Abstract
We have previously reported on a system for large-scale molecular virus screening of clinical samples. As part of an effort to systematically search for unrecognized human pathogens, the technology was applied for virus screening of human respiratory tract samples. This resulted in the identification of a previously unknown polyomavirus provisionally named KI polyomavirus. The virus is phylogenetically related to other primate polyomaviruses in the early region of the genome but has very little homology (<30% amino acid identity) to known polyomaviruses in the late region. The virus was found by PCR in 6 (1%) of 637 nasopharyngeal aspirates and in 1 (0.5%) of 192 fecal samples but was not detected in sets of urine and blood samples. Since polyomaviruses have oncogenic potential and may produce severe disease in immunosuppressed individuals, continued searching for the virus in different medical contexts is important. This finding further illustrates how unbiased screening of respiratory tract samples can be used for the discovery of diverse virus types.
Persistent virus infections are an integrated part of human life. Most humans are persistently infected with one or more herpesviruses, papillomaviruses, polyomaviruses, and anelloviruses and remain healthy. Nevertheless, many of these viruses may occasionally produce severe disease (21, 32, 34, 37). Identification of previously unrecognized viral species is technically difficult. Thus, many potentially medically important persisting human viruses most likely remain undetected.
Polyomaviruses are small DNA viruses capable of persistent infection and having oncogenic potential. They have been found in many mammals and birds worldwide. Two polyomaviruses are known to normally infect humans, JC virus (JCV) and BK virus (BKV), both discovered in 1971 (13, 30). They are genetically closely related to each other, and both viruses show 70 to 80% seroprevalence in adults (23). The routes of acquisition and sites of primary infection are largely unknown, but both viruses can establish a latent infection in the kidneys and, in the case of JCV, also in the central nervous system (31). Persistent replication in the kidneys is evidenced by the fact that JCV, and occasionally also BKV, can be detected in the urine of healthy adults (23). BKV has also been detected in the feces of children (35). JCV and BKV are highly oncogenic in experimental animals, but a role in the development of human tumors has not been established (25). Disease caused by human polyomaviruses has been observed in immunosuppressed individuals. JCV is the causative agent of progressive multifocal leukoencephalopathy, a demyelinating disease of the brain and a feared complication of AIDS (21). This disorder has recently received renewed attention after the occurrence of fatal cases among patients treated with natalizumab for multiple sclerosis (22, 24). BKV has been associated with posttransplantation nephropathy and hemorrhagic cystitis in hematopoietic stem cell transplant (HSCT) recipients (7, 17). In addition to JCV and BKV, there are reports on the presence of the primate polyomavirus simian virus 40 (SV40) in humans, possibly introduced by contaminated poliovirus vaccine produced in monkey cells (4), although other ways of transmission have also been suggested (10, 27). SV40 genomic sequences have been detected in human malignant mesothelioma tumors, but its role in human tumor development remains debated (25).
We have developed a system for large-scale molecular screening of human diagnostic samples for unknown viruses (2). With this technology, we have initiated a systematic search for previously unrecognized viruses infecting humans in order to identify agents that are potentially involved in human disease. We describe here the identification and molecular characterization of a hitherto unknown human polyomavirus, which is only distantly related to the other known primate polyomaviruses. In analogy with the nomenclature of the other human polyomaviruses, we propose the name KI polyomavirus, KIPyV, for the newly discovered virus.
MATERIALS AND METHODS
Molecular virus screening.
As part of a systematic search for unknown viruses in clinical respiratory tract samples, a screening library was constructed from cell-free supernatants of 20 randomly selected nasopharyngeal aspirates made anonymous and submitted to the Karolinska University Laboratory, Stockholm, Sweden, for the diagnosis of respiratory tract infections. The samples were collected from March to June of 2004 and stored at −80°C until analyzed. This study was approved by the Karolinska Institutet local ethics committee. The procedure used for identification of virus nucleic acid sequences, molecular virus screening, has been described previously (2). In brief, samples were pooled and the pool was divided into two aliquots, which were filtered through 0.22- and 0.45-μm-pore-size disc filters (Millex GV/HV; Millipore), respectively. Both aliquots were ultracentrifuged at 41,000 rpm in an SW41 rotor (Beckman) for 90 min. The resulting pellet was recovered, resuspended, and treated with DNase before DNA and RNA were extracted (1). Extracted DNA and RNA were amplified separately by “random PCR” (2, 12). The amplification products were separated on an agarose gel, and fragments between approximately 600 and 1,500 bp in length were cloned. A total of four libraries were generated, derived from DNA or RNA, and filtered through a 0.22- or 0.45-μm filter. Ninety-six clones from each library were sequenced bidirectionally, i.e., a total of 384 clones. A set of specially designed C++ and Perl programs were used for automated quality trimming, clustering, BLAST searches, sorting, and formatting of the sequence reads. The output was a sorted list of the best database hits for nucleotide and translated sequences.
Genomic analysis of the KIPyV genome.
A 4,808-bp-long PCR product reaching around the circular DNA genome was generated by primers directed “outward” from the first cloned fragment (Pol-82R [TTGACTTCTTGGCCTTGTTAG] and Pol-315F [AGATGCTGACACAACTGTATG]) and by using a long-range enzyme mixture (Platinum Taq High Fidelity; Invitrogen). A second PCR product of 500 bp overlapping both ends of the long product and closing the circle was generated by primers PolconF (GGATTTTGTATGTGCTAGAAC) and PolconR (TTAACTAGAGGTACAACAAGC). Both PCR products were directly sequenced in order to obtain a consensus sequence for the complete genome. The same procedure was applied for determining the full-length sequences of three isolates. Putative open reading frames (ORFs) were identified, and sequences were aligned with Clone Manager Suite 6 (version 6.00) and Align Plus (version 4.10) (Scientific and Educational Software, Durham, NC). Prediction of putative binding sites for transcription factors was performed by comparison with consensus sequences and with the help of the AliBaba software (version 2.1) (16).
Phylogenetic analysis.
All sequences were downloaded from GenBank, except those of murine pneumotropic virus, which were based on a corrected sequence (T. Ramqvist, unpublished data). Accession numbers are available upon request. The complete genomes and the amino acid sequences of the early and late proteins, respectively, were aligned and neighbor-joining trees generated with ClustalX version 1.83. The data were bootstrapped with 1,000 replicates, and trees were viewed with NJplot. For whole-genome analysis, the noncoding control regions were removed in accordance with established conventions and the first nucleotide in the T antigens was designated nucleotide 1.
PCR for detection of KIPyV.
PCR experiments for detection of KIPyV were performed in a diagnostic laboratory setting, ensuring that the necessary precautions to avoid contamination were taken. Positive and negative controls were included in each experiment. DNA was extracted by commercially available kits as described under the respective sample type. Five microliters of extracted DNA was used as the template for the nested PCR. The 50-μl reaction mixtures used for the first and second PCRs consisted of 1× GeneAmp PCR buffer II (10 mM Tris-HCl [pH 8.3], 50 mM KCl; Applied Biosystems), 2.5 mM MgCl2, 0.2 mM each deoxynucleoside triphosphate, 2.5 U of AmpliTaq Gold DNA polymerase (Applied Biosystems), and 20 pmol of each of the primers. The first-PCR primers were POLVP1-39F (AAG GCC AAG AAG TCA AGT TC) and POLVP1-363R (ACA CTC ACT AAC TTG ATT TGG). The second-PCR primers were POLVP1-118F (GTA CCA CTG TCA GAA GAA AC) and POLVP1-324R (TTC TGC CAG GCT GTA ACA TAC). The cycling conditions for the first and second PCRs were 10 min at 94°C, followed by 35 cycles of amplification (94°C for 1 min, 54°C for 1 min, and 72°C for 2 min). Products were visualized on an agarose gel. The product size after the second PCR was 207 bp. All PCR products were sequenced in order to confirm that they were specific for KIPyV.
Prevalence study populations. (i) Nasopharyngeal aspirates.
Six hundred thirty-seven stored nasopharyngeal aspirates submitted to the Karolinska University Laboratory for diagnosis of respiratory virus infections from July 2004 to June 2005 were studied. Sampling month, patient's age and sex, and routine diagnostic (immunofluorescence and virus culture) findings were recorded before samples were made anonymous. The median age of the sampled patients was 7 years (range, 0 months to 90 years). Two hundred seventy-one samples came from children <2 years old. Total nucleic acids were extracted from 200-μl samples by the MagAttract Virus Mini M48 kit (QIAGEN), and nucleic acids were eluted in 100 μl. Eluted nucleic acids were initially analyzed in pools of 10 samples, and 5 μl of the pool was used as the template for the PCR. Single samples from PCR-positive pools were analyzed.
(ii) Feces.
One hundred ninety-two fecal samples submitted to the Karolinska University Laboratory for diagnosis of virus infections from 1 July 2005 to 30 November 2005 were studied. Samples were mainly submitted for diagnosis of gastroenteritis. Basic sampling data were recorded before samples were made anonymous. The median age of the sampled patients was 1 year (range, 0 months to 17 years). One hundred nineteen samples came from children <2 years old. Nucleic acids were extracted from 400 μl of a frozen 20% feces suspension by MagAttract Virus Mini M48 kit and the Biorobot M48 instrument (QIAGEN) and eluted in 100 μl, and 5-μl samples were used for subsequent individual PCR assays.
(iii) Urine of HSCT recipients.
One hundred fifty urine samples collected from HSCT recipients for the study of BKV and JCV were analyzed (14). Fifty of the samples were selected on the basis of previous analysis results; 20 were previously shown to be positive for BKV, 8 were positive for JCV, 2 were positive for both BKV and JCV, and 20 were negative for both viruses. JCV and BKV status was unknown for the remaining 100 samples. As described previously, samples were analyzed by PCR without preceding DNA extraction (6).
(iv) Serum of HSCT recipients.
Thirty-three serum samples drawn from 17 HSCT recipients 2 to 6 weeks after transplantation were studied. Total nucleic acids were extracted from 200 μl of serum by QIAamp Virus Spin Kit (QIAGEN) and eluted in 50 μl.
(v) Whole blood.
Whole EDTA blood from 192 healthy volunteer blood donors in Stockholm was analyzed. DNA was extracted from 200-μl samples with the MagAttract DNA Mini M48 kit and the Biorobot M48 instrument (QIAGEN) and eluted in 50 μl.
(vi) Leukocytes.
Ninety-six frozen preparations of Ficoll-separated leukocytes were studied. Samples were originally sent to the laboratory for diagnosis of cytomegalovirus by PCR and virus culture and therefore mainly originated from immunosuppressed patients. DNA was extracted from 105 cells with the MagAttract DNA Mini M48 kit and the Biorobot M48 instrument (QIAGEN) and eluted in 100 μl.
Nucleotide sequence accession numbers.
The sequences reported in this paper have been deposited in the GenBank database under accession no. EF127906 (KIPyV isolate 60), EF127907 (KIPyV isolate 350), and EF127908 (KIPyV isolate 380).
RESULTS
Molecular virus screening of 20 respiratory tract samples.
A virus-enriched DNA-cDNA library was constructed from 20 randomly selected nasopharyngeal aspirate samples by a previously published protocol (2). After vector and low-quality sequences were automatically discarded, sequence reads from 374 (97%) clones remained for database searches. By automated nucleotide and translated BLAST searches (3), the sequences were categorized as likely (expected value, <10−4) human (73%), bacterial (5%), phage (1%), unknown (2%), and virus (20%) sequences. Sixty-nine of the 74 clones with viral sequences matched human rhinovirus or enterovirus species. Reliable discrimination of rhinovirus from enterovirus sequences or type determination could not be performed on the basis of the unassembled sequence reads. Five clones closely matched respiratory syncytial virus. In addition to these virus-like clones, a single clone of 363 bp showed weak amino acid similarity (30% identity, expected value = 0.011) to VP1 of SV40 and was selected for further studies.
Genome analysis of KIPyV.
The source nasopharyngeal aspirate sample containing the SV40-like sequence was identified by PCR analysis of aliquots saved before pooling. The positive sample was named Stockholm 60. A second PCR product reaching around the circular DNA genome was used as a template for determining the complete consensus viral genome sequence. The genome was confirmed to be circular and 5,040 nucleotides in length (accession number EF127906). Two additional isolates that were identified during the subsequent prevalence study (see below) were sequenced by the same approach. (Stockholm 350, accession number EF127907; Stockholm 380, accession number EF127908). The three genomes were highly similar. Both isolates Stockholm 350 and Stockholm 380 differed from the prototype isolate by 10 nucleotide substitutions, and they differed from each other by seven single bases. The variable positions showed some clustering in the regulatory region, but there were also a few isolate-specific amino acid substitutions in the putative proteins.
Overall genome organization.
The genomic organization of KIPyV is typical for a member of the family Polyomaviridae, with an early region encoding regulatory proteins (small t [ST] and large T [LT] antigens) and a late region coding for structural proteins separated by a noncoding regulatory region (Fig. 1). The genome size is within the range of polyomaviruses. Properties of the deduced proteins and their similarities to those of JCV, BKV, and SV40 are shown in Table 1. While the nonstructural proteins have substantial amino acid sequence similarity to those of the other primate polyomaviruses, the structural proteins have a very low degree of similarity to those of other known polyomaviruses.
FIG. 1.
Genome organization of KIPyV. Putative coding regions for VP1 to VP3, ST antigen, and LT antigen are marked by arrows.
TABLE 1.
Putative proteins encoded by KIPyVa
| Protein | Putative coding region(s) | No. of amino acids | Calculated mass (kDa) | Amino acid identity (%) with:
|
||
|---|---|---|---|---|---|---|
| BKV | JCV | SV40 | ||||
| VP1 | 1498-2634 | 378 | 41.6 | 29 | 30 | 29 |
| VP2 | 441-1643 | 400 | 41.8 | 22 | 23 | 22 |
| VP3 | 870-1643 | 257 | 28.2 | 22 | 24 | 22 |
| ST antigen | 4967-4392 | 191 | 23.2 | 37 | 36 | 40 |
| LT antigen | 4967-4716, 4328-2655 | 641 | 74.3 | 48 | 47 | 47 |
Molecular masses were calculated by means of the ProtParam web tool (http://www.expasy.ch/tools/protparam.html).
Regulatory region.
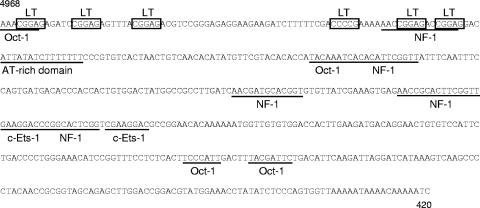
The noncoding regulatory region of polyomaviruses contains the promoters for early and late gene transcription, origin of replication, and transcriptional enhancers. The core origin of replication of KIPyV contains three potential LT antigen binding sites, compared to the four found in most polyomaviruses (Fig. 2). Two of these have the classical sequence GAGGC, while the third has the sequence GGGGC. An A/T-rich domain, probably harboring a TATA box for the early mRNA, lies to the late side of these binding sites. To the early side of the binding sites, polyomaviruses normally contain an imperfect palindrome, followed by additional LT antigen binding sites. KIPyV has three additional potential LT antigen binding sites where expected, but the there is no palindrome pattern. In fact, no long palindromes or repeats were found in the regulatory region of KIPyV. Putative binding sites for transcription factors were predicted in silico. Transcription factors with more than one putative binding site were c-Ets-1, Oct-1, and NF-1 (Fig. 3). In addition, there were transcription factors with a single putative binding site (data not shown). No binding sites for Sp1 could be found.
FIG. 2.
Alignment of the origins of replication of nine polyomavirus species. Putative binding sites for LT antigen are boxed. MPtV, murine pneumotropic virus; BPV, bovine polyomavirus; LPV, lymphotropic papovavirus. This alignment is a modified form of that of Mayer and Dörries (26).
FIG. 3.
The noncoding regulatory region with putative binding sites for transcription factors indicated. Putative LT antigen binding sites are boxed, and other transcription factor binding sites and the A/T-rich domain are underlined.
Early region.
In the early region of the genome, there are putative ORFs for the two regulatory proteins ST antigen and LT antigen (Fig. 1). On the basis of consensus sequences and alignments with LT antigens of other polyomaviruses, we propose a donor splice site for LT antigen at position 4716 and an acceptor site at position 4328. No middle T (MT) antigen seems to be expressed on the basis of the following. (i) Both polyomaviruses that express the MT antigen (murine polyomavirus [MPyV] and hamster polyomavirus) use all three reading frames for synthesis of the ST, LT, and MT antigens, while all other polyomaviruses, KIPyV included, use either one or, in some cases, two. (ii) In both MPyV and hamster polyomavirus, the MT antigen mRNA is produced through splicing, and no corresponding splice sites have been found in KIPyV. Assuming there is no expression of MT antigen, tiny T antigen can most likely not be expressed either (33).
The early proteins show similarities to other members of the polyomavirus family, primarily BKV, JCV, SV40, and simian agent 12 (SA12), and alignment with the LT antigens of other polyomaviruses shows that most regions characteristic of LT antigen are present also in KIPyV. The N-terminal 82 amino acids (aa) of the ST antigen are common to the LT antigen. This region encompasses the J domain carrying the conserved region 1 sequence and the HPDKGG box. In the C-terminal part that is unique to the ST antigen, there is a cysteine- rich domain typical of polyomaviruses. In the LT antigen, the HPDKGG box is followed by a putative Rb binding domain (LRCNE), a nuclear localization signal, a DNA binding domain, a Zn finger region including the zinc finger motif (C-312, C-315, H-327, H-331), and finally an ATPase-p53 binding domain containing the highly conserved GPXXXGKT sequence (aa 434 to 441). Unlike BKV, JCV, SV40, and SA12, the host range domain seems to be missing.
Late region.
In the late region of the genome, there are putative ORFs for capsid proteins VP1, VP2, and VP3 (Fig. 1). As in all polyomaviruses, VP3 is encoded by the same ORF as VP2 by the use of an internal start codon. There is an overlap between the C terminus of VP2/3 and the N terminus of VP1, as is the case in other polyomaviruses. It can be noted that both VP2 and VP3 of KIPyV are large in comparison with those of other members of the polyomavirus family (400 and 257 aa, respectively).
For VP1, there is only one possible start codon, in contrast to the VP1 proteins of BKV, JCV, and SV40. The degree of homology with other VP1 proteins is remarkably low (Table 1). VP1 has only 30% identity with its closest counterparts (those of JCV and MPyV). In KIPyV VP1, the only region that shows a relatively high degree of similarity to those of other polyomaviruses is the sequence that in MPyV VP1 has been shown to bind calcium, corresponding to approximately aa 237 to 248 in VP1 of KIPyV. Otherwise, VP1 of KIPyV has very limited homology to those of other polyomaviruses.
The VP2/VP3 gene showed even lower similarity to its counterparts in other polyomavirus species (Table 1). In fact, neither a nucleotide nor a translated BLAST search with this gene sequence generated any significant matches in the public databases. Thus, the identity of this ORF is only indicated by its position in the genome. VP2 and VP3 of all other polyomaviruses contain a conserved VP1 binding domain (located at approximately aa 281 to 295 in MPyVP2). No corresponding sequence is found in KIPyV.
Several polyomaviruses such as BKV, JCV, and SV40 express an Agno protein from the late mRNA. In KIPyV, the region between the start codons of VP2 and ST/LT, respectively, is large (513 bp) and this could possibly indicate the presence of an agno gene. However, there is no corresponding ORF present in this region.
Phylogenetic analysis.
Phylogenetic trees were constructed on the basis of alignments of the first isolate, Stockholm 60, with known viruses of the Polyomaviridae family. Analysis of early protein genes consistently clustered KIPyV with JCV, BKV, SV40, and SA12 but as an outlier in this clade (Fig. 4). Analysis of the complete genome yielded highly similar results (data not shown). In contrast, analysis of the late protein genes consistently placed KIPyV outside the tree as the most distant group member (Fig. 4).
FIG. 4.
Phylogenetic analysis of LT antigen amino acid sequences (a) and VP1 amino acid sequences (b). Bootstrap values are indicated at each branching point. Analysis of the ST antigen yielded a phylogenetic tree highly similar to that obtained by LT antigen analysis, and whole-genome analysis showed a similar pattern. Analysis of VP2 and VP3 yielded results highly similar to those obtained with VP1. GHPV, goose hemorrhagic polyomavirus; CPyV, crow polyomavirus; FPyV, finch polyomavirus; APV, avian polyomavirus; BPV, bovine polyomavirus; MPtV, murine pneumotropic virus; LPV, lymphotropic papovavirus; HaPyV, hamster polyomavirus.
Prevalence of KIPyV in human samples.
In order to investigate possible replication sites and suitable sample materials for prevalence studies, several sample sets were investigated for the presence of KIPyV DNA by a PCR targeting the gene for VP1 (Table 2). The identities of the PCR products were confirmed by sequencing. The results were also confirmed by a second PCR assay targeting the LT antigen gene. KIPyV was detected in nasopharyngeal aspirates and feces but not in urine, whole-blood, leukocyte, or serum samples (Table 2). Two isolates obtained from nasopharyngeal aspirates were fully sequenced. The ages of the six subjects positive for KIPyV in the nasopharynx ranged from 1 month to 26 years (median, 2 years), and the subject with positive feces was 3 months old. In five of the six nasopharyngeal aspirates, a respiratory virus infection was codetected in the same sample by standard diagnostic tests (immunofluorescence and virus culture) (three respiratory syncytial virus, one human metapneumovirus, and one influenza A virus), suggesting that KIPyV is not likely responsible for the symptoms that prompted nasopharyngeal sampling.
TABLE 2.
Summary of results from the screening of selected human samples for KIPyV by PCR
| Sample type | Source | No. of samples tested | No. (%) positive |
|---|---|---|---|
| Nasopharyngeal aspirate | Consecutive clinical samples, mainly respiratory tract disease | 637 | 6 (1) |
| Urine | HSCT recipients | 150 | 0 (0) |
| Serum | HSCT recipients | 33 | 0 (0) |
| Whole blood | Blood donors | 192 | 0 (0) |
| Feces | Consecutive clinical samples, mainly gastroenteritis | 192 | 1 (0.5) |
| Isolated frozen leukocytes | Samples sent for CMVa diagnosis, mainly from immunosuppressed patients | 96 | 0 (0) |
CMV, cytomegalovirus.
DISCUSSION
The finding of a circular DNA molecule with a genomic organization characteristic for a polyomavirus and with large regions of amino acid sequence homology with polyomaviruses strongly suggests that the discovered DNA molecule is indeed the genome of a previously undescribed polyomavirus. The two previously known human polyomaviruses JCV and BKV were named by the initials of the patients in whom the viruses were first identified (13, 30). This is not the current naming practice. In order to still conform to the previous nomenclature, we propose the provisional name KIPyV for the new polyomavirus. Its recovery from eight human samples confirms that it infects humans, and its partial relatedness to JCV and BKV and its prevalence in children indicate that humans are likely the natural host. However, further studies, in particular, serologic studies, are needed in order to confirm this. Intriguingly, the existence of more human polyomavirus species in addition to JCV and BKV has been suggested earlier by the finding of lymphotropic papovavirus-reactive antibodies in human sera (36). Whether this observation was indeed related to KIPyV remains to be determined. The nucleotide sequence of KIPyV has only limited similarity to that of JCV, BKV, SV40, or any other known polyomavirus. We aligned 18 published primer pairs used for the detection of JCV, BKV, or SV40 with the KIPyV sequence and found that none of them could amplify KIPyV (data not shown). It is thus highly unlikely that the presence of KIPyV would have influenced earlier studies of polyomavirus detection by PCR.
Phylogenetic analysis of the complete genome revealed that KIPyV is clearly separate from all other known polyomaviruses. When the early and late genes were analyzed separately, disparate results were obtained. While the early genes group with JCV, BKV, SV40, and SA12, the late genes form an outlier to the entire polyomavirus family. A possible explanation for this could be that the virus once emerged by recombination of two phylogenetically distant viruses, each contributing half of the genome. Alternatively, the early region may simply be more conserved because of more-rigid functional constraints while the late genes have diverged more rapidly and become very distant from those of its relatives. It is possible that future discoveries of additional polyomavirus species, e.g., in other primates, could make the phylogenetic tree more complete and provide additional clues to the evolution of KIPyV. Several new members of the polyomavirus family besides KIPyV have been discovered in the last few years (18, 19). The unique late region of KIPyV indicates that it may be the first discovered member of a new subfamily of polyomaviruses.
For assignment of nucleotide numbers, two different systems are in use for polyomaviruses. Either the nucleotide adjacent to the start codon of the T antigens, i.e., the first codon in the regulatory region, or a nucleotide in the origin of replication is considered nucleotide 1. The numbering we selected for KIPyV begins within the presumed origin and proceeds clockwise through the late region, as has been done for most primate polyomaviruses, such as JCV, SV40, SA12, and some strains of BKV.
On the basis of the ORF analysis, KIPyV is expected to express VP1 to VP3 and the ST and LT antigens, while the MT antigen and the Agno protein are both missing. The absence of the MT antigen is not surprising, since most polyomaviruses, the primate polyomaviruses included, lack expression of this particular protein. The lack of an ORF for an Agno protein is more interesting, since this protein is expressed by JCV, BKV, SV40, and SA12. The functional implications of this are unclear, since the function of the Agno protein remains to be fully elucidated. However, definitive conclusions about protein expression require further experimental evidence, e.g., in the form of mRNA analysis data.
The previously known primate polyomaviruses are generally not considered to be agents of respiratory tract disease. JCV and BKV have nevertheless been detected in human tonsil tissue, and a respiratory route of transmission of polyomaviruses has been hypothesized (9, 15, 28). BKV has also been found in the feces of children (35). The finding of KIPyV in nasopharyngeal aspirates and feces is consistent with these observations. However, the findings provide few clues to replication or latency sites or to possible disease caused by the virus. The screening of 1,300 clinical samples still provided important data. First, the cloning of a virus infecting humans was confirmed, since KIPyV genomes could be recovered from multiple individuals and since the isolates showed sequence variation. Second, the virus was detected in different age groups. Third, a concomitant finding of a recognized respiratory tract pathogen in most positive persons indicated that KIPyV was likely not the virus responsible for the respiratory tract symptoms.
The prevalence of KIPyV in humans remains unknown. Development of an antibody assay and/or finding relevant material for detecting latent virus is necessary for improved estimates. The findings obtained with nasopharyngeal aspirates suggest that the KIPyV prevalence is at least 1% in our study population. The absence of KIPyV in urine samples suggests that the biology and/or prevalence of KIPyV in kidneys differ significantly from those of JCV and BKV.
The cell type tropism and host range of polyomaviruses stem from both their regulatory regions and the receptor binding characteristics. The existence of multiple predicted c-Ets-1 transcription factor binding sites prompted us to investigate whether KIPyV may possibly replicate in lymphocytes in accordance with lymphotropic papovavirus, which harbors three putative binding sites for this transcription factor (38). This hypothesis is consistent with KIPyV being detected in the nasopharynx during an inflammatory process due to infection by a respiratory virus. However, studies of whole blood of healthy subjects or purified peripheral blood leukocytes of immunosuppressed patients did not support this hypothesis. On the other hand, given the 1% recovery rate in the respiratory tract samples, sample numbers may still be too small for definite conclusions. It must also be recognized that KIPyV is as yet only known by its genome sequence as detected by PCR. The virus has not been replicated in vitro, and no assay for detection of antibodies is available. Such experiments will be important in the further characterization of this virus.
A few newly discovered viruses have still not been associated with disease (20, 29). Nonetheless, the majority of known human viruses are pathogenic in one situation or another and any newly discovered virus must therefore be considered a likely pathogen. A problem with persisting viruses is that they are often discovered out of their symptomatic context, so that establishing their association with a particular disease may require extensive investigation. Historically, this has been the case for hepatitis B virus, Epstein-Barr virus, and parvovirus B19 (5, 8, 11). JCV and BKV are also very prevalent viruses that cause disease only under rare circumstances. Searching for a disease associated with KIPyV will be challenging but may have important medical implications. Primary candidate diseases could include unclear infectious complications in immunocompromised individuals and different types of cancer. Whether JCV, BKV, and SV40 can contribute to tumor development in humans is still a matter of debate, and one could assume that KIPyV will be subject to the same discussion. There are putative binding sites for p53, as well as the Rb family of tumor suppressor proteins, in the LT antigen of KIPyV, which indicates that a role for this virus in tumorigenesis cannot be excluded.
This study reinforces the notion that many human viruses have eluded detection despite more than 100 years of research in virology. Since viruses are likely pathogens, their identification remains an urgent scientific task. This study further illustrates how molecular virus screening of respiratory tract samples can be applied for discovering unknown viruses of different types, and not only agents of respiratory tract disease, thus making it a suitable approach for a “human virome project”.
Acknowledgments
We thank Inga Karlsson, Cecilia Lindau, Hamid Darban, and Daryoush Rahmani for excellent technical assistance and Lisbeth Barkholt and Fredrik Boström for providing samples.
This study was supported by the Torsten and Ragnar Söderberg Foundation, the Swedish Cancer Foundation, Nanna Svartz' Fund, the Gustav Vth Jubilee Society, and the Swedish Society for Clinical Microbiology. T.A. is a fellow of the Swedish Research Council.
Footnotes
Published ahead of print on 7 February 2007.
REFERENCES
- 1.Allander, T., S. U. Emerson, R. E. Engle, R. H. Purcell, and J. Bukh. 2001. A virus discovery method incorporating DNase treatment and its application to the identification of two bovine parvovirus species. Proc. Natl. Acad. Sci. USA 98:11609-11614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allander, T., M. T. Tammi, M. Eriksson, A. Bjerkner, A. Tiveljung-Lindell, and B. Andersson. 2005. Cloning of a human parvovirus by molecular screening of respiratory tract samples. Proc. Natl. Acad. Sci. USA 102:12891-12896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barbanti-Brodano, G., S. Sabbioni, F. Martini, M. Negrini, A. Corallini, and M. Tognon. 2006. BK virus, JC virus and simian virus 40 infection in humans, and association with human tumors. Adv. Exp. Med. Biol. 577:319-341. [DOI] [PubMed] [Google Scholar]
- 5.Blumberg, B. S., H. J. Alter, and S. Visnich. 1965. A “new” antigen in leukemia sera. JAMA 191:541-546. [DOI] [PubMed] [Google Scholar]
- 6.Bogdanovic, G., M. Brytting, P. Cinque, M. Grandien, E. Fridell, P. Ljungman, B. Lönnqvist, and A. L. Hammarin. 1994. Nested PCR for detection of BK virus and JC virus DNA. Clin. Diagn. Virol. 2:211-220. [DOI] [PubMed] [Google Scholar]
- 7.Bogdanovic, G., P. Priftakis, G. Giraud, and T. Dalianis. 2006. A related donor and reduced intensity conditioning reduces the risk of development of BK virus-positive haemorrhagic cystitis in allogeneic haematopoetic stem cell-transplanted patients. Anticancer Res. 26:1311-1318. [PubMed] [Google Scholar]
- 8.Cossart, Y. E., A. M. Field, B. Cant, and D. Widdows. 1975. Parvovirus-like particles in human sera. Lancet i:72-73. [DOI] [PubMed] [Google Scholar]
- 9.Eash, S., R. Tavares, E. G. Stopa, S. H. Robbins, L. Brossay, and W. J. Atwood. 2004. Differential distribution of the JC virus receptor-type sialic acid in normal human tissues. Am. J. Pathol. 164:419-428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Engels, E. A., W. M. Switzer, W. Heneine, and R. P. Viscidi. 2004. Serologic evidence for exposure to simian virus 40 in North American zoo workers. J. Infect. Dis. 190:2065-2069. [DOI] [PubMed] [Google Scholar]
- 11.Evans, A. S., J. C. Niederman, and R. W. McCollum. 1968. Seroepidemiologic studies of infectious mononucleosis with EB virus. N. Engl. J. Med. 279:1121-1127. [DOI] [PubMed] [Google Scholar]
- 12.Froussard, P. 1992. A random-PCR method (rPCR) to construct whole cDNA library from low amounts of RNA. Nucleic Acids Res. 20:2900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gardner, S. D., A. M. Field, D. V. Coleman, and B. Hulme. 1971. New human papovavirus (B.K.) isolated from urine after renal transplantation. Lancet i:1253-1257. [DOI] [PubMed] [Google Scholar]
- 14.Giraud, G., G. Bogdanovic, P. Priftakis, M. Remberger, B. M. Svahn, L. Barkholt, O. Ringden, J. Winiarski, P. Ljungman, and T. Dalianis. 2006. The incidence of hemorrhagic cystitis and BK-viruria in allogeneic hematopoietic stem cell recipients according to intensity of the conditioning regimen. Haematologica 91:401-404. [PubMed] [Google Scholar]
- 15.Goudsmit, J., P. Wertheim-van Dillen, A. van Strien, and J. van der Noordaa. 1982. The role of BK virus in acute respiratory tract disease and the presence of BKV DNA in tonsils. J. Med. Virol. 10:91-99. [DOI] [PubMed] [Google Scholar]
- 16.Grabe, N. 2002. AliBaba2: context specific identification of transcription factor binding sites. In Silico Biol. 2:S1-S15. [PubMed] [Google Scholar]
- 17.Hirsch, H. H. 2002. Polyomavirus BK nephropathy: a (re-)emerging complication in renal transplantation. Am. J. Transplant. 2:25-30. [DOI] [PubMed] [Google Scholar]
- 18.Johne, R., D. Enderlein, H. Nieper, and H. Muller. 2005. Novel polyomavirus detected in the feces of a chimpanzee by nested broad-spectrum PCR. J. Virol. 79:3883-3887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johne, R., W. Wittig, D. Fernandez-de-Luco, U. Hofle, and H. Muller. 2006. Characterization of two novel polyomaviruses of birds by using multiply primed rolling-circle amplification of their genomes. J. Virol. 80:3523-3531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jones, M. S., A. Kapoor, V. V. Lukashov, P. Simmonds, F. Hecht, and E. Delwart. 2005. New DNA viruses identified in patients with acute viral infection syndrome. J. Virol. 79:8230-8236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Khalili, K., J. Gordon, and M. K. White. 2006. The polyomavirus, JCV and its involvement in human disease. Adv. Exp. Med. Biol. 577:274-287. [DOI] [PubMed] [Google Scholar]
- 22.Kleinschmidt-DeMasters, B. K., and K. L. Tyler. 2005. Progressive multifocal leukoencephalopathy complicating treatment with natalizumab and interferon beta-1a for multiple sclerosis. N. Engl. J. Med. 353:369-374. [DOI] [PubMed] [Google Scholar]
- 23.Knowles, W. A. 2006. Discovery and epidemiology of the human polyomaviruses BK virus (BKV) and JC virus (JCV). Adv. Exp. Med. Biol. 577:19-45. [DOI] [PubMed] [Google Scholar]
- 24.Langer-Gould, A., S. W. Atlas, A. J. Green, A. W. Bollen, and D. Pelletier. 2005. Progressive multifocal leukoencephalopathy in a patient treated with natalizumab. N. Engl. J. Med. 353:375-381. [DOI] [PubMed] [Google Scholar]
- 25.Lee, W., and E. Langhoff. 2006. Polyomavirus in human cancer development. Adv. Exp. Med. Biol. 577:310-318. [DOI] [PubMed] [Google Scholar]
- 26.Mayer, M., and K. Dörries. 1991. Nucleotide sequence and genome organization of the murine polyomavirus, Kilham strain. Virology 181:469-480. [DOI] [PubMed] [Google Scholar]
- 27.Minor, P., P. A. Pipkin, K. Cutler, and G. Dunn. 2003. Natural infection and transmission of SV40. Virology 314:403-409. [DOI] [PubMed] [Google Scholar]
- 28.Monaco, M. C., P. N. Jensen, J. Hou, L. C. Durham, and E. O. Major. 1998. Detection of JC virus DNA in human tonsil tissue: evidence for site of initial viral infection. J. Virol. 72:9918-9923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mushahwar, I. K. 2000. Recently discovered blood-borne viruses: are they hepatitis viruses or merely endosymbionts? J. Med. Virol. 62:399-404. [DOI] [PubMed] [Google Scholar]
- 30.Padgett, B. L., D. L. Walker, G. M. ZuRhein, R. J. Eckroade, and B. H. Dessel. 1971. Cultivation of papova-like virus from human brain with progressive multifocal leucoencephalopathy. Lancet i:1257-1260. [DOI] [PubMed] [Google Scholar]
- 31.Randhawa, P., A. Vats, and R. Shapiro. 2006. The pathobiology of polyomavirus infection in man. Adv. Exp. Med. Biol. 577:148-159. [DOI] [PubMed] [Google Scholar]
- 32.Razonable, R. R., and C. V. Paya. 2003. Herpesvirus infections in transplant recipients: current challenges in the clinical management of cytomegalovirus and Epstein-Barr virus infections. Herpes 10:60-65. [PubMed] [Google Scholar]
- 33.Riley, M. I., W. Yoo, N. Y. Mda, and W. R. Folk. 1997. Tiny T antigen: an autonomous polyomavirus T antigen amino-terminal domain. J. Virol. 71:6068-6074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tyler, K. L. 2004. Update on herpes simplex encephalitis. Rev. Neurol. Dis. 1:169-178. [PubMed] [Google Scholar]
- 35.Vanchiere, J. A., R. K. Nicome, J. M. Greer, G. J. Demmler, and J. S. Butel. 2005. Frequent detection of polyomaviruses in stool samples from hospitalized children. J. Infect. Dis. 192:658-664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Viscidi, R. P., and B. Clayman. 2006. Serological cross reactivity between polyomavirus capsids. Adv. Exp. Med. Biol. 577:73-84. [DOI] [PubMed] [Google Scholar]
- 37.zur Hausen, H. 2002. Papillomaviruses and cancer: from basic studies to clinical application. Nat. Rev. Cancer 2:342-350. [DOI] [PubMed] [Google Scholar]
- 38.zur Hausen, H., and L. Gissmann. 1979. Lymphotropic papovaviruses isolated from African green monkey and human cells. Med. Microbiol. Immunol. 167:137-153. [DOI] [PubMed] [Google Scholar]