Abstract
Frequent human immunodeficiency virus type 1 (HIV-1) recombination occurs during DNA synthesis when portions of the two copackaged RNAs are used as templates to generate a hybrid DNA copy. Therefore, the frequency of copackaging of genomic RNAs from two different viruses (heterozygous virion formation) affects the generation of genotypically different recombinants. We hypothesized that the selection of copackaged RNA partners is largely determined by Watson-Crick pairing at the dimer initiation signal (DIS), a 6-nucleotide palindromic sequence at the terminal loop of stem-loop 1 (SL1). To test our hypothesis, we examined whether heterozygous virion formation could be encouraged by manipulation of the DIS. Three pairs of viruses were generated with compensatory DIS mutations, designed so that perfect DIS base pairing could only occur between RNAs derived from different viruses, not between RNAs from the same virus. We observed that vector pairs with compensatory DIS mutations had an almost twofold increase in recombination rates compared with wild-type viruses. These data suggest that heterozygous virion formation was enhanced in viruses with compensatory DIS mutations (from 50% to more than 90% in some viral pairings). The role of the SL1 stem in heterozygous virion formation was also tested; our results indicated that the intermolecular base pairing of the stem sequences does not affect RNA partner selection. In summary, our results demonstrate that the Watson-Crick pairing of the DIS is a major determinant in the selection of the copackaged RNA partner, and altering the base pairing of the DIS can change the proportion of heterozygous viruses in a viral population. These results also strongly support the hypothesis that HIV-1 RNA dimers are formed prior to encapsidation.
Human immunodeficiency virus type 1 (HIV-1), like all retroviruses, packages two copies of its RNA genome into one virus particle. Although each viral RNA contains the complete genetic information, only one provirus is generated from an infection event (17). This feature, referred to as “pseudodiploidy,” has raised much speculation as to the rationale for packaging two complete genomes into each viral particle. It has been proposed that viral RNAs often contain breaks and that packaging two copies of the viral genome allows the reverse transcription complex to switch between the two RNAs, avoiding the breaks, rescuing the genetic information, and allowing the completion of DNA synthesis (9). It is known that frequent recombination occurs during reverse transcription, not between two different incoming viruses but between sequences encoded by the two copackaged RNAs in the same virion (17). Therefore, it has also been proposed that packaging two RNAs allows frequent recombination to occur to reassort viral mutations and increase the total genetic variation in the viral population (17). Regardless of the rationale for packaging two copies of viral RNAs, it is clear that frequent retroviral recombination is closely linked to the dimeric nature of the packaged viral RNA.
Biochemical analyses of the wild-type virion RNAs demonstrate that these RNAs exist as dimers held together by a noncovalent linkage, as heating the virion-associated RNA results in the dissociation of the dimers into monomeric species (11). During or soon after the assembly of HIV-1 particles, viral proteases are activated and cleave Gag and Gag-Pol polyproteins into mature proteins, triggering the virion maturation process (18, 30, 31). During maturation, changes also occur in the viral RNA dimers. RNA isolated from protease-deficient (PR−) virions are found as monomers and fragile, immature dimers; it is thought that the monomeric forms may be the result of dissociated unstable dimers (13, 14, 29). In contrast, RNA isolated from wild-type viruses that have undergone maturation are almost exclusively dimers; furthermore, these dimers are more thermostable than those isolated from immature particles (13, 14).
Electron micrographs of RNA isolated from wild-type virions reveal that the two RNA molecules in the dimer are held together near the 5′ end of the RNA (2, 16, 21). The 5′ untranslated region of the HIV-1 genomic RNA has been predicted to form a number of highly ordered secondary structures (3, 6), one of which, the stem-loop 1 (SL1), plays important roles in efficient RNA encapsidation and dimerization (6, 27). The presence of the SL1 sequence within synthetic RNA molecules results in their spontaneous dimerization in vitro, whereas corresponding RNA molecules without SL1 do not dimerize (8, 22, 27). The SL1 sequence has been demonstrated by nuclear magnetic resonance and X-ray crystallography to form a stem-loop, punctuated by an internal bulge (1, 12). At its apical loop, the SL1 has a highly conserved palindromic sequence called the dimer initiation site or signal (DIS). The palindromic nature of DIS allows for Watson-Crick base pairing to occur between the SL1 structures of two viral RNAs, resulting in the kissing loop (KL) complex, a solution structure for which has also been elucidated (20). The KL complex corresponds to the low-stability dimer observed to spontaneously form in vitro at 37°C. However, upon heating to 55°C, the heat-labile KL dimer alters its conformation into a more thermodynamically stable form referred to as the extended dimer (ED) (24). The ED is proposed to be an extension of intermolecular base pairing from DIS down the stems of each SL1 structure. The transition from KL to ED is also promoted by the addition of the nucleocapsid protein, which has RNA binding and chaperoning activities (23, 32). However, conversion of the KL complex to ED in vivo has not been proven. Furthermore, a role has not been assigned to the ED, nor are the molecular events that lead to the maturation of the dimer fully elucidated.
Although it is clear that HIV-1 packages two copies of RNA, many questions regarding the molecular mechanisms of this feature remain unknown, such as how the two copackaged RNAs are selected. If there are two equally expressed viral RNAs in an infected cell (from virus A and virus B), the following distributions are expected, assuming random assortment of RNAs into the virions. Of the released virions, 50% will contain two different RNAs (heterozygous virions) and 50% will contain two copies of the same RNA (homozygous virions), of which half (25% of total virions) will contain two copies of the A RNA and half (25% of total virions) will contain two copies of the B RNA (Hardy-Weinberg equilibrium). Recombination can occur in all viruses, but only heterozygous viruses can generate genotypically different recombinant progeny. In a system using highly homologous vectors, HIV-1 recombination rates with markers separated by 1, 1.3, and 1.9 kb have been measured. Recombination rates were found to reach the theoretical maximum at a marker distance of 1.3 kb and did not increase further when markers were separated by 1.9 kb (25). The fact that it was possible to reach the theoretical maximum rate of recombination with two equally expressed viruses indicates that heterozygous HIV-1 virions are formed very efficiently, and RNAs from two almost identical viruses are assorted in a random or near-random manner.
We have also observed that although two subtype B viruses or two subtype C viruses can recombine efficiently, recombination between a subtype B and a subtype C virus occurs at a rate ninefold lower than that of intrasubtype recombination (5). The major restrictive factor for efficient intersubtype recombination is the DIS sequence in the SL1 loop. The DIS sequence is GCGCGC for subtype B and GUGCAC for subtype C. Although each DIS is palindromic and, therefore, complementary to RNA from within the same subtype, the subtype B DIS and subtype C DIS are noncomplementary. When the subtype B DIS was changed to subtype C DIS, the recombination rate between the modified B virus and subtype C virus was significantly increased (5). Therefore, a close relationship exists between DIS and the efficiency of heterozygous virion formation, which has a significant impact on recombination rates between two viruses.
Based on these observations, we hypothesize that the DIS sequence in SL1 plays an important role in the selection of copackaged RNAs, thereby influencing the ratios of heterozygous and homozygous viruses. If the ratio of heterozygous virus in a viral population could be reduced due to noncomplementarity of the DIS sequences, then it should be possible to promote the formation of such viruses by targeted manipulation of DIS. To test this hypothesis, we generated various mutants in SL1 and examined the effects of these mutations on viral replication and recombination. Our analyses reveal that the palindromic sequence of DIS is not required for successful viral replication in a single-cycle assay. However, the presence of two nonpalindromic, but complementary, DIS mutant viruses results in enhanced recombination rates, most likely by favoring heterozygous virion formation.
MATERIALS AND METHODS
Nomenclature and plasmid construction.
The names of all plasmids used in this study begin with “p,” the names of the viruses produced from these plasmids do not. The plasmid pHIV-BH0 has been previously described (5); briefly, it encodes the HIV-1 subtype B genomic sequence, derived from pNL4-3, with deletions of env, vpu, vif, and nef. Within the nef reading frame, a dual gene cassette was inserted encoding the murine heat-stable antigen (HSA) gene (hsa) and the green fluorescent protein (GFP) gene (gfp) that contains an inactivating mutation at its 5′ end. The gfp gene is expressed from an internal ribosomal entry site (IRES) from the encephalomyocarditis virus. The SL1 region of pHIV-BH0 was modified by PCR-based site-directed mutagenesis to produce several mutants. PCR products were digested with SpeI and SfoI, and the 806-bp DNA fragments were inserted into pHIV-BH0, replacing its corresponding fragment with the mutated versions. This procedure generated pHIV-BH0-1A, -1B, -2A, -2B, -3A, -3B, -SM, -SM3A, and -SM3B. Plasmid pON-T6 (26) has the same viral sequences as pHIV-BH0, but it carries the mouse Thy1.2 gene (thy) and its gfp gene contains an inactivating mutation at its 3′ end. The AgeI-to-AatII fragment of pHIV-BH0 and its nascent DIS mutant versions were inserted into pON-T6, also digested with AgeI and AatII, to generate pHIV-BT6, pHIV-BT6-1A, -1B, -2A, -2B, -3A, -3B, -SM, -SM3A, and -SM3B. All newly created constructs were characterized by restriction digestion, and the PCR-amplified and mutated regions were verified by DNA sequencing.
Cells, virus production, and infection.
The modified human embryonic kidney cell line 293T and the HeLa-derived HIV-1 reporter cell line TZM were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum (FCS), 1% penicillin-streptomycin, and 1% glutamine. The human T-cell line Hut/CCR5, derived from Hut78 cells but modified to express the HIV-1 entry coreceptor CCR5, was maintained in RPMI 1640 medium, supplemented with 10% FCS, 1% penicillin-streptomycin, 1% glutamine, puromycin (1 μg/ml), and G418 (500 μg/ml). All cultures were maintained at 37°C with 5% CO2.
DNA transfections were performed using 10 μM poly(ethyleneimine), either branched (25 kDa; Sigma) or linear (ExGen500; MBI Fermentas), on 293T cells plated 24 h previously at 4 × 106 cells per 100-mm-diameter dish. For virus production, 10 μg of virus plasmid (pHIV-BH0 or pHIV-BT6 derived) and 5 μg of either pIIINL(AD8)env (10), which expresses the CCR5-tropic HIV-1 Env, or pHCMV-G (33), which encodes the vesicular stomatitis virus glycoprotein, were cotransfected into the 293T cells. Viral supernatants were harvested 48 h posttransfection and clarified through a 0.45-μm-pore-size filter to remove cellular debris prior to use.
Infections of 293T cells were performed in 100-mm-diameter dishes seeded 24 h previously at 1 × 106 cells. Cells were incubated with serial dilutions of virus, supplemented with 50 μg/ml Polybrene, in a final volume of 2 ml. After 1 h, virus was removed and replaced with fresh medium. Cells were harvested 48 h later and processed prior to analysis by flow cytometry. Titrations of the viral supernatants were performed on TZM cells seeded at 4 × 103 cells per well in white flat-bottom 96-well plates. After incubation for 72 h, the medium was replaced with 100 μl of fresh medium without phenol red and 100 μl of brightlite luciferase solution (Perkin Elmer). One minute later, luciferase activity, encoded by the TZM cells under the control of a Tat-responsive promoter, was measured using the LUMIstar Galaxy (BMG) luminometer. For infection of Hut/CCR5 cells, viral supernatant was added to 1 × 106 cells in a six-well plate; the cells were subjected to a spinnoculation procedure (centrifugation at 1,200 × g for 1 h), and analyzed by flow cytometry 72 h later.
Immunostaining and flow cytometry.
To detect viral infection and recombination, cells were stained with r-phycoerythrin-conjugated anti-HSA antibody (0.2 μg/ml) and allophycocyanin-conjugated anti-Thy antibody (2 μg/ml) in phosphate-buffered saline supplemented with 2.5% FCS for 1 h on ice. Cells used for analysis were fixed in 1% paraformaldehyde prior to flow cytometry using a FACSCalibur (BD Biosciences) or CyFlow ML (Partec) apparatus, whereas live cells were sorted on a FACSVantage SE system with the FACSDiVa digital option (BD Biosciences).
Recombination assay.
To measure the recombination rate in one round of viral replication, defined as from provirus to provirus, we harvested virus from dual-infected 293T cell lines, used it to infect Hut/CCR5 target cells, and analyzed the phenotypes of the infected target cells by flow cytometry. Dual-infected virus producer cells were generated by infection of low-passage 293T cells with HSA-encoding virus at a low multiplicity of infection (MOI) of 0.05 to 0.1, followed by cell sorting for HSA-positive cells. These cells were subsequently infected with Thy-encoding virus at a low MOI and sorted twice for double-positive cells. To measure recombination frequency, 4 × 106 cells of each cell line were seeded in 100-mm-diameter dishes and transfected with 4 μg of pIIINL(AD8)env. Forty-eight hours after transfection, virus was harvested, clarified through a 0.45-μm-pore-size filter, and used to transduce the Hut/CCR5 target cell line. Recombination was detected by the recovery of GFP fluorescence in the reporter cells. The numbers of infected cells were converted to MOIs based on Poisson distribution. The recombination frequency was calculated as GFP MOI/infection MOI × 100, where the infection MOI was calculated as log(1 − Zi/Y)/log[(Y − 1)/Y]/Y, the GFP MOI was calculated as log(1 − Zg/Y)/log[(Y − 1)/Y]/Y, Y represents the number of total live cells analyzed during the flow cytometry, Zi represents the number of infected cells, and Zg represents the number of GFP-positive cells.
RNA packaging efficiency.
Viral RNA was extracted from viral particles using the MagMAX viral isolation system (Ambion) according to the manufacturer's protocol. Any DNA contamination was removed by DNase treatment of the RNA samples using TURBO DNA-free (Ambion) according to the manufacturer's protocol. The purified RNA was serially diluted and reverse transcribed into DNA using SuperScript reverse transcriptase (RT; Invitrogen) and random hexadeoxynucleotide primers (Promega). The RNA was reverse transcribed and quantified by real-time PCR using AmpliTaq Gold DNA polymerase, a gag-specific primer-probe set, and an ABI Prism 7700 sequence detector (Applied Biosystems).
Evaluation of the genomic dimerization state within viral particles.
Dual-infected virus producer cell lines were seeded at 4 × 106 cells in three 100-mm-diameter dishes. Cells were plated either in normal medium or in medium supplemented with 1 μM lopinavir, a protease inhibitor used to prevent viral maturation. Twenty-four hours later, fresh medium was added to all cell lines; the medium of the protease-inhibitor-treated cell lines was again supplemented with 1 μM lopinavir. Viral particles were harvested 48 h later, clarified through a 0.45-μm-pore-size filter, and pelleted by centrifugation at 25,000 rpm for 90 min at 4°C using a Beckman SW27 rotor. Viral genomic RNA was extracted from the resultant viral pellet as previously described (14). Nondenaturing Northern blotting analysis was performed on the recovered viral RNA (19). Briefly, RNA was dissolved in R buffer (10 mM Tris-HCl [pH 7.5], 1 mM EDTA, 1% sodium dodecyl sulfate, and 50 mM NaCl) and separated on a nondenaturing 1% agarose gel in 89 mM Tris-HCl (pH 8.3), 89 mM boric acid, 2.5 mM EDTA buffer. After electrophoresis, the gel was incubated with 6% formaldehyde at 65°C for 30 min, and the RNA was transferred to a nylon membrane. The RNA was cross-linked to the wet membrane by using a UV Stratalinker 1800 (Stratagene) prior to hybridization with a gag-specific 32P-radiolabeled RNA riboprobe.
RESULTS
Experimental hypothesis and approach.
Dimerization of HIV-1 RNA is intimately tied to the complementarity of DIS at the apical loop of SL1. We hypothesize that the selection of copackaged RNA partners in HIV-1 can be influenced by the identity of the DIS sequence; furthermore, preferential formation of heterozygous HIV-1 virions can be achieved by manipulating the DIS sequences. To test this hypothesis, we generated three pairs of mutant viruses with their DIS sequences altered to lessen its palindromic nature, thereby decreasing its self-complementarity while allowing each mutated DIS to form perfect base pairing with the DIS of its partner virus. Our hypothesis predicts that rather than random assortment of the RNA from two viruses, the RNA from a mutant virus would preferentially pair with the RNA of its partner virus, thereby favoring heterozygous virion formation. Genotypically different recombinants are generated by heterozygous viruses; therefore, preferential formation of the heterozygous viruses also predicts an increase in observed recombination rates over wild-type viruses.
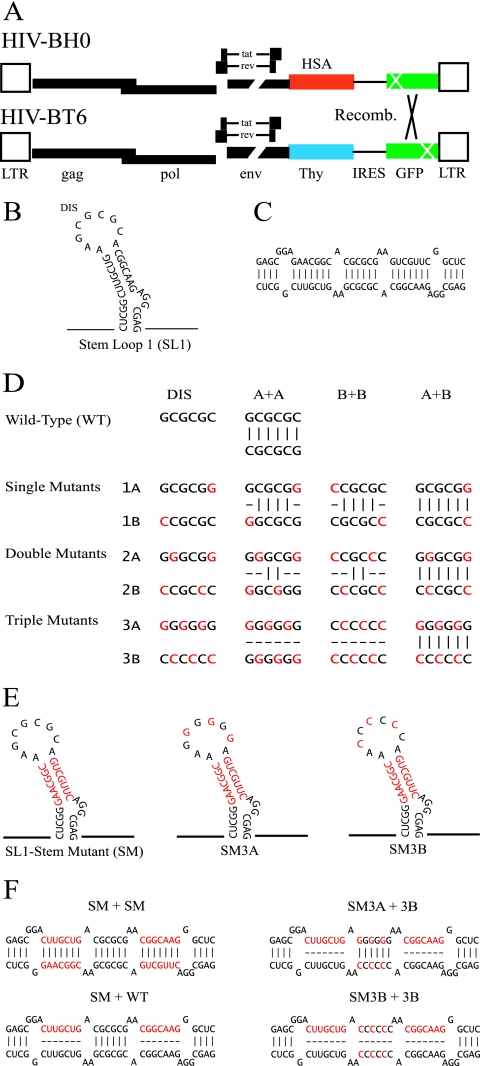
The experimental strategy is illustrated in Fig. 1. By site-directed mutagenesis, the DIS sequence was successively homogenized (Fig. 1D), mutating first one base (virus partners 1A and 1B), then two bases (virus partners 2A and 2B), and finally, three bases, resulting in a G-loop or a C-loop (virus partners 3A and 3B, respectively). Each additional mutation, from the single-mutant viruses to the triple-mutant viruses, led to a gradual loss of predicted Watson-Crick base pairings within the KL complex of homodimers, from the 6 bp of wild-type virus to the 0 bp of virus partners 3A and 3B, while maintaining the 6-bp KL complex of all partner virus heterodimers.
FIG. 1.
Schematic representation of the viruses used in this study and the mutations created within SL1 of the 5′ untranslated region of HIV-1. (A) General structures of the nearly full-length HIV-1 vectors that make up the recombination system. (B) Proposed structure of the HIV-1 SL1. (C) Extension of the KL complex down the stems of SL1 results in a more stable ED conformation. (D) Design of the DIS mutants containing altered sequences in the loop of SL1 and their impact on the base pairing within the KL complex of each virus combination. (E) SL1 SM and the double mutants (SM3A, SM3B) containing both the stem and DIS mutations. (F) Base pairing of the SL1 SM virus with another SL1 SM virus or with the wild-type (WT) virus in either the wild-type DIS or mutant context. Black letters, HIV-1 sequences; red letters, mutated bases. LTR, long terminal repeat.
The KL complex has also been proposed to mature into the more stable ED by base pairing the SL1 stem of the two RNAs (Fig. 1C). The functional significance of this proposed conformational change has not been determined, but presumably, it is to maintain the DIS base pairing and stabilize the RNA dimer. To examine the role of the SL1 stem in partner selection, we swapped the sequences upstream and downstream of the DIS that form the stem to create the SL1 stem-mutant (SM) virus (Fig. 1E). This mutation was also included in viruses with a G-loop or C-loop DIS, creating double-mutant viruses SM3A and SM3B (Fig. 1E). The predicted SL1 stem structure (Fig. 1B) is preserved in all these mutants (Fig. 1E); furthermore, viruses containing the stem mutation were predicted to convert KL dimers to EDs with other SM genomes but not when interacting with wild-type viral genomes (Fig. 1F).
The recombination assay used in this study relies on the establishment of dual-infected cell lines. The two viruses used express gag-pol, tat, and rev. They also harbor different marker genes, hsa or thy, and a gfp gene with different inactivating mutations; the distance between the two mutations in the gfp gene is 588 bp (Fig. 1A). Thus, proviruses derived from virus generated by the dual-infected cells could only carry a functional gfp gene if the two different viral genomes were copackaged and recombination occurred between them, specifically between the two gfp mutations (Fig. 1A). This system allows for the manipulation of each viral genome independently and for the effect of such alterations on recombination to be measured and, therefore, inferences to be made about the efficiency of copackaging and dimerization.
Characterization of the SL1 mutants.
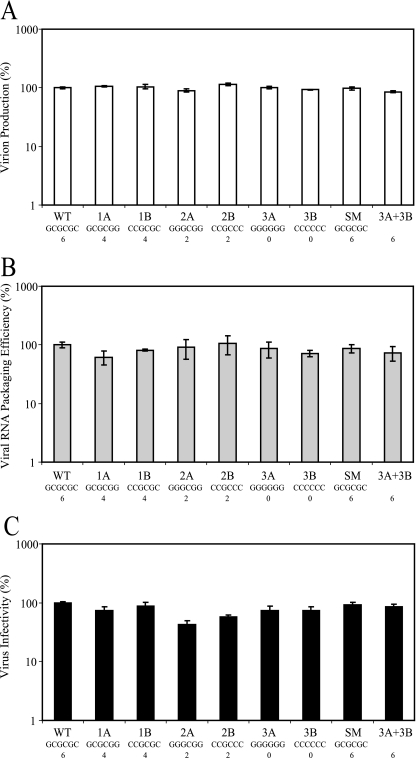
In addition to its critical role in RNA dimerization, SL1 is also an integral part of the HIV-1 packaging signal. Therefore, mutations in SL1, either in the loop (DIS) or in the stem, could cause defects in viral RNA packaging and viral infectivity. To characterize these mutant viruses, we examined their virion production, RNA packaging efficiency, and viral titers. The 293T cells were transfected with a mutant virus plasmid and the pIIINL(AD8) plasmid that expresses CCR5-tropic HIV-1 Env. Because 293T cells do not express CD4, reinfection cannot occur in this system. Viruses were harvested 48 h posttransfection, and virion production levels were determined by an enzyme-linked immunosorbent assay-based p24 assay. Data summarized from three sets of independent experiments demonstrated that all of the mutants produced similar amounts of viruses (Fig. 2A). These p24 values were used to normalize virus particle release in the subsequent RNA analyses and viral titer measurements. The efficiency of RNA packaging was measured by extracting RNA from the viruses, converting it into cDNA, and quantifying the DNA copy numbers by real-time PCR using a gag-specific primer-probe set. Our results demonstrated that the RNAs of all mutants were packaged efficiently, within twofold of the wild-type virus RNA-packaging level (Fig. 2B). The infectivities of the released mutant viruses were measured by infecting the TZM reporter cells with serially diluted virus. TZM cells contain a luciferase gene under the transcriptional regulation of an HIV-1 promoter, thereby providing luciferase detection of Tat expression, a marker of successful infection and proviral integration. Infectivity results are shown in Fig. 2C; of all the mutants, virus 2A had the largest reduction in infectivity, which was approximately 40% of the wild-type virus infectivity. The results of all these assays demonstrated that the aforementioned SL1 mutations, either in DIS or in the stem region, had very limited effects on virus release, RNA packaging, or viral titers in a single-round virus replication assay (Fig. 2).
FIG. 2.
Virus production, RNA packaging efficiency, and virus infectivity of DIS mutants. Virus was produced by transient transfection of 293T cells with wild-type plasmid, pHIVBHO, or the DIS mutant plasmids and pIIINL(AD8)env, encoding HIV-1 Env. (A) Viral particle production as measured by p24 levels. Virus was harvested from the producer cells 48 h posttransfection and assayed for p24 levels by enzyme-linked immunosorbent assay. (B) Efficiency of viral RNA encapsidation. Viral RNA was extracted from a sample of each virus, reverse transcribed, analyzed by real-time PCR using a gag-specific primer-probe set, and normalized to p24 levels. (C) Virion infectivity. Titers of virus were determined on TZM reporter cells containing a Tat-responsive luciferase gene and normalized to p24 levels. All three graphs are plotted as a percentage of the wild-type (WT) levels; shown are the means ± standard errors of results from three independent transfection experiments. Also shown are the DIS sequences and the number of complementary bases within the putative KL complex for each virus assayed.
Testing the hypothesis that altering DIS causes the preferential formation of heterozygous virions and increases the observed recombination rate.
Each DIS mutant was generated in two different vector backgrounds, one containing hsa and gfp with a mutation at the 5′ end and the other containing thy and gfp with a mutation at the 3′ end (Fig. 1A). By sequential infection and cell sorting, we generated two different types of cell lines for each DIS mutant: one type of cell line contained two viruses with the same DIS mutation and another type of cell line contained two viruses with complementary mutations. In general, each cell line was comprised of a cell pool of more than 100,000 independent infection events, and more than 95% of the cells expressed both HSA and Thy markers.
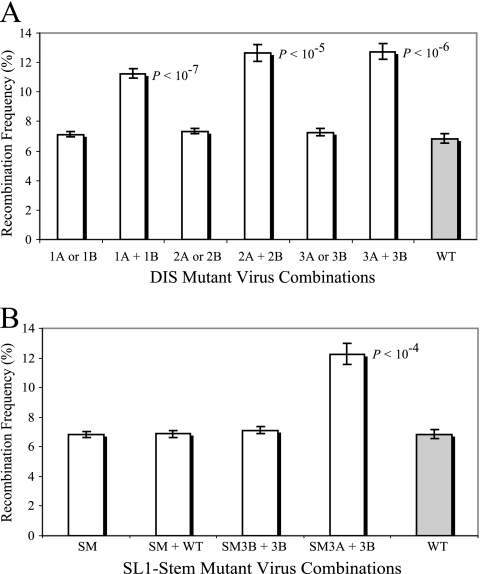
To measure recombination rates between various mutants, these cell lines were transfected with an HIV-1 Env-expressing plasmid. The resulting virus was used to infect a target T cell line, Hut/CCR5; cells were then assayed by immunostaining and flow cytometry for the expression of HSA, Thy, and GFP markers. Recombination rates, calculated as the percentage of infection events that express GFP, are depicted in Fig. 3. The recombination rates between two wild-type HIV-1 subtype B viruses have been previously measured using the same gfp recovery system. When the two gfp mutations were separated by 588 bp, functional gfp was reconstituted through recombination in 6.85% ± 0.3% (mean ± standard error, n = 6) of the infection events; these previously published results are shown for comparison (Fig. 3) (26).
FIG. 3.
Recombination frequency of SL1 mutants. (A) The role of the DIS sequence in RNA partner selection. The recombination rates of the DIS mutant viruses with viruses containing the identical DIS sequence compared with that of two viruses harboring complementary DIS mutations. “A or B” signifies that results were obtained from two cell lines, one containing two A mutant viruses and the other containing two B mutant viruses. For example, “1A or 1B” represents results compiled from a cell line containing two 1A viruses and a cell line containing two 1B viruses. “A + B” signifies that results were obtained from two cell lines both containing an A mutant virus and a B mutant virus. For example, “1A + 1B” represents results from cell lines containing one 1A and one 1B virus. (B) The impact of dimer extension on partner selection. The recombination rate of the SL1 mutant alone or in the presence of wild-type (WT) virus was measured. Also, the double mutants containing a G- or C-loop DIS and the SL1 SM were assayed for recombination with their equivalent DIS mutant with wild-type SL1 stems. Results are depicted as the means ± standard errors (n = 7) for two independent cell lines each transfected and assayed at least three times; P-values, generated by Student's t test, indicate significant deviation from the wild-type recombination rate. The recombination rate of wild-type virus (gray columns) previously measured using the same recombination system is shown for comparison.
The recombination results demonstrate that two viruses with the same DIS mutations can recombine efficiently with each other at levels comparable to that of two wild-type viruses, irrespective of the extent of the DIS mutation or whether the bases were changed to C or G (Fig. 3A). Because recombination requires the presence of two RNAs, these results suggest that two copies of RNAs are packaged into the virion even when the complementarity of the DIS sequence is decreased or abolished between the two RNAs. Furthermore, as long as the mutants contain the same DIS mutations, the two viral RNAs are copackaged randomly, regardless of whether partial complementarity (single or double mutants) or lack of complementarity (triple mutants) exists between the DIS sequences of the two viruses.
In contrast, when virus producer cells coexpressed two partner mutants containing complementary DIS sequences, the recombination rates between these two viruses were significantly higher than that of two wild-type viruses (Fig. 3A). The single-mutant virus pair (1A plus 1B) (Fig. 3A) shows an increase in recombination to 11.3% (P < 10−7, Student's t test), whereas the double and triple mutants (2A plus 2B and 3A plus 3B) (Fig. 3A) both show an 80% increase in recombination, elevating the GFP+ phenotype to 12.6% (P < 10−5) and 12.7% (P < 10−6) of the infection events, respectively.
The increased recombination rates could be caused by preferential formation of heterozygous virions, as we hypothesized, or could be caused by altered template-switching frequencies by RT. To distinguish between these two possibilities, we examined the template-switching frequencies by using the HSA and Thy markers.
Effect of altering the DIS sequence on template-switching frequencies.
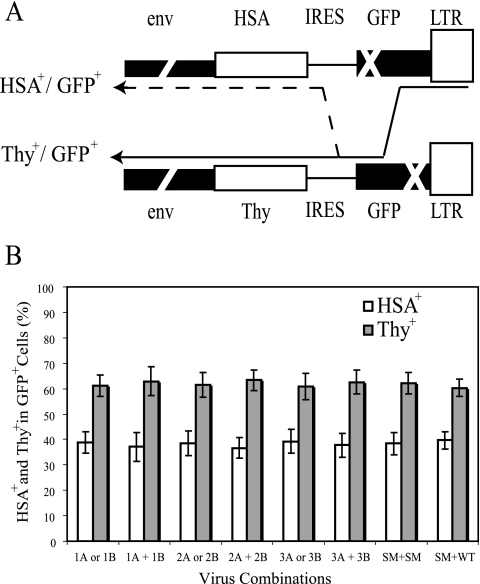
Each virus used in the recombination assay contains a marker gene (hsa or thy) and the mutated gfp gene separated by the same IRES (Fig. 1A). In addition to monitoring the reconstitution of functional gfp, we can also examine the frequency of template switching within the IRES region. To generate a wild-type gfp gene, the RT complex must avoid the 5′ gfp mutation in the hsa-containing viral genome and copy its functional counterpart in the thy-containing viral genome (Fig. 4A). Thus, for progeny with a functional gfp, the RT complex would be on the thy-containing viral genome after completion of reverse transcription of the gfp gene. Without an additional template-switching event in the IRES, the progeny would contain and express thy; however, if template switching occurred within the IRES sequence and RT copied the hsa gene, the progeny would express HSA. Therefore, the frequency of template switching in the IRES region can be measured by examining the percentage of HSA+ cells compared with that of Thy+ cells within the GFP+ population.
FIG. 4.
Effect of SL1 mutations on template-switching frequency. (A) Strategy to measure template switching within the IRES region. The presence of two markers, hsa or thy and gfp, within the viral genomes provides an internal control for the rate of template switching during reverse transcription. After reconstitution of gfp, the RT complex will synthesize the IRES region and then the thy gene. However, an HSA+ GFP+ virus will result if an additional template-switching event occurs within the IRES. (B) Relative levels of HSA+ and Thy+ cells within the GFP+ population of infected cells. LTR, long terminal repeat; WT, wild type.
Our results show that the frequency of template switching was not altered in any of the DIS mutants, either in the noncomplementary setting (A or B combinations) or the complementary setting (A plus B combinations). Therefore, any increase in recombination observed was not due to a concomitant increase in the rate of template switching (Fig. 4B).
Altering the DIS sequences resulted in preferential formation of heterozygous virions.
With the assumptions of random RNA copackaging and maximum possible recombination, theoretically, 12.5% of the infection events would generate a GFP+ phenotype in this system. With two viral RNAs expressed at similar levels and random copackaging, 50% of the viruses would be heterozygous (Hardy-Weinberg equilibrium distribution). Four gfp genotypes can be generated from these heterozygous virions: gfp with a 5′ mutation, gfp with a 3′ mutation, gfp with both mutations, and gfp with no mutation. Of these, only the last genotype can confer the GFP+ phenotype. At the maximum possible recombination level, the four genotypes will be generated at equal frequencies; therefore, the GFP+ phenotype can be generated in 25% of the heterozygous virus population, which is 12.5% of the total infection events. The observed 6.85% GFP+ reconstitution rate between two wild-type HIV-1 viruses corresponds to 55% of the theoretical maximum rate of GFP recovery (6.85%/12.5% = 55%), most likely due to insufficient distance between the two mutations to allow random assortment by template switching to occur during reverse transcription.
Using the Harvey-Weinberg equilibrium distribution and the aforementioned results for wild-type virus, an increase of recombination from 6.85% to 11.3% in the single-mutant pair corresponds to an increase from 50% to 81% of heterozygous virions in the total virus population. Using the same calculation, coexpression of the complementary double or triple mutants resulted in heterozygous virion formation in more than 90% of the total virus population. These results demonstrate the preferential formation of heterozygous particles by viruses containing RNAs with complementary DIS sequences.
Role of the SL1 stem in RNA partner selection.
It was hypothesized that after base pairing of the KL between two RNAs, the HIV-1 RNA dimer is further extended to include the stems of SL1 (Fig. 1C). To examine whether the base pairing of the stem sequences between two RNAs plays a role in RNA partner selection, we have also generated an SL1 SM that has the sequence upstream and downstream of the loop swapped (Fig. 1E). This mutant RNA can form perfect DIS base pairing with a wild-type RNA but cannot extend the dimer into the stems; in contrast, the ED can form when two wild-type viruses (Fig. 1C) or two SM viruses are paired (Fig. 1F). We generated cell lines containing two SM viruses or an SM virus with a wild-type virus and measured the recombination rates of the viruses generated from these two cell lines (Fig. 3B). The recombination rate between the SL1 SM and wild-type virus remained at the wild-type level of 6.85%, as did the rate of GFP reconstitution between the two SL1 SM viruses. Therefore, the intermolecular base pairing of the stem sequences plays limited, if any, role in the RNA partner selection in the context of wild-type DIS sequences. However, in the absence of a complementary DIS, it was unclear whether base pairing of the stem sequences between two RNAs would affect RNA partner selection. To further examine this question, we generated double-mutant viruses, SM3A and SM3B, which harbor both DIS and SL1 stem mutations (Fig. 1E); these viruses were then used in conjunction with DIS mutants containing wild-type SL1 stems. The recombination rate between 3B and SM3B was similar to that of 3B and 3B, demonstrating that even in the absence of a matching DIS or ability to form an ED, two RNAs are packaged at random. Furthermore, the increase in recombination with matching DIS mutations was maintained in the absence of an ED (SM3A plus 3B) (Fig. 3B). These results again indicate that although the SL1 stem is an important element of the RNA packaging signal, intermolecular base pairing of the stem sequences does not play an important role in the selection of copackaged RNA partners.
Analysis of the dimerization state of the virion RNA.
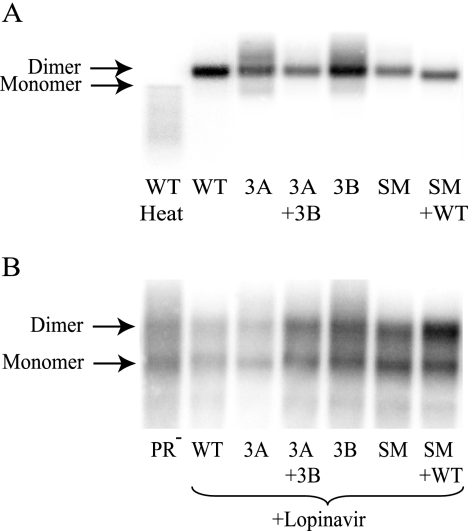
We have generated seven different mutants in SL1, a region known to be involved in RNA dimerization, and yet observed little effects of these mutations on RNA packaging and viral titers. Furthermore, the recombination rate between two viruses containing the same DIS mutation was comparable to that of wild-type viruses. For recombination to occur, more than one copy of genomic RNA must be copackaged. Therefore, virions derived from these SL1 mutants should contain more than one copy of RNA. However, it was unclear whether the copackaged RNA molecules in the virions were in dimeric form or existed as two monomers. To address this question, virus was harvested from a selection of dual infected cell lines, and the viral RNA was extracted, separated by nondenaturing gel electrophoresis, and detected by Northern analysis using a gag-specific riboprobe. As expected, the wild-type viral RNA was detected as a dimeric species (Fig. 5A). All of the mutant virus preparations analyzed (virus 3A, 3A plus 3B, 3B, SM, and SM plus wild type) also contained dimeric RNA as their major genomic species. These dimeric species, from the wild-type virus and the mutants, resolved to monomeric and smaller RNA species when the RNA samples were heated to 85°C prior to Northern analyses (Fig. 5A and data not shown). Our Northern analyses also revealed that the RNA from the noncomplementary viruses (3A or 3B) contained minor aberrant species, which could have resulted from a dimerization or packaging defect. However, the aberrant species were not observed in viruses isolated from producer cells coexpressing the two noncomplementary viruses (3A plus 3B), suggesting that the defect was corrected in the presence of the partner virus.
FIG. 5.
Dimerization state of the SL1 mutant viral genomes. (A) Genomic RNA extracted from virus released from a selection of dual infected cell lines. The RNA was phenol-chloroform extracted from harvested virus and separated by nondenaturing gel electrophoresis. Genomic RNA was identified using a 32P-radiolabeled RNA riboprobe targeting the gag region of HIV-1. Wild-type RNA heat denatured at 85°C was also included as a control. (B) Viral RNA isolated from virions treated with 1 μM lopinavir to block virus maturation. The viral RNA from PR− virus, incapable of maturing its RNA, was also included as a control. WT, wild type.
During and after their release, HIV-1 particles undergo maturation and change from a noninfectious immature particle into a fully infectious virion. In immature HIV-1 virions, both monomer and immature dimer RNA species can be detected; however, during the maturation process, viral RNA takes on a more thermodynamically stable conformation, and only RNA dimers can be detected in mature viruses. To investigate whether the SL1 mutants also have monomer/dimer status similar to wild-type viruses in their immature particles, we examined the RNA from virions treated with the protease inhibitor lopinavir. Upon treatment with 1 μM lopinavir, which is approximately 30-fold higher than the 50% inhibitory concentration, very little Gag cleavage was observed in the viral preparations (data not shown). RNA isolated from lopinavir-treated wild-type virus particles showed a pattern similar to the RNA from PR− particles (Fig. 5B), demonstrating that lopinavir treatment successfully blocked the maturation of the viral RNA dimer. Using this strategy, we examined the RNAs in immature virions generated from cell lines that expressed virus 3A or virus 3B or coexpressed 3A and 3B. We also examined RNA from SL1 SM virus and cell lines that coexpressed SL1 SM and wild-type virus. In all cases, the mutant viruses contained wild-type immature dimers and monomers at ratios similar to that of wild-type and PR− viruses (Fig. 5B). Therefore, the formation of immature dimer was unaffected in these mutants.
DISCUSSION
Multiple stages are involved in the creation of a recombinant retrovirus, including the establishment of a dual-infected cell, the formation of heterozygous virions, and a template-switching event(s) during reverse transcription. In this report, we have shown that the DIS sequence in the loop of SL1 plays a unique role in recombination by influencing the process of RNA partner selection during virion formation. By matching the DIS sequences of two RNAs, we were able to encourage the formation of heterozygous virions, thereby increasing the observed recombination frequencies.
RNA dimerization is an essential step of retroviral replication. It has been shown in vitro that the self-complementary sequence of DIS is essential for genomic RNA dimerization. Moreover, two short RNA molecules containing the same C-loop and G-loop DIS mutations used in our study were shown to not dimerize in vitro (8). However, using in vivo assays, our observations lead us to reach different conclusions. Of the six DIS mutants generated in this report, none had significant defects in viral productivity or RNA packaging in a single-round assay. Northern analyses revealed that the mutant virion RNAs were mostly in dimeric form even when the DIS contained a C-loop or a G-loop. Although minor aberrant species of RNA were also detected in 3A or 3B virus samples, this defect was corrected in the presence of the partner virus. We have also found that dimers isolated from 3A or 3B viruses have thermodynamic stability comparable to that of dimers isolated from wild-type viruses (data not shown). These findings are consistent with other in vivo studies examining mutations in the DIS (7, 15). Furthermore, the recombination rates between viruses containing the same DIS mutations were the same as those of wild-type viruses, indicating that two copies of RNA were packaged in these viruses and both RNAs were contributing to the genetic information. Most importantly, when used in combination, the G-loop and C-loop mutant viruses almost exclusively resulted in dimers in vivo, as observed by an almost doubling of recombination rates. Taken together, the results from our genetic and biochemical studies revealed that even when the self-complementary feature of DIS was abolished, two copies of HIV-1 viral RNA were packaged and these RNAs were mostly in dimeric form. Therefore, although the DIS plays a critical role in the enhancement of dimerization and RNA partner selection, in the absence of a palindromic sequence, other sequences must be present to direct the RNA dimerization process.
Although RNA without a complementary DIS can form dimers, when there is a choice, RNA with matched DIS sequences are preferentially copackaged. This was revealed by our recombination results which demonstrated an increase in recombination rates in matched DIS pairings (1A plus 1B, 2A plus 2B, and 3A plus 3B) compared with the unmatched pairings (1A or 1B, 2A or 2B, 3A or 3B). The same preference was also observed in our previous report demonstrating that the recombination rate between subtype B and subtype C HIV-1 was much lower than the intrasubtype recombination rate, mainly because these two viruses have different DIS sequences, each self-complementary. When the producer cells coexpressed subtype B and subtype C viruses, genomic RNAs with matched DIS were preferentially copackaged, thereby reducing the formation of heterozygous virions and causing the lower recombination rates. This is in contrast to recombination between two viruses containing the same noncomplementary DIS mutation (such as 3A and 3A). In this case, none of the potential RNA partners can perform base pairing at their DIS sequences. Therefore, with no preference, RNA must be copackaged randomly, in the same fashion as RNA from two wild-type viruses, and hence, the recombination rate is similar to that of the wild-type virus.
Across various reports, the effects of DIS mutations on viral infectivity and replication kinetics appear to be somewhat mixed. In a single-round assay, none of our six DIS mutants caused any drastic decrease in viral infectivity. The largest defect was virus 2A, which had 40% of the wild-type virus infectivity. Although the phenotype of our SM is comparable to that of a similar SL1 SM in a previous report, our results with virus 3A are somewhat different from that of a similar DIS mutant (7). In the previous report, a G-loop mutant had a sixfold-lower viral titer than wild-type virus without any associated RNA packaging deficiency (7). The difference between these two observations may be caused by additional mutations inserted at the base of the SL1 of the viruses from the previous study or may be due to inherent experimental variations. However, mutants appearing normal for a single round of replication can have defects in multiple-round assays; therefore, our results do not suggest that DIS is unimportant for viral replication. In the context of replicationcompetent viruses, other reports have shown that deletion or insertion mutants within the DIS result in delayed replication kinetics (4, 28). Intriguingly, it has been shown that the entire SL1 region can be removed without affecting replication in peripheral blood mononuclear cells (15). Therefore, mutations in DIS and SL1 could have varied effects in viral replication depending on the specific mutations and the assay system.
It has been hypothesized that dimerization of viral genomic RNA first initiates in DIS, followed by extension into the stem of SL1; the extended base pairing of the stem sequences is thought to increase the stability of the dimer. We have generated SL1 SMs that preserve the stem structure but prevent intermolecular base pairing of the mutant stem with that of the wild-type virus, hence, blocking the formation of the ED form between these two RNAs. Recombination between an SM and the wild-type virus occurred as frequently as with two wild-type viruses, indicating that heterozygous virions were formed at the same ratios in these two sets of viruses. Because the inability to form the ED did not alter the copackaged partner preference, our data indicate that the proposed KL complex to ED reorganization does not play a role in RNA partner selection. However, our data do not exclude the possibility that the reorganization occurs after RNA partner selection. It is also worth mentioning that although the extension can be observed in vitro with short RNA molecules, no definitive proof of the conversion from KL complex to ED has been shown in vivo.
Although it is known that HIV-1 packages two viral RNAs into each virion, the molecular mechanism(s) of RNA packaging is not completely understood. Two contrasting mechanisms could be responsible for packaging of two RNAs. It is possible that viral RNAs dimerize initially and the resulting dimer is packaged into virions. Alternatively, two monomeric RNAs could be packaged and RNA dimerization occurs during or after virus assembly. We observed that recombination rates between two mutants with complementary DIS sequences were increased significantly without altering the template-switching frequency. The hypothesis that dimeric RNA is packaged can easily explain these results. Under this hypothesis, two RNAs with complementary DIS sequences are preferentially paired and subsequently packaged, thereby increasing the proportion of heterozygous virions and generating the observed increase in recombination rates. If two monomers were packaged into each virion, it is difficult to envision how the DIS sequence could change the ratio of heterozygous virions in the viral population.
An alternative hypothesis is that monomers are packaged, but the identity of the copackaged RNA affects viral infectivity. Under this hypothesis, two monomeric RNAs are packaged, and heterozygous virions are formed in 50% of the viral population. However, the homozygous viruses, having an unpaired DIS region, have decreased infectivity; therefore, most of the detected infection events are from heterozygous viruses, thereby generating the observed higher recombination rates. We observed an 80% increase in the recombination rate and estimated that more than 90% of the virions were heterozygous. Hence, less than 10% of the total infectivity was contributed by homozygous viruses. If the increased recombination rate was caused by a decrease in infectivity of the homozygous virus, then a significant reduction in infectivity of the homozygous virions would be required to account for such large effects on recombination. In this scenario, homozygous virions occupy 50% of the total viral population but account for less than 10% of the viral titer; the infectivity of the homozygous virus must be reduced by at least fivefold compared with the heterozygous virus. Therefore, the inability to base pair at the DIS sequence must cause at least a fivefold decrease in the viral titer. However, our DIS mutants have infectivities similar to that of wild-type virus (Fig. 2C). Virus 2A is the only mutant with a >2-fold difference in viral infectivity (40%) compared with the wild-type virus. These data do not support the hypothesis of random assortment and at least a fivefold reduction in infectivity of the homozygous virus. Therefore, we consider this alternative hypothesis unlikely. Taken together, our results demonstrate that dimerization of HIV-1 genomic RNA occurs prior to its packaging within viral particles.
We have shown that DIS mutants are perfectly capable of packaging two RNAs into their viral particles. However, the RNA packaging mechanism(s) for mutants without a complementary DIS remains unclear. Without a matched DIS sequence, do Gag proteins package two separate RNA monomers? Or do other elements in the viral genome provide a supporting function for dimer formation prior to encapsidation? Further experiments are needed to distinguish between these possibilities.
The work presented here has highlighted the influential role of the DIS sequence in the selection of copackaged RNA partners, whereas the KL to ED conversion has little impact in this process. Only six complementary base pairs in the DIS sequence are capable of directing more than 90% heterozygous virion formation. Intriguingly, in the absence of a complementary DIS sequence, dimers still form and are the majority of packaged RNAs, suggesting that although DIS is an efficient dimerization signal, other redundant mechanisms must exist to direct the dimerization of HIV-1 genomic RNA. This highlights the importance of the dimerization process to the replication cycle of HIV-1, although the rationale of packaging two copies of viral genome into a retroviral particle remains undefined.
Acknowledgments
We thank Anne Arthur for her expert editorial help and Vinay K. Pathak for discussions and intellectual input throughout this project.
This research was supported in part by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research.
Footnotes
Published ahead of print on 31 January 2007.
REFERENCES
- 1.Baba, S., K. Takahashi, S. Noguchi, H. Takaku, Y. Koyanagi, N. Yamamoto, and G. Kawai. 2005. Solution RNA structures of the HIV-1 dimerization initiation site in the kissing-loop and extended-duplex dimers. J. Biochem. (Tokyo) 138:583-592. [DOI] [PubMed] [Google Scholar]
- 2.Bender, W., and N. Davidson. 1976. Mapping of poly(A) sequences in the electron microscope reveals unusual structure of type C oncornavirus RNA molecules. Cell 7:595-607. [DOI] [PubMed] [Google Scholar]
- 3.Berkhout, B., and J. L. van Wamel. 2000. The leader of the HIV-1 RNA genome forms a compactly folded tertiary structure. RNA 6:282-295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berkhout, B., and J. L. van Wamel. 1996. Role of the DIS hairpin in replication of human immunodeficiency virus type 1. J. Virol. 70:6723-6732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chin, M. P., T. D. Rhodes, J. Chen, W. Fu, and W. S. Hu. 2005. Identification of a major restriction in HIV-1 intersubtype recombination. Proc. Natl. Acad. Sci. USA 102:9002-9007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clever, J., C. Sassetti, and T. G. Parslow. 1995. RNA secondary structure and binding sites for gag gene products in the 5′ packaging signal of human immunodeficiency virus type 1. J. Virol. 69:2101-2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clever, J. L., and T. G. Parslow. 1997. Mutant human immunodeficiency virus type 1 genomes with defects in RNA dimerization or encapsidation. J. Virol. 71:3407-3414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clever, J. L., M. L. Wong, and T. G. Parslow. 1996. Requirements for kissing-loop-mediated dimerization of human immunodeficiency virus RNA. J. Virol. 70:5902-5908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coffin, J. M. 1979. Structure, replication, and recombination of retrovirus genomes: some unifying hypotheses. J. Gen. Virol. 42:1-26. [DOI] [PubMed] [Google Scholar]
- 10.Dang, Q., J. Chen, D. Unutmaz, J. M. Coffin, V. K. Pathak, D. Powell, V. N. KewalRamani, F. Maldarelli, and W. S. Hu. 2004. Nonrandom HIV-1 infection and double infection via direct and cell-mediated pathways. Proc. Natl. Acad. Sci. USA 101:632-637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Duesberg, P. H. 1968. Physical properties of Rous sarcoma virus RNA. Proc. Natl. Acad. Sci. USA 60:1511-1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ennifar, E., P. Walter, B. Ehresmann, C. Ehresmann, and P. Dumas. 2001. Crystal structures of coaxially stacked kissing complexes of the HIV-1 RNA dimerization initiation site. Nat. Struct. Biol. 8:1064-1068. [DOI] [PubMed] [Google Scholar]
- 13.Fu, W., R. J. Gorelick, and A. Rein. 1994. Characterization of human immunodeficiency virus type 1 dimeric RNA from wild-type and protease-defective virions. J. Virol. 68:5013-5018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fu, W., and A. Rein. 1993. Maturation of dimeric viral RNA of Moloney murine leukemia virus. J. Virol. 67:5443-5449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hill, M. K., M. Shehu-Xhilaga, S. M. Campbell, P. Poumbourios, S. M. Crowe, and J. Mak. 2003. The dimer initiation sequence stem-loop of human immunodeficiency virus type 1 is dispensable for viral replication in peripheral blood mononuclear cells. J. Virol. 77:8329-8335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hoglund, S., A. Ohagen, J. Goncalves, A. T. Panganiban, and D. Gabuzda. 1997. Ultrastructure of HIV-1 genomic RNA. Virology 233:271-279. [DOI] [PubMed] [Google Scholar]
- 17.Hu, W. S., and H. M. Temin. 1990. Genetic consequences of packaging two RNA genomes in one retroviral particle: pseudodiploidy and high rate of genetic recombination. Proc. Natl. Acad. Sci. USA 87:1556-1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Katoh, I., Y. Yoshinaka, A. Rein, M. Shibuya, T. Odaka, and S. Oroszlan. 1985. Murine leukemia virus maturation: protease region required for conversion from “immature” to “mature” core form and for virus infectivity. Virology 145:280-292. [DOI] [PubMed] [Google Scholar]
- 19.Khandjian, E. W., and C. Meric. 1986. A procedure for Northern blot analysis of native RNA. Anal. Biochem. 159:227-232. [DOI] [PubMed] [Google Scholar]
- 20.Kieken, F., F. Paquet, F. Brule, J. Paoletti, and G. Lancelot. 2006. A new NMR solution structure of the SL1 HIV-1Lai loop-loop dimer. Nucleic Acids Res. 34:343-352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kung, H. J., J. M. Bailey, N. Davidson, M. O. Nicolson, and R. M. McAllister. 1975. Structure, subunit composition, and molecular weight of RD-114 RNA. J. Virol. 16:397-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Laughrea, M., and L. Jette. 1994. A 19-nucleotide sequence upstream of the 5′ major splice donor is part of the dimerization domain of human immunodeficiency virus 1 genomic RNA. Biochemistry 33:13464-13474. [DOI] [PubMed] [Google Scholar]
- 23.Muriaux, D., H. De Rocquigny, B. P. Roques, and J. Paoletti. 1996. NCp7 activates HIV-1Lai RNA dimerization by converting a transient loop-loop complex into a stable dimer. J. Biol. Chem. 271:33686-33692. [DOI] [PubMed] [Google Scholar]
- 24.Muriaux, D., P. Fosse, and J. Paoletti. 1996. A kissing complex together with a stable dimer is involved in the HIV-1 Lai RNA dimerization process in vitro. Biochemistry 35:5075-5082. [DOI] [PubMed] [Google Scholar]
- 25.Rhodes, T., H. Wargo, and W. S. Hu. 2003. High rates of human immunodeficiency virus type 1 recombination: near-random segregation of markers one kilobase apart in one round of viral replication. J. Virol. 77:11193-11200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rhodes, T. D., O. Nikolaitchik, J. Chen, D. Powell, and W. S. Hu. 2005. Genetic recombination of human immunodeficiency virus type 1 in one round of viral replication: effects of genetic distance, target cells, accessory genes, and lack of high negative interference in crossover events. J. Virol. 79:1666-1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Skripkin, E., J. C. Paillart, R. Marquet, B. Ehresmann, and C. Ehresmann. 1994. Identification of the primary site of the human immunodeficiency virus type 1 RNA dimerization in vitro. Proc. Natl. Acad. Sci. USA 91:4945-4949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.St Louis, D. C., D. Gotte, E. Sanders-Buell, D. W. Ritchey, M. O. Salminen, J. K. Carr, and F. E. McCutchan. 1998. Infectious molecular clones with the nonhomologous dimer initiation sequences found in different subtypes of human immunodeficiency virus type 1 can recombine and initiate a spreading infection in vitro. J. Virol. 72:3991-3998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stoltzfus, C. M., and P. N. Snyder. 1975. Structure of B77 sarcoma virus RNA: stabilization of RNA after packaging. J. Virol. 16:1161-1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vogt, V. M. 1996. Proteolytic processing and particle maturation. Curr. Top. Microbiol. Immunol. 214:95-131. [DOI] [PubMed] [Google Scholar]
- 31.Wiegers, K., G. Rutter, H. Kottler, U. Tessmer, H. Hohenberg, and H. G. Krausslich. 1998. Sequential steps in human immunodeficiency virus particle maturation revealed by alterations of individual Gag polyprotein cleavage sites. J. Virol. 72:2846-2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Windbichler, N., M. Werner, and R. Schroeder. 2003. Kissing complex-mediated dimerisation of HIV-1 RNA: coupling extended duplex formation to ribozyme cleavage. Nucleic Acids Res. 31:6419-6427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yee, J. K., T. Friedmann, and J. C. Burns. 1994. Generation of high-titer pseudotyped retroviral vectors with very broad host range. Methods Cell Biol. 43(Pt A):99-112. [DOI] [PubMed] [Google Scholar]