Abstract
A 9.2-kb sequence from a hepatitis C virus (HCV) strain found in southwest France was compared to sequences from reference strains in HCV sequence databases. We found a recombinant virus with genotype 2 at the 5′ end and genotype 5 at the 3′ end. The crossover point was located between genes NS2 and NS3. Recombination between HCV genotypes must now be considered in studies on HCV epidemiology and evolution and in predictions of the virus response to antiviral therapy. Knowing the location of the recombination point may also be useful for constructing infectious chimeric viruses.
Studies of the nucleotide sequences of hepatitis C virus (HCV) variants from different individuals and geographical regions have revealed at least six genotypes and many subtypes (15, 16). The genotype is presently a major factor in the choice of treatment and the prognosis (2, 7). Knowledge of the distributions of genotypes in different parts of the world may help us to understand the epidemiology and evolution of HCV. Until 2001, the evolution of HCV was thought to proceed in a clonal manner, with diversity generated through the accumulation of mutations. However, homologous recombinations have been demonstrated in places as far apart as St. Petersburg (5), Vietnam (10), the Philippines (4), and Peru (1). We have now identified a new natural intergenotypic recombinant. Full-genome sequencing indicated that it was produced by recombination between genotypes 2 and 5.
Identification of a potential recombinant strain of HCV.
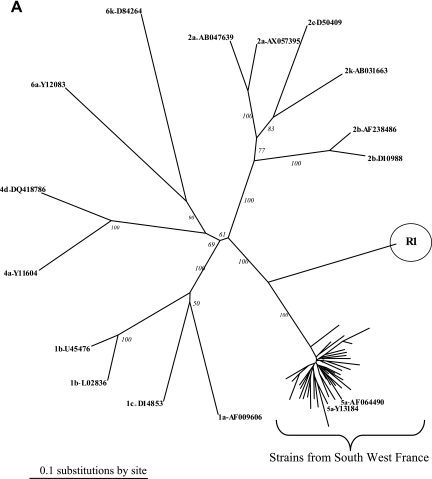
We previously analyzed the responses to treatment of 26 patients infected with HCV genotype 5 (HCV-5) in southwest France and the associated epidemiology (6). Phylogenetic analysis of the NS5B genes of the 26 corresponding strains pointed out a particular strain, R1. Twenty-five local strains formed a cluster with the genotype 5a reference sequences, while the R1 strain formed a distinct branch in the tree (Fig. 1A). The NS5B sequence with accession number AY373486 corresponds to the strain called R1 in the present study. The E2 region of the genome was sequenced to further characterize this strain (12). The MEGA program showed that the R1 sequence belonged to the genotype 2 in the E2 phylogenetic tree (Fig. 1B), but the strain could not be subtyped. Although these preliminary analyses were compatible with a recombinant strain of HCV, they could also indicate a mixed infection with two genotypes. Two genome regions were cloned and sequenced using the Invitrogen TA cloning kit. Phylogenetic analysis indicated that all the clones from the 5′ untranslated region (UTR; n = 19) clustered with genotype 2 and all the clones from the NS5B region (n = 35) clustered with genotype 5 (data not shown). This result excluded a double infection.
FIG. 1.
(A) Phylogenetic analysis of the R1 strain and 25 sequences from HCV-5 from southwestern France, performed with 400 bp within the NS5B region. (B) Phylogenetic analysis of the R1 strain and 137 sequences from HCV-2 from southwestern France, performed with 200 bp within the E2 region. Reference sequences from HCV strains from the Los Alamos HCV sequence database are listed by their accession numbers.
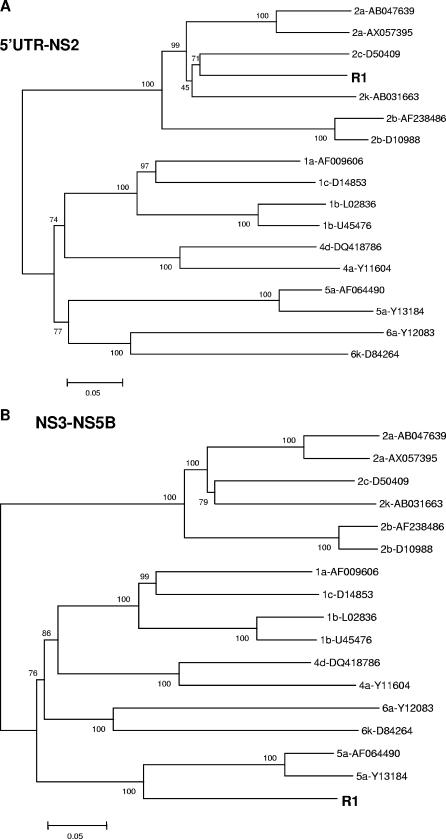
Determination of the entire sequence of the R1 strain.
cDNA was synthesized with reverse transcriptase (Superscript II; Invitrogen) and an RNase inhibitor (RNaseOUT; Invitrogen). A 7,400-nucleotide (nt) fragment was amplified and cloned using the Expand long-template PCR system (Roche Diagnostics, France) with sense primer KS3-2a and antisense primer ENO2 (Table 1). The other parts of the genome were directly sequenced. Almost the whole genome was sequenced (9,246 nt) with the Applied Biosystems Taq polymerase kit. The structure of the HCV genome was checked using the sequence locator tool (http://hcv.lanl.gov/content/hcv-db/LOCATE/locate.html). The various regions of the HCV genome were identified, and no stop codon was identified. The R1 sequences downstream (fragment of 3,430 nt) and upstream (fragment of 5,390 nt) of the putative breakpoint were compared with reference sequences (Fig. 2). The 5′ end of R1, from the 5′ UTR to NS2, corresponded to genotype 2, while the 3′ part of the R1 sequence, from NS3 to NS5B, was closer to that of genotype 5.
TABLE 1.
Primers used to amplify overlapping PCR fragments
| Primer name | Sequence | 5′ positiona | Type |
|---|---|---|---|
| PW11 | GCGTTAGTATGAGTGTCGTACAGCCTCCAG | 1 | Sense |
| KY78 | CTCGCAAGCACCCTATCAGGCAGT | 288 | Antisense |
| ENO1 | TGGGGATCCCGTATGATACCCGCTGCTTTGA | 8,245 | Sense |
| ENO2 | GGCGGAATTCCTGGTCATAGCCTCCGTGAA | 8,645 | Antisense |
| KS3-2a | GCTTGGGATATGATGATGAACTGGTC | 1,296 | Sense |
| LP2-4a | CGGGGTCGRGCAYGRGACATGCTGTGATAAA | 9,246 | Antisense |
| S1-SW-G2 | ACGGCTGGCCGCTGATGGCATCATACTGGG | 2,951 | Sense |
| AS-SW-G2 | CCCTCGGCGCTGGATAGTACATCTGCGTAG | 3,642 | Antisense |
| S1-SW-G5 | TCATAGTGGTGGCTTCAGTATTTCATAGCCC | 2,806 | Sense |
| AS1-SW-G5 | CCCACCAAGAGTCTTTGTCCTACATTAGGTGTACAT | 3,579 | Antisense |
The nucleotide positions are numbered according to the sequence locator tool numbering program from Los Alamos for the strain M67463 subtype 1a.
FIG. 2.
Neighbor-joining tree depicting the phylogenetic relationships among HCV reference sequences and sequences from the R1 strain. Trees were constructed for a 3,430-nucleotide fragment within the 5′ UTR to the NS2 gene (A) and a 5,390-nucleotide fragment within the NS3 region to the NS5B gene (B) by using the MEGA program with the Tamura-Nei model with gamma-corrected rates. Confidence values calculated by bootstrap analysis (100 replicates) are indicated at the major branching points. Reference sequences from HCV strains from the Los Alamos HCV sequence database are listed by their accession numbers.
Identification of the recombination point.
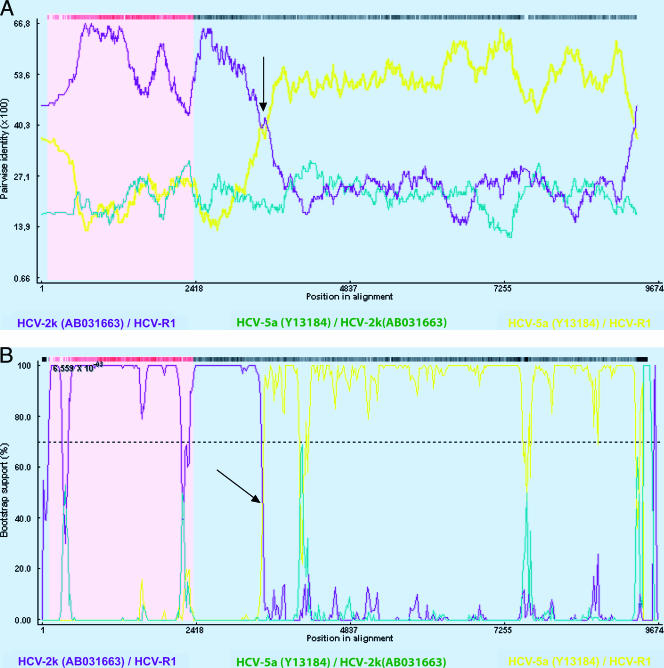
Recombination analysis was performed with the Recombinant Detection Program version 2 (http://darwin.uvigo.es/rdp/rdp.html) (8). This tool provides statistical evidence for the breakpoint site by using six methods (Recombinant Detection Program, Geneconv, BOOTSCAN, maximum chi-square, Chimaera, and sister scanning analyses). Recombination analyses of the R1 sequence and genome 2k (AB0331663), genome 2c (D50409), genome 2a (AB047639 and AX057395), genome 2b (AB030907 and AF238486), and genome 5a (AF064490 and Y13184) reference sequences indicated a crossover point at the beginning of the NS3 region, between nucleotides 3420 and 3440, depending on the method (P value, <0.01 for each method) (Fig. 3). Thus, the analysis by the Recombination Detection Program version 2 corroborated the phylogenetic tree results and showed that the 5′ part of the R1 strain is related to genotype 2k and the 3′ part is related to genotype 5. Further analyses did not reveal recombination with other genotypes.
FIG. 3.
Determination of the recombination point with the Recombinant Detection Program method (A) or the BOOTSCAN method (B) using the Recombinant Detection Program version 2. Plots for comparing the R1 and 2k strains are shown in purple, and plots for comparing the R1 strain and the 5a reference strains are in yellow. An arrow indicates the crossover point. Reference sequences from HCV strains from the Los Alamos HCV sequence database are listed by their accession numbers.
Confirmation of the recombination point.
We ruled out the possibility of artifactual intra-PCR recombination by amplifying by PCR fragments covering part of the E2 regions to the beginning of the NS3 regions of the R1, genotype 2, and genotype 5 strains. The reverse transcription-PCR kit (QIAGEN) was used with 140 μl of serum HCV RNA from the patient infected with the R1 strain. The sets of primers used were as follows: (i) a genotype 2-specific sense primer and a genotype 5-specific antisense primer (S1-SW-G2 and AS1-SW-G5), (ii) genotype 2-specific sense and antisense primers (S1-SW-G2 and AS1-SW-G2), and (iii) genotype 5-specific sense and antisense primers (S1-SW-G5 and AS1-SW-G5) (Table 1). The R1 strain sequence was successfully amplified only when the reverse transcription-PCR used a type 2-specific sense primer and a type 5-specific antisense primer, while the genotype 2 strain sequence was successfully amplified with the type 2-specific primers and the genotype 5 strain sequence with the type 5-specific primers (data not shown). The PCR product obtained with the type 2-specific sense primer and the type 5-specific antisense primer was sequenced; this analysis confirmed the same recombination point.
We have characterized a new HCV-2/5 recombinant genome that was identified because of a difference between the results from E2 and NS5B genotyping. Neither genotype 2 nor genotype 5 strains were found in the serum of the patient infected with R1, suggesting that she was contaminated directly by the R1 strain or that the recombinant strain had replaced the parental strains. The genotype 5-related sequence of the 3′ region from the R1 strain diverges from the other genotype 5 sequences, clustering as subtype 5a. This finding suggests that other HCV-5 subtypes remain to be characterized. Similarly, our analyses of the E2 region or of the sequence from the 5′ UTR to NS2 indicate that the R1 strain belongs to genotype 2, but the subtype is unknown. Our recent work with genotype 2 in our area has shown that this genotype varies greatly. Eight subtypes have been described, and more than 15% of the reported strains are uncharacterized (17). Salemi and Vandamme have demonstrated lower phylogenetic analysis values for the 5′ part of the HCV genome than for the NS5B region (11).
Although recombination may not appear to be common among natural populations of HCV, it must be taken into account as a cause of HCV genetic diversity, which may influence epidemiology and diagnosis. The recombinant strain type 2k/1b is spreading in Europe; 10 strains are described in the HCV database. No data are yet available on the response of this recombinant virus to therapy (5, 9). Genotyping is generally done with small fragments of the HCV genome. The HCV genotype determined directly influences the therapeutic strategy. The widespread existence of recombinant forms would considerably limit the use of genotyping assays that are based on single genome regions, such as the 5′ UTR or the core gene. The genotype may be determined by analyzing regions of interest, like the targets of new inhibitors of the HCV NS5B or NS3 gene.
The entire genome was sequenced, and the recombination site was identified at the beginning of the NS3 region. Several RNA viruses, like the Picornaviridae and the dengue virus, have recombination sites in the region of the genome that codes for the first nonstructural protein (3, 13, 14, 18). The recombination sites of the previously described recombinant HCV strains are also at the NS2/NS3 junction. The NS2/NS3 junction could be a favorable site for generating recombination events, especially with genotype 2 (4, 5, 10).
In conclusion, these data could be most important for vaccine development, diagnosis, and therapeutic strategy.
Nucleotide sequence accession number.
The recombinant sequence determined in this study has been deposited in the EMBL database with accession number AM408911.
Footnotes
Published ahead of print on 31 January 2007.
REFERENCES
- 1.Colina, R., D. Casane, S. Vasquez, L. Garcia-Aguirre, A. Chunga, H. Romero, B. Khan, and J. Cristina. 2004. Evidence of intratypic recombination in natural populations of hepatitis C virus. J. Gen. Virol. 85:31-37. [DOI] [PubMed] [Google Scholar]
- 2.Fried, M. W., M. L. Shiffman, K. R. Reddy, C. Smith, G. Marinos, F. L. Goncales, Jr., D. Haussinger, M. Diago, G. Carosi, D. Dhumeaux, A. Craxi, A. Lin, J. Hoffman, and J. Yu. 2002. Peginterferon alfa-2a plus ribavirin for chronic hepatitis C virus infection. N. Engl. J. Med. 347:975-982. [DOI] [PubMed] [Google Scholar]
- 3.Guillot, S., V. Caro, N. Cuervo, E. Korotkova, M. Combiescu, A. Persu, A. Aubert-Combiescu, F. Delpeyroux, and R. Crainic. 2000. Natural genetic exchanges between vaccine and wild poliovirus strains in humans. J. Virol. 74:8434-8443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kageyama, S., D. M. Agdamag, E. T. Alesna, P. S. Leano, A. M. Heredia, I. P. Abellanosa-Tac-An, L. D. Jereza, T. Tanimoto, J. Yamamura, and H. Ichimura. 2006. A natural inter-genotypic (2b/1b) recombinant of hepatitis C virus in the Philippines. J. Med. Virol. 78:1423-1428. [DOI] [PubMed] [Google Scholar]
- 5.Kalinina, O., H. Norder, S. Mukomolov, and L. O. Magnius. 2002. A natural intergenotypic recombinant of hepatitis C virus identified in St. Petersburg. J. Virol. 76:4034-4043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Legrand-Abravanel, F., K. Sandres-Saune, K. Barange, L. Alric, J. Moreau, P. Desmorat, J. P. Vinel, and J. Izopet. 2004. Hepatitis C virus genotype 5: epidemiological characteristics and sensitivity to combination therapy with interferon-alpha plus ribavirin. J. Infect. Dis. 189:1397-1400. [DOI] [PubMed] [Google Scholar]
- 7.Manns, M. P., J. G. McHutchison, S. C. Gordon, V. K. Rustgi, M. Shiffman, R. Reindollar, Z. D. Goodman, K. Koury, M. Ling, and J. K. Albrecht. 2001. Peginterferon alfa-2b plus ribavirin compared with interferon alfa-2b plus ribavirin for initial treatment of chronic hepatitis C: a randomised trial. Lancet 358:958-965. [DOI] [PubMed] [Google Scholar]
- 8.Martin, D. P., C. Williamson, and D. Posada. 2005. RDP2: recombination detection and analysis from sequence alignments. Bioinformatics 21:260-262. [DOI] [PubMed] [Google Scholar]
- 9.Moreau, I., S. Hegarty, J. Levis, P. Sheehy, O. Crosbie, E. Kenny-Walsh, and L. J. Fanning. 2006. Serendipitous identification of natural intergenotypic recombinants of hepatitis C in Ireland. Virol. J. 3:95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Noppornpanth, S., T. X. Lien, Y. Poovorawan, S. L. Smits, A. D. Osterhaus, and B. L. Haagmans. 2006. Identification of a naturally occurring recombinant genotype 2/6 hepatitis C virus. J. Virol. 80:7569-7577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Salemi, M., and A. M. Vandamme. 2002. Hepatitis C virus evolutionary patterns studied through analysis of full-genome sequences. J. Mol. Evol. 54:62-70. [DOI] [PubMed] [Google Scholar]
- 12.Sandres, K., M. Dubois, C. Pasquier, J. L. Payen, L. Alric, M. Duffaut, J. P. Vinel, J. P. Pascal, J. Puel, and J. Izopet. 2000. Genetic heterogeneity of hypervariable region 1 of the hepatitis C virus (HCV) genome and sensitivity of HCV to alpha interferon therapy. J. Virol. 74:661-668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Santti, J., T. Hyypia, L. Kinnunen, and M. Salminen. 1999. Evidence of recombination among enteroviruses. J. Virol. 73:8741-8749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Simmonds, P. 2004. Genetic diversity and evolution of hepatitis C virus—15 years on. J. Gen. Virol. 85:3173-3188. [DOI] [PubMed] [Google Scholar]
- 15.Simmonds, P., J. Bukh, C. Combet, G. Deleage, N. Enomoto, S. Feinstone, P. Halfon, G. Inchauspe, C. Kuiken, G. Maertens, M. Mizokami, D. G. Murphy, H. Okamoto, J. M. Pawlotsky, F. Penin, E. Sablon, I. T. Shin, L. J. Stuyver, H. J. Thiel, S. Viazov, A. J. Weiner, and A. Widell. 2005. Consensus proposals for a unified system of nomenclature of hepatitis C virus genotypes. Hepatology 42:962-973. [DOI] [PubMed] [Google Scholar]
- 16.Simmonds, P., E. C. Holmes, T. A. Cha, S. W. Chan, F. McOmish, B. Irvine, E. Beall, P. L. Yap, J. Kolberg, and M. S. Urdea. 1993. Classification of hepatitis C virus into six major genotypes and a series of subtypes by phylogenetic analysis of the NS-5 region. J. Gen. Virol. 74:2391-2399. [DOI] [PubMed] [Google Scholar]
- 17.Thomas, F., F. Nicot, K. Sandres-Saune, M. Dubois, F. Legrand-Abravanel, L. Alric, J. M. Peron, C. Pasquier, and J. Izopet. 2007. Genetic diversity of HCV genotype 2 strains in south western France. J. Med. Virol. 79:26-34. [DOI] [PubMed] [Google Scholar]
- 18.Tolou, H. J., P. Couissinier-Paris, J. P. Durand, V. Mercier, J. J. de Pina, P. de Micco, F. Billoir, R. N. Charrel, and X. de Lamballerie. 2001. Evidence for recombination in natural populations of dengue virus type 1 based on the analysis of complete genome sequences. J. Gen. Virol. 82:1283-1290. [DOI] [PubMed] [Google Scholar]