Abstract
The diversity of viral strategies to modulate complement activation indicates that this component of the immune system has significant antiviral potential. One example is the Kaposi's sarcoma-associated herpesvirus (KSHV) complement control protein (KCP), which inhibits progression of the complement cascade. Rhesus rhadinovirus (RRV), like KSHV, is a member of the subfamily Gammaherpesvirinae and currently provides the only in vivo model of KSHV pathobiology in primates. In the present study, we characterized the KCP homologue encoded by RRV, RRV complement control protein (RCP). Two strains of RRV have been sequenced to date (H26-95 and 17577), and the RCPs they encode differ substantially in structure: RCP from strain H26-95 has four complement control protein (CCP) domains, whereas RCP from strain 17577 has eight CCP domains. Transcriptional analyses of the RCP gene (ORF4, referred to herein as RCP) in infected rhesus macaque fibroblasts mapped the ends of the transcripts of both strains. They revealed that H26-95 encodes a full-length, unspliced RCP transcript, while 17577 RCP generates a full-length unspliced mRNA and two alternatively spliced transcripts. Western blotting confirmed that infected cells express RCP, and immune electron microscopy disclosed this protein on the surface of RRV virions. Functional studies of RCP encoded by both RRV strains revealed their ability to suppress complement activation by the classical (antibody-mediated) pathway. These data provide the foundation for studies into the biological significance of gammaherpesvirus complement regulatory proteins in a tractable, non-human primate model.
The complement system bridges the innate and adaptive components of the immune system and provides a rapid response to infecting agents, including viruses. Complement activation is directly lytic, by formation of the membrane attack complex, and results in a number of other functional consequences, including priming and recruiting the adaptive immune system, e.g., through opsonization of foreign surfaces by C3b and release of the anaphylatoxins C3a and C5a. As a consequence, many viruses have evolved means to modulate complement activation. The phylogenetic diversity of these viruses and the different mechanisms by which they regulate complement activation indicate that this immunomodulatory activity has been acquired independently and at different times (reviewed in reference 10).
Kaposi's sarcoma-associated herpesvirus (KSHV) is the etiologic agent of Kaposi's sarcoma (13), primary effusion lymphoma (12), and perhaps multicentric Castleman's disease (38). It encodes a multitude of proteins with demonstrated or putative immunomodulatory activity (for review, see reference 35). The fourth open reading frame (ORF4) of KSHV encodes a protein that inhibits complement activation, as shown by our group and others (29, 39, 41). We named this protein KSHV complement control protein, KCP. KCP is similar in structure and function to a group of human complement inhibitors belonging to the family of regulators of complement activation (RCA). Like these cellular proteins, KCP modulates complement activation by inhibiting the C3 convertases (29, 39), key enzyme complexes that are formed subsequent to complement activation and that determine progression of the complement cascade. ORF4 is a lytic gene (17, 41), and the KCP protein it encodes has been found on the surface of both infected cells and viruses (40, 41). Thus, KCP is likely to help protect virions and infected cells against complement eradication. Moreover, KCP may be multifunctional; we recently showed that, like other human and viral RCAs, it binds to heparin and heparan sulfate (27, 40). On the surface of virions, KCP may therefore aid in viral attachment to host cells, as previously described for the envelope glycoproteins K8.1 and gB (1, 3, 9, 44). Concentrating KSHV on the surface of target cells via binding to heparan sulfate could enhance binding to its other specific receptors (2, 19, 34), a strategy described for other herpes viruses (37).
Kapadia et al. (20) evaluated the in vivo pathogenetic significance of the murine gammaherpesvirus 68 (γHV68) RCA. These authors identified a critical role for the γHV68 RCA in determining virulence, since mutant virus lacking this protein was attenuated. Moreover, mutant virus lacking the γHV68 RCA protein and wild-type γHV68 were equally pathogenic in complement component C3-deficient mice, underscoring the complement dependency of this attenuation. In contrast, these authors found no effect of γHV68 RCA in studies of latency. Such studies of the contribution of KCP to KSHV infection and pathogenesis in the natural host are obviously impossible. However, murine models are being developed that may be amenable to such KSHV work (6, 33, 46). Another putative model for KSHV is the non-human primate gamma2-herpesvirus, rhesus rhadinovirus (RRV). RRV replicates to high titer in cell culture (15) and infects rhesus monkeys, providing an in vivo model of KSHV-like infection since the gene organization and genomic sequences share similarity (5, 7, 25, 36, 45). RRV infection of rhesus macaques is therefore considered a useful animal model to study the relevance of individual genes for rhadinovirus biology in a primate host. Moreover, RRV can yield lymphoid hyperplasia resembling Castleman's disease when combined with simian immunodeficiency virus-induced immunosuppression (45) as well as transient lymphadenopathy in simian immunodeficiency virus-negative macaques (25). The genomes of two RRV isolates, called H26-95 and 17577, have been sequenced to date (GenBank accession numbers AF210726 and AF083501, respectively) (5, 36). Each strain encodes a KCP homologue. This protein has been named RRV complement control protein, RCP, by others (31), in keeping with existing nomenclature for this type of viral complement inhibitor (i.e., vaccinia virus complement control protein and KCP). Thus, as a prelude to investigating the function of a primate gammaherpesvirus complement regulatory protein in vivo, we have performed primary characterization and functional studies on RRV RCP and the protein(s) it encodes. While most gene products from the two different RRV isolates are predicted to share around 98% amino acid sequence similarity (5), the predicted RCP proteins from the two strains are quite different. They are referred to herein as RCP-H (RCP encoded by strain H26-95) and RCP-1 (RCP of strain 17577). RCP-H contains four complement control protein (CCP) domains (TrEMBL accession number Q9J2M6), like the KSHV KCP, while RCP-1 has eight CCP domains (TrEMBL accession number Q9WRU2). Both RCPs have a predicted C-terminal transmembrane region.
Given the structural diversity of RCP-H and RCP-1, we studied both genes and the proteins they encode. Rapid amplification of cDNA ends (RACE) PCR identified the 5′ and 3′ ends of the genes, and their expression in RRV-infected cells was measured by reverse transcriptase PCR (RT-PCR) and Western blotting. RCP-1 is expressed as a full-length mRNA and two alternatively spliced transcripts, while RCP-H RNA is not spliced. Recombinant RCP-H and RCP-1 expressed on the surface of Chinese hamster ovary (CHO) cells inhibited complement activation by the classical (antibody-mediated) pathway, measured by their inhibition of C3b deposition on the cell surface. Further data confirmed the presence of RCP on the surface of RRV, like KSHV-encoded KCP.
MATERIALS AND METHODS
Virus and cell culture.
Cell-free seed stocks of RRV 17577 and H26-95 were grown initially in primary rhesus macaque (Macaca mulatta) fibroblasts (RFB). Thereafter, RRV-infected cells were either primary RFB or RFB that were transduced with a retrovirus expressing human telomerase and referred to as transformed RFB (tRFB). Prior to their inoculation with virus, the cells were treated with polybrene (hexadimethrine bromide; catalogue no. H9268; Sigma, St. Louis, MO) at 2 μg/ml for 60 min. RRV naturally undergoes lytic replication in RFB and tRFB, with cytopathic effect (CPE) detectable by approximately 5 days postinfection. Cells were cultured in Dulbecco's modified Eagle's medium, supplemented with 1% nonessential amino acids, 1% glutamine, and 10% defined fetal bovine serum (catalogue no. SH30070; HyClone South Logan, UT). CHO cells were purchased from ATCC and propagated in RPMI 1640 cell medium (GIBCO) containing 10% fetal calf serum, 4 mM l-glutamine, 50 U/ml penicillin, and 50 μg/ml streptavidin.
Mapping of 5′ and 3′ ends of RCP transcripts.
Primary rhesus fibroblasts were infected with cell-free 17577 or H26-95 RRV. After development of CPE, 5 to 10 days postinfection, total cell RNA was isolated with TRIzol (GIBCO, Invitrogen, Carlsbad, CA), according to the manufacturer's protocol. Transcript 5′ and 3′ ends were mapped with a GeneRacer kit (Invitrogen), according to the manufacturer's instructions. Briefly, 15 to 600 ng of total RNA was used per reaction. An RNA oligonucleotide from the GeneRacer kit was ligated selectively to the mRNAs with an intact 5′ end, and reverse transcription was carried out with oligo(dT) primers and the Superscript III RT. PCRs were then performed on the resulting cDNA with one primer from the GeneRacer kit, annealing either to the sequence originating from the RNA oligonucleotide ligated to the 5′ end or to a 3′ sequence originating from the oligo(dT) primer. Gene-specific primers were designed to anneal around 200 to 250 bp either downstream of the putative initiating ATG or upstream of the putative translation TAA stop codon: 5′-RCP-1, GCAAGTCAGCAGGTGTGGGACATCTT; 3′-RCP-1, CGCCAACGCCAGAAGCACCCAAACCAAA; 5′-RCP-H, GCGTTTGGTGTAGTCCACGTTCCGTTT; and 3′RCP-H, CAGACAACAACCAATCGGCCATCAA.
The sequences of all other primers in the present study are available on request. Specific PCR products were TA cloned into the pCR4-TOPO vector provided with the GeneRacer kit. A total of 10 to 30 colonies of each RACE construct were sequenced. Nucleotide sequences were analyzed manually to identify the consensus DNA sequences, TATA, AATAAA, and YGTGTTYY.
RT-PCR analyses of RCP transcripts.
RNA was isolated from RRV-infected rhesus fibroblasts at the emergence of CPE, as described for the RACE procedure. Contaminating DNA was removed by treatment with DNase I (RQ1 DNase; Promega, Madison, WI) according to the manufacturer's instructions. The RT reaction was carried out using the oligo(dT) primer, Superscript III RT, and transcriptase buffer from the GeneRacer kit (Invitrogen), according to the manufacturer's instructions, followed by RNase H treatment. RCP cDNA was amplified by PCR with sequence-specific primers and Platinum Pfx DNA polymerase (Invitrogen), followed by incubation with Taq DNA polymerase to add 3′ A-overhangs. The PCR products were cloned into either the pCR4-TOPO vector (Invitrogen) or the pPCR-Script Amp SK(+) vector (Stratagene, La Jolla, CA) and sequenced.
Cloning of genomic DNA encoding RCP.
RRV H26-95 or 17577 particles were lysed by heating to 95°C for 30 min. The crude lysate served as a PCR template in a reaction employing Pfu polymerase (Fermentas, Vilnius, Lithuania). 3′ A-overhangs were then added with Taq polymerase (Invitrogen), and the PCR products were ligated into the pCR4-TOPO vector for sequencing (Invitrogen). The inserts were sequenced and found to be identical to the corresponding parts of the published RRV genomes (GenBank accession numbers AF083501 and AF210726).
Production of polyclonal anti-RCP antibodies.
Polyclonal anti-RCP antibodies were produced by the method described previously for antibodies against KCP (26). DNA fragments encoding CCP domains 1 to 2 (CCP1-2) and CCP5-6 from RCP-1 and CCP1-2 from RCP-H were amplified by PCR. The primers introduced sites for the restriction enzymes NdeI and XhoI to enable insertion of the DNA fragments into the prokaryotic expression vector pET-38b(+) (Novagen, EMD Biosciences, Madison, WI) in frame with a C-terminal cellulose binding domain tag from an exoglucanase (CBDcex) and an N-terminal His tag. The proteins were expressed by the Escherichia coli strain BL21-CodonPlus (DE3)-RIPL and accumulated in inclusion bodies. The inclusion bodies were isolated after lysis of the bacteria with lysozyme treatment (200 μg/ml in phosphate-buffered saline [PBS]) and sonication, washed in PBS, and then dissolved by sonication in 6 M guanidine HCl, 10 mM reduced glutathione, and 20 mM Tris-HCl (pH 8). Recombinant protein was purified by a nickel-nitrilotriacetic acid Superflow column (QIAGEN, Venlo, The Netherlands). The protein was refolded, when bound to the nickel matrix, by application of a linear gradient starting with 6 M guanidine HCl, 10 mM reduced glutathione, and 20 mM Tris-HCl (pH 8) and ending in 50 mM Tris-HCl, pH 8, followed by washing in 50 mM Tris-HCl, pH 8, plus 20 mM imidazole. The protein was then eluted with 0.7 M imidazole. Eluted protein was a mixture of monomeric RCP molecules and aggregates, as determined by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis and Coomassie brilliant blue staining (data not shown). To separate these species, the protein was dialysed against 50 mM Tris, pH 8.5, and then applied to a MonoQ column (GE Health Care, Fairfield, CT) equilibrated in the same buffer. Proteins were eluted with a salt gradient ranging from 0 to 0.5 M NaCl in the equilibration buffer. Fractions containing unaggregated protein were dialysed against PBS and used for immunization of rabbits according to a standard protocol at Cardiff University. The resulting sera are referred to as polyclonal antibodies (pAb) throughout the text.
Western blotting.
Cell lysates were prepared from either RRV-infected tRFB demonstrating significant CPE or from CHO cells stably transfected to express recombinant forms of RCP. Cells were detached from the culture flask by an EDTA-containing buffer to ensure cell surface protein integrity and then dissolved in a solubilization buffer (1% Triton X-100, 20 mM Tris-HCl, pH 8.0, 0.15 M NaCl, 5 mM EDTA, 1% Trasylol, and 2 mM phenylmethylsulfonyl fluoride [PMSF]). SDS-polyacrylamide gel electrophoresis was performed with a 12% acrylamide resolving gel. Denatured lysates were prepared by adding an equal volume of 2× SDS loading buffer (100 mM Tris-HCl, pH 6.8, 4% [wt/vol] SDS, 20% [vol/vol] glycerol, 3% [vol/vol] 2-mercaptoethanol, 0.05% [wt/vol] bromophenol blue) and heating to 100°C for 2 min. A total of 5 μg of each lysate was loaded, as determined by a Bradford assay. Cell culture fluid of RRV 17577-infected tRFB was also analyzed by Western blotting, after being concentrated threefold through a YM-10 Centriplus filter device (Millipore) by centrifugation (3,200 × g for 16 h at 4°C). Both the concentrate and flowthrough were analyzed.
Western blotting was performed according to the method of Towbin et al. (42). Resolved proteins were transferred to a 0.45-μm-pore-size Immobilon-P membrane (Millipore, Billerica, MA) by electrophoresis. Proteins were detected with primary antibody at a dilution of 1:250 and in turn detected by horseradish peroxidase (HRP)-conjugated anti-rabbit immunoglobulin G (IgG) secondary antibody (Dako, Glostrup, Denmark). The antibody-protein complex was visualized by enhanced chemiluminescence. Equal volumes of solution 1 (2.5 mM luminol, 0.4 mM p-coumaric acid, 100 mM Tris-HCl, pH 8.5) and solution 2 (0.0192% [vol/vol] H2O2, 100 mM Tris-HCl, pH 8.5) were mixed and added to the membrane. After about 1 min, excess solution was blotted off, and the membrane was exposed to autoradiographic film.
Chemical deglycosylation.
Aliquots of tRFB cytosol containing 1 mg of total protein were lyophilized overnight. Deglycosylation was performed on the freeze-dried samples using a GlycoProfile IV chemical deglycosylation kit (Sigma) as instructed by the manufacturer. The samples were then dialysed against PBS using tubing with a molecular size cutoff of 12,000 kDa.
IEM.
RRV for immune electron microscopy (IEM) was prepared from the culture fluid of infected tRFB showing CPE. Cells and debris were removed by centrifugation (1,900 × g for 5 min at 4°C), and virions were then concentrated (13,000 × g for 180 min at 4°C) before being resuspended in serum-free medium by overnight incubation at 4°C. The IEM procedure was performed essentially as described for staining KSHV virion-associated KCP (40). Briefly, RRV was applied to Parlodion-coated 200-mesh nickel grids (Agar Scientific, Stansted, United Kingdom). RCP was detected by staining with polyclonal anti-RCP antibody pAb H2612. The grids were washed in PBS, and bound antibody was detected with goat anti-rabbit antibody conjugated to 5-nm gold (British Biocell, Cardiff, United Kingdom). Preparations were negatively stained with phosphotungstic acid, pH 7 (Agar Scientific, United Kingdom), and viewed with a Jeol 1200 electron microscope (80kV; magnification, ×40,000). As a negative control, preimmune serum was used instead of pAb H2612.
Eukaryotic cell surface expression of RCP and C3b deposition assay.
In order to assess RCP regulation of complement activation, membrane-bound RCP was expressed with a glycosylphosphatidylinositol (GPI) anchor from human decay-accelerating factor (DAF; CD55) in the C-terminal part of the protein, as described previously for KCP functional studies (41). To achieve this expression, DNA encoding the soluble parts of RCP-1 and RCP-H, ending approximately 50 amino acids after the most C-terminal CCP domain, was amplified by PCR with appropriate primers from the respective full-length RCP genes in pCR4-TOPO. These fragments were cloned with the restriction enzymes XbaI and NotI into a derivative of the pDR2ΔEF1 vector (41) that contained the cDNA signal for GPI anchor addition and transfected with Lipofectamine (Invitrogen) into CHO cells. Following selection with hygromycin B (Roche, Basel, Switzerland), cell surface RCP expression on stable transfectants was confirmed by flow cytometric analyses with our polyclonal antibodies against RCP-H and RCP-1. For this analysis, the cells were detached with Versene (Invitrogen) and incubated with serum from the RCP-immunized animals [diluted 1:100 in binding buffer (10 mM HEPES, 140 mM NaCl, 5 mM KCl, 1 mM MgCl2, 2 mM CaCl2, pH 7)], for 1 h at room temperature, followed by incubation for 45 min with fluorescein isothiocyanate (FITC)-labeled anti-rabbit antibodies (catalogue no. F0205; DAKO), diluted 1:75.
C3b deposition was quantified on the surface of the stable CHO transfectants, expressing either RCP or KCP (39), or on mock-transfected cells. The cells were detached with Versene and washed twice with binding buffer. Between 200,000 and 500,000 cells per sample were sensitized with anti-hamster lymphocyte serum (catalogue no. H-4769; Sigma, Poole, United Kingdom), diluted 1:40, and increasing concentrations of fresh-frozen normal human serum diluted in DGVB++ (2.5 mM veronal buffer, pH 7.35, 72 mM NaCl, 140 mM glucose, 0.1% gelatin, 1 mM MgCl2, and 0.15 mM CaCl2).
C3b was quantified by washing the cells with binding buffer and then incubating them with rabbit anti-human C3c FITC-conjugated antibodies (catalogue no. F0201, diluted 1:100 in binding buffer; DAKO) for 30 min at room temperature. Fluorescence-activated cell sorting buffer (50 mM HEPES, 100 mM NaCl, 30 mM NaN3, 1% bovine serum albumin, pH 7.4) was added, and the cells were then analyzed with a Becton Dickinson FACSort flow cytometer (BD Biosciences, Franklin Lake, NJ). The mean fluorescence intensity (MFI) provided a measurement of the extent of C3b deposition. The experiments were carried out in triplicate, and data were acquired on 5,000 cells.
Detection of anti-RCP antibodies in rhesus serum.
Serum was drawn from a rhesus monkey experimentally infected with RRV 17577 and from a noninfected control. Both animals were housed at the Oregon National Primate Research Center, Beaverton, OR, and were cared for according to institutional guidelines. Serum was also obtained from two RRV H26-95-infected monkeys, animals Mm 122-96 and Mm 193-96, from the New England Regional Primate Research Center, Southborough, MA. Serum proteins were precipitated with (NH4)2SO4 at 60% of saturation and then dissolved in and dialysed against PBS. The preparations from each animal were adjusted to contain equal amounts of protein, as determined by UV spectroscopy. Microtiter plates (Maxisorp; Nunc) were coated with RCP-tagged CBDcex expressed in bacteria (described above) by incubation with a 10 μg/ml solution of the protein in 75 mM Na2CO3, pH 9.6, at 4°C overnight. The wells were blocked by incubation with quench buffer (3% fish gelatin, 10 mM Tris HCl, 75 mM NaCl, 0.05% Tween 20, pH 8.0) for 1 h at room temperature. The plates were then incubated with the rhesus serum proteins, diluted in quench buffer for 90 min at room temperature, followed by incubation with rabbit anti-human serum that cross-reacts with rhesus Ig (our data not shown, data found in reference 32, and data from the manufacturer). These sera were anti-human IgG (catalogue no. A0423; DAKO) and anti-human IgM (catalogue no. A0425; DAKO), diluted 1:1,000 in quench buffer for 1 h at room temperature. An HRP-conjugated anti-rabbit antibody (catalogue no. P0448; DAKO), diluted 1:1,000 in quench buffer, was then added for 1 h at room temperature. Between each incubation step, the microtiter plates were washed with 10 mM Tris HCl, 75 mM NaCl, 0.05% Tween 20, pH 8.0. Bound antibodies were detected using OPD (o-phenylenediamine dihydrochloride) tablets (DAKO) dissolved in H2O and supplemented with H2O2.
RESULTS
Mapping the 5′ and 3′ ends of the RCP transcripts.
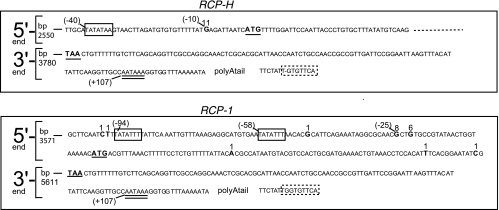
To identify the 5′ and 3′ ends of the RCP genes, the RACE procedure was performed on RNA prepared from RRV-infected primary RFB demonstrating CPE. Between 10 and 30 cDNA PCR clones derived from each transcript were sequenced, and the results are presented in Fig. 1. For RCP-H, the mRNA start site was unambiguously mapped to the position 10 nucleotides upstream of the predicted ATG start codon. The most preferred initiation start site for RCP-1 was 25 nucleotides upstream of the ATG start codon (−25; eight clones), but the transcription initiation was less stringently controlled than for RCP-H, since a further six clones identified an initiation site at position −22. Another six clones revealed putative transcription start sites to be scattered about this region of the gene. However, since no more than one clone identified each site, the sites specified by these six clones are deduced to be of minor relevance and may be artifacts arising due to RNA degradation during sample treatment.
FIG. 1.
The 5′ and 3′ ends of the RCP genes. Parts of the genomes of RRV 17577 (GenBank accession no. AF083501) and RRV H26-95 (AF210726) at the 5′ and 3′ ends of the RCP genes are displayed. The predicted RCP start (ATG) and stop (TAA) codons are in boldface and underlined. The detected mRNA start sites, as found by 5′ RACE in the present study, are in boldface, and the number above the nucleotide indicates the number of sequenced clones in which each site was found. For orientation, the number in parentheses indicates the position upstream (−) of the ATG start codon or downstream (+) of the TAA stop codon. The mRNA stop site and location of the poly(A) tail were determined by 3′ RACE. The position in the genome is indicated at the beginning of each sequence. Putative TATA boxes are enclosed by a rectangle, the consensus AATAAA box is underlined with two lines, and putative YGTGTTYY boxes are enclosed in rectangles with dashed lines, where Y stands for pyrimidine. The lack of a G residue in the RCP-H YGTGTTYY box compared to that of RCP-1 is indicated with a minus sign.
Examination of the sequences about the identified 5′ and 3′ ends of the genes revealed conserved motifs for the promoter (TATA) (11) and for polyadenylation (AATAAA and YGTGTTYY) (28, 43).
Primary sequence analysis of RCP proteins and comparison with KCP.
To identify similarities and differences between RCP from the two strains and KCP, their amino acid sequences were compared. CCP domains are predicted for all three proteins by sequence analysis tools (41; also SMART at http://smart.embl.de/). A CCP domain consists of approximately 60 amino acid residues that are arranged into antiparallel β-sheets, forming a β-sandwich arrangement stabilized by four cysteine residues that form two disulfide bonds in a 1-3,2-4 pattern, and conserved hydrophobic and aromatic residues that form a hydrophobic core (30).
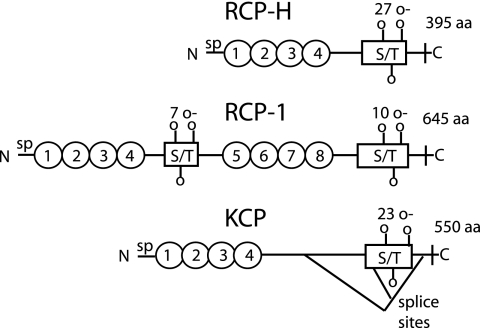
These sequence analyses predict that RCP-1 contains eight CCP domains, whereas RCP-H has four, like KCP (Fig. 2). All three proteins have a transmembrane domain in the C terminus. KCP exists in three isoforms due to splicing of its mRNA, where both shorter forms retain the transmembrane region as well as the four CCP domains (41). In between the last CCP domain and the transmembrane region, all three proteins have a region that is predicted to contain several O-glycosylated serine and threonine residues. This type of sequence is present also in the region between CCP4-5 in RCP-1.
FIG. 2.
Analysis and comparison of predicted structures of RCP and KCP. All three proteins have a signal peptide (sp) in the N terminus. The ovals symbolize CCP domains. S/T boxes are areas with predicted O-glycosylated serine and threonine residues, where the number of O-glycosylation sites is indicated above the boxes. In the C terminus there is a transmembrane region followed by a single intracellular amino acid. Three isoforms of KCP have been identified due to splicing events, for which the two donor and the single acceptor sites have been identified (41).
Pairwise alignment of the amino acid sequences in the CCP domains was performed by BLAST analyses (for which RCP-1 was divided into CCP1-4 and CCP5-8) (Table 1). The blocks of four CCP domains ranged in similarity from 58 to 63% (41 to 47% identity), with the exception of RCP-H CCP1-4 aligned with RCP-1 CCP5-8, which had much higher similarity (80% similarity and 64% identity).
TABLE 1.
A comparison of the identity and similarity of CCP domains in RCP-H, RCP-1, and KCP
| Aligned sequencesa | Identity (%) | Similarity (%) | |
|---|---|---|---|
| RCP-H CCP1-4 | RCP-1 CCP5-8 | 64 | 80 |
| RCP-H CCP1-4 | RCP-1 CCP1-4 | 47 | 63 |
| RCP-1 CCP1-4 | RCP-1 CCP5-8 | 45 | 64 |
| RCP-1 CCP1-4 | KCP CCP1-4 | 45 | 63 |
| RCP-1 CCP5-8 | KCP CCP1-4 | 45 | 58 |
| RCP-H CCP1-4 | KCP CCP1-4 | 41 | 58 |
The following amino acid residues were used (numbering refers to full-length proteins containing their signal peptide): 27 to 270 for RCP-H CCP1-4 (Q9J2M6); 23 to 265 for RCP-1 CCP1-4 (Q9WRU2); 320 to 562 for RCP-1 CCP5-8 (Q9WRU2); and 25 to 266 for KCP CCP1-4 (P88903). The TrEMBL accession numbers are in parentheses. Alignment was performed at http://www.ncbi.nlm.nih.gov/BLAST/bl2seq/wblast2.cgi, using the BLOSUM62 matrix algorithm.
Multiple splice forms of RCP-1 mRNA and a single RCP-H mRNA.
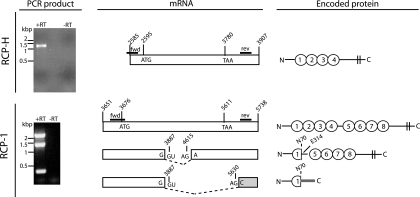
To determine whether the RCP transcript of either RRV strain is alternatively spliced, like KSHV KCP (41), RT-PCR analyses of primary RFB infected with RRV were performed. PCR primers were designed to anneal downstream of the transcription start and upstream of the transcription stop sites, determined from our RACE transcript mapping. A single, unspliced mRNA was detected for RCP-H. However, three different RCP-1 mRNA species were identified (Fig. 3, left). The largest mRNA was not spliced and encodes the predicted RCP-1. The two shorter mRNAs arise due to splicing events. There is one donor site and two acceptor sites (Fig. 3, middle); all obey the consensus G(U/A)G rule. The second largest mRNA encodes a predicted protein in which the first CCP domain is truncated (Fig. 3, right): the splice event causes loss of the last 10 amino acids of CCP1, including the last cysteine residue. CCP2-4 are also predicted to be absent, while CCP5-8 and the C-terminal region are present and in frame. The predicted size of this gene product is 40 kDa. The smallest mRNA encodes a protein with the truncated first CCP domain and a C-terminal end of 17 amino acids. Its molecular size is predicted to be 8 kDa. The splice acceptor site is downstream of the RCP stop codon, and the C-terminal domain is frameshifted compared to the N-terminal part. No evidence of a transmembrane domain in the C terminus of this putative protein was found by sequence analysis using the TMpred tool at EMBnet (http://www.ch.embnet.org).
FIG. 3.
Splicing pattern of RCP mRNA. RNA was extracted from rhesus fibroblasts that were infected with RRV. Reverse transcription generated cDNA, which was amplified with RCP-specific primers. The positions of the primers are indicated in the middle panel, as lines called fwd and rev. The resulting PCR products are shown in the left panel. The presence (+) or absence (−) of RT during the cDNA reaction is indicated above the lanes. One PCR product was obtained for RCP-H and three for RCP-1. These products were subcloned and sequenced. The cartoon in the middle panel interprets the sequence data (not to scale), with the transcript ends identified by RACE (Fig. 1). The PCR product from RCP-H represents the entire predicted ORF. For RCP-1, the largest PCR product represents the full-length gene. The nucleotides of certain key features, such as the protein start (ATG) and stop (TAA) codons, are given with their positions in the viral genome. The gray shading indicates a frameshift with respect to the full-length protein. The panel to the right describes the predicted proteins that are encoded by the different splice variants. Ovals represent CCP domains, vertical double lines are the transmembrane regions, and the bold line in gray indicates the frameshift. Amino acids and their positions in the full-length protein are shown at the splice border.
Expressed RCP proteins.
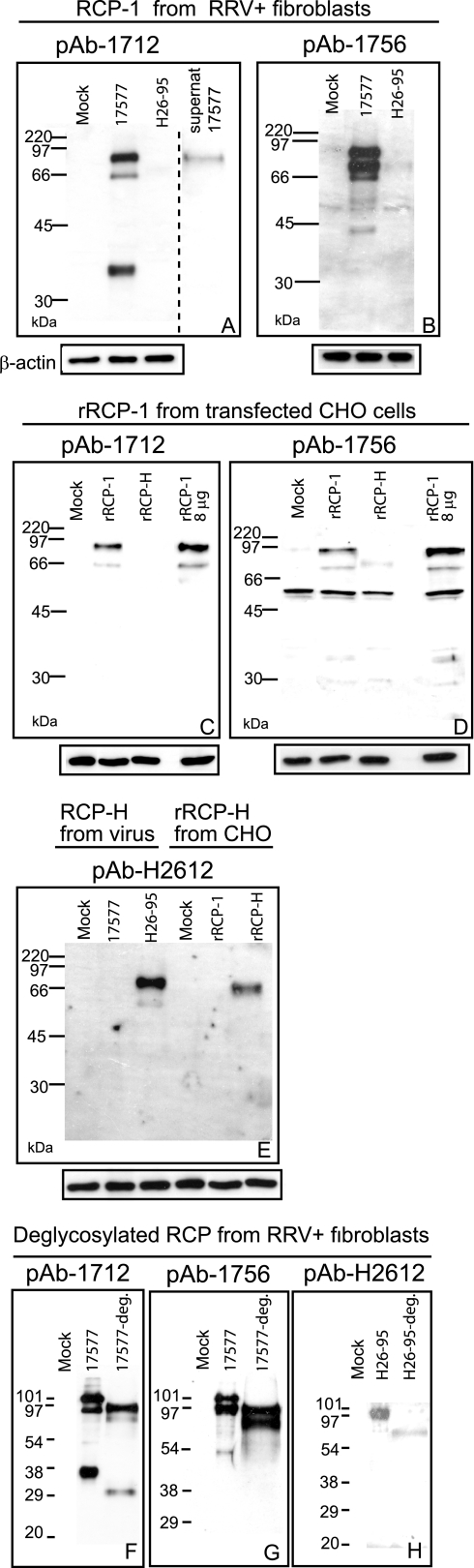
To determine if the identified transcription profiles of RCP-1 and RCP-H mRNA coincided with the expression profiles of the cognate proteins, Western blot analyses of RRV-infected tRFB were performed. In addition, these analyses were performed on CHO cells engineered to express recombinant RCP (rRCP) in which the protein was bound to the cell membrane via a GPI anchor derived from human DAF and inserted in frame approximately 50 amino acids downstream of the most C-terminal CCP domain.
Polyclonal antibody pAb-1712, raised against recombinant CCP1-2 of RCP-1, bound to proteins of an approximate size of 90, 66, and 40 kDa in cell lysates of 17577-infected RFB cells (Fig. 4A). This antibody also detected 90- and 66-kDa proteins from lysates of CHO cells expressing GPI-anchored recombinant forms of rRCP-1 (Fig. 4C). The 90-kDa and 66-kDa bands are inferred to be glycosylated forms of the proteins encoded by the full-length and middle size mRNAs of RCP-1, respectively, which encode proteins with masses of 69 and 40 kDa before posttranslational modification (see Discussion). Indeed, chemical deglycosylation resulted in RCP isoforms with a lower apparent molecular size (Fig. 4F and G), confirming that the expressed RCP-1 isoforms are glycosylated; the slight discrepancy in the apparent molecular sizes of the protein bands in Fig. 4F, G, and H compared with the sizes in Fig. 4A to E is due to differences in the molecular size markers used in the deglycosylation study. Antibody pAb-1712 also detected only the 90-kDa species in virion-rich, cell-free supernatant, even after longer exposures of the photographic film during Western blot analyses (Fig. 4A). The 66-kDa protein was detected at lower efficiency than the 90-kDa protein under all conditions by pAb-1712 (Fig. 4A and C), including in the virion-rich supernatant (Fig. 4A). This reduced detection could be due either to the predicted loss of cognate epitopes within CCP2 and 10 amino acid residues of CCP1 missing in this isoform, since pAb-1712 only recognizes CCP1-2 or to lower expression of the 66-kDa protein. The identification of the RCP-1 40-kDa protein is ongoing and no RCP-1 8-kDa protein was detected (see Discussion).
FIG. 4.
Native RCP expression in RRV-infected rhesus macaque fibroblasts and rRCP expression in engineered CHO cells. Cell lysates (5 μg of total protein) of either RRV-infected tRFB or CHO cells transfected with plasmid expressing rRCP were loaded in each lane. Exceptions are the supernatant loaded in panel A and samples with 8 μg of total protein in panels C and D. The uppermost text indicates which form of RCP was detected and from which cell-type. “Mock” refers to either noninfected rhesus fibroblasts or CHO cells transfected with an empty expression vector. Membranes were probed with antibodies indicated above the blots. These antibodies were directed to either RCP-1 CCP1-2 (pAb 1712) or RCP-1 CCP5-6 (pAb 1756) or RCP-H CCP1-2 (pAb H2612). Next, the membranes were incubated with anti-goat anti-rabbit HRP-conjugated antibody. Bound antibody was detected by chemiluminescence, and a number of different lengths of film exposure were processed. Equal loading of lysates was verified by measuring β-actin levels (lower panels). The dashed line in panel A indicates the boundary between two separate blots. Panels A to E present results of unmodified, denatured proteins. In panels F to H, chemically deglycosylated RCP (−deg) from RRV-infected tRFBs was compared to unmodified RCP from the same cells.
Staining RRV 17577-infected cell lysates with polyclonal antibody pAb-1756 (Fig. 4B), raised against CCP5-6 of RCP-1, identified the two bands that correspond in size and staining intensity to the 90- and 66-kDa bands of Fig. 4A. There is also a band slightly smaller than the 90-kDa species, of approximately 75 kDa. This protein is likely a more glycosylated variant of the 66-kDa protein, since it does not appear in the deglycosylated sample (Fig. 4G). Three minor bands in the size range 40 to 60 kDa are also detected with pAb-1756 in RRV 17577-infected cells upon long exposure (Fig. 4B); the middle-sized protein (ca. 55 kDa) is also detected in mock-infected cells, indicating some cross-reactivity of this antibody with one or more cellular proteins. Antibody pAb-1756 also detected the equivalent 90- and 66-kDa recombinant forms in transfected CHO cells expressing rRCP-1 (Fig. 4D). Since very weak reactivity with a 70-kDa band was observed for H26-95-infected RFB (Fig, 4B) and slightly stronger reactivity to this protein was detected in transfected CHO cells expressing rRCP-H (Fig. 4D), pAb1756 is most likely cross-reacting with the RCP-H protein that is the most similar in sequence to RCP-1 (Table 1). Antibody pAb-1756 also cross-reacted with the cellular protein of 55 kDa in untransfected CHO controls (Fig. 4D), detected in mock-infected cells (Fig. 4B), as well as demonstrating mild reactivity with other bands (Fig. 4B and D).
Polyclonal antibody pAb-H2612, raised against CCP1-2 of RCP-H, bound a protein of approximately 70 kDa in lysates from H26-95-infected RFB cells (Fig. 4E), the presumptive glycosylated variant of the full-length protein encoded by RCP-H, as confirmed by chemical deglycosylation studies (Fig. 4H). Antibody pAb-H2612 also detected the GPI-anchored rRCP-H in transfected CHO cells (Fig. 4E). The GPI-anchored rRCP-H is slightly smaller than the virion-expressed RCP-H due to a reduced number of amino acid residues. Unlike studies with pAb-1756, no cross-reactivity with the other RCP (RCP-1) was noted for pAb-H2612 (Fig. 4E).
RCP detection on RRV virions.
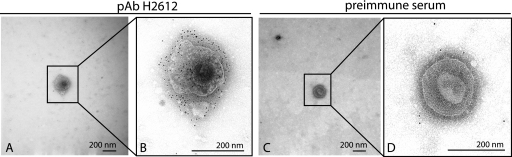
We have shown previously that KCP is located on the virion envelope and the surface of complete virions (40). Therefore, we reasoned that RCP would be expressed on the surface of RRV, and this supposition was confirmed by IEM. Figure 5A and B show RCP-H expression on the surface of H26-95 virions detected with pAb-H2612 and gold-conjugated secondary antibodies but not with preimmune rabbit serum (Fig. 5C and D).
FIG. 5.
RCP is present on the surface of RRV virions. (A and B) RRV H26-95 particles were purified and stained with pAb H2612 that was detected with gold-conjugated secondary antibodies, visible as black dots. Panel B shows an enlargement of the field of view shown in panel A. (C and D) As a negative control, RRV H26-95 particles were stained with preimmune serum instead of pAb H2612. Panel D is an enlargement of the field of view shown in panel C.
RCP antibodies in rhesus macaque monkeys infected with RRV.
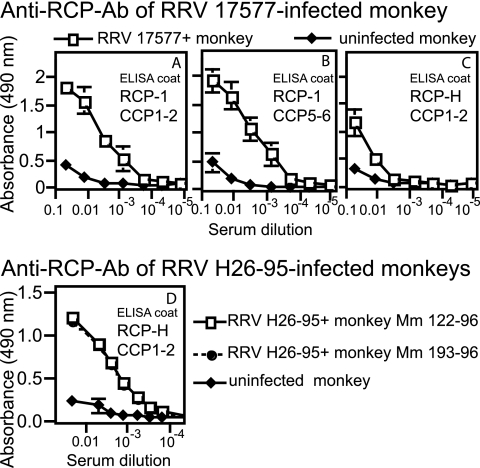
The presence of anti-RCP antibodies in the serum was determined with a modified enzyme-linked immunosorbent assay in which the assay wells were coated with recombinant proteins containing two CCP domains of either RCP-1 or RCP-H. Serum was drawn from one rhesus monkey experimentally infected with the 17577 strain of RRV and two infected with RRV H26-95 (animals Mm 122-96 and Mm 193-96). The serum from the RRV-infected monkeys contained substantial antibody titers against the respective variants of RCP (Fig. 6). Very low levels of RCP antibodies were detected in the noninfected control monkey and were of the IgM isotype (data not shown). Some cross-reactivity was demonstrated by antibodies from the RRV 17577-infected monkey to RCP-H.
FIG. 6.
Sera from RRV-infected rhesus monkeys contain antibodies against RCP. (A to C) Serum from a 17577-positive monkey was compared to that of a noninfected monkey. Microtiter plates were coated with CCP1-2 (A) or CCP5-6 (B) from RCP-1 or CCP1-2 from RCP-H (C) expressed in E. coli. Different dilutions of serum from the 17577-positive or noninfected monkey were added to the wells, and bound antibodies were captured by a mixture of goat anti-human IgG and IgM antibodies. (D) Microtiter plates were coated with CCP1-2 from RCP-H. Different dilutions of serum from either of two RRV H26-95-infected monkeys or a noninfected monkey were added, and bound antibodies were detected as described for panels A to C. ELISA, enzyme-linked immunosorbent assay.
RCP at the surface of CHO cells protects against complement-mediated lysis.
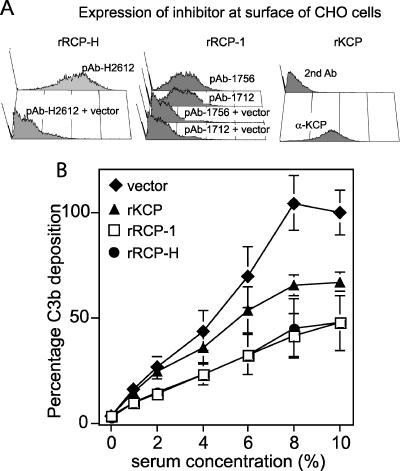
The capacity of RCP to regulate complement activation was determined according to the method by which we first showed KCP to inhibit complement activation (41). The CHO cells engineered to express recombinant GPI-anchored rRCP-1 or rRCP-H contained similar isoforms as RRV-infected rRFB cells (Fig. 4C to E) and were examined by flow cytometry with the polyclonal antibodies to ensure cell surface expression (Fig. 7A). CHO cells expressing KCP were also included as a positive control. The cells were sensitized with anti-CHO antibodies to activate the classical complement cascade and thereafter incubated with increasing concentrations of human serum. Resultant C3b deposition mediated by classical complement pathway activation was quantified by flow cytometry, staining with an anti-C3b antibody directly conjugated to FITC. Recombinant RCP from both RRV strains protected the cells against human C3b deposition in a manner comparable to KCP (39).
FIG. 7.
rRCP-H and rRCP-1 inhibit C3b deposition. CHO cells were transfected with constructs encoding either recombinant membrane-bound rRCP or rKCP. Negative control cells were transfected with an empty vector. (A) The levels of rRCP and rKCP expression by the transfected cells were determined by staining with polyclonal antibodies directed against CCP1-2 of RCP-H, CCP1-2 or CCP5-6 of RCP-1, and CCP1-2 of KCP. For rRCP expression, the negative controls were stained in the same manner as the rRCP-transfected cells, as indicated. For rKCP, the negative control is rKCP-expressing cells stained with only the secondary antibody. (B) C3b deposition was measured by flow cytometry. Cells were incubated with anti-hamster antibodies and different concentrations of human serum to activate complement and deposit C3b on the cell surface. The cells were then stained with an anti-C3b antibody. The experiment was performed twice in triplicate (or duplicate for rRCP-H). The geometric MFI obtained for the vector-only control cells in 10% serum was set as 100% C3b deposition. The C3b deposition percentages were calculated for all of the individual samples. The average and standard deviations of these values are plotted. A 100% C3b deposition corresponded to a geometric MFI of 195 or 125.
DISCUSSION
KSHV encodes the complement inhibitor KCP. The effect of KCP on the complement system has been well characterized in vitro, but the study of the biological impact of KCP for KSHV is confounded by the difficulty of culturing the virus in cell culture and the lack of an animal host. New small-animal model systems may help to alleviate this deficiency (6, 33, 46), although they are not primate based. RRV infection of rhesus macaques provides the most promising animal model in this regard to study pathophysiology and therapeutic interventions in the primate setting.
Thus, to ascertain the function and evaluate the mechanism of action of the RRV complement regulatory protein, we have performed primary characterization of the RCP genes of the two sequenced strains H26-95 and 17577. These data enabled cloning of the genes from both strains and, thereby, the expression of the proteins for functional studies and the generation of antibodies.
The sites of RCP transcription initiation and termination were mapped (Fig. 1). Transcription initiation occurred either 25 nucleotides (RCP-1) or 10 nucleotides (RCP-H) upstream of the putative translation initiation codons, increasing their credibility. Putative TATA boxes were found 33 (RCP-1) and 30 (RCP-H) bp upstream of the identified transcription initiation start sites, consistent with their normal situation for mammalian genes (11). An additional candidate TATA box positioned 69 bp upstream of RCP-1 is less ideally situated. Although the TATA boxes follow the consensus rules (11), they are not identical for RCP-1 and RCP-H, and neither of them is identical to the TATA box identified upstream from KCP (41). Both RCP genes have a Kozak element containing the important residue A three nucleotides upstream of the first methionine codon, but they lack G at position +4, suggesting that the context of these methionine codons is reasonably favorable (22, 23).
The 3′ ends of the RCP genes are identical. The polyadenylation site is 127 bp downstream of the stop codon. This site is preceded by the consensus AATAAA signal, present in most transcripts transcribed by RNA polymerase II (43). Approximately 30 bp downstream of the AATAAA box is a sequence adhering well to the YGTGTTYY consensus sequence found in certain herpesvirus genes and two-thirds of surveyed mammalian genes and thought to be involved in efficient 3′ RNA processing (28). Taken together, this analysis indicates that RCP-1 and RCP-H maintain a conventional eukaryotic gene organization.
Splicing of KSHV genes is well documented (for review, see reference 47), and the KSHV ORF4, or KCP, encoding KCP is expressed as a full-length transcript along with two alternatively spliced mRNAs; all three mRNA species specify membrane-bound KCP isoforms (41). Therefore, after the 5′ and 3′ ends of the RCP genes were mapped, the transcripts of RRV-infected cells were analyzed by RT-PCR for alternative splicing. Three splice variants of the RCP-1 mRNA from strain 17577 were identified (Fig. 3). The largest, unspliced RCP-1 transcript encodes the full-length RCP-1 protein, with eight CCP domains and a transmembrane region. The two smaller mRNAs arise through alternative splicing, where the splice donor sites are conserved and the acceptor sites differ. Each spliced transcript species specifies an RCP-1 isoform that is truncated in CCP1, resulting in the deletion of the carboxyl terminal 10 amino acid residues, including the fourth cysteine. This cysteine residue forms one of the two disulfide bonds that contribute to the tertiary structure of a CCP domain. It is therefore not clear what kind of conformation this truncated CCP domain adopts. Nevertheless, the full-length and middle-sized mRNAs of RCP-1 are expressed; proteins with the approximate apparent sizes of 90 and 66 kDa are detected with pAb-1712 and pAb-1756 (directed against CCP1-2 and CCP5-6 of RCP-1, respectively) in Western blot analyses of cell lysates of both RRV 17577-infected RFB as well as CHO cells expressing recombinant RCP-1 (Fig. 4). The predicted masses of the proteins encoded by the full-length and middle-sized splice forms are 69 and 40 kDa, respectively. The increased apparent molecular mass is likely due to glycosylation on any of the predicted N- and O-glycosylation sites found within the protein (http://www.cbs.dtu.dk/services/NetNGlyc/and http://www.cbs.dtu.dk/services/NetOGlyc/, respectively) (18). Chemical deglycosylation did, indeed, decrease the apparent molecular size of the RCP isoforms (Fig. 4F-H).
The smallest RCP-1 isoform may also be expressed, since we consistently detected by Western blotting a protein of ca. 40 kDa in RRV 17577-infected cell lysates (Fig. 4A) but not in RRV H26-95-infected or noninfected cells. This protein was not detected with pAb-1756, indicating that it lacks epitopes in CCP5-6, consistent with its being translated from the smallest mRNA variant. However, the predicted size of the protein encoded by this smallest isoform is ca. 8 kDa, and it does not contain any putative N- or O-glycosylation sites that could explain a size of 40 kDa. We have not detected an 8-kDa protein from RRV 17577-infected cells. Explanations include the possibility that the 40-kDa protein could be a very stable multimer of the smallest isoform, although the proteins were separated under reducing conditions. Alternatively, it could be the 8-kDa protein with covalently attached ubiquitin groups, targeting it for intracellular degradation due to misfolding (16). This possibility agrees with the absence of this protein in the supernatant of RRV 17577-infected cells (Fig. 4A); it is not secreted, despite having features of a secreted protein, including a signal peptide but no additional transmembrane domain. The 40-kDa protein was not present in lysates of CHO cells expressing recombinant RCP-1 (Fig. 4C and D). In this construct, the C-terminal transmembrane segment was replaced by a GPI anchor link from human DAF. Therefore, the most 5′ acceptor site in the RCP-1 mRNA is missing. The 40-kDa protein could also be derived from an as yet unidentified spliced isoform of RCP-1, and we cannot exclude the possibility that it and, indeed, the other small RCP-1 isoform are generated by proteolytic processing. Nevertheless, studies to determine the identity of the 40-kDa protein are in progress.
Another example of a viral RCA with a partial CCP domain, like the RCP-1 isoforms derived from spliced transcripts, is MOPICE (monkeypox inhibitor of complement enzymes) encoded by the monkeypox virus (24), of which only the first cysteine residue of the fourth CCP domain is present. The reason for the existence of alternative RCP-1 isoforms is unclear, as it is for KCP. Herpesvirus saimiri encodes a complement regulatory protein, expressed as a membrane-bound protein from full-length mRNA and a soluble form from spliced transcripts (4), but neither the KCP isoforms nor the two largest RCP-1 isoforms are expected to be soluble; they are all predicted to contain a transmembrane domain. Western blot analysis of concentrated supernatant from RRV 17577-infected cultures revealed low levels of the largest RCP-1 isoform, containing a membrane-spanning domain (Fig. 4A). The manner by which this protein is released into the supernatant is not known; possibly, it is cleaved from the cell surface, as was proposed for the γHV68 RCA (21). Alternatively, its presence in the supernatant is due to its virion-association. The other isoforms may also be present in the supernatant associated with virions but at levels too low to detect by our assay.
RCP-H contains four CCP domains, similar to previously described viral RCAs, while RCP-1 has eight CCP. The pairwise alignment performed for the CCP encoding sequences, with RCP-1 divided into CCP1-4 and CCP5-8, revealed the highest similarity between RCP-1 CCP5-8 and RCP-H (Table 1). If the eight CCP domains had arisen through a recent gene duplication of a four-CCP precursor, as the presence of a short S/T region between CCP4 and CCP5 suggests, a greater degree of similarity between CCP1-4 and CCP5-8 would be expected. However, although unlikely, a recombination event between viral strains might explain the higher similarity between CCP5-8 of RCP-1 and CCP1-4 of RCP-H. S/T-rich regions, predicted to be heavily O-glycosylated, are commonly found between the last CCP domain and the membrane, regardless of whether attachment to the membrane is via a glycolipid anchor or a transmembrane region and proposed to project the functional CCP domains to a suitable location above the plasma membrane (14). The presence of an S/T region between CCP domains, as identified in RCP-1, is unique compared to all other complement regulators identified (host and virus), and its significance is under investigation.
IEM analyses indicated that RCP-H encoded by RRV H26-95 is incorporated into the virion envelope. These data are consistent with the primary structure of RCP-H, which contains a membrane-spanning domain, and with our observations of KCP on the surface of KSHV virions (40). Moreover, our data are consistent with the studies of O'Connor et al. (31), who analyzed the protein composition of the different compartments of RRV H26-95 virions by mass spectometry and found RCP-H in the lipid envelope at a level indicating that RCP is more abundant than glycoprotein gL and as abundant as gN. Taken together, several lines of evidence support the presence of RCP-H on the virion surface, where it likely defends the virus against complement-mediated lysis by the membrane attack complex, the product of the terminal components of the complement cascade. Whether RCP-H contributes to cell binding, as KCP does for KSHV (40), is currently under investigation.
That RCP modulates complement activation was demonstrated by expressing the protein at the surface of CHO cells on the DAF GPI anchor. Here, both RCP-1 and RCP-H reduced deposition of C3b on the cell surface, following complement activation by the classical pathway (Fig. 7). These data indicate that RCP inhibits the classical C3 convertase, as does KCP. KCP exerts this inhibition both by acting as a cofactor for the serine protease factor I, enabling it to cleave C4b and C3b, and by directly accelerating the decay of the classical C3 convertase (29, 41). However, the mechanism(s) by which RCP regulates the classical C3 convertase activity remains to be determined. The C3b deposition assay was performed with human serum, suggesting that RCP cross-reacts with human complement components.
RRV-infected monkeys develop antibodies against RRV proteins, with seroconversion detectable within a few weeks after infection (25). Here, we show that antibodies can be generated in vivo against RCP (Fig. 6), indicating that RCP is expressed during the course of infection. Antibodies from RRV-infected animals neutralize infectivity in a complement-independent manner (8). Whether antibodies against RCP contribute to this neutralization is currently being determined. They may do so, for example, by either confounding a putative role for RCP in mediating virus-cell attachment or interfering with RCP complement regulation.
In conclusion, the present study adds another virus, RRV, to the growing list of those known to modulate complement activity (10). It serves to underline, therefore, the importance of complement to the biology of viruses of diverse phylogeny and emphasizes the role that the study of viruses plays in elucidating the function and significance of the immune response. These data also provide the framework for further studies on the nature of the molecular interactions between RCP and the complement system in vitro and in vivo in the non-human primate.
Acknowledgments
This study was supported by a FEBS short-term fellowship (L.M.), funds made available through the Tom Owen Memorial Fund (O.B.S.) and grants from Cancer Research UK (C7934 to D.J.B. and O.B.S.), the Swedish Research Council (A.B.), the Swedish Foundation for Strategic Research (A.B.), and Cancerfonden (A.B.).
The authors thank Adrienne Edkins for help in preparing RNA from RRV-infected primary rhesus macaque fibroblasts and Ronald Desrosiers and John Billelo for providing sera from H26-95-infected macaques.
Footnotes
Published ahead of print on 27 February 2007.
REFERENCES
- 1.Akula, S. M., N. P. Pramod, F. Z. Wang, and B. Chandran. 2001. Human herpesvirus 8 envelope-associated glycoprotein B interacts with heparan sulfate-like moieties. Virology 284:235-249. [DOI] [PubMed] [Google Scholar]
- 2.Akula, S. M., N. P. Pramod, F. Z. Wang, and B. Chandran. 2002. Integrin α3β1 (CD 49c/29) is a cellular receptor for Kaposi's sarcoma-associated herpesvirus (KSHV/HHV-8) entry into the target cells. Cell 108:407-419. [DOI] [PubMed] [Google Scholar]
- 3.Akula, S. M., F. Z. Wang, J. Vieira, and B. Chandran. 2001. Human herpesvirus 8 interaction with target cells involves heparan sulfate. Virology 282:245-255. [DOI] [PubMed] [Google Scholar]
- 4.Albrecht, J. C., and B. Fleckenstein. 1992. New member of the multigene family of complement control proteins in herpesvirus saimiri. J. Virol. 66:3937-3940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alexander, L., L. Denekamp, A. Knapp, M. R. Auerbach, B. Damania, and R. C. Desrosiers. 2000. The primary sequence of rhesus monkey rhadinovirus isolate 26-95: sequence similarities to Kaposi's sarcoma-associated herpesvirus and rhesus monkey rhadinovirus isolate 17577. J. Virol. 74:3388-3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.An, F. Q., H. M. Folarin, N. Compitello, J. Roth, S. L. Gerson, K. R. McCrae, F. D. Fakhari, D. P. Dittmer, and R. Renne. 2006. Long-term-infected telomerase-immortalized endothelial cells: a model for Kaposi's sarcoma-associated herpesvirus latency in vitro and in vivo. J. Virol. 80:4833-4846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bergquam, E. P., N. Avery, S. M. Shiigi, M. K. Axthelm, and S. W. Wong. 1999. Rhesus rhadinovirus establishes a latent infection in B lymphocytes in vivo. J. Virol. 73:7874-7876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bilello, J. P., J. S. Morgan, B. Damania, S. M. Lang, and R. C. Desrosiers. 2006. A genetic system for rhesus monkey rhadinovirus: use of recombinant virus to quantitate antibody-mediated neutralization. J. Virol. 80:1549-1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Birkmann, A., K. Mahr, A. Ensser, S. Yaguboglu, F. Titgemeyer, B. Fleckenstein, and F. Neipel. 2001. Cell surface heparan sulfate is a receptor for human herpesvirus 8 and interacts with envelope glycoprotein K8.1. J. Virol. 75:11583-11593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blue, C. E., O. B. Spiller, and D. J. Blackbourn. 2004. The relevance of complement to virus biology. Virology 319:176-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bucher, P. 1990. Weight matrix descriptions of four eukaryotic RNA polymerase II promoter elements derived from 502 unrelated promoter sequences. J. Mol. Biol. 212:563-578. [DOI] [PubMed] [Google Scholar]
- 12.Cesarman, E., Y. Chang, P. S. Moore, J. W. Said, and D. M. Knowles. 1995. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in AIDS-related body-cavity-based lymphomas. N. Engl. J. Med. 332:1186-1191. [DOI] [PubMed] [Google Scholar]
- 13.Chang, Y., E. Cesarman, M. S. Pessin, F. Lee, J. Culpepper, D. M. Knowles, and P. S. Moore. 1994. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi's sarcoma. Science 266:1865-1869. [DOI] [PubMed] [Google Scholar]
- 14.Coyne, K. E., S. E. Hall, S. Thompson, M. A. Arce, T. Kinoshita, T. Fujita, D. J. Anstee, W. Rosse, and D. M. Lublin. 1992. Mapping of epitopes, glycosylation sites, and complement regulatory domains in human decay accelerating factor. J. Immunol. 149:2906-2913. [PubMed] [Google Scholar]
- 15.Desrosiers, R. C., V. G. Sasseville, S. C. Czajak, X. Zhang, K. G. Mansfield, A. Kaur, R. P. Johnson, A. A. Lackner, and J. U. Jung. 1997. A herpesvirus of rhesus monkeys related to the human Kaposi's sarcoma-associated herpesvirus. J. Virol. 71:9764-9769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gao, G., and H. Luo. 2006. The ubiquitin-proteasome pathway in viral infections. Can. J. Physiol. Pharmacol. 84:5-14. [DOI] [PubMed] [Google Scholar]
- 17.Jenner, R. G., M. M. Alba, C. Boshoff, and P. Kellam. 2001. Kaposi's sarcoma-associated herpesvirus latent and lytic gene expression as revealed by DNA arrays. J. Virol. 75:891-902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Julenius, K., A. Molgaard, R. Gupta, and S. Brunak. 2005. Prediction, conservation analysis, and structural characterization of mammalian mucin-type O-glycosylation sites. Glycobiology 15:153-164. [DOI] [PubMed] [Google Scholar]
- 19.Kaleeba, J. A., and E. A. Berger. 2006. Kaposi's sarcoma-associated herpesvirus fusion-entry receptor: cystine transporter xCT. Science 311:1921-1924. [DOI] [PubMed] [Google Scholar]
- 20.Kapadia, S. B., B. Levine, S. H. Speck, and H. W. t. Virgin. 2002. Critical role of complement and viral evasion of complement in acute, persistent, and latent gamma-herpesvirus infection. Immunity 17:143-155. [DOI] [PubMed] [Google Scholar]
- 21.Kapadia, S. B., H. Molina, V. van Berkel, S. H. Speck, and H. W. t. Virgin. 1999. Murine gammaherpesvirus 68 encodes a functional regulator of complement activation. J. Virol. 73:7658-7670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kozak, M. 1991. An analysis of vertebrate mRNA sequences: intimations of translational control. J. Cell Biol. 115:887-903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kozak, M. 1991. Structural features in eukaryotic mRNAs that modulate the initiation of translation. J. Biol. Chem. 266:19867-19870. [PubMed] [Google Scholar]
- 24.Liszewski, M. K., M. K. Leung, R. Hauhart, R. M. Buller, P. Bertram, X. Wang, A. M. Rosengard, G. J. Kotwal, and J. P. Atkinson. 2006. Structure and regulatory profile of the monkeypox inhibitor of complement: comparison to homologs in vaccinia and variola and evidence for dimer formation. J. Immunol. 176:3725-3734. [DOI] [PubMed] [Google Scholar]
- 25.Mansfield, K. G., S. V. Westmoreland, C. D. DeBakker, S. Czajak, A. A. Lackner, and R. C. Desrosiers. 1999. Experimental infection of rhesus and pig-tailed macaques with macaque rhadinoviruses. J. Virol. 73:10320-10328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mark, L., W. H. Lee, O. B. Spiller, D. Proctor, D. J. Blackbourn, B. O. Villoutreix, and A. M. Blom. 2004. The Kaposi's sarcoma-associated herpesvirus complement control protein mimics human molecular mechanisms for inhibition of the complement system. J. Biol. Chem. 279:45093-45101. [DOI] [PubMed] [Google Scholar]
- 27.Mark, L., W. H. Lee, O. B. Spiller, B. O. Villoutreix, and A. M. Blom. 2006. The Kaposi's sarcoma-associated herpesvirus complement control protein (KCP) binds to heparin and cell surfaces via positively charged amino acids in CCP1-2. Mol. Immunol. 43:1665-1675. [DOI] [PubMed] [Google Scholar]
- 28.McLauchlan, J., D. Gaffney, J. L. Whitton, and J. B. Clements. 1985. The consensus sequence YGTGTTYY located downstream from the AATAAA signal is required for efficient formation of mRNA 3′ termini. Nucleic Acids Res. 13:1347-1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mullick, J., J. Bernet, A. K. Singh, J. D. Lambris, and A. Sahu. 2003. Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) open reading frame 4 protein (kaposica) is a functional homolog of complement control proteins. J. Virol. 77:3878-3881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Norman, D. G., P. N. Barlow, M. Baron, A. J. Day, R. B. Sim, and I. D. Campbell. 1991. Three-dimensional structure of a complement control protein module in solution. J. Mol. Biol. 219:717-725. [DOI] [PubMed] [Google Scholar]
- 31.O'Connor, C. M., and D. H. Kedes. 2006. Mass spectrometric analyses of purified rhesus monkey rhadinovirus reveal 33 virion-associated proteins. J. Virol. 80:1574-1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pachner, A. R., K. Amemiya, M. Bartlett, H. Schaefer, K. Reddy, and W. F. Zhang. 2001. Lyme borreliosis in rhesus macaques: effects of corticosteroids on spirochetal load and isotype switching of anti-Borrelia burgdorferi antibody. Clin. Diagn. Lab. Immunol. 8:225-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Parsons, C. H., L. A. Adang, J. Overdevest, M. O'Connor, C. J. R. Taylor, D. Camerini, and D. H. Kedes. 2006. KSHV targets multiple leukocyte lineages during long-term productive infection in NOD/SCID mice. J. Clin. Investig. 116:1963-1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rappocciolo, G., F. J. Jenkins, H. R. Hensler, P. Piazza, M. Jais, L. Borowski, S. C. Watkins, and C. R. Rinaldo, Jr. 2006. DC-SIGN is a receptor for human herpesvirus 8 on dendritic cells and macrophages. J. Immunol. 176:1741-1749. [DOI] [PubMed] [Google Scholar]
- 35.Rezaee, S. A., C. Cunningham, A. J. Davison, and D. J. Blackbourn. 2006. Kaposi's sarcoma-associated herpesvirus immune modulation: an overview. J. Gen. Virol. 87:1781-1804. [DOI] [PubMed] [Google Scholar]
- 36.Searles, R. P., E. P. Bergquam, M. K. Axthelm, and S. W. Wong. 1999. Sequence and genomic analysis of a rhesus macaque rhadinovirus with similarity to Kaposi's sarcoma-associated herpesvirus/human herpesvirus 8. J. Virol. 73:3040-3053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shukla, D., and P. G. Spear. 2001. Herpesviruses and heparan sulfate: an intimate relationship in aid of viral entry. J. Clin. Investig. 108:503-510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Soulier, J., L. Grollet, E. Oksenhendler, P. Cacoub, D. Cazals-Hatem, P. Babinet, M. F. d'Agay, J. P. Clauvel, M. Raphael, L. Degos, et al. 1995. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in multicentric Castleman's disease. Blood 86:1276-1280. [PubMed] [Google Scholar]
- 39.Spiller, O. B., D. J. Blackbourn, L. Mark, D. Proctor, and A. M. Blom. 2003. Functional activity of the complement regulator encoded by Kaposi's sarcoma-associated herpesvirus. J. Biol. Chem. 278:9283-9289. [DOI] [PubMed] [Google Scholar]
- 40.Spiller, O. B., L. Mark, C. E. Blue, D. G. Proctor, J. A. Aitken, A. M. Blom, and D. J. Blackbourn. 2006. Dissecting the regions of virion-associated Kaposi's sarcoma-associated herpesvirus complement control protein required for complement regulation and cell binding. J. Virol. 80:4068-4078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Spiller, O. B., M. Robinson, E. O'Donnell, S. Milligan, B. P. Morgan, A. J. Davison, and D. J. Blackbourn. 2003. Complement regulation by Kaposi's sarcoma-associated herpesvirus ORF4 protein. J. Virol. 77:592-599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Towbin, H., T. Staehelin, and J. Gordon. 1979. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc. Natl. Acad. Sci. USA 76:4350-4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wahle, E., and U. Ruegsegger. 1999. 3′ End processing of pre-mRNA in eukaryotes. FEMS Microbiol. Rev. 23:277-295. [DOI] [PubMed] [Google Scholar]
- 44.Wang, F. Z., S. M. Akula, N. P. Pramod, L. Zeng, and B. Chandran. 2001. Human herpesvirus 8 envelope glycoprotein K8.1A interaction with the target cells involves heparan sulfate. J. Virol. 75:7517-7527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wong, S. W., E. P. Bergquam, R. M. Swanson, F. W. Lee, S. M. Shiigi, N. A. Avery, J. W. Fanton, and M. K. Axthelm. 1999. Induction of B cell hyperplasia in simian immunodeficiency virus-infected rhesus macaques with the simian homologue of Kaposi's sarcoma-associated herpesvirus. J. Exp. Med. 190:827-840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wu, W., J. Vieira, N. Fiore, P. Banerjee, M. Sieburg, R. Rochford, W. Harrington, Jr., and G. Feuer. 2006. KSHV/HHV-8 infection of human hematopoietic progenitor (CD34+) cells: persistence of infection during hematopoiesis in vitro and in vivo. Blood 108:141-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zheng, Z. M. 2003. Split genes and their expression in Kaposi's sarcoma-associated herpesvirus. Rev. Med. Virol. 13:173-184. [DOI] [PMC free article] [PubMed] [Google Scholar]