Abstract
The E6 protein from high-risk human papillomavirus types interacts with and degrades several PDZ domain-containing proteins that localize to adherens junctions or tight junctions in polarized epithelial cells. We have identified the tight junction-associated multi-PDZ protein PATJ (PALS1-associated TJ protein) as a novel binding partner and degradation target of high-risk types 16 and 18 E6. PATJ functions in the assembly of the evolutionarily conserved CRB-PALS1-PATJ and Par6-aPKC-Par3 complexes and is critical for the formation of tight junctions in polarized cells. The ability of type 18 E6 full-length to bind to, and the subsequent degradation of, PATJ is dependent on its C-terminal PDZ binding motif. We demonstrate that the spliced 18 E6* protein, which lacks a C-terminal PDZ binding motif, associates with and degrades PATJ independently of full-length 18 E6. Thus, PATJ is the first binding partner that is degraded in response to both isoforms of 18 E6. The ability of E6 to utilize a non-E6AP ubiquitin ligase for the degradation of several PDZ binding partners has been suggested. We also demonstrate that 18 E6-mediated degradation of PATJ is not inhibited in cells where E6AP is silenced by shRNA. This suggests that the E6-E6AP complex is not required for the degradation of this protein target.
Human papillomaviruses (HPVs) are DNA tumor viruses that infect basal cells of the mucosal or cutaneous epithelium, where they cause proliferative lesions on genital tissues, the upper respiratory tract, and skin. Papillomas induced by high-risk mucosal HPV types, of which 16, 18, 31, and 45 are predominant, are inherently oncogenic and associated with the vast majority of cervical cancers (64) and many oral cancers (9). In contrast, low-risk types, such as 6 and 11, are rarely associated with carcinogenesis (10). HPV-associated carcinogenesis is attributed to the high-risk specific activities of the viral gene products E6 and E7, and introduction of these genes from HPV types 16 or 18 can induce immortalization of primary human keratinocytes (16, 35). Studies using transgenic mice that express 16 E6 and/or 16 E7 have revealed that E7 initiates tumor formation, while E6 induces tumor promotion and progression, in the skin and cervix of transgenic animals (13, 47, 58).
High-risk E6 proteins are multifunctional and associate with functionally diverse cellular proteins. Degradation of p53 by the E6-E6AP complex (17) is a property of high-risk E6 types, although this activity alone is not sufficient for E6-dependent immortalization activity in tissue culture or tumorigenesis in transgenic animals. For example, certain E6 mutant proteins that fail to degrade p53 retain their ability to immortalize primary cells (30, 45). Moreover, the phenotypes of E6 transgenic mice are not observed in p53-null mice (59).
A 4-amino-acid PDZ binding motif (X-S/T-X-V/L, where X is any amino acid) is present at the carboxy termini of all known high-risk mucosal HPV E6 proteins. The conservation of this motif among high-risk, but not low-risk, mucosal E6 types suggests the importance of PDZ domain protein interactions for E6-induced oncogenesis (24). The PDZ binding motif is required for the transformation activity of E6 in certain established and immortalized cell lines (24, 66) and for certain stages of tumorigenesis, such as spontaneous tumor formation and the development of epithelial hyperplasia, in 16 E6 transgenic mice (38, 56). Recently, the PDZ binding motif of E6 was shown to be required for the early stages of the life cycle of HPV type 31, specifically, for cell proliferation and maintenance of episomal genomes (26).
In cervical cancer-derived cell lines and tissues that contain high-risk HPV, the majority of HPV RNAs do not encode full-length (FL) E6 (4, 52, 55, 57). In the context of the complete HPV type 16 and 31 genomes, several alternatively spliced E6 RNAs, designated E6*, have been detected (4, 18). In cell lines that contain integrated HPV 18 DNA, a single spliced 18 E6 RNA, 18 E6*, and its protein product were described (51, 52). High-risk E6, expressed ectopically, has the potential to encode either one, in the case of types 18 (52) and 31 (18), or two, in the case of type 16 (57), alternatively spliced isoforms. The E6* proteins are thought to negatively regulate the immortalization activities of E6 and E7 (53). In particular, 18 E6* binds 16 E6, 18 E6, and E6AP, a ubiquitin ligase, and inhibits the 18 E6-mediated degradation of p53 (43, 44). The gene products of 16 E6* and 18 E6* have C termini that differ from their full-length counterparts and lack the canonical PDZ binding motif. However, based on the ability of E6* to bind E6, it remains possible that this high-risk specific E6 isoform forms a complex with E6 and its PDZ binding partners.
The PDZ binding domain of high-risk E6 types mediates its interaction with seven PDZ domain-containing proteins: Dlg (24), Scribble (36), MUPP1 (27), MAGI1 (11), MAGI2 and -3 (63), and TIP-2/GIPC (6). In general, the multiple PDZ domain proteins with which E6 interacts localize to cell junctions, where they are believed to assemble multiprotein complexes that regulate cell growth and polarity (5). Disruption of the Drosophila melanogaster homologs of these PDZ proteins leads to loss of polarity and overgrowth of epithelial tissue (reviewed in reference 50). Additionally, reduced levels of Dlg, Scrib, and MUPP1 in human tumors is associated with increased dysplasia and invasiveness (8, 31, 37, 65).
While the tumor suppressor properties of the PDZ interaction partners of E6 suggest an intriguing link to E6-associated oncogenesis, the biological consequences of the associations between high-risk E6 types and PDZ proteins remain unclear. Similar to other high-risk specific E6 interaction partners, these PDZ proteins are targeted for degradation as a result of E6 expression in tissue culture systems (7, 11, 27, 36, 63). However, the stability of only certain PDZ proteins is reduced in 16 E6 transgenic mice (39, 56). These findings raise the possibilities that high-risk E6 has alternative effects on its PDZ interaction partners or that the E6 PDZ binding site contributes to the HPV life cycle and E6-associated carcinogenesis via interactions with alternative PDZ proteins.
In this study, we describe the interaction between high-risk E6 types 16 and 18 and PATJ (PALS1-associated tight junction). PATJ is the first PDZ binding partner of E6 shown to be required for the formation of tight junctions (TJ) in mammalian epithelial cells (54). The association of PATJ with PALS1 is required for the proper TJ localization of PALS1 (49) and, in turn, the formation of two evolutionarily conserved TJ apical complexes, CRB-PALS1-PATJ and Par6-aPKC-Par3 (19). Loss of PATJ leads to mislocalization of TJ components and overall defects in cell polarity (34, 54). Recent evidence suggests that PATJ is a common interaction partner for PDZ binding viral oncoproteins (25). The ability of adenovirus type 9 E4-ORF1 to cause mislocalization of PATJ correlates with its tumorigenic potential (25). PATJ contains a series of PDZ domains and an amino-terminal L27 domain (49), all of which are protein-protein interaction domains. PDZ domain 6 mediates its interaction with the peripheral TJ protein ZO-3 and, consequently, its trafficking to TJ (48). Here, we report that PATJ binds to the PDZ binding site of 18 E6 via its fourth and fifth PDZ domains. We provide evidence that PATJ is a novel degradation target of both 16 E6 and 18 E6 and that E6AP is not required for 18 E6-mediated reduction of PATJ levels. The ability of 18 E6* to associate with and reduce the stability of PATJ defines a novel property of the spliced isoform of 18 E6.
MATERIALS AND METHODS
Plasmids. (i) HPV E6 plasmids.
The E6 constructs pAS2-1-18 E6, pGEX-2T-6 E6, and 18 E6 for bacterial expression of glutathione-S-transferase (GST) fusion proteins pMycL.1-6 E6 and 18 E6 were previously described (3). To clone 16 E6 into pMycL.1, 16 E6 was amplified from pAS2-1-16 E6 (3) using the primers 5′-GTCGACCATGCACCAAAAGAGAACTGC-3′ and 5′-CAGGTCGACGGATCCTTACAGC-3′ and inserted between the SalI and BamHI sites of pMycL.1 The 18 E6 clones containing the V158L and ΔETQV mutation were created by PCR amplification from the pMycL.1-18 E6 construct with the primers 5′-CTGAGGAAGATCTCA-3′ and either 5′-CCATGGTTATAGTTGTGTTTCTCTGCGTC-3′ or 5′-CCATGGTTATCTGCGTCGTTGGAG-3′, respectively. The PCR products were inserted between the SalI and NcoI sites of pMycL.1. 18 E6 V158A was released from pAS2-1-18 E6 V158A by NcoI digestion, treated with Klenow, and inserted into the PmeI site of pMycL.1. pAS2-1-18 E6V158A was created by QuikChange site-directed mutagenesis (Stratagene, La Jolla, CA) using pCRII-18 E6 as template and the primers 5′-CGCAGAGAAACACAAGCATAATATTAAGTATCCATG-3′ and 5′-CATGGATACTTAATATTATGCTTGTGTTTCTCTGCG-3′. The 18 E6 V158A open reading frame (ORF) was subcloned as an NcoI fragment from pCRII into pAS2-1. The 18 E6* cDNA was obtained from reverse transcription-PCR (RT-PCR) and subcloned between the SalI and NcoI sites of pMycL.1.
(ii) PATJ plasmids.
A clone of pGAD10 that expressed amino acids (aa) 535 to 882 of PATJ was identified in a yeast two-hybrid screen of a human foreskin keratinocyte library (Clontech Laboratories, Mountain View, CA). A clone of the alternatively spliced isoform of PATJ encoding 1,552 aa in pCDNAII was a gift from Stephan Philipp (42). The PATJ fragments and full-length PATJ were cloned into the Gal4 AD vector pACT2 (Clontech) as follows. The primers 5′-CTGTGGGAATTCAGTCAGGATGC-3′ and 5′-CAACTCCATGTCGACTGAGGGATCTTG-3′ were used to amplify a PATJ fragment encoding aa 1 to 883, which was inserted between the EcoRI and SalI sites of pACT2. A PATJ fragment encoding aa 1 to 318 was amplified by PCR using the primers 5′-GAGAATGGATCCAAATGCCTGAAAATCCTGCTAC-3′ and 5′-CCTGACTGAATTCCCACAGTTCC-3′ and subcloned between the BamHI and EcoRI sites of pACT2. The PATJ fragments encoding aa 535 to 887 and aa 680 to 887 were amplified by PCR with the primers 5′-GGGATACAACTCCATGGATGGTGAGGG-3′ and either 5′-CCAGATGGGAAACCATGGTGGTCCTGATTATG-3′ or 5′-GATGGGGAATTAGCCATGGGGTCCCCTGAAGTC-3′, respectively, and inserted into the NcoI site of pACT 2-2. The PATJ fragments encoding aa 535 to 775 and aa 680 to 775 were amplified by PCR with the primers 5′-CTTCATTGAATTCCACCAAAGGCTTACAGATGCCAAGG-3′ and either 5′-CCAGATGGGAAACCATGGTGGTCCTGATTATG-3′ and 5′-GATGGGGAATTAGCCATGGGGTCCCCTGAAGTC-3′, respectively, and inserted between the NcoI and EcoRI sites of pACT2. pACT2-aa 885-1552 was created by subcloning the NcoI-XhoI fragment of pCDNAII-PATJ into pACT2. pACT2-aa 535-1552 was created by inserting the NcoI fragment from pACT2-aa 535-887 into pACT2-aa 885-1552. pACT2-PATJ FL was obtained by subcloning the EcoRI-XhoI fragment of pCDNAII-PATJ into pACT2-aa 1-318.
For in vitro transcription and translation, PATJ was cloned in pET21d (EMD Biosciences, San Diego, CA). pET21d-aa 1-318, pET21d-aa 535-887, pET21d-aa 885-1552, and pET21d-PATJ FL were cloned as described for pACT2-aa 1-318, pACT2-aa 535-887, pACT2-aa 885-1552, and pACT2-PATJ FL.
For expression of PATJ with an N-terminal Flag epitope tag in mammalian cells, PATJ encoding the 1,552-aa isoform was cloned into pFlag-CMV-2 (Eastman Kodak, New Haven, CT). First, the EcoRI-XhoI fragment from pcDNA2-PATJ was inserted between the EcoRI and SalI sites of pFlag-CMV-2. The remaining 5′ sequence of PATJ was amplified by PCR from pcDNA2-PATJ using the primers 5′-GAAGAGAATCGGCCGGAATGCCTGAAAATCCTG-3′ and 5′-CCTGACTGAATTCCCACAGTTCC-3′ and inserted between the NotI and EcoRI sites of pFlag-CMV-2-PATJ-nt 951-3′. The additional 3′ sequence of PATJ encoded by the alternatively spliced isoform was subcloned in pFlag-CMV-2-PATJ aa 1 to 1552 from an IMAGE consortium EST clone (EST BG768372; ATCC, Manassas, VA) to generate pFlag-CMV-2-PATJ aa 1-1801.
(iii) Other constructs.
The pSUPER plasmid was provided by David Sayah. To create the E6AP-specific shRNA expression constructs E6AP-1 and E6AP-2, the oligo pairs 5′-GATCCCCCCTCAGTCTGACGACATTGTTCAAGAGACAATGTCGTCAGACTGAGGTTTTTGGAAA-3′/5′-AGCTTTTCCAAAAACCTCAGTCTGACGACATTGTCTCTTGAACAATGTCGTCAGACTGAGGGGG-3′ and 5′-GATCCCCGAGCCCATCCCTGAGTCCATTCAAGAGATGGACTCAGGGATGGGCTCTTTTTGGAAA-3′/5′-AGCTTTTCCAAAAAGAGCCCATCCCTGAGTCCATCTCTTGAATGGACTCAGGGATGGGCTCGGG-3′, respectively, were annealed and inserted between the BglII and HindIII sites of pSUPER (2). pSUPER-LaminA/C was created by using the oligo pair 5′-GATCCCCTGGACTTCCAGAAGAACATTCAAGAGATGTTCTTCTGGAAGTCCAAGTCAGTTTTTGGAAA-3′ and 5′-AGCTTTTCCAAAAACTGGACTTCCAGAAGAACATCTCTTGAATGTTCTTCTGGAAGTCCAGGG-3′. The p53 mammalian expression plasmid, pCB6-p53, was a gift from Wei Gu.
RT-PCR.
Cytoplasmic RNA was collected from 293T cells transfected with empty pMycL.1 or the pMycL.1-18 E6 wild-type (WT) or splice donor (SD) constructs using the QIAGEN RNeasy mini-prep kit (Valencia, CA). For first-strand cDNA synthesis, a 10-μl mixture that contained 2 μg of RNA template, 2.5 mM deoxynucleoside triphosphates, and 1 μl of a gene-specific primer (10 μM) that binds to RNA transcribed from the pMyc vector sequence downstream of the 18 E6 insert (5′-GTAACCATTATAAGCTGCAATAAAC-3′) was incubated at 65°C for 10 min and then chilled on ice. To synthesize the first cDNA strand, the RNA-DNA mixtures with deoxynucleoside triphosphates were incubated with 1 U SuperScript III reverse transcriptase and RT buffer (Invitrogen), 5 mM MgCl2, 0.01 mM dithiothreitol, and 40 U RNasin RNase inhibitor (Promega, Madison, WI) for 20 min at 42°C and 30 min at 50°C. The reactions were stopped by incubation at 85°C for 15 min. Two μl of these reactions was used as template for PCRs. The primers 5′-CTGAGGAAGATCTCA-3′ and 5′-CCATGGTTATAGTTGTGTTTCTCTGCGTC-3′ were used; they bind to pMycL.1 and the 3′pMycL.1-18E6 junction, respectively.
Yeast strains and β-Gal assays.
The Saccharomyces cerevisiae strain YGH1 was cultured and transformed as previously described (3). To measure the production of β-galactosidase (β-Gal) in the cotransformed yeast, three individual colonies from each transformation were grown to log phase. Equal amounts of yeast culture, equivalent to 1 optical density at 600 nm (OD600) unit, were lysed in 50 μl of Y-PER yeast protein extraction reagent (Pierce Biotechnology, Inc., Rockford, IL). The yeast lysates were centrifuged to remove cell debris. One ml of Z-buffer was added to the supernatant following centrifugation to remove cell debris, and the samples were preincubated for 5 min at 30°C. The β-Gal reactions were performed as previously described (3). The units of β-Gal activity were calculated as follows: 1,000 × OD420/(Δt × OD equivalent), where Δt is the length of time, in minutes, from the addition of o-nitrophenyl-β-d-galactopyranoside to quenching the reaction and the OD equivalent is 1.
In vitro transcription/translation.
The pET21-PATJ constructs were used as template for in vitro transcription and translation as previously described (3).
GST protein purification and GST pull-down assays. (i) GST protein purification.
The GST fusion proteins were expressed in the Escherichia coli BL21 strain and purified as previously described (3). The purity and amount of the GST fusion proteins were determined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Coomassie blue staining. For antibody production, the GST-PATJ peptide fusion was eluted from beads in 5 mM of reduced glutathione (Sigma). Protein concentration was then measured by the Bradford assay (Bio-Rad, Hercules, CA).
(ii) GST pull-down assays.
Equivalent amounts of GST-6 E6 or GST-18 E6 fusion proteins, or GST alone, were incubated with in vitro-translated [35S]PATJ FL or the [35S]PATJ peptides in 1 ml binding buffer as previously described (3). After binding, the complexes were washed five times at 4°C for 10 min each in binding buffer with 50, 150, or 250 mM KCl. After the last wash, the bound protein was released from the beads by boiling in 15 μl 2× SDS LB (1× SDS LB is l0 mM Tris-HCl pH 6.8, 2% [wt/vol] SDS, 0.12% [wt/vol] bromophenol blue, 10% [wt/vol] glycerol, 100 mM dithiothreitol) for 7 min. The 35S-labeled proteins were detected by autoradiography.
Mammalian cell culture and transfection.
Human 293T fibroblasts and H1299 lung carcinoma cells were maintained as monolayer cultures in Dulbecco's modified Eagle medium (DMEM; GIBCO-BRL, Grand Island, NY) supplemented with 8% fetal clone II and 2% fetal bovine serum (HyClone, Ogden, UT), 100 U/ml penicillin, and 100 μg/ml streptomycin (GIBCO-BRL) at 37°C in a 5% CO2 atmosphere. The cervical carcinoma-derived cell lines C33A, CaSki, and HeLa were also cultured under these conditions. Twenty-four hours prior to transfection, cells were seeded into either 25-mm or 60-mm tissue culture dishes. Cells were transfected using Lipofectamine Plus in Opti-MEM medium according to the manufacturer's recommendations (Invitrogen, Carlsbad, CA). Cells were processed for analysis 48 h posttransfection. For the RNA inhibition studies in 293A or H1299 cells, the pSUPER plasmids were transfected twice, on days 1 and 4. On day 4, the PATJ and green fluorescent protein (GFP) or p53 expression plasmids and E6 constructs were cotransfected with the pSUPER plasmids. Cells were processed on day 6.
Chemicals and drugs.
Mammalian cells were treated with 10 μM MG132 (EMD Biosciences) from a 10 mM stock in dimethyl sulfoxide (DMSO; Sigma), or 10 μM epoxomicin (EMD Biosciences) from a 1 mM stock in DMSO for 5 hours prior to harvesting.
Mammalian cell protein extraction.
Confluent cell monolayers in six-well or 60-mm tissue culture dishes were washed in chilled phosphate-buffered saline and resuspended in 200 μl or 400 μl, respectively, of chilled RIPA lysis buffer (50 mM Tris-HCl pH 8.0, 150 mM NaCl, 1% [wt/vol] NP-40, 0.5% [wt/vol] sodium deoxycholate, 0.1% [wt/vol] SDS, 50 mM NaF) plus Complete protease inhibitor cocktail (Roche, Indianapolis, IN) and incubated on ice for 30 min. Cell extracts were clarified by centrifugation at 22,500 × g for 15 min at 4°C in a high-speed refrigerated microcentrifuge. Total protein concentration was measured using the Bio-Rad protein assay. A 5× stock of SDS LB was added to the samples to a final concentration of 1× prior to boiling for 5 min and analysis by SDS-PAGE.
Pulse-chase assay.
Cells were transfected in sets of four 60-mm dishes for each cotransfection. Each dish was chased for a different period of time. After 48 h, cells were washed three times in modified DMEM lacking methionine and cysteine (GIBCO-BRL). Prior to metabolic labeling, cells were incubated in this starvation medium supplemented with 10% dialyzed calf serum for 30 min. Medium was then replaced with starvation medium containing 10% dialyzed calf serum and a total of 300 μCi/ml of a [35S]methionine and [35S]cysteine mixture (ICN, Irvine, CA). Cells were labeled for 45 min. After the labeling period, all dishes were washed three times in normal DMEM with 10% FBS without label, incubated at 37°C in DMEM 10% FBS for the indicated times, and then rinsed in chilled phosphate-buffered saline. Cells were lysed in 400 μl of chilled RIPA lysis buffer plus Complete protease inhibitor cocktail. Cell extracts were clarified by centrifugation at 22,500 × g for 15 min in a refrigerated microcentrifuge. Equal amounts of radiolabeled protein, based on trichloroacetic acid-precipitable counts, were incubated with Flag or Myc affinity matrix. Following extensive washing, the immunoprecipitated complexes were boiled off the affinity matrix, subjected to SDS-PAGE, and visualized by autoradiography.
Immunoprecipitation.
The anti-Flag M2 agarose matrix (Sigma) and anti-Myc agarose matrix (Sigma) were washed and resuspended in RIPA lysis buffer to make a 50% slurry. One hundred μl of the 50% slurry was incubated overnight by rotating at 4°C with equal amounts of total protein for the coimmunoprecipitation experiment or equal amounts of incorporated radioactivity for the pulse-chase assay. The proteins bound to the Flag or Myc affinity matrix were washed three to four times for 5 min each with 1 ml RIPA lysis buffer plus Complete protease inhibitor. For the coimmunoprecipitation experiment, proteins that remained bound were eluted off the beads by rotating overnight at 4°C with 50 μl of 150 μg/ml Flag peptide (Sigma) in RIPA buffer. The eluates were boiled in 1× SDS LB for 5 min and subjected to SDS-PAGE. For the pulse-chase assay, 35S-labeled proteins that remained bound were boiled off the beads in 2× SDS LB.
Antibodies. (i) Antibodies to PATJ.
A fusion protein consisting of GST fused to the N terminus of a PATJ peptide encompassing aa 509 to 1181 served as the antigen for production of antibodies to PATJ in rabbits (Cocalico Biologicals, Inc., Reamstown, PA). Antibodies to PATJ were purified from the exsanguinated bleed on a PATJ affinity column containing PATJ peptide aa 509 to 1181 bound to cyanogen bromide (CNBr)-activated Sepharose 4B beads (GE Healthcare, Piscataway, NJ).
(ii) Other antibodies.
Antibody to Flag was purchased from Stratagene (no. 200471; La Jolla, CA). Santa Cruz Biotechnology supplied the mouse monoclonal antibody to Myc (clone 9E10), p53 (clone DO1), and Dlg (2D11). Mouse monoclonal antibodies to E6AP (clone E6AP-330) and actin (clone AC-74) were purchased from Sigma. Mouse anti-GFP was purchased from Invitrogen (Carlsbad, CA). Goat anti-mouse and goat anti-rabbit antibodies conjugated to horseradish peroxidase were purchased from Kierkegaard and Perry Laboratories (Gaithersburg, MD).
RESULTS
Interaction of PATJ peptides with 18 E6 in the yeast two-hybrid system.
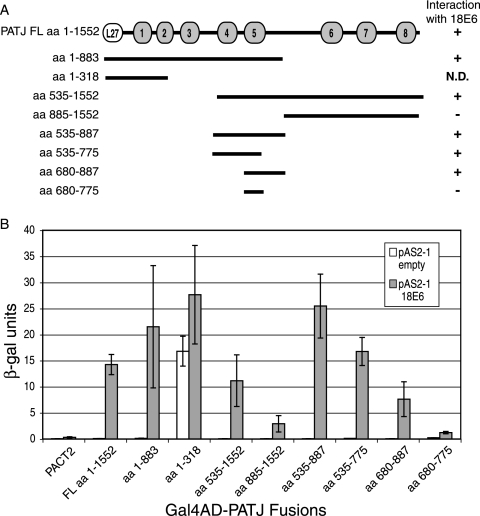
A yeast two-hybrid screen for proteins that interact with high-risk type 18 E6 identified a fragment of PATJ encoding aa 535 to 887. This region spans PDZ domains 4 and 5 of PATJ. PATJ sequence was not identified from a similar screen using low-risk type 6 E6 as bait. To confirm the 18 E6 interaction with PATJ, an alternatively spliced isoform of FL PATJ that encodes 1,552 amino acids and eight PDZ domains (42) and a series of PATJ peptides that encompass different PDZ domains were tested (Fig. 1A). Expression of the PATJ peptides was demonstrated by Western blotting (not shown). The ability of FL PATJ and PATJ peptides to interact with 18 E6 is summarized in Fig. 1A. The N-terminal peptide aa 1-883 that spans the L27 domain and PDZ domains 1 to 5 scored positive for interaction with 18 E6 (Fig. 1B). The ability of the smaller peptide aa 1-318 to interact with 18 E6 could not be determined in this system because it transactivated the reporter gene in the absence of 18 E6. The C-terminal fragment aa 535-1552, encompassing PDZ domains 4 to 8, interacted with 18 E6. However, a C-terminal peptide (aa 885-1552) that lacks PDZ domains 4 and 5 mediated only low levels of reporter activity in the presence of 18 E6. The ability of the peptide aa 535-887 to interact with 18 E6 reinforces the association of18 E6 with PDZ domains 4 and 5 of PATJ. Both peptides aa 535-775, containing PDZ 4 and only part of PDZ 5, and aa 680-887, containing only PDZ 5, associated with 18 E6, demonstrating that PDZ 4 and PDZ 5 are each able to interact. A peptide (aa 680-775) that lacks the complete PDZ 5 domain failed to interact. This deletion analysis did not exclude the possibility that 18 E6 also binds PDZ domains 1, 2, and 3 of PATJ.
FIG. 1.
PDZ domains 4 and 5 of PATJ are sufficient to interact with 18 E6. (A) Schematic representation of FL PATJ, amino acids 1 to 1552, identifying the position of its L27 domain and eight PDZ domains. PATJ deletion mutants, named for the amino acids of PATJ that they encode, were cloned in the Gal4AD plasmid pACT2. They are shown as lines below the FL PATJ schematic. The ability of PATJ and the PATJ peptides to interact with 18 E6 in the yeast two-hybrid system is indicated to the right of the line schematic. (B) Yeast strain YGH1 was cotransformed with 1 μg of either the Gal4DBD vector pAS2-1 or pAS2-1-18 E6 and 1 μg of either Gal4AD vector pACT2 or the Gal4AD-PATJ fusions. Liquid β-Gal assays were performed on the extracts from cotransformed yeast colonies.
In vitro binding of PATJ to GST-18 E6.
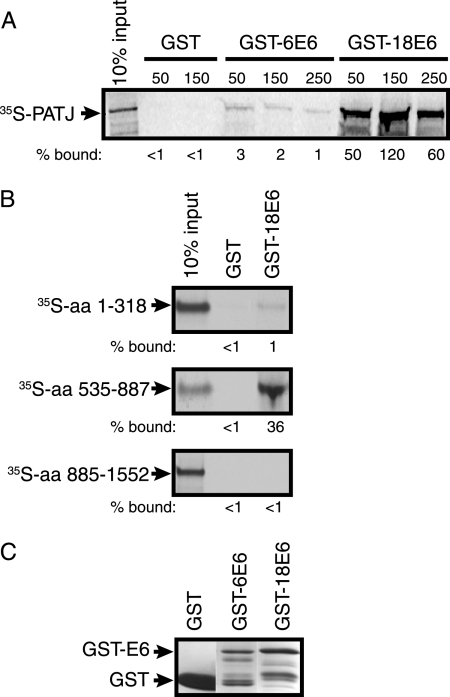
To determine if full-length PATJ binds specifically to high-risk E6 and to confirm the region of PATJ that mediates binding, GST pull-down assays were performed. PATJ cDNAs encoding aa 1 to 1552, 1 to 318, 535 to 887, and 885 to 1552 were incubated in a coupled transcription and translation reaction in the presence of [35S]methionine. The amount and purity of the GST, GST-6 E6, and GST-18 E6 proteins were determined by staining with Coomassie brilliant blue (Fig. 2C). 35S-labeled PATJ FL was incubated with purified GST or equal amounts of GST-6 E6 or GST-18 E6 bound to glutathione beads in the presence of 50 mM KCl (Fig. 2A). The complexes were washed in binding buffer containing increasing concentrations of KCl. The background level of PATJ bound to GST was below the limit of detection. The amount bound to GST-6 E6 was negligible compared to GST alone and reduced from 3% to 1% by washing at increased salt concentrations. In contrast, the PATJ-GST-18 E6 complexes were stable at all salt concentrations tested, and approximately 50% remained bound to GST-18 E6 at 250 mM salt. Thus, the interaction between PATJ and E6 is stable and specific to high-risk 18 E6. Truncated, 35S-labeled PATJ polypeptides were incubated with either GST or GST-18 E6, and the bound complexes were washed with 50 mM KCl. Only 1% of [35S]peptide aa 1-318 remained bound to GST-18 E6, excluding the possibility that PDZ domains 1 and 2 bind to 18 E6. In agreement with the yeast two-hybrid data (Fig. 2B), a peptide containing aa 535 to 887 bound to 18 E6, while the C-terminal peptide containing aa 885 to 1552 did not. This GST pull-down assay confirms the results of the yeast two-hybrid analyses and provides biochemical evidence for the association of 18 E6 with PDZ 4 and 5 of PATJ.
FIG. 2.
PATJ binds to GST-18 E6 but not GST-6 E6 in vitro. (A) Purified GST, GST-6 E6, or GST-18 E6 protein bound to glutathione beads was incubated with in vitro-translated 35S-labeled PATJ aa 1 to 1552. The complexes were washed with buffer containing 50, 150, or 250 mM KCl as indicated above the lanes. The bound 35S-labeled protein was resolved by SDS-PAGE and detected by autoradiography. The percentage of 35S-PATJ that remained bound, shown below each lane, was based on the intensity of the band representing 10% of the input. Band intensity was quantified using ImageJ (http://rsb.info.nih.gov/ij/). The arrow points to the position of [35S]PATJ. (B) 35S-labeled fragments of PATJ encompassing aa 1 to 318, 535 to 887, and 885 to 1552 were incubated with either GST or GST-18 E6. The complexes were incubated and washed in 50 mM KCl. The percentage of each fragment that bound to GST and GST-18 E6 was determined as described for panel A. (C) GST, GST-6 E6, and GST-18 E6 proteins used in the pull-down experiment were resolved by SDS-PAGE and visualized by staining with Coomassie brilliant blue. Arrows identify the bands representing GST and GST-6 E6 or GST-18 E6.
In vivo association of Flag-PATJ with E6 and PDZ binding site mutant E6 proteins.
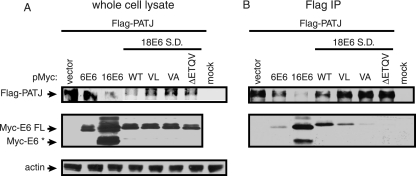
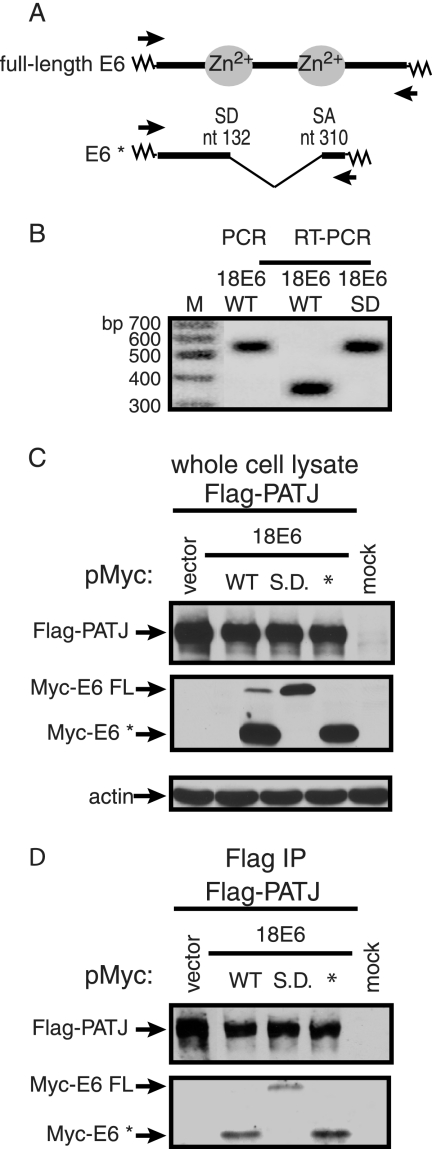
The E6 interaction with PATJ in vivo was addressed by coimmunoprecipitation experiments. 293T cells were transiently transfected with a Flag-tagged expression plasmid encoding PATJ with 10 PDZ domains (49) to determine if the longer isoform associates with 18 E6. Cells were cotransfected with Myc-tag vector or Myc-tagged E6 expression constructs that encoded low-risk type 6 E6, high-risk types 16 or 18 E6, or 18 E6 PDZ binding site mutant proteins. The 18 E6 clones contained a point mutation in the SD site to achieve higher levels of FL 18 E6. Comparable levels of the various Myc-E6 proteins were expressed (Fig. 3A). The transfected cells were treated with MG132, a proteasome inhibitor, for 5 h prior to lysis to stabilize Flag-PATJ levels because, as presented in Fig. 5, below, the accumulation of Flag-PATJ is reduced by types 16 and 18 E6. Despite proteasome inhibition, lower levels of Flag-PATJ accumulated in cells with 16 E6 and 18 E6 WT than in those without E6 or with 6 E6 (Fig. 3A). The reduced accumulation of Flag-PATJ in these samples resulted in lower levels of Flag-PATJ bound to the Flag affinity beads (Fig. 3B). However, even these low levels of Flag-PATJ were sufficient to coimmunoprecipitate FL 16 E6 and what is presumed to be an alternatively spliced isoform of 16 E6, 16 E6*. Flag-PATJ also associated with 18 E6 FL. The V158L mutation changed the PDZ binding site of 18 E6 (ETQV) to that of 16 E6 (ETQL) and reduced the amount of 18 E6 that interacted with Flag-PATJ. This result demonstrates that, while both types 16 and 18 E6 associate with Flag-PATJ, the 16 E6 PDZ binding site, in the context of 18 E6, associates with Flag-PATJ with lower affinity. Both the V158A point mutation and ΔETQV deletion mutation in the PDZ binding site of FL 18 E6 disrupted the association with Flag-PATJ. The level of low-risk 6 E6 that associated with Flag-PATJ was similar to that of 18 E6 PDZ proteins with a disrupted PDZ binding site and probably not significant. Coimmunoprecipitation experiments performed with the shorter PATJ isoform yielded similar results (data not shown). Thus, the ability to interact with PATJ is shared by E6 from the predominant high-risk types 16 and 18 but not low-risk type 6 E6.
FIG. 3.
In vivo association of Flag-PATJ with E6 is dependent on the presence of a PDZ binding site. Approximately 2 × 106 293T cells were transiently transfected with 2 μg of Flag-PATJ expression plasmid and 3 μg of pMycL.1 plasmid or Myc-E6 expression constructs. Mock-transfected cells were incubated with transfection reagent in the absence of plasmid DNA. Cells were collected at 48 h posttransfection. (A) To determine the level of protein accumulation, equal amounts of protein from whole-cell lysates were subjected to Western blot analysis with antibody to Flag, Myc, and actin. (B) Equal amounts of protein were incubated with Flag affinity beads. Following extensive washing, the bound complexes were eluted and subjected to Western blot analysis with antibody to Flag and Myc to determine the levels of Flag-PATJ and Myc-E6. Arrows point to Flag-PATJ, Myc-E6 FL, Myc- E6*, and actin.
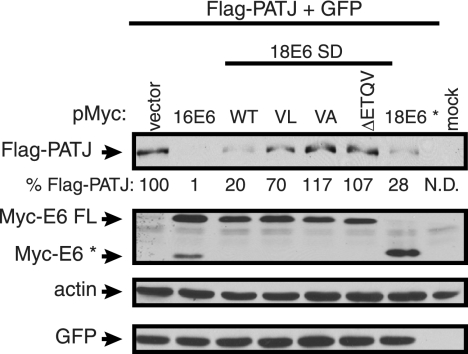
FIG. 5.
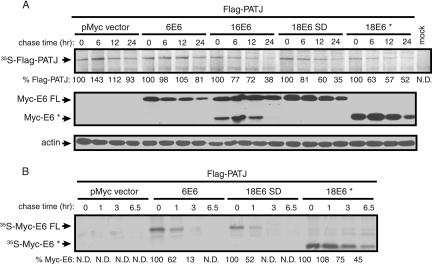
The steady-state level of Flag-PATJ is reduced by 16 E6, 18 E6, and 18 E6*, but not 18 E6 PDZ binding site mutants. Approximately 1 × 106 293T cells were cotransfected with 1 μg of pFlag-CMV-2-PATJ and 3 μg of pMycL.1 empty or pMycL.1 encoding 16 E6, 18 E6 S.D., 18 E6 SD, PDZ binding site mutants (VL, VA, and ΔETQV), or 18 E6*. All cotransfection mixtures contained 0.2 μg of a GFP expression vector. The mock lane contained lysate from cells incubated with Lipofectamine reagent in the absence of plasmid DNA. Forty-eight hours posttransfection, cell lysates were collected, and equal amounts of protein from whole-cell lysates were resolved by SDS-PAGE. Western blot assays were performed using antibody to the Flag or Myc epitopes, actin, and GFP. Arrows identify the positions of Flag-PATJ, Myc-E6 FL, Myc-E6*, actin, and GFP. Flag-PATJ levels, shown below the lanes, were determined by dividing the absolute band intensity by the intensity of the actin band in the sample.
In vivo association of Flag-PATJ with an alternatively spliced isoform of 18 E6.
The Myc-18 E6 WT construct contains an SD and acceptor (SA) site that if used produce RNA encoding Myc-tagged 18 E6* (Fig. 4A). We confirmed that 18 E6 RNA expressed from the Myc-tagged 18 E6 WT construct is internally spliced by performing RT-PCR on total RNA extracted from 293T cells transiently transfected with 18 E6 WT or SD constructs. The 5′ primer bound to the Myc vector, and the 3′ primer bound to the junction between 18 E6 and the Myc vector. A PCR using Myc-18 E6 WT plasmid DNA as template was included as a size standard for unspliced 18 E6 cDNA (Fig. 4B). An RT-PCR product the same size as the 18 E6 PCR amplimer, corresponding to an unspliced cDNA of approximately 500 bp, was obtained from cells transfected with the 18 E6 SD expression construct (Fig. 4B). However, in cells transfected with 18 E6 WT DNA a 300-bp RT-PCR product predominated, suggesting the presence of the spliced product (Fig. 4B). DNA sequence analysis of this product confirmed that the 18 E6 intron was removed. The failure to detect the larger RT-PCR product from cells transfected with 18 E6 WT could result from less efficient amplification of the larger 18 E6 cDNA or the presence of lower levels of full-length 18 E6 RNA. To determine if Flag-PATJ associates with 18 E6* in the presence and absence of 18 E6 FL, 293T cells were cotransfected with plasmids expressing Flag-PATJ and a Myc-tag vector or Myc-tagged E6 constructs expressing 18 E6 WT, SD, or 18 E6*. After treatment with MG132 for 5 h to enhance PATJ accumulation, cells were lysed. The levels of Flag-PATJ were comparable in the presence of the different species of E6 (Fig. 4C). Although Myc-18 E6 WT expressed predominantly Myc-18 E6*, low levels of Myc-18 E6 FL were detected. Myc-18 E6 SD and 18E6* expressed only the full-length and * proteins, respectively. In agreement with prior findings, 18 E6 FL interacts with Flag-PATJ (Fig. 4D). 18 E6* also coimmunoprecipitated with Flag-PATJ in cell lysates that contained low levels of 18 E6 FL. Moreover, when only 18 E6* was expressed, it associated with Flag-PATJ, revealing that the association between Flag-PATJ and 18 E6* does not require 18 E6 FL as a bridge molecule. This result was surprising, considering that the unique carboxy terminus of 18 E6* lacks a classical PDZ binding motif. Instead, the association between 18 E6* and Flag-PATJ appears to be mediated through other regions of 18 E6*, or may be indirect and mediated by a cell protein.
FIG. 4.
18 E6* is expressed and associates in vivo with Flag-PATJ. (A) Alignment of FL 18 E6 and 18 E6* ORFs. Splicing of 18 E6 removes an intron between nucleotides (nt) 132 and 310 and results in an alternative 3′ reading frame. The zig-zag lines represent DNA from the Myc-tag vector. The arrows identify the binding sites for the 5′ and 3′ primers used for RT-PCR. (B) RT-PCR was performed on total RNA collected from approximately 1.5 × 106 293T cells transiently transfected with 1 μg of pMycL.1-18 E6 WT or 18 E6 SD construct. PCR was also performed using pMycL.1-18 E6 WT vector DNA and the same primers. The PCR products were resolved on an agarose gel and detected by ethidium bromide staining. The sizes of the DNA molecular weight markers (M) are indicated. Approximately 2 × 106 293T cells were transiently transfected with 2 μg of Flag-PATJ expression plasmid and 3 μg of pMycL.1 plasmid or pMycL.1 constructs containing 18 E6 WT, SD, or E6*. (C and D) Equal amounts of protein, extracted after 48 h, were either resolved directly (C) or bound to Flag affinity beads (D). The complexes bound to Flag affinity beads were washed extensively and eluted. Following SDS-PAGE, the whole-cell lysates and Flag-bound complexes were probed with antibody to Flag, Myc, and actin. Arrows identify Flag-PATJ, Myc-E6 FL, Myc-E6*, and actin.
Steady-state level of Flag-PATJ in the presence of high-risk E6, 18 E6*, and PDZ binding site mutant proteins.
Like many high-risk E6 protein interaction partners, the PDZ binding partners are targeted for degradation by high-risk E6 (6, 7, 11, 27, 36, 63). The steady-state level of Flag-PATJ was examined in the presence of 16 E6 and 18 E6 with either a WT or mutated PDZ binding site. Because we found that 18 E6* associates with Flag-PATJ, the effect of this protein on the accumulation of Flag-PATJ was also tested. 293T cells were cotransfected with the Flag-PATJ expression plasmid, a Myc-tag vector, or the various Myc-tagged E6 expression constructs and a GFP expression plasmid as a control for plasmid DNA uptake. Equal amounts of total protein from cell lysates were subjected to Western blot analysis with the indicated antibodies. The steady-state level of Flag-PATJ was reduced in the presence of 16 E6 and 18 E6 FL by 99% and 80%, respectively (Fig. 5). The effects of the 18 E6 mutant proteins on the level of Flag-PATJ correlated with their ability to bind Flag-PATJ (Fig. 3B). The V158L mutation reduced the ability of 18 E6 to bind to and reduce the level of Flag-PATJ, and the V158A and ΔETQV mutant proteins failed to bind to or reduce the level of Flag-PATJ. This correlation demonstrates that, for 18 E6 FL, the PDZ binding site-mediated interaction with Flag-PATJ is required to reduce the level of Flag-PATJ. Similar to 18 E6 FL, 18 E6* reduced the level of Flag-PATJ by 72%. Immunoblot assays using antibody to actin and GFP confirmed that comparable amounts of total protein were loaded, and the cells took up the same amounts of DNA.
Half-life of Flag-PATJ in the presence of low-risk E6, high-risk E6, and 18 E6*.
Pulse-chase experiments were performed to confirm that the E6-dependent decrease of Flag-PATJ resulted from reduced protein stability. 293T cells transiently expressing Flag-PATJ with or without Myc-6 E6, 16 E6, 18 E6 FL, or 18 E6* were metabolically labeled and chased for the indicated times (Fig. 6A, top panel). Flag-PATJ is a stable protein that decayed only slightly after 24 h and 48 h (not shown) in the absence of E6. The stability of Flag-PATJ was not affected in the presence of low-risk 6 E6. Quantification of the level of [35S]Flag-PATJ revealed that 93% remained after 24 h in the absence E6 and 81% remained after 24 h in the presence of 6 E6. In contrast, less PATJ remained after 6 h in the presence of 16 E6, 18 E6 FL, and 18 E6* than after 24 h in the absence of E6. The half-life of [35S]Flag-PATJ was reduced to between 12 and 24 h in the presence of 16 E6, 18 E6 FL, or 18 E6*. Therefore, the stability of PATJ is reduced in the presence of these E6 forms. To confirm the presence of Myc-E6 in the samples, equal amounts of total protein were subjected to Western blot analysis with antibody to Myc. Between the 0- and 12-h time points, the levels of the different Myc-tagged E6 proteins were similar (Fig. 6A, middle panel). The reduced level of E6 at 24 h is probably from decreased cell viability. Analysis of actin levels confirmed that similar amounts of total protein were loaded (Fig. 6A, bottom panel). We also examined the half-life of 18 E6*, because its stability, unlike 18 E6 FL, is not known. Considering that 6 E6 (23) and 18 E6 FL (61) are relatively unstable proteins, data from earlier time points were collected from the pulse-chase experiments described above. As expected, the decay rates of 6 E6 FL and 18 E6 were rapid (Fig. 6B). Quantification of the levels of 35S-labeled Myc-6 E6 and 18 E6 FL revealed that only 50% of each remained after 1 h. The half-life of Myc-18 E6* was approximately 6 h. Thus, 18 E6* is more stable than 18 E6 FL.
FIG. 6.
The half-life of Flag-PATJ is reduced in the presence of 16 E6, 18 E6 FL, and 18 E6*, but not 6 E6. Approximately 2 × 106 293T cells were transfected with 2 μg of Flag-PATJ expression construct, 6 μg of empty pMycL.1, or pMycL.1 encoding 6 E6, 16 E6, 18 E6 FL, or E6*. Forty-eight hours later, cells were metabolically labeled with Trans35S-label and chased for the times indicated above each lane. (A) Equal amounts of trichloroacetic acid-precipitable counts were bound to Flag affinity beads. The captured complexes were washed and resolved by SDS-PAGE. 35S-labeled Flag-PATJ was visualized by autoradiography and is identified by the arrow (top panel). The percent remaining 35S-Flag-PATJ at each chase time point, shown below each lane, was normalized to the relative background intensity in the lane. Quantifications were performed using ImageJ. The expression level of the Myc-tagged E6 types was determined after equal amounts of protein from whole-cell lysates were resolved by SDS-PAGE and immunoblotted with antibody to the Myc epitope (middle panel). Equal loading was confirmed by analyzing the levels of actin (bottom panel). (B) The half-life of the Myc-tagged E6 proteins was determined as for Flag-PATJ, except that earlier time points were analyzed and equal amounts of total extracted protein were incubated with Myc affinity beads. 35S-labeled Myc-6 E6, 18 E6 FL, or 18 E6* is identified by the arrows. The level of the 35S-labeled Myc-E6 proteins that remained at the different times was quantified using ImageJ.
Requirement of E6AP for 18 E6-dependent reduction of Flag-PATJ.
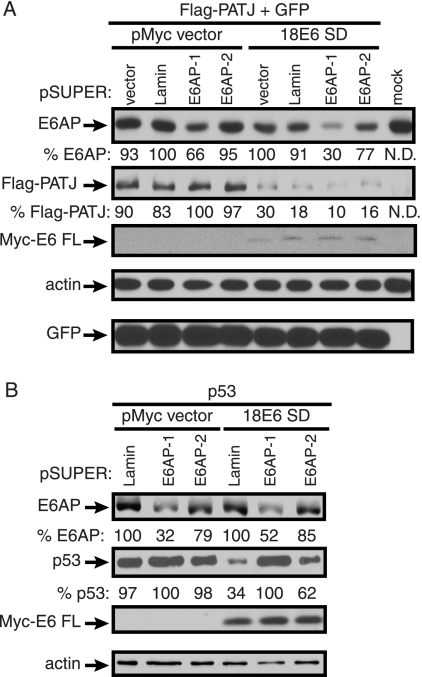
We wished to determine if the ubiquitin ligase E6AP is involved in the degradation of PATJ by 18 E6. Many binding partners of E6, including the PDZ proteins Dlg and Scrib, are stimulated for degradation by the E6-E6AP complex (1, 33, 36). However, data indicate that high-risk E6 can also mediate degradation of the PDZ targets Dlg and MAGI-1, -2, and -3 in the absence of E6AP (14, 46). To determine if E6AP is required for the E6-dependent destabilization of Flag-PATJ, expression of E6AP was inhibited by E6AP-specific shRNA. pSUPER plasmids expressing E6AP-specific shRNA molecules E6AP-1 or E6AP-2, lamin A/C shRNA (41) as a nonspecific control, or empty pSUPER plasmid were introduced into 293T cells. Cells were cotransfected with the Flag-PATJ expression plasmid, with or without the Myc-18 E6 SD construct and, to monitor transfection efficiency, a GFP expression plasmid. Western blots of lysates were probed with antibody to E6AP to determine if the E6AP targeting constructs diminished the level of E6AP. In the absence of 18 E6, pSUPER plasmids expressing E6AP-1 or -2 shRNA reduced the level of E6AP by 34% and 5%, respectively (Fig. 7A). In the presence of E6, the basal level of E6AP was reduced, suggesting that 18 E6, like 16 E6, decreases E6AP stability (22). It is conceivable that because of differences in the synthesis of E6AP in the presence of Myc-18 E6, the levels of E6AP were more susceptible to E6AP shRNA. In cells cotransfected with the Myc-18 E6 SD construct, the E6AP-1 or -2 targeting constructs reduced the accumulation of E6AP by 70% and 23%, respectively, compared to the corresponding cotransfection with empty pSUPER. Analysis of Western blot assays of these lysates demonstrated that Flag-PATJ was degraded when Myc-18 E6 FL was expressed in the presence of control and E6AP-targeting pSUPER plasmids. Thus, the 70% reduction of the E6AP level did not affect 18 E6-dependent reduction of Flag-PATJ. Western blotting with antibodies to actin and GFP confirmed that comparable amounts of cell and exogenously expressed proteins were loaded. 18 E6-mediated reduction of p53 in the presence of the pSUPER targeting constructs was tested to confirm that E6AP activity was dampened in cells treated with shRNAs. The experiment could not be performed in 293T cells, because the resident adenoviral proteins and simian virus 40 (SV40) T-antigen result in high levels of stabilized p53. Instead, a p53 expression plasmid was introduced into p53-null H1299 cells with or without the Myc-18 E6 SD construct and the control or E6AP targeting constructs. In the absence of 18 E6, expression of E6AP-1 or -2 shRNA reduced the level of E6AP by 68% and 21%, respectively (Fig. 7B). In the presence of 18 E6, the level of E6AP was similarly reduced by E6AP-1 or -2 by 48% and 15%, respectively, compared to the corresponding cotransfection with the lamin-targeting construct. In the presence of 18 E6 and lamin shRNA, the accumulation of p53 was reduced by 76%. Both of the E6AP-targeting constructs decreased the level of 18 E6-mediated degradation of p53. E6AP-1 was more effective than E6AP-2 in reducing E6AP levels, and it completely inhibited 18 E6-mediated degradation of p53. The E6AP-targeting constructs interfered with 18 E6-mediated degradation of p53, despite their incomplete reduction of E6AP. This indicates that residual E6AP is not available for 18 E6-mediated substrate degradation. Thus, the E6AP-specific shRNAs extinguish functionality of the E6-E6AP complex.
FIG. 7.
shRNA-mediated E6AP silencing does not affect 18 E6-dependent reduction of Flag-PATJ. (A) On day 1, approximately 0.75 × 106 293T cells were cotransfected with 2.5 μg of either empty pSUPER vector, lamin A/C, E6AP-1, or E6AP-2 targeting constructs. On day 4, the same amount of pSUPER constructs and 0.5 μg of Flag-PATJ expression plasmid, 0.125 μg of GFP expression plasmid, and 1.5 μg of empty Myc vector or Myc-18 E6 SD construct were introduced into these same cells. Cells were harvested on day 6. Equal amounts of protein were analyzed with antibodies to E6AP, the Flag and Myc epitopes, actin, and GFP. Arrows identify E6AP, Flag-PATJ, Myc-E6 FL, actin, and GFP. The levels of E6AP and Flag-PATJ, shown below the images, were quantified using ImageJ and normalized to the corresponding levels of actin. (B) The effect of E6AP silencing on 18 E6-dependent p53 reduction was determined as described for panel A, except that H1299 cells were used and cells were cotransfected with p53 instead of Flag-PATJ, expression plasmid, empty Myc vector, or Myc-18 E6 SD, and lamin A/C, E6AP-1 or E6AP-2 targeting constructs. Cell lysates were probed with antibodies to E6AP, p53, the Myc epitope, and actin. Arrows identify E6AP, p53, Myc-E6 FL, and actin.
Effect of proteasome inhibition on PATJ, Dlg, and p53 accumulation in cervical cancer-derived cell lines.
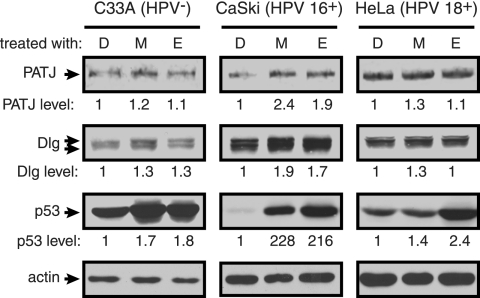
The reduced stability of Flag-PATJ in the presence of types 16 and 18 E6 and 18 E6* led us to ask what the levels of endogenous PATJ are in cell lines derived from cervical cancers. CaSki and HeLa cells continually express 16 and 18 E6, respectively. C33A cells are derived from HPV-negative cervical cancer tissue. These cell lines were treated for 6 h with the proteasome inhibitors MG132 or epoxomicin or DMSO alone prior to lysis. Equal amounts of protein from these treated cells were analyzed by Western blotting with antibody to PATJ, Dlg, p53, and actin. The levels of p53 are high in C33A cells, as reported, and increased by approximately twofold after treatment with proteasome inhibitors (Fig. 8). The levels of PATJ and the PDZ target Dlg are not altered in response to the inhibitors in C33A cells. In contrast, the levels of both PATJ and Dlg are increased approximately twofold in CaSki cells. Moreover, the level of p53 increases dramatically in CaSki cells. Thus, PATJ, Dlg, and p53 are regulated by the proteasome in an HPV 16+ cell line. In HeLa cells, treatment with proteasome inhibitors had a modest effect on the level of p53 and no effect on the levels of PATJ and Dlg. This phenomenon was previously reported for other PDZ interaction partners of E6: Dlg, Scribble, and MUPP1 were stabilized significantly in CaSki cells, but not HeLa cells, following proteasome inhibition (32).
FIG. 8.
Proteasome regulation of PATJ, Dlg, and p53 in cervical cancer-derived cell lines. C33A, HPV 16+ CaSki, and HPV 18+ HeLa cells were grown to confluence and incubated with medium containing DMSO alone (D), MG132 (M), or epoxomicin (E) at 10 μg/ml final concentration. Cells were incubated with proteasome inhibitors for 6 hours before lysis in RIPA buffer. Seventy μg of C33A protein, 28 μg of CaSki protein, and 111 μg of HeLa protein were analyzed by Western blotting using antibodies to PATJ (described in Materials and Methods), Dlg, p53, and actin. The levels of PATJ, Dlg, and p53 were quantified using ImageJ and normalized to the level of actin. The difference in PATJ, Dlg, and p53 levels, shown below each lane, was determined based on their levels in the corresponding DMSO-treated cell.
DISCUSSION
In this study, we describe the association of high-risk types 16 and 18 E6 with the tight junction-associated protein PATJ. The interaction was identified in a yeast two-hybrid screen and verified by GST pull-down and coimmunoprecipitation assays. Similar to the interaction between E6 and other PDZ proteins, the interaction with PATJ is specific for high-risk types 16 and 18 E6. In the context of full-length 18 E6, expressed from a gene with a splice donor site mutation, the C-terminal PDZ binding motif is required for binding to PATJ. PATJ shares certain properties with the known PDZ binding partners of E6, such as its multiple PDZ domains (49), and membrane localization upon the formation of cell-cell contacts (28). While MUPP1 (27) and MAGI-1 (11) localize to tight junctions, only PATJ is known to be required for the localization of TJ plaque proteins and overall cell polarity (34, 54). The localization and functions of PATJ at TJ are likely to depend on its protein associations. Through analysis of peptide fragments of PATJ in the yeast two-hybrid system and GST pull-down assays, the region of PATJ that interacts with 18 E6 was mapped to PDZ domains 4 and 5. Because this region of PATJ is distinct from those that mediate its associations with PALS1 (49), ZO-3, and Claudin-1 (48), we predict that association with 18 E6 is unlikely to interfere directly with binding of the known cellular interaction partners of PATJ.
We have provided evidence that PATJ is a novel degradation substrate of high-risk types 16 and 18 E6. The reduction in the steady-state level of PATJ in the presence of full-length E6 is dependent on the PDZ binding motif of 18 E6. Pulse-chase assays revealed that PATJ is a stable protein that was reduced by less than 10% after 24 h. While low-risk 6 E6 did not reduce the stability of PATJ, the half-life of PATJ was decreased to between 12 and 24 h in the presence of 16 E6 and 18 E6 FL. Because the abundance of tight junction proteins is thought to be directly related to junction tightness (reviewed in reference 12) and a reduction of PATJ leads to loss of its associated TJ complexes (54), the decrease in PATJ level in the presence of high-risk E6 is predicted to reduce TJ function. Endogenous PATJ, like Dlg and p53, was stabilized by treatment with proteasome inhibitors in an HPV 16+ (CaSki) cervical cancer-derived cell line. The levels of PATJ, Dlg, and p53 were not significantly altered by proteasome inhibition in an HPV− (C33A) or HPV 18+ (HeLa) cell line. Massimi et al. (32) also found that proteasome inhibition resulted in increased accumulation of PDZ binding partners of E6, including Dlg in CaSki cells compared with HeLa cells. Together, these data suggest that these PDZ proteins are regulated by the proteasome in cervical cancer-derived cell lines that harbor high-risk E6 but that certain cell lines, like HeLa cells, have acquired differences that affect the stability of 18 E6 degradation targets.
The E6-dependent degradation of Dlg (46) and MAGI-1, -2, and -3 was suggested to occur independently of the ubiquitin ligase E6AP (14, 60). We found that shRNA-mediated silencing of E6AP did not interfere with 18 E6-dependent degradation of PATJ. The ability of the E6AP-specific shRNAs to rescue p53 from 18 E6-dependent degradation demonstrated that the function of the E6-E6AP heterodimer was inhibited. These findings point to the association of E6 with a non-E6AP ubiquitin ligase for the destruction of PATJ and possibly other degradation targets.
The findings presented here reveal that 18 E6* associates with PATJ to effect its degradation. The majority of high-risk HPV RNAs in cervical lesions and cell lines contain spliced E6 (4, 55). 18 E6* is the only E6 spliced product detected in HPV 18+ cervical cancer-derived cell lines (52), and its protein product is present in cervical carcinomas grown in nude mice (51). Our pulse-chase analysis shows that 18 E6* is stable relative to the full-length E6 species. The only described function of 18 E6* is the negative regulation of E6-dependent p53 degradation. This activity of 18 E6* is probably responsible for its antiproliferative effect in HPV 16+ and 18+ cervical carcinoma-derived cells (43, 44). Considering that 18 E6* lacks a C-terminal PDZ binding motif and has a unique 14-amino-acid C terminus, we predict that a cellular protein bridges the in vivo association between 18 E6* and Flag-PATJ. Alternatively, a direct interaction between 18 E6* and PATJ could occur and involve a different region of 18 E6* and, possibly, PATJ. Based on the recently solved partial structure of 16 E6, it is suggested that the E6* structure resembles that of the N terminus of full-length E6 (40). Thus, if the 18 E6* interaction with PATJ is direct, we hypothesize that it is not mediated by the N terminus, which is predicted to resemble that of 18 E6, but rather sequence within its unique C terminus. It would be of considerable interest to perform in vitro interaction assays to determine if 18 E6* and PATJ interact directly. Similar to 18 E6 FL, 18 E6* reduced the accumulation and stability of PATJ. It will be of interest to determine if this form of 18 E6 affects the stability of other PDZ binding partners of FL high-risk E6 types. Overexpression of 18 E6* in HPV 16+ cells did not affect the membrane localization of Dlg and Scribble (15), suggesting that E6* does not affect an identical set of PDZ proteins as E6 FL. This prediction explains the requirement for the C-terminal PDZ binding site in FL E6 for the proliferation of HPV 31-transfected keratinocytes (26) and the transformation activity of 16 E6 (24) and 18 E6 (66). The identification of spliced isoforms of 16 E6 (4, 57) and 31 E6 (18) raises the possibility that their putative protein products similarly regulate PATJ levels.
In summary, PATJ, an evolutionarily conserved polarity determinant, is a new member of the class of PDZ proteins whose interaction with E6 could contribute to the life cycle and carcinogenesis of high-risk HPV types. Our data demonstrate that PATJ is degraded by E6 and may thus be an important target of E6. It remains speculative how the regulation of PATJ by E6 could affect the cell. A link between PATJ and control of cell signaling and cell growth is suggested by the finding that PATJ is required for assembly and maintenance of the TJ-associated complex Par6-aPKC-Par3 (34, 54). This complex regulates the kinase activity of aPKC and the Cdc42 GTPase (21, 29). High-risk E6 binds to the adherens junction (AJ)-localized PDZ proteins Dlg (24) and Scribble (36). They have been proposed to act as negative regulators of the cell cycle (20, 62) and may thereby alter cell growth control. MUPP1 is a paralog of PATJ, based on its sequence homology and its ability to bind PALS1 (49). The apparent functional redundancy between PATJ and MUPP1, like Dlg and Scrib, suggests that E6 associates with these proteins to effectively ablate the functions of the same or similar membrane-associated complexes. In both Drosophila and mammalian cells, an interplay between the AJ complex involving Dlg and Scribble and the TJ complex involving PATJ and MUPP1 regulates the segregation of apical and basolateral membrane compartments during cell polarization (reviewed in reference 50). As a result, mutations in the Drosophila homologs of both AJ and TJ components exhibit phenotypes that are less severe than individual defects. It is thus tempting to speculate that E6 targets PDZ proteins to disrupt cell growth control but, because it affects both AJ- and TJ-associated PDZ proteins, it does not necessarily prevent the epithelial differentiation necessary to complete the HPV life cycle.
Acknowledgments
We thank Yan Degenhardt and the members of the Silverstein lab for their support and insight. We are also grateful to Matt Evans for his critical comments on the manuscript.
Footnotes
Published ahead of print on 7 February 2007.
REFERENCES
- 1.Brimer, N., C. Lyons, and S. B. Vande Pol. 2006. Association of E6AP (UBE3A) with human papillomavirus type 11 E6 protein. Virology 358:303-310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brummelkamp, T. R., R. Bernards, and R. Agami. 2002. A system for stable expression of short interfering RNAs in mammalian cells. Science 296:550-553. [DOI] [PubMed] [Google Scholar]
- 3.Degenhardt, Y. Y., and S. J. Silverstein. 2001. Gps2, a protein partner for human papillomavirus E6 proteins. J. Virol. 75:151-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Doorbar, J., A. Parton, K. Hartley, L. Banks, T. Crook, M. Stanley, and L. Crawford. 1990. Detection of novel splicing patterns in a HPV16-containing keratinocyte cell line. Virology 178:254-262. [DOI] [PubMed] [Google Scholar]
- 5.Fanning, A. S., and J. M. Anderson. 1999. PDZ domains: fundamental building blocks in the organization of protein complexes at the plasma membrane. J. Clin. Investig. 103:767-772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Favre-Bonvin, A., C. Reynaud, C. Kretz-Remy, and P. Jalinot. 2005. Human papillomavirus type 18 E6 protein binds the cellular PDZ protein TIP-2/GIPC, which is involved in transforming growth factor beta signaling and triggers its degradation by the proteasome. J. Virol. 79:4229-4237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gardiol, D., C. Kuhne, B. Glaunsinger, S. S. Lee, R. Javier, and L. Banks. 1999. Oncogenic human papillomavirus E6 proteins target the discs large tumour suppressor for proteasome-mediated degradation. Oncogene 18:5487-5496. [DOI] [PubMed] [Google Scholar]
- 8.Gardiol, D., A. Zacchi, F. Petrera, G. Stanta, and L. Banks. 2006. Human discs large and scrib are localized at the same regions in colon mucosa and changes in their expression patterns are correlated with loss of tissue architecture during malignant progression. Int. J. Cancer 119:1285-1290. [DOI] [PubMed] [Google Scholar]
- 9.Gillison, M. L., and K. V. Shah. 2001. Human papillomavirus-associated head and neck squamous cell carcinoma: mounting evidence for an etiologic role for human papillomavirus in a subset of head and neck cancers. Curr. Opin. Oncol. 13:183-188. [DOI] [PubMed] [Google Scholar]
- 10.Gissmann, L., E. M. deVilliers, and H. zur Hausen. 1982. Analysis of human genital warts (condylomata acuminata) and other genital tumors for human papillomavirus type 6 DNA. Int. J. Cancer 29:143-146. [DOI] [PubMed] [Google Scholar]
- 11.Glaunsinger, B. A., S. S. Lee, M. Thomas, L. Banks, and R. Javier. 2000. Interactions of the PDZ-protein MAGI-1 with adenovirus E4-ORF1 and high-risk papillomavirus E6 oncoproteins. Oncogene 19:5270-5280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gonzalez-Mariscal, L., A. Betanzos, P. Nava, and B. E. Jaramillo. 2003. Tight junction proteins. Prog. Biophys. Mol. Biol. 81:1-44. [DOI] [PubMed] [Google Scholar]
- 13.Griep, A. E., R. Herber, S. Jeon, J. K. Lohse, R. R. Dubielzig, and P. F. Lambert. 1993. Tumorigenicity by human papillomavirus type 16 E6 and E7 in transgenic mice correlates with alterations in epithelial cell growth and differentiation. J. Virol. 67:1373-1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grm, H. S., and L. Banks. 2004. Degradation of hDlg and MAGIs by human papillomavirus E6 is E6-AP-independent. J. Gen. Virol. 85:2815-2819. [DOI] [PubMed] [Google Scholar]
- 15.Guccione, E., D. Pim, and L. Banks. 2004. HPV-18 E6*I modulates HPV-18 full-length E6 functions in a cell cycle dependent manner. Int. J. Cancer 110:928-933. [DOI] [PubMed] [Google Scholar]
- 16.Hudson, J. B., M. A. Bedell, D. J. McCance, and L. A. Laiminis. 1990. Immortalization and altered differentiation of human keratinocytes in vitro by the E6 and E7 open reading frames of human papillomavirus type 18. J. Virol. 64:519-526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huibregtse, J. M., M. Scheffner, and P. M. Howley. 1993. Cloning and expression of the cDNA for E6-AP, a protein that mediates the interaction of the human papillomavirus E6 oncoprotein with p53. Mol. Cell. Biol. 13:775-784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hummel, M., J. B. Hudson, and L. A. Laimins. 1992. Differentiation-induced and constitutive transcription of human papillomavirus type 31b in cell lines containing viral episomes. J. Virol. 66:6070-6080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hurd, T. W., L. Gao, M. H. Roh, I. G. Macara, and B. Margolis. 2003. Direct interaction of two polarity complexes implicated in epithelial tight junction assembly. Nat. Cell Biol. 5:137-142. [DOI] [PubMed] [Google Scholar]
- 20.Ishidate, T., A. Matsumine, K. Toyoshima, and T. Akiyama. 2000. The APC-hDLG complex negatively regulates cell cycle progression from the G0/G1 to S phase. Oncogene 19:365-372. [DOI] [PubMed] [Google Scholar]
- 21.Joberty, G., C. Petersen, L. Gao, and I. G. Macara. 2000. The cell-polarity protein Par6 links Par3 and atypical protein kinase C to Cdc42. Nat. Cell Biol. 2:531-539. [DOI] [PubMed] [Google Scholar]
- 22.Kao, W. H., S. L. Beaudenon, A. L. Talis, J. M. Huibregtse, and P. M. Howley. 2000. Human papillomavirus type 16 E6 induces self-ubiquitination of the E6AP ubiquitin-protein ligase. J. Virol. 74:6408-6417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kehmeier, E., H. Ruhl, B. Voland, M. C. Stoppler, E. Androphy, and H. Stoppler. 2002. Cellular steady-state levels of “high risk” but not “low risk” human papillomavirus (HPV) E6 proteins are increased by inhibition of proteasome-dependent degradation independent of their p53- and E6AP-binding capabilities. Virology 299:72-87. [DOI] [PubMed] [Google Scholar]
- 24.Kiyono, T., A. Hiraiwa, M. Fujita, Y. Hayashi, T. Akiyama, and M. Ishibashi. 1997. Binding of high-risk human papillomavirus E6 oncoproteins to the human homologue of the Drosophila discs large tumor suppressor protein. Proc. Natl. Acad. Sci. USA 94:11612-11616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Latorre, I. J., M. H. Roh, K. K. Frese, R. S. Weiss, B. Margolis, and R. T. Javier. 2005. Viral oncoprotein-induced mislocalization of select PDZ proteins disrupts tight junctions and causes polarity defects in epithelial cells. J. Cell Sci. 118:4283-4293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee, C., and L. A. Laimins. 2004. Role of the PDZ domain-binding motif of the oncoprotein E6 in the pathogenesis of human papillomavirus type 31. J. Virol. 78:12366-12377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee, S. S., B. Glaunsinger, F. Mantovani, L. Banks, and R. T. Javier. 2000. Multi-PDZ domain protein MUPP1 is a cellular target for both adenovirus E4-ORF1 and high-risk papillomavirus type 18 E6 oncoproteins. J. Virol. 74:9680-9693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lemmers, C., E. Medina, M. H. Delgrossi, D. Michel, J. P. Arsanto, and A. Le Bivic. 2002. hINADl/PATJ, a homolog of discs lost, interacts with crumbs and localizes to tight junctions in human epithelial cells. J. Biol. Chem. 277:25408-25415. [DOI] [PubMed] [Google Scholar]
- 29.Lin, D., A. S. Edwards, J. P. Fawcett, G. Mbamalu, J. D. Scott, and T. Pawson. 2000. A mammalian PAR-3-PAR-6 complex implicated in Cdc42/Rac1 and aPKC signalling and cell polarity. Nat. Cell Biol. 2:540-547. [DOI] [PubMed] [Google Scholar]
- 30.Liu, Y., J. J. Chen, Q. Gao, S. Dalal, Y. Hong, C. P. Mansur, V. Band, and E. J. Androphy. 1999. Multiple functions of human papillomavirus type 16 E6 contribute to the immortalization of mammary epithelial cells. J. Virol. 73:7297-7307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martin, T. A., G. Watkins, R. E. Mansel, and W. G. Jiang. 2004. Loss of tight junction plaque molecules in breast cancer tissues is associated with a poor prognosis in patients with breast cancer. Eur. J. Cancer 40:2717-2725. [DOI] [PubMed] [Google Scholar]
- 32.Massimi, P., N. Gammoh, M. Thomas, and L. Banks. 2004. HPV E6 specifically targets different cellular pools of its PDZ domain-containing tumour suppressor substrates for proteasome-mediated degradation. Oncogene 23:8033-8039. [DOI] [PubMed] [Google Scholar]
- 33.Matsumoto, Y., S. Nakagawa, T. Yano, S. Takizawa, K. Nagasaka, K. Nakagawa, T. Minaguchi, O. Wada, H. Ooishi, K. Matsumoto, T. Yasugi, T. Kanda, J. M. Huibregtse, and Y. Taketani. 2006. Involvement of a cellular ubiquitin-protein ligase E6AP in the ubiquitin-mediated degradation of extensive substrates of high-risk human papillomavirus E6. J. Med. Virol. 78:501-507. [DOI] [PubMed] [Google Scholar]
- 34.Michel, D., J. P. Arsanto, D. Massey-Harroche, C. Beclin, J. Wijnholds, and A. Le Bivic. 2005. PATJ connects and stabilizes apical and lateral components of tight junctions in human intestinal cells. J. Cell Sci. 118:4049-4057. [DOI] [PubMed] [Google Scholar]
- 35.Munger, K., W. C. Phelps, V. Bubb, P. M. Howley, and R. Schlegel. 1989. The E6 and E7 genes of the human papillomavirus type 16 together are necessary and sufficient for transformation of primary human keratinocytes. J. Virol. 63:4417-4421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nakagawa, S., and J. M. Huibregtse. 2000. Human scribble (Vartul) is targeted for ubiquitin-mediated degradation by the high-risk papillomavirus E6 proteins and the E6AP ubiquitin-protein ligase. Mol. Cell. Biol. 20:8244-8253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nakagawa, S., T. Yano, K. Nakagawa, S. Takizawa, Y. Suzuki, T. Yasugi, J. M. Huibregtse, and Y. Taketani. 2004. Analysis of the expression and localisation of a LAP protein, human scribble, in the normal and neoplastic epithelium of uterine cervix. Br. J. Cancer 90:194-199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nguyen, M. L., M. M. Nguyen, D. Lee, A. E. Griep, and P. F. Lambert. 2003. The PDZ ligand domain of the human papillomavirus type 16 E6 protein is required for E6's induction of epithelial hyperplasia in vivo. J. Virol. 77:6957-6964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nguyen, M. M., M. L. Nguyen, G. Caruana, A. Bernstein, P. F. Lambert, and A. E. Griep. 2003. Requirement of PDZ-containing proteins for cell cycle regulation and differentiation in the mouse lens epithelium. Mol. Cell. Biol. 23:8970-8981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nomine, Y., M. Masson, S. Charbonnier, K. Zanier, T. Ristriani, F. Deryckere, A. P. Sibler, D. Desplancq, R. A. Atkinson, E. Weiss, G. Orfanoudakis, B. Kieffer, and G. Trave. 2006. Structural and functional analysis of E6 oncoprotein: insights in the molecular pathways of human papillomavirus-mediated pathogenesis. Mol. Cell 21:665-678. [DOI] [PubMed] [Google Scholar]
- 41.Paul, C. P., P. D. Good, I. Winer, and D. R. Engelke. 2002. Effective expression of small interfering RNA in human cells. Nat. Biotechnol. 20:505-508. [DOI] [PubMed] [Google Scholar]
- 42.Philipp, S., and V. Flockerzi. 1997. Molecular characterization of a novel human PDZ domain protein with homology to INAD from Drosophila melanogaster. FEBS Lett. 413:243-248. [DOI] [PubMed] [Google Scholar]
- 43.Pim, D., and L. Banks. 1999. HPV-18 E6*I protein modulates the E6-directed degradation of p53 by binding to full-length HPV-18 E6. Oncogene 18:7403-7408. [DOI] [PubMed] [Google Scholar]
- 44.Pim, D., P. Massimi, and L. Banks. 1997. Alternatively spliced HPV-18 E6* protein inhibits E6 mediated degradation of p53 and suppresses transformed cell growth. Oncogene 15:257-264. [DOI] [PubMed] [Google Scholar]
- 45.Pim, D., A. Storey, M. Thomas, P. Massimi, and L. Banks. 1994. Mutational analysis of HPV-18 E6 identifies domains required for p53 degradation in vitro, abolition of p53 transactivation in vivo and immortalisation of primary BMK cells. Oncogene 9:1869-1876. [PubMed] [Google Scholar]
- 46.Pim, D., M. Thomas, R. Javier, D. Gardiol, and L. Banks. 2000. HPV E6 targeted degradation of the discs large protein: evidence for the involvement of a novel ubiquitin ligase. Oncogene 19:719-725. [DOI] [PubMed] [Google Scholar]
- 47.Riley, R. R., S. Duensing, T. Brake, K. Munger, P. F. Lambert, and J. M. Arbeit. 2003. Dissection of human papillomavirus E6 and E7 function in transgenic mouse models of cervical carcinogenesis. Cancer Res. 63:4862-4871. [PubMed] [Google Scholar]
- 48.Roh, M. H., C. J. Liu, S. Laurinec, and B. Margolis. 2002. The carboxyl terminus of zona occludens-3 binds and recruits a mammalian homologue of discs lost to tight junctions. J. Biol. Chem. 277:27501-27509. [DOI] [PubMed] [Google Scholar]
- 49.Roh, M. H., O. Makarova, C. J. Liu, K. Shin, S. Lee, S. Laurinec, M. Goyal, R. Wiggins, and B. Margolis. 2002. The Maguk protein, Pals1, functions as an adapter, linking mammalian homologues of Crumbs and Discs Lost. J. Cell Biol. 157:161-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Roh, M. H., and B. Margolis. 2003. Composition and function of PDZ protein complexes during cell polarization. Am. J. Physiol. Renal Physiol. 285:F377-F387. [DOI] [PubMed] [Google Scholar]
- 51.Schneider-Gadicke, A., S. Kaul, E. Schwarz, H. Gausepohl, R. Frank, and G. Bastert. 1988. Identification of the human papillomavirus type 18 E6 and E6 proteins in nuclear protein fractions from human cervical carcinoma cells grown in the nude mouse or in vitro. Cancer Res. 48:2969-2974. [PubMed] [Google Scholar]
- 52.Schneider-Gadicke, A., and E. Schwarz. 1986. Different human cervical carcinoma cell lines show similar transcription patterns of human papillomavirus type 18 early genes. EMBO J. 5:2285-2292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sedman, S. A., M. S. Barbosa, W. C. Vass, N. L. Hubbert, J. A. Haas, D. R. Lowy, and J. T. Schiller. 1991. The full-length E6 protein of human papillomavirus type 16 has transforming and trans-activating activities and cooperates with E7 to immortalize keratinocytes in culture. J. Virol. 65:4860-4866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shin, K., S. Straight, and B. Margolis. 2005. PATJ regulates tight junction formation and polarity in mammalian epithelial cells. J. Cell Biol. 168:705-711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shirasawa, H., H. Tanzawa, T. Matsunaga, and B. Simizu. 1991. Quantitative detection of spliced E6-E7 transcripts of human papillomavirus type 16 in cervical premalignant lesions. Virology 184:795-798. [DOI] [PubMed] [Google Scholar]
- 56.Simonson, S. J., M. J. Difilippantonio, and P. F. Lambert. 2005. Two distinct activities contribute to human papillomavirus 16 E6's oncogenic potential. Cancer Res. 65:8266-8273. [DOI] [PubMed] [Google Scholar]
- 57.Smotkin, D., and F. O. Wettstein. 1986. Transcription of human papillomavirus type 16 early genes in a cervical cancer and a cancer-derived cell line and identification of the E7 protein. Proc. Natl. Acad. Sci. USA 83:4680-4684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Song, S., A. Liem, J. A. Miller, and P. F. Lambert. 2000. Human papillomavirus types 16 E6 and E7 contribute differently to carcinogenesis. Virology 267:141-150. [DOI] [PubMed] [Google Scholar]
- 59.Song, S., H. C. Pitot, and P. F. Lambert. 1999. The human papillomavirus type 16 E6 gene alone is sufficient to induce carcinomas in transgenic animals. J. Virol. 73:5887-5893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sterlinko Grm, H., M. Weber, R. Elston, P. McIntosh, H. Griffin, L. Banks, and J. Doorbar. 2004. Inhibition of E6-induced degradation of its cellular substrates by novel blocking peptides. J. Mol. Biol. 335:971-985. [DOI] [PubMed] [Google Scholar]
- 61.Stewart, D., S. Kazemi, S. Li, P. Massimi, L. Banks, A. E. Koromilas, and G. Matlashewski. 2004. Ubiquitination and proteasome degradation of the E6 proteins of human papillomavirus types 11 and 18. J. Gen. Virol. 85:1419-1426. [DOI] [PubMed] [Google Scholar]
- 62.Takizawa, S., K. Nagasaka, S. Nakagawa, T. Yano, K. Nakagawa, T. Yasugi, T. Takeuchi, T. Kanda, J. M. Huibregtse, T. Akiyama, and Y. Taketani. 2006. Human scribble, a novel tumor suppressor identified as a target of high-risk HPV E6 for ubiquitin-mediated degradation, interacts with adenomatous polyposis coli. Genes Cells 11:453-464. [DOI] [PubMed] [Google Scholar]
- 63.Thomas, M., R. Laura, K. Hepner, E. Guccione, C. Sawyers, L. Lasky, and L. Banks. 2002. Oncogenic human papillomavirus E6 proteins target the MAGI-2 and MAGI-3 proteins for degradation. Oncogene 21:5088-5096. [DOI] [PubMed] [Google Scholar]
- 64.Walboomers, J. M., M. V. Jacobs, M. M. Manos, F. X. Bosch, J. A. Kummer, K. V. Shah, P. J. Snijders, J. Peto, C. J. Meijer, and N. Munoz. 1999. Human papillomavirus is a necessary cause of invasive cervical cancer worldwide. J. Pathol. 189:12-19. [DOI] [PubMed] [Google Scholar]
- 65.Watson, R. A., T. P. Rollason, G. M. Reynolds, P. G. Murray, L. Banks, and S. Roberts. 2002. Changes in expression of the human homologue of the Drosophila discs large tumour suppressor protein in high-grade premalignant cervical neoplasias. Carcinogenesis 23:1791-1796. [DOI] [PubMed] [Google Scholar]
- 66.Watson, R. A., M. Thomas, L. Banks, and S. Roberts. 2003. Activity of the human papillomavirus E6 PDZ-binding motif correlates with an enhanced morphological transformation of immortalized human keratinocytes. J. Cell Sci. 116:4925-4934. [DOI] [PubMed] [Google Scholar]