. Bound MBP-PreS fusion proteins were resolved by SDS-PAGE and analyzed by Western blotting. An anti-PreS1 monoclonal antibody (MAb), 125E11 (21, 47), and an anti-MBP MAb (Santa Cruz Biotechnology, Santa Cruz, CA) were used as primary antibodies in Western blotting. Signals were visualized using the enhanced chemiluminescence method with a horseradish peroxidase-labeled rabbit anti-mouse immunoglobulin (Dako, Carpenteria, CA).
Determination of peptide sequences.
Peptide sequences were deduced from sequences of the inserts of randomly picked individual phagemids. Alignment of the peptides were performed with the Vector NTI Suite (InforMax) with manual adjustment. Peptide-protein BLAST searching was performed online (http://www.ncbi.nlm.nih.gov/BLAST/) using the program of “short, nearly exact matches”.
cDNA clones and expression plasmids.
Total RNA was extracted with the TRIzol reagent (Invitrogen) from human adipose tissues (Ruijin Hospital, Shanghai, China) and used as the template for random-primed reverse transcription with Superscript II (Invitrogen). The full-length cDNA of human LPL was assembled by overlapping PCR using gene-specific primers and inserted into pcDNA3 (Invitrogen) in frame and upstream of the FLAG (DYKDDDDK) tag. Mutations resulting in Trp393Trp394→AlaAla were introduced by overlapping PCR with the internal primers containing the mutations. The coding sequence of the carboxyl domain of LPL (LPLc, amino acids [aa] 313 to 448) was amplified by PCR and inserted into pGEX-2T (Amersham Pharmacia) to create a GST-LPLc expression plasmid. For the expression plasmid of FLAG-LPLc, the GST fragment in pGEX-LPLc was replaced by FLAG.
cDNAs for the extracellular domains of signal-regulatory protein b2 (SIRP-b2) and prolactin receptor were obtained from human liver and adipose tissue by reverse transcription-PCR with gene-specific primers (SIRP-b2, 174f [5′-AAGTGGCAGGTGAGGAGGAG-3′] and 1163B/r [5′-ATAGGATCCAGGGGTAGCATCTGAGC-3′]; prolactin receptor, 292f [5′-AACTTCTGATACATTTCCTG-3′] and 1013B/r [5′-ATAGGATCCACACGGTTGTATC-3′]) and inserted into pcDNA3 (Invitrogen) in frame and upstream of a FLAG-coding sequence.
For expression of GST-fused peptide, an oligonucleotide duplex encoding the peptide was inserted into pGEX-2T in frame with GST. All of the above-mentioned constructs were verified by sequencing. pcDNA3-SHBs contains the coding sequence for HBV small envelope protein (subtype adr-1) driven by the cytomegalovirus early promoter in pcDNA3.
Transient transfections and eukaryotic expression.
HepG2 and COS cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum at 37°C and 5% CO2. To prepare HBV virions and subviral particles composed of only SHBs, p3.6II (17, 24) or pcDNA3-SHBs was transfected into HepG2 cells by the calcium phosphate precipitation method (24). At 72 h posttransfection, the medium supernatants were collected and concentrated with Centricon YM-100 (Millipore). HBsAg production was assessed by enzyme-linked immunosorbent assay (ELISA) with an HBsAg diagnostic kit (SABC, Shanghai, China). For the production of LPL, COS cells were transfected with expression plasmid for FLAG-tagged wild-type or mutant LPL. At 72 h posttransfection, cell lysate was prepared by sonication on ice and concentrated with Centricon YM-10 (Millipore). The production of LPL was verified by Western blotting with an anti-FLAG MAb (Sigma, St. Louis, MO).
ELISAs.
For phage enzyme-linked immunosorbent assay (ELISA), about 1011 phage were applied to microwells coated with purified thioredoxin or Thio-PreS protein (1 μg/well) and incubated for 1 h at room temperature. After washing with PBS containing 0.05% Tween 20, bound phage were detected with an HRP-labeled anti-M13 MAb (Amersham Pharmacia).
In virus capture assays, N-biotinylated consensus (pC, biotin-S-G-S-G-W-T-N-W-W-S-T) and mutant (pM, biotin-S-G-S-G-W-T-N-A-A-S-T [mutated residues are underlined]) peptides were synthesized (GL Biochem, Shanghai, China). Twenty micrograms of each peptide was dissolved in 10 μl of dimethyl sulfoxide and diluted with 1 ml PBS containing 0.1% Tween 20. Biotin-conjugated peptides (100 μl/well) were immobilized on streptavidin-coated plates (Roche, Mannheim, Germany). PBS-diluted HBV serum samples (viral titer determined by real-time PCR; provided by Ruijin Hospital, Shanghai, China) were mixed with an equal volume of an HRP-labeled anti-HBs (SABC) according to the instructions of the manufacturer and incubated in the peptide- or LPL-coated wells for 1 h at 37°C, followed by washing with PBS containing 0.05% Tween 20. Color was developed using the TMB method as described previously (15). For experiments on peptide-HBV interaction, concentrated medium containing HBV particles or SHBs-only subviral particles was added to peptide-coated wells (∼10 μg HBsAg/well) together with the HRP-labeled anti-HBs. In virus capture assay with LPL-FLAG, the recombinant proteins in cell lysates were immobilized on streptavidin wells precoated with biotinylated anti-FLAG (Sigma). In virus capture assay with purified bovine LPL (bLPL) (Sigma), bLPL in PBS was directly applied to microwells.
HBV attachment studies.
The human monocyte cell line THP-1 was cultured in RPMI 1640 supplemented with 10% (vol/vol) fetal calf serum. For macrophage differentiation (16, 26), cells were suspended in 1.6 × 10−7 M phorbol 12-myristate 13-acetate (Sigma) in growth medium and plated in 24-well Falcon multidishes (2 × 105 to 5 × 105 cells/well). After 24 h of induction with phorbol 12-myristate 13-acetate, 10 nM dexamethasone (Sigma) was added to the medium and incubated for 48 h to stimulate the production of LPL (26).
For the detection of LPL expression on the cell surface, we used anti-LPL MAb 5D2 (10 μg/ml; kindly provided by J. D. Brunzell) (8). Heparin treatment (10 mg/ml) was used to remove LPL from the cell surface. For studies of HBV-cell interaction via LPL, THP-1 macrophages were washed with PBS and incubated with 1.5 ml/well of prechilled RPMI 1640 medium containing 30 to 50 μl of HBV-positive serum (containing approximately 1 × 107 to 2 × 107 copies of HBV DNA) for 4 h at 4°C. Cells were washed and fixed with 4% paraformaldehyde in PBS, and the virus bound to the cell surface was detected by incubating cells for 45 min at room temperature with anti-HBs MAb (1:500; MAb 3E7; Dako) or fluorescein isothiocyanate (FITC)-labeled polyclonal anti-HBs antibody (1:50; ab32914; Abcam). In the competition assay, cells were incubated with HBV in the presence of anti-LPL MAb 5D2 (10 μg/ml) prior to detection with FITC-labeled polyclonal anti-HBs. Cells were mounted in Vectashield mounting medium (Vector Laboratories, United Kingdom) and examined by fluorescence microscopy. The staining by FITC-labeled anti-HBs was directly visualized. The bound primary MAb was detected with Alexa 488-labeled rabbit anti-mouse immunoglobulin G (Molecular Probes).
RESULTS
Identification of PreS-binding peptides by screening a phage display random peptide library.
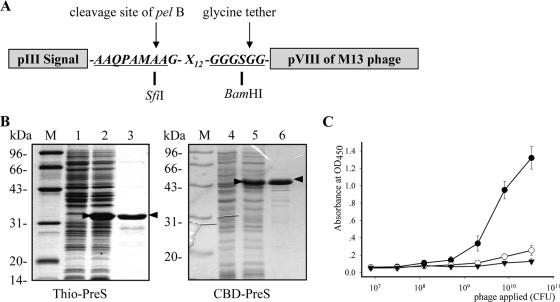
For the screening of PreS-binding peptides, an M13-based phage display library of linear 12-mer random peptides was constructed. The peptides were fused in frame between a signal peptide derived from M13 protein pIII and the major surface protein pVIII (Fig. 1A). In this way, the peptides can be displayed on the phage surface in multiple copies, since a single M13 phage contains several thousand copies of pVIII. Such a multivalent display of the peptide may facilitate the identification of weak peptide-bait interactions. The library was estimated to contain at least 5 × 108 independent clones. The randomness of the library was verified by sequencing the inserts of a phagemid pool, which indicated a sufficient diversity of peptides (data not shown).
FIG. 1.
Library construction, screening, and verification of PreS-binding phages. (A) A phage library was constructed for the surface display of linear 12-mer peptides (X12). The pIII secretion signal facilitates the transport of the fusion peptide to the phage surface. The arrows indicate the cleavage site for the signal peptide by pelB and the glycine tether for enhanced flexibility for the peptide. (B) Recombinant Thio-PreS (left) and CBD-PreS (right) were analyzed by SDS-PAGE. Lanes 1 and 4, E. coli lysate before IPTG (isopropyl-β-d-thiogalactopyranoside) induction; lanes 2 and 5, E. coli lysate after IPTG induction; lanes 3 and 6, the affinity-purified products. PreS fusions are indicated by arrowheads. (C) The PreS-binding property of the enriched phage pool after the final round of screening was assessed by phage-ELISA. Closed circles, assay with Thio-PreS; open circles, assay with thioredoxin; triangles, phages from the original library assayed with Thio-PreS. OD450, optical density at 450 nm. Error bars indicate standard deviations.
Meanwhile, efforts were made to produce the PreS domain (PreS1 plus PreS2) as the bait for the screening. PreS alone produced in E. coli is usually vulnerable to degradation and mainly insoluble (data not shown). Therefore, several fusion partners were tested with PreS to achieve better stability and solubility. Thio-PreS and CBD-PreS turned out to be stable and soluble and were subsequently purified (Fig. 1B) (15).
Thio-PreS and CBD-PreS were used alternatively as baits in the screening to minimize the possibility of enriching phages that bind only to the fusion proteins or tags. After five rounds of screening, the phage pool showed a significantly enhanced capability of binding to the immobilized Thio-PreS proteins than the thioredoxin proteins (Fig. 1C), indicating the enrichment of PreS-binding phages. A total of 26 clones were randomly picked from the final phage pool and propagated. The PreS-binding specificity of each phage on Thio-PreS-coated microwells was assessed by phage ELISA (data not shown), and that on CBD-PreS-coupled chitin beads was assessed by directly determining the titer of the bound phages (Table 1). Phages showing a high background interaction with fusion tags (CBD-PreS/CBD ratio of <5) were excluded. Consequently, 13 phages were verified to possess specific PreS-binding activity. The coding sequences of peptides were determined, and the corresponding peptide sequences were deduced. Among them, p5 and p18 occurred twice and four times, respectively (Table 1). An interesting feature of these peptides is the frequent presence of tryptophan residue.
TABLE 1.
Peptide sequences of PreS-binding phages derived from screening a phage display library of linear 12-mer random peptides
| Peptide | Sequence | Frequencya | CBD-PreS/CBD ratiob |
|---|---|---|---|
| p1 | G-G-W-T-Q-W-W-W-T-A-F-Y | 1/13 | 10 |
| p2 | N-N-W-W-Y-W-W-D-T-L-V-N | 1/13 | 12 |
| p5 | G-L-W-R-F-W-F-G-D-F-L-T | 2/13 | 10 |
| p12 | Q-M-M-L-T-L-L-W-A-F-W-Y | 1/13 | 9 |
| p15 | M-S-E-A-L-W-T-A-W-T-Q-W | 1/13 | 9 |
| p17 | M-T-G-R-L-I-S-W-W-W-S-L | 1/13 | 5 |
| p18 | W-T-D-M-F-T-A-W-W-S-T-P | 4/13 | 16 |
| p19 | G-L-W-R-F-W-F-G-D-F-L-T | 1/13 | 5 |
| p20 | W-V-E-Y-M-Y-S-W-I-P-T-A | 1/13 | 6 |
Occurrence among the total of 13 specific PreS-binding clones.
Number of bound phages eluted from CBD-PreS-coupled beads divided by that eluted from CBD-coupled beads.
The peptides bind to residues 21 to 47 of PreS1.
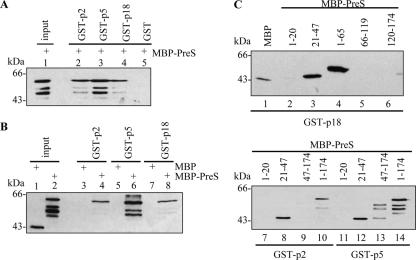
Peptides p2, p5, and p18 were chosen for further characterization due to their relatively higher PreS-binding specificities (Table 1). To verify whether these peptides truly interact with PreS, in vitro binding assays were performed. The GST-fused peptides were produced in E. coli. To avoid the fusion tags employed in screening, MBP-PreS was used in binding assays. Bound MBP-PreS proteins were detected with a monoclonal antibody against PreS1 (Fig. 2A) or MBP (Fig. 2B). As shown in Fig. 2A, all the GST-peptides (lanes 2 to 4) but not GST (lane 5) could bind to MBP-PreS. Moreover, these GST-peptides did not bind to MBP (Fig. 2B, lanes 3, 5, and 7). These results indicate that p2, p5, and p18 can bind to the PreS domain of LHBs.
FIG. 2.
GST pull-down assays for the verification of PreS-binding properties of the peptides. (A) Western blot with anti-PreS1 MAb 125E11. An equal amount of MBP-PreS was applied to beads conjugated with GST-p2 (lane 2), GST-p5 (lane 3), GST-p18 (lane 4), and GST (lane 5). Lane 1 shows the input of MBP-PreS. (B) Western blot with anti-MBP MAb. Comparable amount of MBP-PreS (lanes 4, 6, and 8) and MBP (lanes 3, 5, and 7) were applied to beads conjugated with GST-p2 (lanes 3 and 4), GST-p5 (lanes 5 and 6), and GST-p18 (lanes 7, 8). Lanes 1 and 2 show the MBP and MBP-PreS inputs, respectively. (C) Mapping of the peptide binding site within PreS. Comparable amount of indicated MBP-PreS fragments were applied to beads conjugated with GST-p18, GST-p2, or GST-p5. PreS fragments as MBP fusion proteins are denoted above each lane.
To determine the precise site where these peptides bind within PreS, several PreS fragments (aa 1 to 20, 21 to 47, 47 to 174, 1 to 65, 66 to 119, and 120 to 174) were fused to MBP and used in GST pull-down assays (Fig. 2C). The results indicate that residues 21 to 47 of PreS (PreS121-47) was recognized by these peptides (lanes 3, 8, and 12). A higher background of nonspecific binding was observed with p5 (Fig. 2C, lanes 13 and 14, and data not shown), likely owing to there being more hydrophobic residues present in this peptide. The results of the mapping experiment are interesting, since the full-length PreS was used in the screening but apparently the peptides interact almost exclusively with PreS121-47. It is noteworthy that these peptides bound more stably to residues 1 to 65 of PreS (PreS1-65) (Fig. 2C, lane 4, and data not shown), which covers PreS121-47. Consequently, PreS1-65 was used in subsequent binding assays.
Identification and mutational analysis of the consensus sequence for PreS-binding peptides.
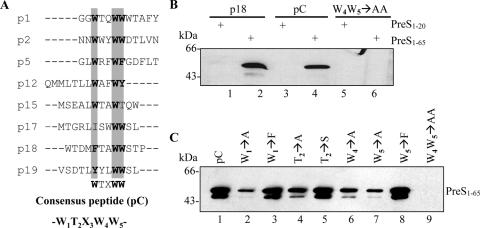
Since the selected peptides all bind to PreS121-47, there might be a common PreS-interacting motif. In addition, we noticed a high frequency of tryptophan in all enriched peptides, which implies a common feature required for PreS binding among these peptides. Alignment of the peptides revealed, with the exception of p20, a putative PreS-binding consensus pentapeptide, W1T2X3W4W5 (Fig. 3A). The three tryptophan residues are highly conserved. Residues with hydroxyl side chains, such as threonine or serine, were observed at the second position of this motif in several peptides. On the other hand, the residue at the third position seems to be relatively more flexible.
FIG. 3.
Identification and mutational analysis of a consensus PreS-binding motif. (A) Peptide alignment. Highly conserved residues are shaded and in bold. The deduced consensus sequence (pC) is W1T2X3W4W5. (B) GST pull-down assays. MBP-PreS11-65 was used for the binding assays (lanes 2, 4, and 6). Parallel controls were performed with MBP-PreS11-20 (lanes 1, 3, and 5). Lanes 1 and 2, GST-p18; lanes 3 and 4, GST-pC, with the sequence W1T2N3W4W5; lanes 5 and 6, W4W5→AA mutant of GST-pC. (C) GST pull-down assays with A, F, or S residue substitutions. The substitutions are denoted above each lane.
To determine whether this consensus peptide can bind to PreS and to define the residues critical for PreS binding, mutational analysis was performed. GST-fused wild-type and mutant peptides were incubated with MBP-PreS1-65. As shown in Fig. 3B, the wild-type GST-consensus peptide (pC) could bind to MBP-PreS1-65 (lane 4) but not to MBP-PreS1-20 (lane 3), while alanine-substituted mutant GST-peptides either were unable to bind (Fig. 3B, lane 6, and C, lane 9) or showed a much reduced binding (Fig. 3C, lanes 2, 4, 6, and 7). Phenylalanine is a hydrophobic residue with a side chain of a benzene ring and might be a potential substitution to tryptophan in the consensus peptide. In fact, both FTXWW and WTXWF were found in some enriched peptides (p5 and p18). Thus, the first and last tryptophans were replaced with phenylalanine, respectively. No apparent difference in the interaction with MBP-PreS1-65 was observed using phenylalanine-substituted GST-peptides (Fig. 3C, lanes 3 and 8). Moreover, threonine at the second position could be replaced by serine without affecting PreS binding (Fig. 3C, lane 5).
HBV particles can be captured by the consensus peptide.
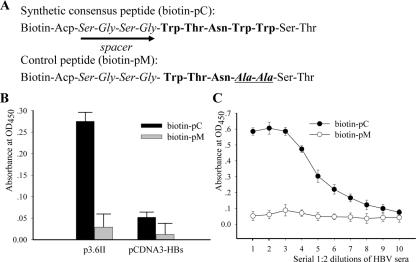
To investigate whether the consensus peptide is able to bind to HBV particles, virus capture assays were performed. To facilitate peptide coating on a solid surface, biotin was conjugated to the N terminus of the peptide during synthesis, separated by a flexible short spacer (G-S-G-S) (Fig. 4A). As shown in Fig. 4B, the wild-type peptide but not the mutant peptide showed a strong binding to viral particles produced by HepG2 cells transfected with p3.6II, which contains a terminally redundant, replication-competent HBV genome capable of producing both HBV virions and subviral particles (9, 17, 24). To assess whether the wild-type consensus peptide may bind to small envelope protein which lacks PreS, subviral particles composed of SHBs were prepared from HepG2 cells transfected with pcDNA3-SHBs. The results of capture assay showed that the wild-type consensus peptide bound poorly to SHBs (Fig. 4B). Therefore, there is little interaction between the wild-type peptide and SHBs. Taken together with the GST pull-down results, this indicates that the consensus peptide can capture viral particles via its interaction with the PreS1 domain of LHBs. Furthermore, virus samples (adr or adw serotype) from chronic hepatitis B patients were serially diluted and incubated in peptide-coated wells. After extensive washing, the bound viral particles were revealed by anti-HBs ELISA. The results showed that only the wild-type consensus peptide could capture HBV particles (Fig. 4C).
FIG. 4.
Capture of HBV particles by the synthetic consensus peptide. (A) Sequences of the biotin-conjugated synthetic consensus peptide (pC) and mutant peptide (pM). Pentapeptide sequences are in bold. Mutated residues in pM are indicated in bold italics. The spacers in both peptides are depicted by arrows. (B) Medium of HepG2 culture transfected with p3.6II (left) or pcDNA3-SHBs (right) was concentrated and applied to biotin-peptide-immobilized streptavidin-coated wells. Bound particles were revealed by detection of HBs by ELISA. Comparable HBs loads (normalized by HBs) were assayed. (C) Serially 1:2 diluted HBV sera with an initial 106 viral DNA copies were applied, and bound viral particles were detected by anti-HBs antibody. OD450, optical density at 450 nm. Error bars indicate standard deviations.
The pentapeptide as a motif to search for potential HBV binding proteins.
The pentapeptide can serve as a motif to identify potential HBV binding proteins. By performing a BLAST search in GenBank with the sequence W/F1T2X3W4W/F5, many proteins bearing such a motif were found. More hits were obtained from the search with WTXWF. However, most proteins in this case are intracellularly located. Therefore, we focused mainly on proteins obtained from the search with W/FTXWW, while we picked a few membrane or extracellular proteins containing the WTXWF sequence (Table 2). In view of the potential properties of a putative HBV carrier(s) or receptor(s), possible HBV-interacting proteins were first selected based on cellular location (extracellular or cell surface) and liver distribution or accessibility. Next, the topological location of the pentapeptide PreS-binding motif in each protein was analyzed based on known structural information or predicted via an online analysis (http://www.ch.embnet.org/software/TMPRED_form.html#). A few liver-enriched or -specific membrane proteins are excluded because their PreS-binding motifs are embedded in the lipid bilayer (reduced folate carrier protein and multispecific organic anion transporter-C) or within a signal peptide (renal dipeptidase). A few proteins are new or hypothetical, so that little information on their biochemical properties is available. These proteins will await further characterization. Consequently, LPL, prolactin receptor, and SIRP-b2 were selected for further investigation.
TABLE 2.
Proteins with the potential PreS-binding motifa
| Proteins | Gene | Potential motif (aa) | Cellular location | Tissue distribution |
|---|---|---|---|---|
| Renal dipeptidase | DPEP1 | MWSGWWL (1-7) | Extracellular | Kidney |
| Reduced folate carrier protein | SLC19A1 | LWSLWWV (269-275) | Membrane | Ubiquitous |
| LPL | LPL | SWSDWWS (416-422) | Extracellular | Adipose, cardiac and skeletal muscle, mammary gland |
| Multispecific organic anion transporter-C, isoform 1 | ABCC5 | AFSTWWL (874-880) | Membrane | Heart, brain, muscle, kidney |
| Interleukin-22 receptor α2, isoform 1b | IL22RA2 | RFTPWWE (151-157) | Extracellular | Spleen, placenta |
| Prolactin receptor | PRLR | TFTCWWR (43-49) | Membrane | Abundant in uterus, breast, kidney, and liver |
| SIRP-b2 isoform 1 | SIRPG | NWTSWFL (309-315) | Membrane | Widely distributed, abundant in liver |
| Mucin 5 subtype B | MUC5B | QWTEWFD (1511-1517) | Extracellular | Lung, stomach |
| Cartilage intermediate-layer protein | CILP | EWTTWFN (55-61) | Membrane | Articular cartilage |
| Elongation of very-long-chain fatty acids | ELOVL1 | PWSWWWG (149-155) | Membrane | Ubiquitous |
| Solute carrier family 19, member 2 | SLC19A2 | CWSVWWA (297-303) | Membrane | Intestinal and renal epithelia |
| KIAA0821 protein | KIAA0821 | RWTGWWS (58-64) | Membrane | NDc |
| Hypothetical protein LOC401260 | LOC401260 | AWTRWWR (71-77) | Membrane | ND |
| PRP8 pre-mRNA processing factor 8 homolog | PRPF8 | RFTLWWS (1533-1539) | Nucleus | Ubiquitous |
| Small membrane protein 1 | TMEM50A | FFTGWWI (32-38) | Membrane | ND |
| Interleukin-12B precursor | IL12B | RFTCWWL (139-145) | Extracellular | Macrophages, dendritic cells |
BLAST search score, >19.
All three isoforms contain the PreS-binding motif. Only isoform 1 is listed.
ND, not determined.
SIRP-b2 and prolactin receptor are proposed to be single-pass and membrane-anchored proteins (33, 41) that are particularly abundant in human liver and variably expressed in other tissues. The pentapeptide motif in SIRP-b2 is predicted to be at its long extracellular domain (33). The extracellular portion of prolactin receptor encompasses about 210 amino-terminal residues (41). cDNAs of these two proteins were cloned from human adipose tissue (prolactin receptor) and liver (SIRP-b2) by reverse transcription-PCR. The extracellular domain of prolactin receptor or SIRP-b2 was expressed in cultured mammalian cells and used in virus capture assays. However, specific viral particle capture was not observed (data not shown).
LPL binds to HBV.
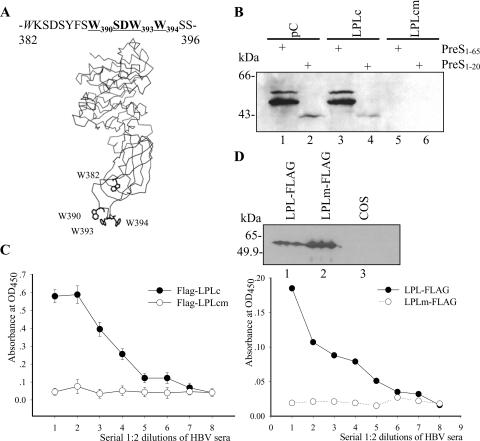
LPL protein is organized into two structurally distinct regions (27, 37), an amino-terminal domain (aa 1 to 312) and a smaller carboxyl-terminal domain (aa 313 to 448). Interestingly, the potential PreS-binding motif is located in a carboxyl-terminal loop exposed on the protein surface, according to a molecular modeling analysis based on the structure of human pancreatic lipase (Fig. 5A) (44).
FIG. 5.
LPL is a novel HBV binding protein. (A) Schematic view of the three-dimensional model of human LPL (25). The PreS-binding motif is exposed in a surface loop, residues 382 to 396 between the stacked sheets in the C-terminal domain. (B) GST-pC (lanes 1 and 2), GST-LPLc (lanes 3 and 4), and GST-LPLcm (W393W394→AA) (lanes 5 and 6) were assessed for binding to PreS11-65 (lanes 1, 3, and 5) and PreS11-20 (lanes 2, 4, and 6). (C) Virus capture assay with FLAG-tagged LPLc. HBV particles from serum samples were applied to FLAG-LPLc-immobilized streptavidin-coated wells. OD450, optical density at 450 nm. Error bars indicate standard deviations. (D) Virus capture by recombinant LPL produced in COS cells. Upper panel, recombinant LPL protein with a C-terminal FLAG tag (lane 1) and the corresponding mutant (lane 2) from lysates of transfected COS cells were verified by Western blotting with anti-FLAG MAb. Lane 3, COS cell lysate without transfection. Lower panel, LPL proteins from cell lysates were applied to anti-FLAG-coated wells and used to capture virus particles.
The cDNA of LPL was cloned from human adipose tissue. To examine whether LPL is able to interact with PreS and HBV particles, the wild-type (LPLc) and mutant (LPLcm) carboxyl domains of LPL were expressed as GST fusions in E. coli and used in pull-down assays. As shown in Fig. 5B, GST-LPLc could interact with MBP-PreS11-65 (lane 3) but not PreS11-20 (lane 4). The PreS-binding ability was lost in GST-LPLcm, where two tryptophan residues (aa 393 and 394) in the PreS-binding motif were mutated (lane 5). Furthermore, recombinant FLAG-tagged LPLc was able to capture HBV particles (Fig. 5C). Moreover, recombinant full-length LPL with a C-terminal FLAG tag and its mutant form was prepared from COS cells and examined by Western blotting (Fig. 5D, upper panel). Cell lysates were used in virus capture assay. As shown in Fig. 5D (lower panel), LPL-FLAG was capable of capturing viral particles, while the corresponding mutant was not.
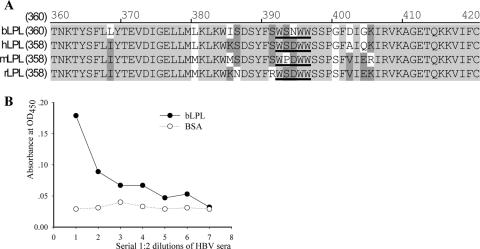
There is a high homology of LPL protein sequences among mammalian species (>90%) (Fig. 6A) (36). We further demonstrated that purified bovine LPL had the capacity to capture HBV via the PreS-binding motif WSNWW, which is well matched to the consensus motif (bLPL) (Fig. 6B). In this assay, bLPL was directly applied to microwells, which might lead to an attenuated binding compared with biotin-labeled peptide. Nevertheless, these in vitro data demonstrate that LPL is a novel binding protein of HBV.
FIG. 6.
Conservation of the PreS-binding motif in non-human lipoprotein lipases. (A) Sequence alignment around the PreS-binding motif. The active sites are in bold and underlined. hLPL, human LPL; mLPL, mouse LPL; rLPL, rat LPL; bLPL, bovine LPL. (B) HBV capture by bLPL. Purified bLPL was directly applied to microwells. BSA, bovine serum albumin as a negative control. OD450, optical density at 450 nm.
LPL bridges HBV virions to the cell surface.
LPL is synthesized in extrahepatic tissues (27, 37) and circulates in the blood in association with lipoproteins (45). LPL has been proposed to play an important role in the clearance of triacylglycerol-rich lipoproteins and chylomicron remnants by hepatocytes through bridging lipoproteins to cell surface receptors (6, 7, 38, 46). In view of the in vivo biological relevance of LPL, we addressed the question of whether LPL could play a role as an extrahepatic carrier in bridging HBV virions to the cell surface during viral infection.
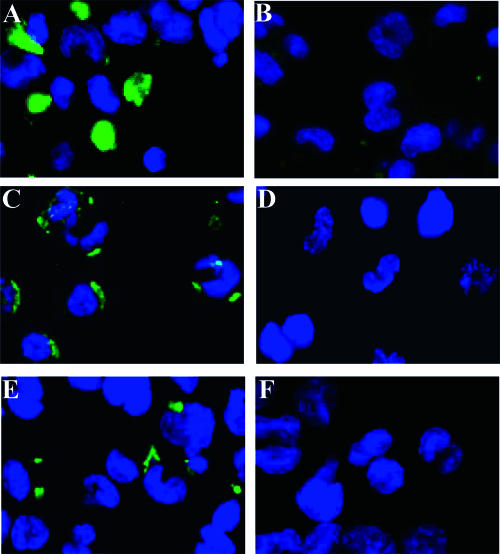
Because hepatocytes and hepatic cell lines such as HepG2 can effectively bind to HBV (13, 14, 29) in the absence of LPL (data not shown), we used the human THP-1 monocyte cell line to investigate the potential LPL-virus interaction on cell surface. The production and surface presentation of LPL proteins by THP-1 monocytes after differentiation into macrophages are well documented (1, 16, 20, 23, 26, 49). We first confirmed previous reports that dexamethasone enhances the expression of LPL by THP-1-derived macrophages (Fig. 7A) (16), while heparin treatment removes LPL from the cell surface (Fig. 7B) (16, 26). Accordingly, the immunofluorescence staining showed clearly the binding of HBV from patient serum on the surface of LPL-expressing THP-1-derived macrophages (Fig. 7C), which was greatly reduced by heparin treatment (Fig. 7D). Moreover, anti-LPL MAb 5D2, which targets the PreS-binding motif in LPL (8), was able to block the binding of HBV to LPL-expressing cells (Fig. 7F). Collectively, these results suggest that LPL can mediate the attachment of HBV to cells.
FIG. 7.
HBV is bridged to the surface of THP-1 cells by LPL. Differentiated THP-1 cells were incubated with 10 nM dexamethasone to stimulate the production of LPL (16). (A and B) Immunostaining of LPL expressed on the cells with anti-LPL MAb 5D2 without (A) or with (B) heparin pretreatment. (C and D) Immunostaining of HBV bound to the cells by anti-HBs MAb without (C) or with (D) heparin pretreatment. (E and F) Competition assay using FITC-labeled anti-HBs polyclonal antibody for the detection of HBV binding. Cells were coincubated without (E) or with (F) the anti-LPL MAb 5D2. DAPI (4′,6′-diamidino-2-phenylindole)-stained nuclei are in blue. Signals of FITC- or Alexa 488-labeled antibody are in green.
DISCUSSION
A group of HBV PreS-binding peptides were identified in this study by screening a phage display library of random peptides. Alignment of the peptides led to the discovery of a short consensus sequence, W/FTXWW/F, with PreS- and HBV-binding activities. Among 13 identified peptides, four (31%) with a relatively higher PreS-binding activity in phage ELISA are identical (represented by p18) (Table 1), which implies that the enrichment was successful. Analysis of the p18 sequence revealed two consecutive PreS-binding motifs (WTDMFTAWWSTP) (underlined and in bold), which may account for its preferential selection in the screening. A remarkable feature of these peptides is the high frequency of the tryptophan residue, which mediates interaction of proteins with lipid/water interfaces due to its amphipathic nature. Its structure, with a large hydrophobic double ring and a nitrogen atom, allows both hydrophobic and hydrophilic interactions. In this study, the in vitro binding assays with mutant peptides suggest that all the four residues examined in the consensus peptide (positions 1, 2, 4, and 5) contribute to PreS binding. Among the forces contributing to peptide-PreS interaction, hydrophobic interaction may play an important role, since replacement of the first or the last tryptophan with phenylalanine does not impair PreS binding.
Another important feature of the selected peptides is that they bind mainly to the region from aa 21 to 47 in the PreS1 domain. It is noteworthy that our search for PreS-binding peptides is not exhaustive, and sequencing more PreS-binding peptides may reveal new binding sites within the PreS region. Nevertheless, this remarkable specificity suggests that aa 21 to 47 of the PreS1 domain may form a prominent or stable conformation which accounts for the role of this region as the major cell attachment site of HBV. Given its crucial role in HBV infection, PreS121-47 is considered a target for antiviral drug development. Since the peptides identified in this study are able to bind HBV, they might have an inhibitory effect on HBV infection by impairing or blocking virus attachment to hepatocytes. Actually, preincubation of HBV-positive serum samples with the consensus peptide reduced the binding of HBV to LPL-expressing THP-1 cells (data not shown). Whether these peptides can block the infection of primary human hepatocytes by HBV awaits further study. Thus, the PreS-binding peptides can be useful tools for development of specific inhibitors of HBV infection.
The putative cellular receptor of HBV is still the puzzle in the field of HBV research. Although more than 10 proteins have been demonstrated to interact with HBV PreS or S proteins in vitro and thus might be potential receptor candidates, none of them has been experimentally proved to be a bona fide HBV receptor. In addition, the molecular mechanisms of these interactions are largely unknown. Previous strategies of searching for HBV binding proteins, including yeast two-hybrid screening and affinity purification, are based on the interaction of HBV envelope proteins, in particular the PreS domain, with cellular proteins. In this study, we adopted a novel approach to search for putative HBV binding proteins. The deduced consensus peptide presents a motif for an effective interaction with PreS, providing useful biochemical information for identification of novel HBV-interacting proteins. Interestingly, none of the previously found candidates harbors the WTXWW motif or a similar one. It is noteworthy that although we have delineated a single consensus PreS-binding motif, our results by no means rule out other potential interactions which may involve PreS121-47 and other sites, linear or conformational, on the PreS or S proteins. Given that some enveloped viruses such as HCV often interacts with different proteins (receptor and coreceptors) to mediate cell entry (2), it is very likely that HBV may also employ a similar strategy of multiple interacting proteins for cell infection.
We found several proteins containing the consensus PreS-binding motif by BLAST searching. Regarding the putative carrier or receptor of HBV, several biochemical properties, including cellular location (membrane spanning or extracellular), tissue distribution (liver distribution or accessibility), and the topology of the PreS-binding motif were taken into account to further define the potential candidates. It is noteworthy that our analysis was not comprehensive regarding the proteins found from the search with WTXWF, due to the large number of hits. A more thorough analysis is currently under way. It also remains to be seen whether a few completely new or hypothetical proteins (Table 2), for which there is currently no information on their biochemical properties, will be able to interact with HBV. These proteins are under further characterization.
LPL (EC 3.1.1.34) is a key enzyme in lipid homeostasis in vertebrates, providing intravascular release of fatty acids from circulating triacylglycerol-rich lipoproteins. LPL is produced by heart, adipose tissue, and muscle as well as in small amounts by many other tissues (27, 37). Although LPL is synthesized in extrahepatic tissues, it circulates in blood in association with lipoproteins and is cleared by the liver (45, 46). Several studies have shown that LPL enhances the binding and uptake of lipoproteins by cultured hepatoma cells and hepatocytes (6, 7, 38) and enhances the uptake of lipoproteins and lipid emulsions in perfused rat livers (40). LPL can bind to at least three plasma membrane receptors (6): (i) heparan sulfate proteoglycans, the most abundant and widely expressed LPL-binding molecules present at the surfaces of most cells; (ii) the α2-macroglobulin receptor/low-density lipoprotein receptor-related protein (LRP); and (iii) other members of the LDL receptor family, such as the LDL receptor, the VLDL receptor, and GP330/LRP2. Molecular modeling of LPL structure based on the structure of pancreatic lipase reveals an exposed loop sensitive to chymotrypsin located in the C-terminal domain (44). Cleavage of the peptide bonds between F388-S389 and W390-S391 generates a truncated LPL, whose catalytic activity against relatively soluble substrates is retained while activity against chylomicrons is lost (25). Our study demonstrates that this exposed loop contains a PreS1-binding motif which can interact with PreS1 and HBV viral particles. Moreover, the LPL-HBV interaction can occur on LPL-expressing or -binding cells. Therefore, LPL in vitro bears at least two necessary features of a potential HBV receptor, the PreS and viral particle-binding activity and the targeting to hepatocytes. The role of LPL in virus transportation or cell infection with HBV has to await functional studies in vivo or with an animal model.
Only humans and chimpanzees are the natural hosts of HBV. Studies with rat hepatoma cells transfected with the HBV genome and of transgenic mice with an integrated HBV genome indicate that HBV is capable of replicating once it bypasses the attachment and entry steps of infection, suggesting that the absence of a specific cellular receptor(s) most probably constitutes the barrier to HBV infection of nonhuman hepatocytes (9). In this regard, the interaction between LPL and HBV particles does not show species specificity. In fact, the binding motifs in rodent or bovine LPL are highly similar to those in human LPL and bLPL can indeed bind to HBV particles. Thus, LPL is unlikely to be the sole HBV receptor that determines the species specificity of the virus. Accordingly, although LPL can bridge HBV to human cells, when added as exogenous ligand or overexpressed in HepG2 cells, no infection of HepG2 cells was observed (data not shown). We propose that HBV infection is a multiple-step process which requires both species-dependent and -independent proteins or receptors. Our findings suggest that the role of LPL in HBV infection in vivo is worthy of further investigation.
Acknowledgments
This work is supported by the Basic Research Program from MOST of China (grant 2005CB522902), the Shanghai municipal government (grant 044107019), and an agreement between Shanghai Institutes for Biological Science and INSERM of France. Q. Deng holds a fellowship from the Ministry of Foreign Affairs of France.
The anti-PreS1 MAb 125E11 was a kind gift from Zhu-Chuan Zhang. We thank Jie-Hong Jiang for preparing HBV sera, Huai-Dong Song for providing human adipose and liver tissues, J. D. Brunzell for the 5D2 anti-LPL MAb, and Ursula Andero and Patrick Maillard for help with the immunofluorescence staining assay.
Footnotes
Published ahead of print on 27 December 2006.
REFERENCES
- 1.Auwerx, J. H., S. Deeb, J. D. Brunzell, G. Wolfbauer, and A. Chait. 1989. Lipoprotein lipase gene expression in THP-1 cells. Biochemistry. 28:4563-4567. [DOI] [PubMed] [Google Scholar]
- 2.Bartosch B., and F. L. Cosset. 2006. Cell entry of hepatitis C virus. Virology 348:1-12. [DOI] [PubMed] [Google Scholar]
- 3.Budkowska, A., P. Bedossa, F. Groh, A. Louise, and J. Pillot. 1995. Fibronectin of human liver sinusoids binds hepatitis B virus: identification by an anti-idiotypic antibody bearing the internal image of the pre-S2 domain. J. Virol. 69:840-848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Budkowska, A., C. Quan, F. Groh, P. Bedossa, P. Dubreuil, J. P. Bouvet, and J. Pillot. 1993. Hepatitis B virus (HBV) binding factor in human serum: candidate for a soluble form of hepatocyte HBV receptor. J. Virol. 67:4316-4322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burritt, J. B., C. W. Bond, K. W. Doss, and A. J. Jesaitis. 1996. Filamentous phage display of oligopeptide libraries. Anal. Biochem. 238:1-13. [DOI] [PubMed] [Google Scholar]
- 6.Casaroli-Marano, R. P., R. Garcia, E. Vilella, G. Olivecrona, M. Reina, and S. Vilaro. 1998. Binding and intracellular trafficking of lipoprotein lipase and triacylglycerol-rich lipoproteins by liver cells. J. Lipid Res. 39:789-806. [PubMed] [Google Scholar]
- 7.Chang, S., N. Maeda, and J. Borensztajn. 1996. The role of lipoprotein lipase and apoprotein E in the recognition of chylomicrons and chylomicron remnants by cultured isolated mouse hepatocytes. Biochem. J. 318:29-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chang, S. F., B. Reich, J. D. Brunzell, and H. Will. 1998. Detailed characterization of the binding site of the lipoprotein lipase-specific monoclonal antibody 5D2. J. Lipid Res. 39:2350-2359. [PubMed] [Google Scholar]
- 9.Chisari, F. V. 1995. Hepatitis B virus transgenic mice: insights into the virus and the disease. Hepatology 22:1316-1325. [DOI] [PubMed] [Google Scholar]
- 10.Choi, Y. H., W. S. Rho, N. D. Kim, S. J. Park, D. H. Shin, J. W. Kim, S. H. Im, H. S. Won, C. W. Lee, C. B. Chae, and Y. C. Sung. 2001. Short peptides with induced beta-turn inhibit the interaction between HIV-1 gp120 and CD4. J. Med. Chem. 44:1356-1363. [DOI] [PubMed] [Google Scholar]
- 11.Cooper, A., N. Paran, and Y. Shaul. 2003. The earliest steps in hepatitis B virus infection. Biochim. Biophys. Acta 1614:89-96. [DOI] [PubMed] [Google Scholar]
- 12.Dash, S., K. V. Rao, B. Joshi, N. C. Nayak, and S. K. Panda. 1991. Significance of natural polymerized albumin and its receptor in hepatitis B infection of hepatocytes. Hepatology 13:134-142. [PubMed] [Google Scholar]
- 13.Dash, S., K. V. Rao, and S. K. Panda. 1992. Receptor for pre-S1(21-47) component of hepatitis B virus on the liver cell: role in virus cell interaction. J. Med. Virol. 37:116-121. [DOI] [PubMed] [Google Scholar]
- 14.De Falco, S., M. G. Ruvoletto, A. Verdoliva, M. Ruvo, A. Raucci, M. Marino, S. Senatore, G. Cassani, A. Alberti, P. Pontisso, and G. Fassina. 2001. Cloning and expression of a novel hepatitis B virus-binding protein from HepG2 cells. J. Biol. Chem. 276:36613-36623. [DOI] [PubMed] [Google Scholar]
- 15.Deng, Q., Y. Y. Kong, Y. H. Xie, and Y. Wang. 2005. Expression and purification of the complete preS region of hepatitis B virus. World J. Gastroenterol. 11:3060-3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Domin, W. S., A. Chait, and S. S. Deeb. 1991. Transcriptional activation of the lipoprotein lipase gene in macrophages by dexamethasone. Biochemistry 30:2570-2574. [DOI] [PubMed] [Google Scholar]
- 17.Feng, Y., Y. Y. Kong, Y. Wang, and G. R. Qi. 2001. Inhibition of hepatitis B virus by hammerhead ribozyme targeted to the poly(A) signal sequence in cultured cells. Biol. Chem. 382:655-660. [DOI] [PubMed] [Google Scholar]
- 18.Franco, A., M. Paroli, U. Testa, R. Benvenuto, C. Peschle, F. Balsano, and V. Barnaba. 1992. Transferrin receptor mediates uptake and presentation of hepatitis B envelope antigen by T lymphocytes. J. Exp. Med. 175:1195-1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gong, Z. J., S. De Meyer, J. van Pelt, K. Hertogs, E. Depla, A. Soumillion, J. Fevery, and S. H. Yap. 1999. Transfection of a rat hepatoma cell line with a construct expressing human liver annexin V confers susceptibility to hepatitis B virus infection. Hepatology 29:576-584. [DOI] [PubMed] [Google Scholar]
- 20.Huang, Y., M. J. Ghosh, and M. F. Lopes-Virella. 1997. Transcriptional and post-transcriptional regulation of LDL receptor gene expression in PMA-treated THP-1 cells by LDL-containing immune complexes. J. Lipid Res. 38:110-120. [PubMed] [Google Scholar]
- 21.Hui, J., M. Mancini, G. Li, Y. Wang, P. Tiollais, and M. L. Michel. 1999. Immunization with a plasmid encoding a modified hepatitis B surface antigen carrying the receptor binding site for hepatocytes. Vaccine 17:1711-1718. [DOI] [PubMed] [Google Scholar]
- 22.Imai, M., Y. Yanase, T. Nojiri, Y. Miyakawa, and M. Mayumi. 1979. A receptor for polymerized human and chimpanzee albumins on hepatitis B virus particles co-occurring with HBeAg. Gastroenterology 76:242-247. [PubMed] [Google Scholar]
- 23.Li, L., M. C. Beauchamp, and G. Renier. 2002. Peroxisome proliferator-activated receptor alpha and gamma agonists upregulate human macrophage lipoprotein lipase expression. Atherosclerosis 16:101-110. [DOI] [PubMed] [Google Scholar]
- 24.Li, M., Y. H. Xie, Y. Y. Kong, X. Wu, L. Zhu, and Y. Wang. 1998. Cloning and characterization of a novel human hepatocyte transcription factor, hB1F, which binds and activates enhancer II of hepatitis B virus. J. Biol. Chem. 273:29022-29031. [DOI] [PubMed] [Google Scholar]
- 25.Lookene, A., N. B. Groot, J. J. Kastelein, G. Olivecrona, and T. Bruin. 1997. Mutation of tryptophan residues in lipoprotein lipase. Effects on stability, immunoreactivity, and catalytic properties. J. Biol. Chem. 272:766-772. [DOI] [PubMed] [Google Scholar]
- 26.Makoveichuk, E., S. Castel, S. Vilaro, and G. Olivecrona. 2004. Lipoprotein lipase-dependent binding and uptake of low density lipoproteins by THP-1 monocytes and macrophages: possible involvement of lipid rafts. Biochim. Biophys. Acta 1686:37-49. [DOI] [PubMed] [Google Scholar]
- 27.Mead, J. R., S. A. Irvine, and D. P. Ramji. 2002. Lipoprotein lipase: structure, function, regulation, and role in disease. J. Mol. Med. 80:753-769. [DOI] [PubMed] [Google Scholar]
- 28.Mehdi, H., M. J. Kaplan, F. Y. Anlar, X. Yang, R. Bayer, K. Sutherland, and M. E. Peeples. 1994. Hepatitis B virus surface antigen binds to apolipoprotein H. J. Virol. 68:2415-2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Neurath, A. R., S. B. Kent, N. Strick, and K. Parker. 1986. Identification and chemical synthesis of a host cell receptor binding site on hepatitis B virus. Cell 46:429-436. [DOI] [PubMed] [Google Scholar]
- 30.Neurath, A. R., N. Strick, and P. Sproul. 1992. Search for hepatitis B virus cell receptors reveals binding sites for interleukin 6 on the virus envelope protein. J. Exp. Med. 175:461-469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Neurath, A. R., and N. Strick. 1990. Antigenic mimicry of an immunoglobulin A epitope by a hepatitis B virus cell attachment site. Virology 178:631-634. [DOI] [PubMed] [Google Scholar]
- 32.Ostapchuk, P., P. Hearing, and D. Ganem. 1994. A dramatic shift in the transmembrane topology of a viral envelope glycoprotein accompanies hepatitis B viral morphogenesis. EMBO. J. 13:1048-1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Piccio, L., W. Vermi, K. S. Boles, A. Fuchs, C. A. Strader, F. Facchetti, M. Cella, and M. Colonna. 2005. Adhesion of human T cells to antigen-presenting cells through SIRP{beta}2-CD47 interaction costimulates T-cell proliferation. Blood 105:2421-2427. [DOI] [PubMed] [Google Scholar]
- 34.Pontisso, P., M. G. Ruvoletto, G. Tiribelli, W. H. Gerlich, A. Ruol, and A. Alberti. 1992. The preS1 domain of hepatitis B virus and IgA cross-react in their binding to the hepatocyte surface. J. Gen. Virol. 73:2041-2045. [DOI] [PubMed] [Google Scholar]
- 35.Prange, R., and R. E. Streeck. 1995. Novel transmembrane topology of the hepatitis B virus envelope proteins. EMBO J. 14:247-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Raisonnier, A., J. Etienne, F. Arnault, D. Brault, L. Noe, J. C. Chuat, and F. Galibert. 1995. Comparison of the cDNA and amino acid sequences of lipoprotein lipase in eight species. Comp. Biochem. Physiol. B 111:385-398. [DOI] [PubMed] [Google Scholar]
- 37.Santamarina-Fojo, S., and K. A. Dugi. 1994. Structure, function and role of lipoprotein lipase in lipoprotein metabolism. Curr. Opin. Lipidol. 5:117-125. [DOI] [PubMed] [Google Scholar]
- 38.Sehayek, E., T. Olivecrona, G. Bengtsson-Olivecrona, I. Vlodavsky, H. Levkovitz, R. Avner, and S. Eisenberg. 1995. Binding to heparan sulfate is a major event during catabolism of lipoprotein lipase by HepG2 and other cell cultures. Atherosclerosis 114:1-8. [DOI] [PubMed] [Google Scholar]
- 39.Sidhu, S. S. 2000. Phage display in pharmaceutical biotechnology. Curr. Opin. Biotechnol. 11:610-616. [DOI] [PubMed] [Google Scholar]
- 40.Skottova, N., R. Savonen, A. Lookene, M. Hultin, and G. Olivecrona. 1995. Lipoprotein lipase enhances removal of chylomicrons and chylomicron remnants by the perfused rat liver. J. Lipid Res. 36:1334-1344. [PubMed] [Google Scholar]
- 41.Somers, W., M. Ultsch, A. M. De Vos, and A. A. Kossiakoff. 1994. The X-ray structure of a growth hormone-prolactin receptor complex. Nature 372:478-481. [DOI] [PubMed] [Google Scholar]
- 42.Stefas, I., M. Rucheton, A. D. D'Angeac, C. Morel-Baccard, J. M. Seigneurin, J. P. Zarski, M. Martin, M. Cerutti, J. P. Bossy, D. Misse, H. Graafland, and F. Veas. 2001. Hepatitis B virus Dane particles bind to human plasma apolipoprotein H. Hepatology 33:207-217. [DOI] [PubMed] [Google Scholar]
- 43.Treichel, U., K. H. Meyer zum Buschenfelde, R. J. Stockert, T. Poralla, and G. Gerken. 1994. The asialoglycoprotein receptor mediates hepatic binding and uptake of natural hepatitis B virus particles derived from viraemic carriers. J. Gen. Virol. 75:3021-3029. [DOI] [PubMed] [Google Scholar]
- 44.van Tilbeurgh, H., A. Roussel, J. M. Lalouel, and C. Cambillau. 1994. Lipoprotein lipase. Molecular model based on the pancreatic lipase X-ray structure: consequences for heparin binding and catalysis. J. Biol. Chem. 269:4626-4633. [PubMed] [Google Scholar]
- 45.Vilella, E., J. Joven, M. Fernandez, S. Vilaro, J. D. Brunzell, T. Olivecrona, and G. Bengtsson-Olivecrona. 1993. Lipoprotein lipase in human plasma is mainly inactive and associated with cholesterol-rich lipoproteins. J. Lipid Res. 34:1555-1564. [PubMed] [Google Scholar]
- 46.Wallinder, L., J. Peterson, T. Olivecrona, and G. Bengtsson-Olivecrona. 1984. Hepatic and extrahepatic uptake of intravenously injected lipoprotein lipase. Biochim. Biophys. Acta 795:513-524. [DOI] [PubMed] [Google Scholar]
- 47.Yang, X., W. Hu, F. Li, H. Xia, and Z. Zhang. 2005. Gene cloning, bacterial expression, in vitro refolding, and characterization of a single-chain Fv antibody against pre-S1(21-47) fragment of HBsAg. Protein Expr. Purif. 41:341-348. [DOI] [PubMed] [Google Scholar]
- 48.Zhang, X. X., Q. Deng, S. Y. Zhang, J. Liu, Q. Cai, Z. M. Lu, and Y. Wang. 2003. Broadly cross-reactive mimotope of hypervariable region 1 of hepatitis C virus derived from DNA shuffling and screened by phage display library. J. Med. Virol. 71:511-517. [DOI] [PubMed] [Google Scholar]
- 49.Zimmermann, R., P. Sartipy, R. Winkler, R. Zechner, E. Hurt-Camejo, and G. M. Kostner. 2000. Endogenously produced glycosaminoglycans affecting the release of lipoprotein lipase from macrophages and the interaction with lipoproteins. Biochim. Biophys. Acta 1484:316-324. [DOI] [PubMed] [Google Scholar]