Abstract
Overcoming senescence signals in somatic cells is critical to cellular immortalization and carcinogenesis. High-risk human papillomavirus (HPV) can immortalize epithelial cells in culture through degradation of the retinoblastoma protein by HPV E7 and activation of hTERT transcription, the catalytic subunit of telomerase, by the heterodimer HPV E6/E6-associated protein (E6AP). Recent work in our laboratory identified a novel repressor of hTERT transcription, NFX1-91, which is targeted for ubiquitin-mediated degradation by HPV type 16 (HPV16) E6/E6AP. In contrast, NFX1-123, a splice variant NFX1, increased expression from an hTERT promoter that was activated by HPV16 E6/E6AP. Here, we show that HPV16 E6 bound both NFX1-91 and NFX1-123 through the common central domain of NFX1 in the absence of E6AP. NFX1-123 positively regulated hTERT expression, as its knockdown decreased hTERT mRNA levels and telomerase activity and its overexpression increased telomerase activity. We identified new protein partners of NFX1-123, including several cytoplasmic poly(A) binding proteins (PABPCs) that interacted with NFX1-123 through its N-terminal PAM2 motif, a protein domain characteristic of other PABPC protein partners. Furthermore, NFX1-123 and PABPCs together had a synergistic stimulatory effect on hTERT-regulated reporter assays. The data suggest that NFX1-123 is integral to hTERT regulation in HPV16 E6-expressing epithelial cells and that the interaction between NFX1-123 and PABPCs is critical to hTERT activity.
Normally, somatic cells undergo a finite series of population doublings before entering cellular senescence (24, 25). A critical marker of a cell's age is the length of its telomeric DNA (1); with each cellular division, up to 200 nucleotides of DNA are lost at the ends of chromosomes (23, 41). Cells that require infinite replicative potential, such as stem cells, protect their telomeric DNA from erosion by constitutively expressing telomerase, a ribonucleoprotein complex that extends telomeric DNA, and thus, these cells avoid senescence. Tumors also overcome cellular senescence in order to continue their growth (22), and many activate telomerase through up-regulation of hTERT, the catalytic subunit of telomerase (63). Thus, hTERT expression and telomerase activity are critical in cellular immortalization and carcinogenesis.
Various proteins have been shown to be important regulators of hTERT. They include those that act as transcriptional repressors, including p53, p73, AP-1, and Menin (45, 59, 62, 67), as well as transcriptional activators, such as N-terminally truncated p73, c-Myc, and Sp1 (5, 56, 57, 68, 70, 74). c-Myc and Sp1 have been shown to bind to the core hTERT promoter and increase hTERT mRNA levels (56, 57, 70, 74), although Sp1 and Sp3 can also recruit histone deactylase to the hTERT promoter to repress expression (73). c-Myc and Sp1 have been found to affect hTERT, but their relative protein levels do not always correlate with the downstream hTERT mRNA and protein expression levels (17, 57, 69). Other important factors have also been identified as hTERT regulators, such as BRCA1 and Wilms' tumor 1, but their roles appear to be cell type specific (42, 55, 77). While most studies have focused on regulation of hTERT at the transcriptional level, one study has also identified posttranscriptional regulation of hTERT (8).
Viral regulators of hTERT have also been identified. The hepatitis B virus genome has been found to integrate into the hTERT promoter and, in turn, can drive its expression through the viral promoter in liver cancer (27). High-risk human papillomavirus (HPV) E6 protein can heterodimerize with E6AP and activate transcription of hTERT (17, 46), and this activation of telomerase is a critical step in cellular immortalization of keratinocytes in anogenital cancers (35). It is still unclear how HPV E6 activates hTERT expression. Studies have shown E6 binding to the hTERT promoter at E boxes as a transcriptional activator with E6AP and c-Myc (46, 69). An increase in histone acetylation (31; W. Luo, M. Xu, D. Elzi, C. Grandori, and D. Galloway, submitted for publication) and dimethylation of the hTERT promoter in the presence of HPV E6/E6AP (Luo et al., submitted) have also been found.
Recent work from our laboratory identified a transcriptional repressor, NFX1-91, which binds to a proximal X1 box in the hTERT promoter and is targeted for ubiquitin-mediated degradation by HPV E6/E6AP (18). NFX1 was identified more than 10 years ago by Song et al. as a major histocompatibility complex class II gene repressor (65). Two splice variant isoforms, NFX1-91 and NFX1-123, named for their respective masses (in kilodaltons), are expressed in epithelial cells (Fig. 1A). The two isoforms share a common N terminus and central domain, including a PHD/RING domain that functions as an E3 ubiquitin ligase and a zinc finger region that is the putative DNA binding domain of NFX1 (18, 65). This central domain is homologous to the Drosophila homolog of NFX1, shuttle craft (66). NFX1-91 has a novel C terminus that is lysine rich, is important for binding to the X1 box sequences of the hTERT promoter, and is the site for ubiquitination by HPV16 E6/E6AP (18). The novel C terminus of NFX1-123 contains two additional zinc fingers and an R3H domain, which is involved in single-strand nucleic acid binding. Although NFX1-91 represses hTERT expression in HPV type 16 (HPV16) E6- or c-Myc-expressing keratinocytes, NFX1-123 was found to augment expression in hTERT promoter reporter assays (18).
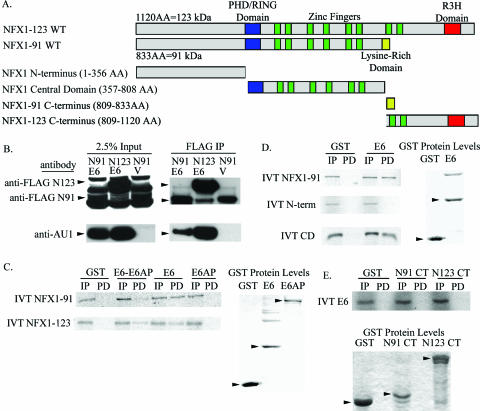
FIG. 1.
HPV16 E6 interacted with NFX1 at its central domain. (A) Map of NFX1-91 and NFX1-123 wild-type (WT) constructs, including common and unique domains, and the truncated constructs used in GST pull-down assays. (B) FLAG-tagged NFX1-91 (N91) or FLAG-tagged NFX1-123 (N123) was coexpressed with AU1-tagged HPV16 E6 (E6) or vector control (V) in 293T cells. Twenty-four hours later, the cells were lysed and FLAG-tagged protein was immunopurified. AU1-tagged HPV16 E6 coimmunoprecipitated with both NFX1-91 and NFX1-123. (C) In vitro-translated (IVT) NFX1-91 and NFX1-123, both pulled down with GST HPV16 E6 with and without GST E6AP present. GST alone did not interact with either NFX1 isoform. (D) In vitro-translated NFX1 central domain (CD) interacted with GST HPV16 E6. (E) GST-tagged novel C termini (CT) of NFX1-91 and NFX1-123 did not interact with in vitro-translated HPV16 E6. All films were scanned in through Adobe Photoshop. IP, 5% in vitro-translated input; PD, pull down. The GST protein levels shown represent 100% input. The arrowheads indicate GST proteins.
To understand the dichotomous effect of the NFX1 isoforms and to define how NFX1-123 specifically could function to increase the expression of hTERT in HPV16 E6 epithelial cells, we examined the effect of NFX1-123 expression and knockdown on hTERT and identified novel protein partners of NFX1-123. Our findings indicate that NFX1-123 augmented telomerase activity in HPV16 E6-expressing cells and that decreasing the cellular level of NFX1-123 reduced hTERT expression. NFX1-123 was found to interact with cytoplasmic poly(A) binding proteins (PABPCs), and together they synergistically augmented expression from the hTERT promoter when activated by HPV16 E6. When NFX1-123 was mutated to decrease or lose its binding capacity for PABPCs, its ability to augment hTERT promoter-driven expression was also lost. This effect adds a new level of control of hTERT by HPV-expressing cells and expands our understanding of how telomerase can be deregulated in cellular immortalization and cancer progression.
MATERIALS AND METHODS
Plasmids.
NFX1-123 was cloned into the pZome-1-C vector (Cellzome, Heidelberg, Germany) by adding BglII linkers and deleting the stop codon using the forward primer 5′GAAGATCTTCGCCACCATGGCGGAGGCGCCTC 3′ and the reverse primer 5′ CTAGATCTGTCCTGGACGTCAAAATAG 3′. The PCR products were ligated into pGEM-T Easy vector (Promega, Madison, WI), restriction digested by BglII, and ligated into the pZome-1-C BamHI site.
Truncated constructs of NFX1 for in vitro translation were cloned into the gateway vectors pDONR201 and pDEST12.2 (Invitrogen, Carlsbad, CA) by nested PCR using FLAG-tagged NFX1-91 pDEST12.2 as a template (18). First-round primers for the NFX1 common N terminus were forward, 5′ AAAAAGCAGGCTATGGCGGAGGCGCCTC 3′, and reverse, 5′ AGAAGGCTGGGTCTAGTATTTTTCTGTTGTTAGTTG 3′; those for the NFX1 central domain were forward, 5′ AAAAAGCAGGCTATGGAGTGCATGGTGTGCTG 3′, and reverse, 5′ AGAAAGCTGGGTCTACTCATGCTTGCCCATGC 3′. The second-round universal primers were forward, 5′ GGGGACAAGTTTGTACAAAAAAGCAGGCT 3′, and reverse, 5′ GGGGACCACTTTGTACAAGAAAGCTGGGT 3′.
Glutathione S-transferase (GST)-tagged truncated constructs of NFX1 were cloned into pGEX2T by PCR amplification and restriction enzyme digestion. The NFX1 common N terminus was cloned with the forward primer 5′ ATAGAATTCATATGGCGGAGGCGCCTC 3′ and the reverse primer 5′ GATGAATTCCGTATTTTTCTGTTGTTAG 3′, using the NFX1-91 EcoRI silent-mutant pGEM-T Easy vector as a template, and ligated into the pGEX2T EcoRI site (Luo et al. submitted). The NFX1 central domain was cloned by QuikChange site-directed mutagenesis (71) with the forward primer 5′ CAGAAGTGGTGCATGGGCAAGCATGAGGGAATCCATCGTGACTGACTGACGATC 3′ and the reverse primer 5′ GATCGTCAGTCAGTCACGATGGATTCCCTCATGCTTGCCCATGCACCACTTCTG 3′, using the NFX1-91 central domain and C terminus pGEX2T vector as a template (Luo et al., submitted). The NFX1-123 C terminus was cloned with the forward primer 5′ GCCGGATCCTTTCGGAGCAACATCCCC 3′ and the reverse primer 5′ GCCGAATTCCTATAAACATGCGTGTGCAGGTAT 3′, using the NFX1-123 EcoRI silent-mutant pGEM-T Easy vector as a template, and ligated into the pGEX2T BamHI and EcoRI sites. The NFX1-91 C terminus was constructed with phosphorylated and annealed forward and reverse oligonucelotides 5′ GATCCCAGTCCCACTACTGGGCGTCTACCCAGAAGAAAAGAAGTCATTATATGAAAAAGATACCTGCACACGCATGTTTATAGG 3′ and 5′ AATTCCTATAAACATGCGTGTGCAGGTATCTTTTTCATATAATGACTTCTTTTCTTCTGGGTAGACGCCCAGTAGTGGGACTGG 3′, which were ligated into the pGEX2T BamHI and EcoR1 sites. The NFX1 common N terminus PAM2 mutant was constructed by QuikChange site-directed mutagenesis (71) using the forward primer 5′ CAGATGCTGCTGAGGCCATTCCTCAGGAG 3′ and the reverse primer 5′ CTCCTGAGGAATGGCCTCAGCAGCATCTG 3′ and the NFX1 common N terminus pGEX2T vector as a template.
The FLAG-NFX1-123 PAM2 mutant was constructed by QuikChange site-directed mutagenesis (71) using the same primers mentioned above and FLAG-NFX1-123 pDONR201 (18) (Invitrogen, Carlsbad, CA) as a template and then cloned into pDEST12.2 vector.
FLAG-NFX1-123 ΔPAM2, with the first 25 amino acids of NFX1-123 deleted, was cloned into the gateway vectors pDONR201 and pDEST12.2 (Invitrogen, Carlsbad, CA) by nested PCR using NFX1-123 pGEM-T Easy vector as a template. The first-round primers were 5′ AAAAAGCAGGCTTTATGGACTACAAAGACGACGACGACAAAAAAAATTCTGGTCTAAATTGTGGG 3′ and 5′AGAAAGCTGGGTTTAGTCCTGGACGTCAAAATAG 3′. The second-round primers were as described above.
The GST HPV16 E6 (34) and E6AP (30) vectors were published previously. PABPC4, named pBShiPABP, was kindly provided by G. J. Goodall with permission from T. Lindsten (64, 75). PABPC4 was hemagglutinin (HA) tagged at its 3′ end and cloned into the gateway vectors pDONR201, pDEST12.2, and pGEXgtwy by nested PCR with the first-round primers forward, 5′ AAAAAGCAGGCTATGAACGCTGCGGCCAG 3′, and reverse, 5′ AGAAAGCTGGGTCTAAGCATAATCTGGAACATCATATGGATAAGCATAATCTGGAACATCATATGGATAAGAGGTAGCAGCAGCAAC 3′, and second-round universal primers as mentioned above.
PABPC1 was purchased from ATCC, IMAGE Clone no. 3940309, and cloned into vectors pDONR201 and pDEST12.2 by gateway cloning.
The NFX1-123 short hairpin was designed specifically for the novel C terminus of NFX1-123, utilizing the program at http://www.ambion.com/techlib/misc/siRNA_finder.html. The scramble short hairpin was designed with equivalent nucleotide distribution as the short hairpin for NFX1-123. Both sequences were BLAST searched at http://www.ensembl.org and confirmed to align with only NFX1-123 and no known gene, respectively. The NFX1-123 short hairpin and scramble were made with the phosphorylated and annealed oligonucleotides sh forward, 5′ GATCCAACCGAAGCGCAATGTGGTGGTTCAAGAGACCACCACATTGCGCTTCGGTTTTTTTTGG3′, and sh reverse, 5′ AATTCCAAAAAAAACCGAAGCGCAATGTGGTGGTCTCTTGAACCACCACATTGCGCTTCGGTTG 3′, and scramble forward, 5′ GATCCTCGAACGGTAGGACTGCGAGATTCAAGAGATCTCGCAGTCCTACCGTTCGATTTTTTGG 3′, and scramble reverse 5′ AATTCCAAAAAATCGAACGGTAGGACTGCGAGATCTCTTGAATCTCGCAGTCCTACCGTTCGAG 3′, which were ligated into the C-FUGW vector BamHI and EcoRI sites (48).
Tissue culture.
Primary human foreskin keratinocytes (HFKs) and 293T cells were cultured as described previously (18). Briefly, HFKs were derived from neonatal foreskins and grown in EpiLife medium supplemented with calcium chloride (60 μM), human keratinocyte growth supplement (Cascade Biologics, Portland, OR), and penicillin-streptomycin. 293T cells were grown in Dulbecco's modified Eagle's medium (GIBCO-BRL, Carlsbad, CA) containing 10% fetal calf serum and penicillin-streptomycin.
Retrovirus production and infection.
Retroviruses were produced either in established viral producer cell lines (PA317 or PG13) or in 293T cells by a transient vesicular stomatitis virus G-pseudotyped virus production protocol as previously described (4). Lentivirus was produced as previously published (48). Briefly, short-hairpin NFX1-123 (shNFX1-123) c-FUGW or scramble c-FUGW constructs were cotransfected with cytomegalovirus-vesicular stomatitis virus G and Δ8.9 plasmids into 293T cells using FuGENE6 (Roche, Alameda, CA), and retrovirus was collected. Retrovirus was concentrated by ultracentrifuge, mixed with Polybrene (8 μg/ml), and incubated with HFKs at 50 to 60% confluence. Three hours later, the Epilife medium (Cascade Biologics, Portland, OR) was replaced; 24 h later, the cells were expanded, and 48 h later, the cells were placed under neomycin/G418 selection (50 μg/ml) or puromycin selection (0.5 μg/ml). All lentivirus infections were also confirmed by green fluorescent protein expression.
Telomerase activity.
Telomerase activity was detected using the TRAPeze detection kit (Chemicon International, Temecula, CA) as described previously (18).
Coimmunoprecipitation and Western blot assays.
Coimmunoprecipitation assays were conducted as previously published (18). Western blots were probed with antibodies: anti-FLAG M2 antibody (1:2,000; Sigma, St. Louis, MO), anti-AU1 antibody (1:2,000; Bethyl, Montgomery, TX), anti-HA antibody (1:1,000; Roche, Alameda, CA), anti-p53 antibody Ab-6 (1:1,000; Calbiochem, San Diego, CA), Nucleolin C23 (1:1,000; Santa Cruz Biotechnology, Santa Cruz, CA), and anti-PABP1 antibody (1:1,000 overnight at 4°C; Cell Signaling, Danvers, MA).
Tandem-affinity purification protocol.
The tandem-affinity purification protocol was conducted as published previously (10) with the following modifications. Plates (15 cm2) of 293T cells were transiently transfected at 50% confluence with NFX1-123 pZome-1-C or vector alone as a control, using FuGENE6 (Roche, Alameda, CA). The total cell count for NFX1-123 pZome-1-C-transfected cells was 2 × 106 cells, and for vector-transfected cells it was 8 × 107 cells. Twenty-four hours later, each plate was washed twice with 10 ml cold phosphate-buffered saline (PBS) and lysed with 1 ml of NP-40 lysis buffer (150 mM sodium chloride, 0.1% NP-40 alternative, 50 mM Tris-HCl, pH 8.0, COMPLETE mini protease inhibitor tablet EDTA free [Roche, Alameda, CA], double-distilled H2O [ddH2O]), with occasional rocking on ice for 30 min. Lysates were collected, freeze-thawed for three cycles, and clarified by pelleting the cellular debris. The clarified lysate was diluted 1:4 with IPP150 buffer (10 mM Tris-HCl, pH 8.0, 150 mM sodium chloride, 0.1% NP-40 alternative, COMPLETE mini protease inhibitor tablet EDTA-free, 0.1 mM phenylmethylsulfonyl fluoride, ddH2O) and incubated with 20 μl equilibrated immunoglobulin G (IgG) Sepharose 6 Fast-Flow beads for lysate from each 15-cm2 plate (Amersham Biosciences, Piscataway, NJ) with rocking overnight at 4°C. The IgG beads were gently pelleted, washed three times with IPP150 buffer and once with TEV cleavage buffer (10 mM Tris-HCl, pH 8.0, 150 mM sodium chloride, 0.1% NP-40 alternative, 0.5 mM EDTA, ddH2O, and freshly added 1 M dithiothreitol diluted to 1 μl/ml). One milliliter TEV cleavage buffer was then added to 80 to 100 μl pooled IgG beads. Ten microliters of AcTEV protease (Invitrogen, Carlsbad, CA) was added, and the beads were rotated at 4°C for 4 hours. The IgG beads were pelleted and then washed with additional TEV cleavage buffer, and the supernatants were collected serially for a total 1.2-ml volume per 80 to 100 μl pooled IgG beads; 3.3 μl 2 M calcium chloride, 575 μl calmodulin binding buffer (10 mM Tris-HCl, pH 8.0, 150 mM sodium chloride, 1 mM magnesium acetate, 1 mM imidazole, 2 mM calcium chloride, ddH2O, freshly added COMPLETE mini tablet protease inhibitor EDTA-free, 0.1 mM phenylmethylsulfonyl fluoride, and beta-mercaptoethanol diluted to 0.697 μl/ml), and 40 to 50 μl equilibrated calmodulin affinity resin (Stratagene, La Jolla, CA) were then added to each 1.2 ml supernatant and rocked for 4 hours at 4°C. The resin was washed three times with calmodulin binding buffer, and all like resins were pooled. Proteins were serially eluted at 25°C with 50 to 100 μl calmodulin elution buffer (10 mM Tris-HCl, pH 8.0, 150 mM sodium chloride, 1 mM magnesium acetate, 1 mM imidazole, 50 mM EGTA, ddH2O, and freshly added beta-mercaptoethanol diluted to 0.697 μl/ml) every 5 minutes. A total of six elutions were collected. A portion of each elution was run on a 4 to 12% NuPAGE Tris-Bis gradient gel (Invitrogen, Carlsbad, CA), silver stained (GelCode SilverSNAP Stain Kit II; Pierce, Rockford, IL), and dried (DryEase Mini-gel Drying System; Invitrogen, Carlsbad, CA) to identify purified tagged NFX1-123 and other proteins interacting with NFX1-123 compared to vector alone. Samples collected throughout the protein purification process were analyzed by Western blotting to ensure the recovery of tagged NFX1-123 protein (anti-NFX1 antibody; 1:500) (18).
The sample elutions were proteolytically digested by adding 2 μg of sequencing-grade trypsin (Promega, Madison, WI) and incubating them at 37°C for 16 h. The sample was concentrated and desalted using a C18micro ZipTip (Millipore, Billerica, MA), following the manufacturer's instructions, and dried by vacuum centrifugation. The samples were then resuspended in 7 μg of 0.1% formic acid and analyzed by liquid chromatography/electrospray ionization mass spectrometry (MS)/MS with an LCQ DECA XP mass spectrometer (Thermo Electron, Waltham, MA) using an instrument configuration described by Gatlin et al. (15). Data were collected in a data-dependent mode in which an MS scan was followed by MS/MS scans of the three most abundant ions from the preceding MS scan. Proteins were identified from MS data with the protein database search algorithm COMET (Institute for Systems Biology), using a human protein database from the National Cancer Institute Advanced Biomedical Computing Center (a human subset from a nonredundant protein database downloaded on 1 December 2004). Protein identification results were filtered by using raw scores greater than 200 for +1 ions, 300 for +2 ions, and 300 for +3 ions; Z scores greater than or equal to 4; percent ions greater than or equal to 15%; and unique peptides greater than or equal to 2; also, the identification did not appear in the control sample elution.
In vitro translation.
All proteins were in vitro translated using the TNT Coupled Wheat Germ Extract system (Promega, Madison, WI).
GST protein expression and pull-down assays.
All pGEX2T or pGEXgtwy constructs were transformed into either BL21 codon+ cells or Rosetta2 cells (Novagen, San Diego, CA), grown in 500-ml cultures, and induced at logarithmic growth with 0.5 mM isopropyl-β-d-thiogalactopyranoside for 2 hours. GST-tagged NFX1 constructs were induced at a lower optical density (0.1 to 0.3 versus 0.6) and with 0.1 mM isopropyl-β-d-thiogalactopyranoside to improve protein solubility and recovery. Bacteria were pelleted, resuspended in 25 ml PBS-P (20 μM zinc chloride for GST-tagged NFX1 constructs or 50 mM EDTA, pH 8.0, for all others, 0.1 mM phenylmethylsulfonyl fluoride, COMPLETE tablets EDTA-free, PBS), and run twice through a microfluidizer for lysis or sonicated on ice with four 30-second bursts separated by 30-second cooling periods, with increasing duty cycles (20 to 40%) and amplitudes (20 to 40%). Cellular debris was pelleted, and the clarified lysate was incubated with 500 μl equilibrated glutathione Sepharose 4B beads (Roche, Alameda, CA) for 1 hour at 4°C. The glutathione beads were gently pelleted and washed four times with PBS-P. GST-tagged protein was eluted from the beads with 20 mM reduced glutathione in 50 mM Tris-HCl, pH 8.0, and placed in a Slide-A-Lyzer (Pierce, Rockford, IL) for dialysis overnight in dialysis buffer (0.1% NP-40 alternative, 0.1 mM phenylmethylsulfonyl fluoride, PBS) at 4°C. Protein expression and recovery were confirmed by Western blotting (anti-GST antibody; 1:500; sc-138; Santa Cruz Biotechnology, Santa Cruz, CA) and GelCode Blue stain reagent (Pierce, Rockford, IL).
For GST pull-down assays, equal amounts of GST-tagged proteins were incubated with in vitro-translated proteins in binding buffer (1% NP-40 alternative, 2 mM dithiothreitol, COMPLETE mini protease inhibitor tabs EDTA-free, PBS) at a final volume of 200 μl and rocked for 1 hour at 4°C. Thirty microliters of equilibrated glutathione Sepharose 4B beads were added to each pull down and rocked for 2 hours at 4°C. The beads were washed three times in binding buffer, and bound proteins were recovered by boiling them in sample buffer. Samples were run on sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis gels, dried, and exposed to Biomax film (Kodak, Rochester, NY) at −80°C.
Luciferase assays.
Luciferase assays were performed as described previously (17, 18). Briefly, HFKs were grown to 50 to 60% confluence in six-well plates and transfected with a pGL3-hTERT reporter-based plasmid, including 710 nucleotides upstream of the transcriptional start site (710 hTERT). A total of 2 μg of DNA was transfected into each well, using a 1:3 DNA-FuGENE6 (Roche, Alameda, CA) ratio. The cells were incubated for 24 h, washed with cold PBS, and lysed in the well by freeze-thawing with 100 to 150 μl of reporter lysis buffer (Promega, Madison, WI). Cellular debris was pelleted, and luminescence was quantitated with 20 μl of lysate mixed with luciferase assay buffer (Promega, Madison, WI) in a Lumat LB953 luminometer. Each experiment was done in triplicate and normalized for total protein concentration.
Real-time PCR.
HFK cells were lysed with 1 ml TRIzol reagent (Invitrogen, Carlsbad, CA) and purified by chloroform extraction. One microgram of total RNA was used to synthesize cDNA using random hexamers and a Superscript II reverse transcriptase system (Invitrogen, Carlsbad, CA). Real-time PCR of hTERT and 36B4 using SYBR green was conducted on an ABIPrism 9700 and analyzed with SDS 2.2.2 software. Samples were amplified in triplicate and normalized to 36B4. Error bars represent 95% confidence intervals of the quotient hTERT/36B4. Sequences for hTERT and 36B4 primers were previously described (17).
RESULTS
HPV16 E6 interacts with the central domain of NFX1.
NFX1-123 was initially identified as a protein partner of E6/E6AP in a yeast two-hybrid screen coexpressing HPV16 E6 and E6AP (18). NFX1-91 was then found to interact with E6/E6AP by coimmunoprecipitation, and both isoforms are expressed in epithelial cells (18). The binding site of E6/E6AP to either NFX1 isoform was not known, although it was presumed to be in a shared region. Additionally, it was not known whether NFX1 bound to E6, E6AP, or both. To address these issues, we coexpressed FLAG-tagged NFX1-123 or NFX1-91 with AU1-tagged HPV16 E6 in 293T cells. By coimmunoprecipitation, HPV16 E6 bound both NFX1-91 and NFX1-123 (Fig. 1B). To determine whether NFX1 interacted with E6, E6AP, or both, GST pull-down assays with GST-tagged HPV16 E6 and E6AP were conducted with in vitro-translated full-length NFX1-91 and NFX1-123 using wheat germ extracts that did not contain endogenous E6AP (29). Both NFX1-91 and NFX1-123 interacted with GST-tagged HPV16 E6 alone or when E6AP was present, but not with E6AP alone (Fig. 1C). The reduced interaction seen in the presence of both E6 and E6AP may be because less of each protein was used in the combined GST pull-down reaction or because E6AP competes with NFX1 for binding to E6.
To define the region of NFX1 to which HPV16 E6 bound, the N terminus and central domain shared by NFX1-91 and NFX1-123 were tested, as well as their unique C termini (depicted in Fig. 1A). HPV16 E6 bound the common central domain of NFX1 (Fig. 1D). Again, this interaction with the central domain of NFX1 occurred whether or not E6AP was included (data not shown). No interaction was seen between HPV16 E6 and the GST-tagged novel C terminus of NFX1-91 or NFX1-123 (Fig. 1E). Similar results were obtained when the experiment was performed in the opposite orientation using GST-tagged truncated NFX1 constructs and in vitro-translated HPV16 E6 and E6AP (data not shown).
NFX1-123 affects telomerase in vivo.
Previous studies in our laboratory showed that NFX1-123 augmented the expression of luciferase driven by an hTERT promoter when activated by HPV16 E6 or c-Myc (18). To determine whether NFX1-123 affected hTERT and telomerase activities in vivo, HFKs expressing HPV16 E6 or vector control (pBABE [pB]) were established and then transduced with either NFX1-123 or a control (LXSN) (Fig. 2C). E6 increased telomerase activity by TRAP assay threefold over background; coexpression of NFX1-123 more than tripled telomerase activity in E6 cells but had no effect in the absence of E6 (pB HFKs) (Fig. 2A).
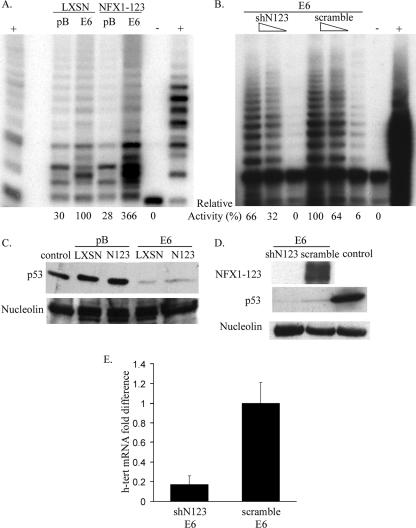
FIG. 2.
NFX1-123 affected hTERT mRNA levels and telomerase activity. (A) TRAP assay of HFKs expressing HPV16 E6 (E6) and NFX1-123 or vector controls, LXSN and pB. Telomerase activity was seen in cells expressing HPV16 E6, and NFX1-123 increased that activity by more than threefold. Two micrograms of protein lysate was used for each sample. −, negative control, CHAPS {3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate} lysis buffer; +, positive controls, telomerase plus cells and TSR8 synthetic template for PCR. (B) TRAP assay of HFKs expressing HPV16 E6 (E6) and shRNA against NFX1-123 (shN123) or scramble oligonucleotide as a control (scramble). Telomerase activity was reduced in cells with knockdown NFX1-123 protein. One microgram, 0.5 μg, and 0.1 μg of protein lysate were used for each sample. −, negative control, CHAPS lysis buffer; +, telomerase plus cells. (C) Western blots of cell lysates used in panel A. Expression of HPV16 E6 (E6) degraded p53 compared to vector control cells (pB) and uninfected HFKs (control). Nucleolin is shown as a loading control. (D) Western blots of cell lysates used in panel B. Expression of HPV16 E6 (E6) degraded p53 compared to uninfected HFKs (control), and shRNA against NFX1-123 (shN123) knocked down protein levels in HFKs compared to the scrambled oligonucleotide (scramble). Nucleolin is shown as a loading control. (E) Real-time PCR of NFX1-123 knockdown cells showed an 80% reduction in endogenous hTERT mRNA levels compared to scramble control cells. Error bars, 95% confidence interval. All films were scanned in through Adobe Photoshop.
To determine whether NFX1-123 was required for hTERT expression, a short hairpin against NFX1-123 or a scrambled oligonucleotide control was expressed in HPV16 E6 HFKs. NFX1-123 protein levels were knocked down in shNFX1-123 cells (Fig. 2D), and TRAP activity was reduced by one-half to one-third compared to the equivalent scramble control (Fig. 2B). By real-time PCR, the mRNA levels of hTERT in shNFX1-123 HPV16 E6 cells were reduced by 80% compared to the scramble control (Fig. 2E). Thus, overexpression of NFX1-123 increased telomerase activity, and a reduction in the endogenous NFX1-123 protein levels, in turn, reduced telomerase activity.
NFX1-123 interacts with poly(A) binding proteins.
To begin to understand how NFX1-123 augments hTERT levels, additional protein partners of NFX1-123 were sought using a tandem-affinity purification of NFX1-123-bound protein and MS. Elution of proteins copurified with NFX1-123 or vector control were evaluated by SDS-polyacrylamide gel electrophoresis silver staining (Fig. 3) and were identified in a shotgun approach by MS (Table 1). Multiple proteins associated with NFX1-123, including two PABPCs, PABPC1 and PABPC4. PABPC1, also known as PABP1, and PABPC4, also known as the inducible poly(A) binding protein, are approximately 65 to 70 kDa in size and function to stabilize mRNA (33, 39). PABPCs bind to the 3′ poly(A) tail of mRNA and directly interact with 5′ cap proteins, forming a closed loop of RNA (49). PABPCs increase expression from mRNA transcripts through nuclear-to-cytoplasmic shuttling of RNA, recruitment and maintenance of translation machinery on RNA, and prevention of deadenylation of the poly(A) tail (39, 49). Other proteins involved in RNA stabilization, RNA processing and splicing, and protein deubiquitination were also identified by MS (Table 1). Only background IgG, calmodulin, and trypsin, used in the protein purification steps, were identified in the vector control elutions by MS.
FIG. 3.
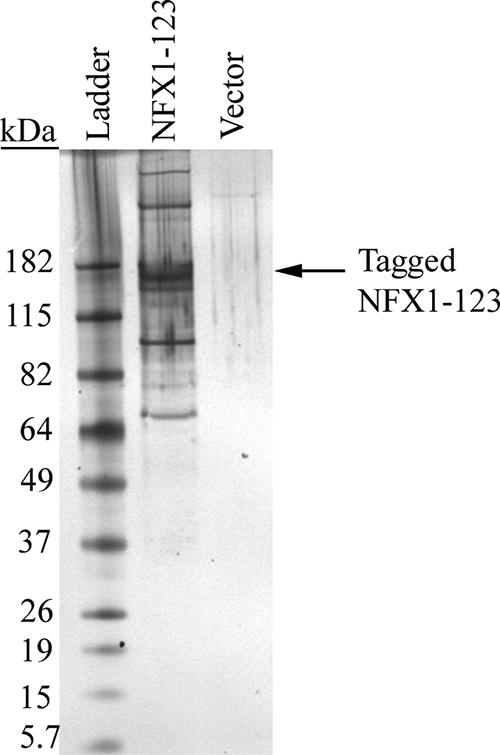
NFX1-123 interacted with poly(A) binding proteins. A silver-stained gel of eluted tagged NFX1-123 in 293T cells showed multiple proteins copurifying with NFX1-123. No protein bands were seen in the vector elution alone. The gel was scanned in through Adobe Photoshop.
TABLE 1.
NFX1-123 and copurified proteins identified by MS in tandem-affinity purification of 293T cellsa
| Name | Accession no. | No. of peptides | No. unique |
|---|---|---|---|
| NFX1 | U15306, NM_002504 | 33 | 18 |
| Calmodulin | 15 | 5 | |
| USP/FAFX | X98296 | 10 | 10 |
| Inducible PABP, PABP4 | NM_002568 | 9 | 8 |
| Trypsin | 8 | 4 | |
| Keratin | 7 | 6 | |
| Elongation factor 1-α2 | NM_001958 | 6 | 5 |
| Matrin 3 | NM_018834 | 6 | 6 |
| Calmodulin 2 | 5 | 2 | |
| PABP3 | NM_030979 | 5 | 5 |
| Heat shock protein 70 protein 1 | M59828 | 4 | 4 |
| Heterogeneous nuclear ribonucleoprotein m | L03532 | 4 | 4 |
| Heterogeneous nuclear ribonucleoprotein u | NM_031844 | 4 | 4 |
| 60S ribosomal protein L13a | X64707 | 3 | 2 |
| Splicing factor, arginine/serine-rich 4 | L14076 | 3 | 2 |
| PABP1 | NM_002568 | 3 | 3 |
| Histone H2A type 1 | M60752 | 3 | 3 |
| Keratin | 3 | 3 | |
| 60S ribosomal protein 17a | X06705 | 3 | 3 |
| 40S ribosomal protein S2 | BC001759 | 3 | 3 |
| KIAA0731 | NM_015315 | 3 | 3 |
| Dead box protein p68; dead box protein 5 | NM_004396 | 2 | 2 |
| Dystrophin | M18533 | 2 | 2 |
| Keratin | 2 | 2 | |
| Keratin | 2 | 2 | |
| IgG | 2 | 2 | |
| Nucleolin | NM_005381 | 2 | 2 |
| 60S ribosomal protein L13a | X56932 | 2 | 2 |
| 60S ribosomal protein L17 | X53777 | 2 | 2 |
| 60S ribosomal protein L22 | X59357 | 2 | 2 |
| 60S ribosomal protein L3 | X73460 | 2 | 2 |
| 60S ribosomal protein L6 | X69391 | 2 | 2 |
| Heterogeneous nuclear ribonucleoproteins A2/B1 | NM_031243 | 2 | 2 |
| Heterogeneous nuclear ribonucleoprotein H | L22009 | 2 | 2 |
| 40S ribosomal protein S19 | M81757 | 2 | 2 |
| 40S ribosomal protein S3A | M77234 | 2 | 2 |
| Splicing factor, arginine/serine-rich 3 | NM_003017 | 2 | 2 |
| APCL protein | NM_005883 | 2 | 2 |
| Nuclease-sensitive element binding protein 1 | BC002411 | 2 | 2 |
| His1h4f protein | CR542189 | 2 | 2 |
NFX1 was the most abundant protein identified by MS. Two poly(A) binding proteins were identified in two independent purification studies (boldface). Other RNA-processing proteins and deubiquitinating protein were also identified. Accession numbers for proteins copurified with NFX1-123 are listed.
Other proteins that interact with PABPCs contain defined PABP-interacting motifs called PAM1 or PAM2 (36, 37). The scansite website (http://scansite.mit.edu) (53) was used to search the NFX1-123 amino acid sequence. Indeed, the NFX1-123 sequence encoded a putative PAM2 motif from residues 9 to 26. The canonical PAM2 motif is 12 to 15 amino acids in length and contains 10 amino acids in its consensus sequence, 4 of which are most critical for binding to a PABC domain (36). The PAM2 motif found in NFX1-123 had 6 of the 10 consensus amino acids, with 3 amino acid substitutions of similar size or charge (Fig. 4A).
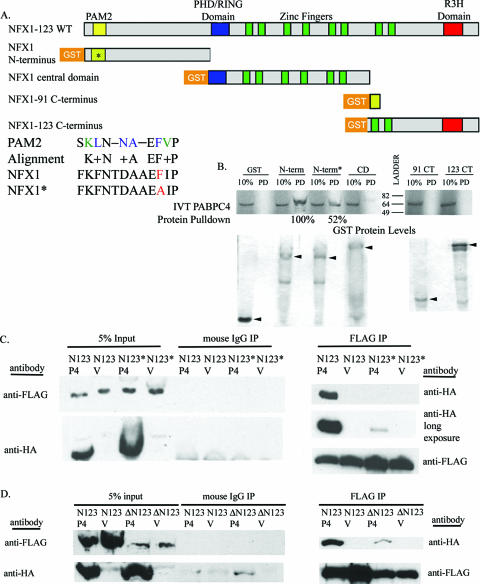
FIG. 4.
NFX1-123 contains a PAM2 motif, and it bound PABPCs via this domain. (A) Maps of NFX1-123, including the PAM2 motif, and the GST-tagged truncated or mutated constructs used in GST pulldowns. (*, single mutation in PAM2 motif). An alignment of the PAM2 canonical motif with NFX1 sequence is shown. Blue, amino acids most critical to PAM2-PABC interactions (36); green, common but not consensus amino acids; +, amino acids of similar charge or hydrophobicity. The phenylalanine mutated to alanine is shown in red. (B) GST pull down of truncated and mutated constructs of NFX1 showed only interaction with the N termini of NFX1 and PABPC4. A single amino acid mutation in the PAM2 motif decreased this interaction by half. 10%, 10% in vitro-translated input; PD, pulldown; N-term, GST NFX1 N terminus; N-term*, GST NFX1 N terminus PAM2 mutant; CD, GST NFX1 central domain; 91 CT, GST NFX1-91 C terminus; 123 CT, GST NFX1-123 C terminus. The GST protein levels shown represent 50% input. The arrowheads indicate GST proteins. (C) FLAG-tagged NFX1-123 (N123) or a FLAG-tagged NFX1-123 PAM2 mutant (N123*) was coexpressed with HA-tagged PABPC4 (P4) or vector control (V) in 293T cells; 24 h later, the cells were lysed and the FLAG-tagged protein was immunopurified. HA-PABPC4 coimmunoprecipitated with wild-type NFX1-123, but its interaction with the PAM2 mutant of NFX1-123 was dramatically reduced. (D) FLAG-tagged NFX1-123 (N123) or FLAG-tagged NFX1-123 with PAM2 deleted (ΔN123) was coexpressed with HA-tagged PABPC4 (P4) or vector control (V) in 293T cells. HA-PABPC4 coimmunoprecipitated with wild-type NFX1-123, and its interaction with NFX1-123 with PAM2 deleted was similar to background with mouse IgG immunoprecipitate. All films were scanned in through Adobe Photoshop.
To determine whether NFX1-123 bound PABPCs via the PAM2 motif, GST-tagged truncated constructs of NFX1 were incubated with in vitro-translated PABPC4 and PABPC1. PABPC4 interacted only with the N terminus of NFX1-123 (Fig. 4B), which encompasses the putative PAM2 motif. A single amino acid mutation (F to A) was created in the NFX1-123 N terminus PAM2 motif (Fig. 4A), since this residue in the PAM2 motif of the poly(A) binding protein regulatory protein, Paip2, had been shown to decrease PAM2-PABC domain interactions (36). With a single amino acid change in the PAM2 motif, we observed that the binding of PABPC4 to the NFX1 N terminus was reduced by half (Fig. 4B). PABPC1 also bound to the N terminus of NFX1, and its binding was similarly reduced with the single mutation in the PAM2 site (data not shown).
To confirm that full-length NFX1-123 interacted with PABPCs in vivo via the PAM2 motif, 293T cells were transiently transfected with FLAG-tagged NFX1-123 and HA-tagged PABPC4. By FLAG tag immunoprecipitation, NFX1-123 interacted with HA-PABPC4 (Fig. 4C). By contrast, NFX1-123 with the F-to-A mutation in the PAM2 motif (N123*) was stably expressed, but it lost nearly all detectable interaction with PABPC4. Only a long exposure of the film revealed any PABPC4 protein interaction with N123* (Fig. 4C).
The first 25 amino acids of NFX1-123 were also deleted to fully remove the PAM2 motif to which PABPCs bind. Equivalent amounts of NFX1-123 or NFX1-123 proteins with PAM2 deleted were immunoprecipitated with an anti-FLAG antibody (Fig. 4D, right), and again by coimmunoprecipitation, NFX1-123 interacted with HA-PABPC4. The NFX1-123 protein with PAM2 deleted interacted only with HA-PABPC4 (right), as well as the background amount seen in the mouse IgG immunoprecipitation (middle). Endogenously expressed PABPC1 also coimmunoprecipitated with NFX1-123, whereas the mutation of the PAM2 motif in N123* fully abolished this protein interaction (data not shown). Thus, the PAM2 motif was critical for NFX1-123 to bind PABPCs in vivo.
NFX1-123 augments the activation of hTERT expression through interactions with PABPCs.
As shown previously, NFX1-123 augmented expression in hTERT promoter-driven assays when c-Myc or HPV16 E6 was coexpressed (18). We wanted to determine whether the ability of NFX1-123 to augment expression required interaction with PABPCs. HPV16 E6 increased luciferase expression regulated by the hTERT promoter in HFKs approximately fivefold (Fig. 5A), and coexpression of NFX1-123 increased luciferase levels 10- to 15-fold (18). Of note, coexpression of PABPC4 and NFX1-123 increased expression 150-fold over the control and 10-fold over NFX1-123 plus HPV16 E6 alone (Fig. 5A). In contrast to wild-type NFX1-123, NFX1-123 with the single amino acid mutation in the PAM2 motif reduced the enhancement of luciferase expression by half. PABPC4 did not affect hTERT promoter-driven luciferase levels without NFX1-123.
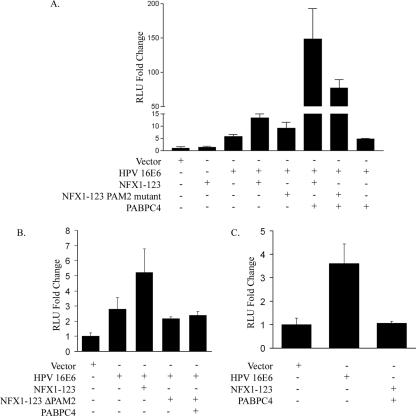
FIG. 5.
NFX1-123 and PABPC4 synergistically augmented hTERT promoter-regulated luciferase expression through the PAM2 motif in HFKs. (A) By luciferase assay, HFKs expressing HPV16 E6 activated expression from the hTERT promoter, and NFX1-123 augmented that expression only when HPV16 E6 was present. NFX1-123 and PABPC4 coexpressed could further increase expression over NFX1-123 alone, and the PAM2 mutant of NFX1-123 decreased this effect by half. PABPC4 did not increase expression without NFX1-123 present. (B) NFX1-123 augmented activation of HPV16 E6, but NFX1-123 with PAM2 deleted (NFX1-123 ΔPAM2) did not. (C) NFX1-123 and PABPC4 did not activate hTERT promoter-driven expression without HPV16 E6. RLU, relative light units. The error bars indicate 95% confidence intervals.
To determine whether the PAM2 motif is required for the augmentation of hTERT-driven expression seen with NFX1-123, HFKs expressing HPV16 E6 and NFX1-123 or NFX1-123 ΔPAM2 were compared by luciferase assay. NFX1-123 ΔPAM2 was unable to augment luciferase expression, and PABPC4 had no effect with NFX1-123 ΔPAM2 (Fig. 5B). Finally, we wanted to determine whether NFX1-123 and PABPC4 together could have an effect on hTERT promoter-driven luciferase levels without HPV16 E6. As with NFX1-123 alone (Fig. 5A), NFX1-123 and PABPC4 together did not increase expression (Fig. 5C).
DISCUSSION
The results of our study provide evidence that NFX1-123 is integral to hTERT regulation in HPV16 E6-expressing cells and that it may occur at a posttranscriptional level. Central to our findings were our observations that PABPCs copurify with NFX1-123, that a PAM2 motif is present in NFX1, and this motif and the PABPCs are important in the enhancement of hTERT activity by NFX1-123.
We found that NFX1-123 increased hTERT and telomerase activities in vivo. Previous work in our laboratory showed that NFX1-123 did not itself activate an hTERT promoter-driven reporter; however, when NFX-123 was coexpressed with c-Myc or HPV16 E6, expression was tripled (18). Now, we have shown that when NFX1-123 is overexpressed in vivo in HPV16 E6-expressing cells, telomerase activity is increased. Conversely, when NFX1-123 is knocked down, telomerase activity and hTERT mRNA levels both decrease. Finally, in coimmunoprecipitation and luciferase assays, the PAM2 motif of NFX1-123 is critical for its in vivo interaction with PABPCs and for its effect on hTERT promoter-driven expression.
Most of the proteins we identified by tandem-affinity purification of NFX1-123 support the role of NFX1-123 as a posttranscriptional regulator. RNA splicing factors, helicases, and ribonucleoproteins all have roles downstream of transcription (51) and were found to be associated with NFX1-123. Arginine/serine-rich splicing factors are involved in pre-mRNA splicing and processing in the nuclei of cells (2, 3, 6, 9, 28, 76), and DEAD box proteins are helicases found to unwind RNA during splicing (2, 14, 21, 26, 43, 44, 47, 54, 60). Heterogeneous ribonucleoproteins function in splicing and in pre-mRNA binding (7, 11, 16, 32, 38, 40, 52).
Interestingly, some of the proteins that were associated with NFX1-123 have roles in both RNA regulation and transcriptional regulation. The DEAD box protein p68 can activate transcription through interactions with other coactivators that have histone acetyltransferase activities (12, 61, 72, 72). Heterogeneous ribonucleoprotein u interacts with scaffold proteins that can recruit the histone acetyltransferase p300/CBP to activate gene expression (13, 19, 50). Thus, these proteins may function dually with NFX1-123, at the hTERT promoter and additionally on the hTERT mRNA, to affect its possible stability or translational efficiency.
Posttranscriptional regulation can occur at multiple steps (reviewed in reference 51). Controlling mRNA splicing can affect which isoforms predominate within a cell. Poly(A) tail formation and 5′ cap and poly(A) binding protein recruitment can affect RNA stability in the nucleus and cytoplasm (20, 39, 49). Shuttling from the nucleus to the cytoplasm for translation and recruiting translation machinery each can affect protein expression levels (33, 49). Finally decapping, deadenylation, and degradation of RNA at P bodies removes transcripts from further use (58). There was one prior study which described posttranscriptional regulation of hTERT. Cerezo et al. found that treatment of HaCaT cells with transforming growth factor β1 blocked the transcription of full-length hTERT and the active α splice variant, allowing only the inactive β transcript to be expressed (8). It is interesting to hypothesize that NFX1-123 also functions to affect hTERT mRNA transcripts.
To determine how NFX1-123 could be recruited to regulate hTERT, we studied how the NFX1 isoforms, HPV16 E6, and E6AP interacted. We have shown that the common central domain of NFX1 can interact with HPV16 E6, consistent with the results from our previous yeast two-hybrid screen that identified NFX1-123 as interacting with E6/E6AP and the fact that NFX1-91 coimmunoprecipitated with E6/E6AP. The independence from E6AP binding had not been tested directly, and this may clarify how NFX1-123 is affected by HPV16 E6. Unlike NFX1-91, which is degraded by E6/E6AP to derepress hTERT, NFX1-123 may be brought to the hTERT promoter or transcript by HPV16 E6 alone to increase expression.
NFX1-91 and NFX1-123 share a common N terminus, and both contain a PAM2 motif. Therefore, we wonder why NFX1-91 does not increase hTERT expression. NFX1-91 is quite unstable and undergoes rapid turnover in cells (18), so NFX1-123 is more likely to associate with PABPCs. Also, NFX1-91 is primarily in the nucleus, whereas NFX1-123 showed substantial cytoplasmic staining (M. Xu and D. A. Galloway, unpublished data). Finally, NFX1-123 has an R3H domain in its novel C terminus that may specify its role in RNA binding and posttranscriptional stabilization.
In summary, NFX1-123 increased telomerase expression in epithelial cells expressing HPV16 E6, its knockdown decreased telomerase activity and hTERT expression, and NFX1-123 with PABPCs synergistically augmented expression in gene reporter assays. Studies are ongoing to further explore the mechanisms by which NFX1-123 and PABPCs regulate hTERT and whether it is through a classic transcriptional activator mechanism or occurs downstream of promoter regulation, as suggested by our findings of PABPCs as interaction partners of NFX1-123.
Acknowledgments
We thank G. J. Goodall for providing the PABPC4 expression vector with T. Lindsten's permission.
This work was supported by STD/AIDS Research Training grant T32 AI07140 (R.A.K.), by Child Health Research Center grant K12 HD043376 (R.A.K. and P.V.-G.), by R01 CA064795 to D.A.G., and by Viral Oncology Training grant T32 CA09229 (E.M.E.).
Footnotes
Published ahead of print on 31 January 2007.
REFERENCES
- 1.Allsopp, R. C., H. Vaziri, C. Patterson, S. Goldstein, E. V. Younglai, A. B. Futcher, C. W. Greider, and C. B. Harley. 1992. Telomere length predicts replicative capacity of human fibroblasts. Proc. Natl. Acad. Sci. USA 89:10114-10118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barnard, D. C., J. Li, R. Peng, and J. G. Patton. 2002. Regulation of alternative splicing by SRrp86 through coactivation and repression of specific SR proteins. RNA 8:526-533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barnard, D. C., and J. G. Patton. 2000. Identification and characterization of a novel serine-arginine-rich splicing regulatory protein. Mol. Cell. Biol. 20:3049-3057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bartz, S. R., and M. A. Vodicka. 1997. Production of high-titer human immunodeficiency virus type 1 pseudotyped with vesicular stomatitis virus glycoprotein. Methods 12:337-342. [DOI] [PubMed] [Google Scholar]
- 5.Beitzinger, M., C. Oswald, R. Beinoraviciute-Kellner, and T. Stiewe. 2006. Regulation of telomerase activity by the p53 family member p73. Oncogene 25:813-826. [DOI] [PubMed] [Google Scholar]
- 6.Blencowe, B. J., R. Issner, J. A. Nickerson, and P. A. Sharp. 1998. A coactivator of pre-mRNA splicing. Genes Dev. 12:996-1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burd, C. G., M. S. Swanson, M. Gorlach, and G. Dreyfuss. 1989. Primary structures of the heterogeneous nuclear ribonucleoprotein A2, B1, and C2 proteins: a diversity of RNA binding proteins is generated by small peptide inserts. Proc. Natl. Acad. Sci. USA 86:9788-9792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cerezo, A., H. Kalthoff, M. Schuermann, B. Schafer, and P. Boukamp. 2002. Dual regulation of telomerase activity through c-Myc-dependent inhibition and alternative splicing of hTERT. J. Cell Sci. 115:1305-1312. [DOI] [PubMed] [Google Scholar]
- 9.Chaudhary, N., C. McMahon, and G. Blobel. 1991. Primary structure of a human arginine-rich nuclear protein that colocalizes with spliceosome components. Proc. Natl. Acad. Sci. USA 88:8189-8193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cox, D. M., M. Du, X. Guo, K. W. Siu, and J. C. McDermott. 2002. Tandem affinity purification of protein complexes from mammalian cells. BioTechniques 33:267-270. [DOI] [PubMed] [Google Scholar]
- 11.Datar, K. V., G. Dreyfuss, and M. S. Swanson. 1993. The human hnRNP M proteins: identification of a methionine/arginine-rich repeat motif in ribonucleoproteins. Nucleic Acids Res. 21:439-446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Endoh, H., K. Maruyama, Y. Masuhiro, Y. Kobayashi, M. Goto, H. Tai, J. Yanagisawa, D. Metzger, S. Hashimoto, and S. Kato. 1999. Purification and identification of p68 RNA helicase acting as a transcriptional coactivator specific for the activation function 1 of human estrogen receptor alpha. Mol. Cell. Biol. 19:5363-5372. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 13.Fackelmayer, F. O., K. Dahm, A. Renz, U. Ramsperger, and A. Richter. 1994. Nucleic-acid-binding properties of hnRNP-U/SAF-A, a nuclear-matrix protein which binds DNA and RNA in vivo and in vitro. Eur. J. Biochem. 221:749-757. [DOI] [PubMed] [Google Scholar]
- 14.Ford, M. J., I. A. Anton, and D. P. Lane. 1988. Nuclear protein with sequence homology to translation initiation factor eIF-4A. Nature 332:736-738. [DOI] [PubMed] [Google Scholar]
- 15.Gatlin, C. L., G. R. Kleemann, L. G. Hays, A. J. Link, and J. R. Yates III. 1998. Protein identification at the low femtomole level from silver-stained gels using a new fritless electrospray interface for liquid chromatography-microspray and nanospray mass spectrometry. Anal. Biochem. 263:93-101. [DOI] [PubMed] [Google Scholar]
- 16.Gattoni, R., D. Mahe, P. Mahl, N. Fischer, M. G. Mattei, J. Stevenin, and J. P. Fuchs. 1996. The human hnRNP-M proteins: structure and relation with early heat shock-induced splicing arrest and chromosome mapping. Nucleic Acids Res. 24:2535-2542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gewin, L., and D. A. Galloway. 2001. E box-dependent activation of telomerase by human papillomavirus type 16 E6 does not require induction of c-myc. J. Virol. 75:7198-7201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gewin, L., H. Myers, T. Kiyono, and D. A. Galloway. 2004. Identification of a novel telomerase repressor that interacts with the human papillomavirus type-16 E6/E6-AP complex. Genes Dev. 18:2269-2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gohring, F., and F. O. Fackelmayer. 1997. The scaffold/matrix attachment region binding protein hnRNP-U (SAF-A) is directly bound to chromosomal DNA in vivo: a chemical cross-linking study. Biochemistry 36:8276-8283. [DOI] [PubMed] [Google Scholar]
- 20.Gorlach, M., C. G. Burd, and G. Dreyfuss. 1994. The mRNA poly(A)-binding protein: localization, abundance, and RNA-binding specificity. Exp. Cell Res. 211:400-407. [DOI] [PubMed] [Google Scholar]
- 21.Guil, S., R. Gattoni, M. Carrascal, J. Abian, J. Stevenin, and M. Bach-Elias. 2003. Roles of hnRNP A1, SR proteins, and p68 helicase in c-H-ras alternative splicing regulation. Mol. Cell. Biol. 23:2927-2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hanahan, D., and R. A. Weinberg. 2000. The hallmarks of cancer. Cell 100:57-70. [DOI] [PubMed] [Google Scholar]
- 23.Harley, C. B., A. B. Futcher, and C. W. Greider. 1990. Telomeres shorten during ageing of human fibroblasts. Nature 345:458-460. [DOI] [PubMed] [Google Scholar]
- 24.Hayflick, L. 1965. The limited in vitro lifetime of human diploid cell strains. Exp. Cell Res. 37:614-636. [DOI] [PubMed] [Google Scholar]
- 25.Hayflick, L., and P. S. Moorhead. 1961. The serial cultivation of human diploid cell strains. Exp. Cell Res. 25:585-621. [DOI] [PubMed] [Google Scholar]
- 26.Hirling, H., M. Scheffner, T. Restle, and H. Stahl. 1989. RNA helicase activity associated with the human p68 protein. Nature 339:562-564. [DOI] [PubMed] [Google Scholar]
- 27.Horikawa, I., and J. C. Barrett. 2001. cis-Activation of the human telomerase gene (hTERT) by the hepatitis B virus genome. J. Natl. Cancer Inst. 93:1171-1173. [DOI] [PubMed] [Google Scholar]
- 28.Huang, Y., and J. A. Steitz. 2001. Splicing factors SRp20 and 9G8 promote the nucleocytoplasmic export of mRNA. Mol. Cell 7:899-905. [DOI] [PubMed] [Google Scholar]
- 29.Huibregtse, J. M., M. Scheffner, and P. M. Howley. 1991. A cellular protein mediates association of p53 with the E6 oncoprotein of human papillomavirus types 16 or 18. EMBO J. 10:4129-4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huibregtse, J. M., M. Scheffner, and P. M. Howley. 1993. Localization of the E6-AP regions that direct human papillomavirus E6 binding, association with p53, and ubiquitination of associated proteins. Mol. Cell. Biol. 13:4918-4927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.James, M. A., J. H. Lee, and A. J. Klingelhutz. 2006. HPV16-E6 associated hTERT promoter acetylation is E6AP dependent, increased in later passage cells and enhanced by loss of p300. Int. J. Cancer 119:1878-1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kafasla, P., M. Patrinou-Georgoula, J. D. Lewis, and A. Guialis. 2002. Association of the 72/74-kDa proteins, members of the heterogeneous nuclear ribonucleoprotein M group, with the pre-mRNA at early stages of spliceosome assembly. Biochem. J. 363:793-799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kahvejian, A., Y. V. Svitkin, R. Sukarieh, M. N. M'Boutchou, and N. Sonenberg. 2005. Mammalian poly(A)-binding protein is a eukaryotic translation initiation factor, which acts via multiple mechanisms. Genes Dev. 19:104-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Keen, N., R. Elston, and L. Crawford. 1994. Interaction of the E6 protein of human papillomavirus with cellular proteins. Oncogene 9:1493-1499. [PubMed] [Google Scholar]
- 35.Kiyono, T., S. A. Foster, J. I. Koop, J. K. McDougall, D. A. Galloway, and A. J. Klingelhutz. 1998. Both Rb/p16INK4a inactivation and telomerase activity are required to immortalize human epithelial cells. Nature 396:84-88. [DOI] [PubMed] [Google Scholar]
- 36.Kozlov, G., G. De Crescenzo, N. S. Lim, N. Siddiqui, D. Fantus, A. Kahvejian, J. F. Trempe, D. Elias, I. Ekiel, N. Sonenberg, M. O'Connor-McCourt, and K. Gehring. 2004. Structural basis of ligand recognition by PABC, a highly specific peptide-binding domain found in poly(A)-binding protein and a HECT ubiquitin ligase. EMBO J. 23:272-281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kozlov, G., J. F. Trempe, K. Khaleghpour, A. Kahvejian, I. Ekiel, and K. Gehring. 2001. Structure and function of the C-terminal PABC domain of human poly(A)-binding protein. Proc. Natl. Acad. Sci. USA 98:4409-4413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kozu, T., B. Henrich, and K. P. Schafer. 1995. Structure and expression of the gene (HNRPA2B1) encoding the human hnRNP protein A2/B1. Genomics 25:365-371. [DOI] [PubMed] [Google Scholar]
- 39.Kuhn, U., and E. Wahle. 2004. Structure and function of poly(A) binding proteins. Biochim. Biophys. Acta 1678:67-84. [DOI] [PubMed] [Google Scholar]
- 40.Kumar, A., K. R. Williams, and W. Szer. 1986. Purification and domain structure of core hnRNP proteins A1 and A2 and their relationship to single-stranded DNA-binding proteins. J. Biol. Chem. 261:11266-11273. [PubMed] [Google Scholar]
- 41.Levy, M. Z., R. C. Allsopp, A. B. Futcher, C. W. Greider, and C. B. Harley. 1992. Telomere end-replication problem and cell aging. J. Mol. Biol. 225:951-960. [DOI] [PubMed] [Google Scholar]
- 42.Li, H., T. H. Lee, and H. Avraham. 2002. A novel tricomplex of BRCA1, Nmi, and c-Myc inhibits c-Myc-induced human telomerase reverse transcriptase gene (hTERT) promoter activity in breast cancer. J. Biol. Chem. 277:20965-20973. [DOI] [PubMed] [Google Scholar]
- 43.Liew, C. C., and H. C. Smith. 1989. Immunological evidence for the role of phosphoprotein p68/pI = 7.3 in premessenger RNA splicing. FEBS Lett. 248:101-104. [DOI] [PubMed] [Google Scholar]
- 44.Lin, C., L. Yang, J. J. Yang, Y. Huang, and Z. R. Liu. 2005. ATPase/helicase activities of p68 RNA helicase are required for pre-mRNA splicing but not for assembly of the spliceosome. Mol. Cell. Biol. 25:7484-7493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lin, S. Y., and S. J. Elledge. 2003. Multiple tumor suppressor pathways negatively regulate telomerase. Cell 113:881-889. [DOI] [PubMed] [Google Scholar]
- 46.Liu, X., H. Yuan, B. Fu, G. L. Disbrow, T. Apolinario, V. Tomaic, M. L. Kelley, C. C. Baker, J. Huibregtse, and R. Schlegel. 2005. The E6AP ubiquitin ligase is required for transactivation of the hTERT promoter by the human papillomavirus E6 oncoprotein. J. Biol. Chem. 280:10807-10816. [DOI] [PubMed] [Google Scholar]
- 47.Liu, Z. R. 2002. p68 RNA helicase is an essential human splicing factor that acts at the U1 snRNA-5′ splice site duplex. Mol. Cell. Biol. 22:5443-5450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lois, C., E. J. Hong, S. Pease, E. J. Brown, and D. Baltimore. 2002. Germline transmission and tissue-specific expression of transgenes delivered by lentiviral vectors. Science 295:868-872. [DOI] [PubMed] [Google Scholar]
- 49.Mangus, D. A., M. C. Evans, and A. Jacobson. 2003. Poly(A)-binding proteins: multifunctional scaffolds for the post-transcriptional control of gene expression. Genome Biol. 4:223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Martens, J. H., M. Verlaan, E. Kalkhoven, J. C. Dorsman, and A. Zantema. 2002. Scaffold/matrix attachment region elements interact with a p300-scaffold attachment factor A complex and are bound by acetylated nucleosomes. Mol. Cell. Biol. 22:2598-2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mata, J., S. Marguerat, and J. Bahler. 2005. Post-transcriptional control of gene expression: a genome-wide perspective. Trends Biochem. Sci. 30:506-514. [DOI] [PubMed] [Google Scholar]
- 52.Matunis, M. J., J. Xing, and G. Dreyfuss. 1994. The hnRNP F protein: unique primary structure, nucleic acid-binding properties, and subcellular localization. Nucleic Acids Res. 22:1059-1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Obenauer, J. C., L. C. Cantley, and M. B. Yaffe. 2003. Scansite 2.0: proteome-wide prediction of cell signaling interactions using short sequence motifs. Nucleic Acids Res. 31:3635-3641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ogilvie, V. C., B. J. Wilson, S. M. Nicol, N. A. Morrice, L. R. Saunders, G. N. Barber, and F. V. Fuller-Pace. 2003. The highly related DEAD box RNA helicases p68 and p72 exist as heterodimers in cells. Nucleic Acids Res. 31:1470-1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Oh, S., Y. Song, J. Yim, and T. K. Kim. 1999. The Wilms' tumor 1 tumor suppressor gene represses transcription of the human telomerase reverse transcriptase gene. J. Biol. Chem. 274:37473-37478. [DOI] [PubMed] [Google Scholar]
- 56.Oh, S., Y. H. Song, U. J. Kim, J. Yim, and T. K. Kim. 1999. In vivo and in vitro analyses of Myc for differential promoter activities of the human telomerase (hTERT) gene in normal and tumor cells. Biochem. Biophys. Res. Commun. 263:361-365. [DOI] [PubMed] [Google Scholar]
- 57.Oh, S. T., S. Kyo, and L. A. Laimins. 2001. Telomerase activation by human papillomavirus type 16 E6 protein: induction of human telomerase reverse transcriptase expression through Myc and GC-rich Sp1 binding sites. J. Virol. 75:5559-5566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Parker, R., and H. Song. 2004. The enzymes and control of eukaryotic mRNA turnover. Nat. Struct. Mol. Biol. 11:121-127. [DOI] [PubMed] [Google Scholar]
- 59.Racek, T., N. Mise, Z. Li, A. Stoll, and B. M. Putzer. 2005. C-terminal p73 isoforms repress transcriptional activity of the human telomerase reverse transcriptase (hTERT) promoter. J. Biol. Chem. 280:40402-40405. [DOI] [PubMed] [Google Scholar]
- 60.Rossler, O. G., P. Hloch, N. Schutz, T. Weitzenegger, and H. Stahl. 2000. Structure and expression of the human p68 RNA helicase gene. Nucleic Acids Res. 28:932-939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rossow, K. L., and R. Janknecht. 2003. Synergism between p68 RNA helicase and the transcriptional coactivators CBP and p300. Oncogene 22:151-156. [DOI] [PubMed] [Google Scholar]
- 62.Shats, I., M. Milyavsky, X. Tang, P. Stambolsky, N. Erez, R. Brosh, I. Kogan, I. Braunstein, M. Tzukerman, D. Ginsberg, and V. Rotter. 2004. p53-dependent down-regulation of telomerase is mediated by p21waf1. J. Biol. Chem. 279:50976-50985. [DOI] [PubMed] [Google Scholar]
- 63.Shay, J. W., and S. Bacchetti. 1997. A survey of telomerase activity in human cancer. Eur. J. Cancer 33:787-791. [DOI] [PubMed] [Google Scholar]
- 64.Sladic, R. T., C. A. Lagnado, C. J. Bagley, and G. J. Goodall. 2004. Human PABP binds AU-rich RNA via RNA-binding domains 3 and 4. Eur. J. Biochem. 271:450-457. [DOI] [PubMed] [Google Scholar]
- 65.Song, Z., S. Krishna, D. Thanos, J. L. Strominger, and S. J. Ono. 1994. A novel cysteine-rich sequence-specific DNA-binding protein interacts with the conserved X-box motif of the human major histocompatibility complex class II genes via a repeated Cys-His domain and functions as a transcriptional repressor. J. Exp. Med. 180:1763-1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Stroumbakis, N. D., Z. Li, and P. P. Tolias. 1996. A homolog of human transcription factor NF-X1 encoded by the Drosophila shuttle craft gene is required in the embryonic central nervous system. Mol. Cell. Biol. 16:192-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Takakura, M., S. Kyo, M. Inoue, W. E. Wright, and J. W. Shay. 2005. Function of AP-1 in transcription of the telomerase reverse transcriptase gene (TERT) in human and mouse cells. Mol. Cell. Biol. 25:8037-8043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Toh, W. H., S. Kyo, and K. Sabapathy. 2005. Relief of p53-mediated telomerase suppression by p73. J. Biol. Chem. 280:17329-17338. [DOI] [PubMed] [Google Scholar]
- 69.Veldman, T., X. Liu, H. Yuan, and R. Schlegel. 2003. Human papillomavirus E6 and Myc proteins associate in vivo and bind to and cooperatively activate the telomerase reverse transcriptase promoter. Proc. Natl. Acad. Sci. USA 100:8211-8216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wang, J., L. Y. Xie, S. Allan, D. Beach, and G. J. Hannon. 1998. Myc activates telomerase. Genes Dev. 12:1769-1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wang, W., and B. A. Malcolm. 1999. Two-stage PCR protocol allowing introduction of multiple mutations, deletions and insertions using QuikChange Site-Directed Mutagenesis. BioTechniques 26:680-682. [DOI] [PubMed] [Google Scholar]
- 72.Watanabe, M., J. Yanagisawa, H. Kitagawa, K. Takeyama, S. Ogawa, Y. Arao, M. Suzawa, Y. Kobayashi, T. Yano, H. Yoshikawa, Y. Masuhiro, and S. Kato. 2001. A subfamily of RNA-binding DEAD-box proteins acts as an estrogen receptor alpha coactivator through the N-terminal activation domain (AF-1) with an RNA coactivator, SRA. EMBO J. 20:1341-1352. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 73.Won, J., J. Yim, and T. K. Kim. 2002. Sp1 and Sp3 recruit histone deacetylase to repress transcription of human telomerase reverse transcriptase (hTERT) promoter in normal human somatic cells. J. Biol. Chem. 277:38230-38238. [DOI] [PubMed] [Google Scholar]
- 74.Wu, K. J., C. Grandori, M. Amacker, N. Simon-Vermot, A. Polack, J. Lingner, and R. Dalla-Favera. 1999. Direct activation of TERT transcription by c-MYC. Nat. Genet. 21:220-224. [DOI] [PubMed] [Google Scholar]
- 75.Yang, H., C. S. Duckett, and T. Lindsten. 1995. iPABP, an inducible poly(A)-binding protein detected in activated human T cells. Mol. Cell. Biol. 15:6770-6776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zahler, A. M., W. S. Lane, J. A. Stolk, and M. B. Roth. 1992. SR proteins: a conserved family of pre-mRNA splicing factors. Genes Dev. 6:837-847. [DOI] [PubMed] [Google Scholar]
- 77.Zhang, Y., S. Fan, Q. Meng, Y. Ma, P. Katiyar, R. Schlegel, and E. M. Rosen. 2005. BRCA1 interaction with human papillomavirus oncoproteins. J. Biol. Chem. 280:33165-33177. [DOI] [PubMed] [Google Scholar]