Abstract
The major protein constituents of the filoviral envelope are the matrix protein VP40 and the surface transmembrane protein GP. While VP40 is recruited to the sites of budding via the late retrograde endosomal transport route, GP is suggested to be transported via the classical secretory pathway involving the endoplasmic reticulum, Golgi apparatus, and trans-Golgi network until it reaches the plasma membrane where most filoviral budding takes place. Since both transport routes target the plasma membrane, it was thought that GP and VP40 join there to form the viral envelope. However, it was recently shown that, upon coexpression of both proteins, GP is partially recruited into peripheral VP40-enriched multivesicular bodies, which contained markers of the late endosome. Accumulation of GP and VP40 in this compartment was presumed to play an important role in the formation of the filoviral envelope. Using a domain-swapping approach, we were able to show that the transmembrane domain of GP was essential and sufficient for (i) partial recruitment of chimeric glycoproteins into VP40-enriched multivesicular bodies and (ii) incorporation into virus-like particles (VLPs) that were released upon expression of VP40. Only those chimeric glycoproteins which were targeted to VP40-enriched endosomal multivesicular bodies were subsequently recruited into VLPs. These data show that the transmembrane domain of GP is critical for the mixing of VP40 and GP in multivesicular bodies and incorporation of GP into the viral envelope. Results further suggest that trapping of GP in the VP40-enriched late endosomal compartment is important for the formation of the viral envelope.
The assembly of viral envelopes represents a unique model to study functional interactions between membrane proteins, to monitor the sorting of proteins to specific sites in a target membrane, and to identify targeting signals in the respective proteins. Many targeting signals, interaction domains, and signals for incorporation into progeny virions have been identified in the cytoplasmic domain of viral transmembrane glycoproteins (5, 8, 9, 13, 17, 25, 28, 36, 41, 42). Likewise, the transmembrane domain of viral surface proteins can contain signals for their incorporation into the viral envelope (1), and in some cases, amino acid sequences that are important for efficient virus assembly are detectable in the ectodomain of viral transmembrane proteins (30).
Marburg virus (MARV), a filovirus, causes severe hemorrhagic fever in humans and nonhuman primates (23). During the latest and largest outbreak of MARV in Angola, the case fatality rate reached 85% (39, 40). Until now, neither a vaccine nor an effective treatment for the filovirus-induced hemorrhagic fever is available for human use.
The filamentous MARV particle is composed of seven structural proteins. Four proteins (NP, VP35, L, and VP30) constitute the nucleocapsid complex, which encapsidates the single-stranded negative-sense viral RNA genome (4). Two matrix proteins, the most abundant viral protein VP40 and VP24, are located between the nucleocapsid complex and the lipid envelope in the viral particle (2, 4). Homotrimers of the single surface protein GP, a type I transmembrane protein, are integrated into the viral envelope (3, 12, 32). During its transport along the secretory pathway to the cell surface, GP is subjected to glycosylation and phosphorylation at its ectodomain (12, 16, 31) and acylation at two cysteine residues at the boundary between the transmembrane and the cytoplasmic domain (14). GP is cleaved by the prohormone convertase furin in the trans-Golgi network into two disulfide-bond linked subunits, GP1 (170 kDa) and GP2 (46 kDa) (37).
The MARV matrix protein VP40 is transported to the cell surface by the retrograde late endosomal pathway (20). VP40 accumulates in multivesicular bodies (MVBs), a subcompartment of the late endosome, and is finally released from the cell surface by inducing the formation of filamentous virus-like particles (VLPs) that resemble progeny virions (21, 34). Since VP40 is necessary and sufficient to induce budding of VLPs, it is presumed to represent the key protein in the assembly and budding of viral particles.
The characteristic endoplasmic reticulum (ER)/Golgi localization pattern of GP changes in the presence of VP40. Upon coexpression, both proteins are detectable in peripheral clusters that also contain markers of the late endosome (21). Coexpression of GP and VP40 also results in an increased release of VLPs that contain both proteins (21). It has been presumed that, as VP40 and GP accumulate in MVBs, the late endosome serves as a platform for the assembly of the viral envelope and the budding of viral particles (21). Until now, the domain(s) of GP that is essential for both its efficient integration into the viral envelope and its accumulation in the late endosomal compartment has remained unidentified. In the present study, we analyzed the prerequisites for the accumulation of GP in VP40-positive MVBs and its incorporation into VP40-induced VLPs.
We show that the transmembrane domain of GP is essential and sufficient to mediate accumulation of GP in VP40-positive MVBs. Accumulation of GP in VP40-positive MVBs, in turn, is the prerequisite for its incorporation into VLPs.
MATERIALS AND METHODS
Cell lines.
HUH-7, a hepatoma cell line, and human embryonic kidney (HEK) 293 cells were cultured in Dulbecco's modified Eagle medium supplemented with 10% fetal calf serum, 2% l-glutamine, and a penicillin-streptomycin solution. The cells were cultivated in an incubator at 37°C under 5% CO2. HUH-7 cells were used for immunofluorescence (IF) analysis, while HEK 293 cells were used for the investigation of VLP release.
Molecular cloning.
The open reading frames of MARV VP40 and GP (MARV strain Musoke) (for reference, see EMBL Nucleotide Sequence Database accession number Z12132) were amplified by PCR and cloned into the mammalian expression vector pCAGGS using the restriction enzymes SmaI and NotI. The resultant plasmids contained sequences of MARV VP40 or GP under the control of a chicken β-actin promoter, as was verified by DNA sequencing. The open reading frame of the full-length Lassa virus glycoprotein (LASV GPC) (Lassa virus strain Josiah) was cloned into the β-actin promoter-driven pCAGGS vector via the restriction enzymes EcoRI and BglII (10). Using recombinant PCR, the chimeric glycoproteins, composed of different domain combinations of LASV GPC and MARV GP, were constructed (18) (see Fig. 2B). The resulting constructs were cloned into the pCAGGS vector using the restriction enzymes SmaI and SacI. Using pTM1-MARV-GP as a template, the mutant MARV-GPΔCD was generated and subcloned into the pCAGGS vector. In MARV-GPΔCD, the entire cytoplasmic domain starting from amino acid residue 674 (arginine) was removed by PCR. To this end, the template plasmid was amplified using primers flanking the cytoplasmic domain. Subsequently, the blunt ends of the amplicons were internally ligated. All constructs were verified by DNA sequencing. Primer sequences are available on request.
FIG. 2.
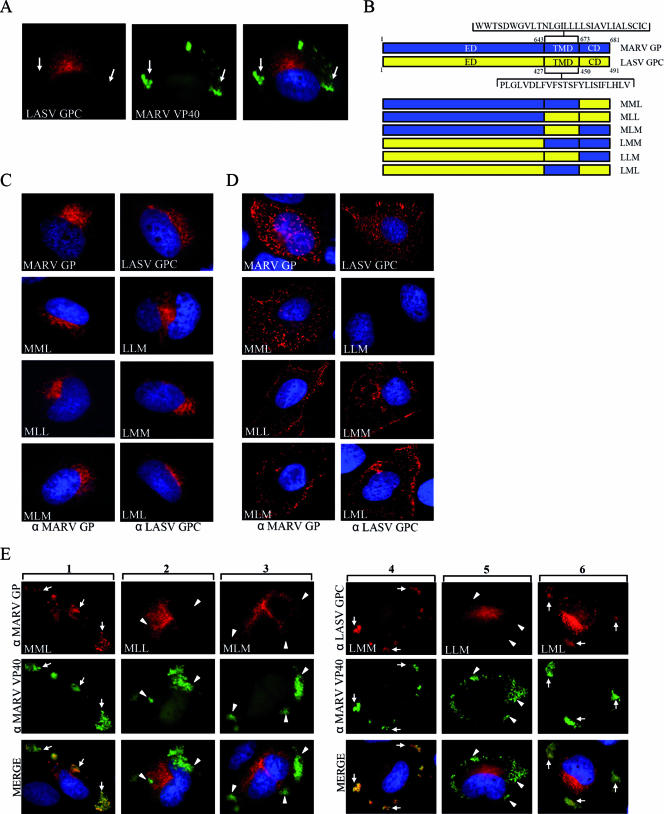
The transmembrane domain of MARV GP mediates accumulation of GP in MVBs upon coexpression with MARV VP40. HUH-7 cells were transfected with plasmids encoding VP40 and LASV GPC (A), chimeric glycoproteins (D), schematically presented in panel B, or chimeric glycoproteins and VP40 (E). Nuclei were counterstained with DAPI (4′,6′-diamidino-2-phenylindole). (A) HUH-7 cells were transfected with plasmids encoding LASV GPC and MARV VP40. At 24 h posttransfection, cells were fixed with 4% paraformaldehyde and immunostained with a rabbit anti-GPC and a monoclonal mouse anti-VP40 antibody. Bound antibodies were detected using secondary goat anti-rabbit IgG conjugated with rhodamine and goat anti-mouse IgG conjugated with FITC. The arrows show peripheral MARV VP40-positive clusters characterized as MVBs, which did not colocalize with LASV GPC. (B) Schematic presentation of MARV and LASV glycoproteins composed of the N-terminal ectodomain (ED), the hydrophobic membrane-spanning transmembrane domain (TMD) and the C-terminal cytoplasmic domain (CD). Chimeric proteins constructed by recombinant PCR are shown below and designated using a three letter code. The first letter of the construct's name indicates the origin of the ED (M, MARV; L, LASV), the second letter indicates the origin of the TMD, and the third letter indicates the origin of the CD. (C) Chimeric glycoproteins were expressed in HUH-7 cells and subjected to IF analysis as described above. Cells were immunostained with a rabbit anti-MARV GP or a rabbit anti-LASV GPC IgG depending on the origin of the ED. Bound antibodies were detected by a goat anti-rabbit antibody conjugated with rhodamine. (D) HUH-7 cells were transfected with plasmids encoding the chimeric glycoproteins and stained with goat anti-MARV IgG or rabbit anti-LASV GPC IgG at 4°C for 1 h. Cells were fixed but not permeabilized (note the difference from cells shown in panel C), and bound antibodies were detected using a donkey anti-goat IgG or a goat anti-rabbit IgG, both conjugated with rhodamine. (E) HUH-7 cells were cotransfected with plasmids encoding the chimeric glycoproteins and MARV VP40. At 24 h posttransfection, cells were fixed and immunostained with an anti-MARV VP40 monoclonal antibody and a rabbit anti-MARV GP (columns 1 to 3) or a rabbit anti-LASV GPC (columns 4 to 6) IgG. As secondary antibodies, a goat anti-mouse IgG conjugated with FITC and a goat anti-rabbit conjugated with rhodamine were used. Arrows indicate colocalization of VP40 and GP; arrowheads indicate VP40 located in peripheral MVBs showing no colocalization with expressed glycoproteins.
Antibodies.
For the identification of MARV VP40, a mouse monoclonal antibody (kindly provided by M. C. Georges-Courbot, Lyon, France) was used (dilution for IF, 1:50). For the detection of MARV GP, an affinity-purified rabbit serum (dilutions: IF, 1:100; immunoelectron microscopy [IEM], 1:100) and an anti-MARV goat serum were used (dilutions: IF, 1:100; Western blotting [WB], 1:2,500). The identification of LASV GPC was performed using a mouse monoclonal antibody (dilutions: WB, 1:200; IEM, 1:50; kindly provided by M.C. Georges-Courbot, Institut Pasteur, Lyon, France) or a rabbit serum (dilutions: IF, 1:100; WB, 1:2,000), which was raised by immunization of a rabbit with a chemically synthesized peptide that was homologous to the N terminus of GP2 (amino acid positions 259 to 279). Secondary antibodies conjugated to rhodamine or fluorescein isothiocyanate (FITC) (Dianova, Hamburg, Germany) were used for IF analysis (dilution, 1:200). Secondary antibodies conjugated to horseradish peroxidase (Dako, Copenhagen, Denmark) were used for WB (dilution 1:40,000). Secondary antibodies conjugated to 10-nm or 12-nm colloidal gold particles (Dianova, Hamburg, Germany or BB International, Cardiff, United Kingdom) were used for IEM (dilution, 1:30).
Transfection of cells.
HEK 293 cells were transfected using Lipofectamine 2000 reagent (Invitrogen, Karlsruhe, Germany). HUH-7 cells were transfected using Lipofectamine PLUS reagent (Invitrogen, Karlsruhe, Germany) or FUGENE 6 transfection reagent (Roche, Mannheim, Germany) according to the manufacturers' instructions. Transfected cells were incubated at 37°C under 5% CO2 for 24 or 48 h. HUH-7 and HEK 293 cells were cotransfected with plasmids encoding MARV VP40 and chimeric glycoproteins under the control of the β-actin promoter (pCAGGS). The optimal proportion of pCAGGS-MARV-VP40 and GP that resulted in an expression level similar to the one in MARV-infected cells was 4:1 (our unpublished data). This ratio was also used for coexpression of VP40 and GP mutants or chimeric glycoproteins.
Purification of vesicular and filamentous particles extracted from cellular supernatant.
At 48 h posttransfection, cellular supernatant from HEK 293 cells was harvested and pelleted in an SW41 rotor through a 20% sucrose cushion at 35,000 rpm for 3 h at 4°C. The pellet was resuspended in TNE buffer (10 mM Tris/HCl [pH 7.5], 150 mM NaCl, 1 mM EDTA [pH 8]), laid on a Nycodenz step gradient, and centrifuged in a SW60 rotor at 16,000 rpm for 15 min at 4°C. This Nycodenz gradient was composed of seven steps from 30% to 2.5% Nycodenz, increasing from the top to the bottom. Fractions (500 μl) were collected from the top, whereas fractions 1 to 3 (vesicular particles) and fractions 4 to 6 (filamentous particles) were pooled. To concentrate membranes and membrane-associated proteins, pooled fractions were centrifuged in a TLA45 rotor at 45,000 rpm for 2 h at 4°C. The resultant pellet was resuspended in 30 μl phosphate-buffered saline (PBS)/1% paraformaldehyde for IEM analysis.
Electrophoresis and immunoblot analysis.
WB analysis was carried out as previously described (21). The antibodies are listed in the figure legends.
Indirect IF analysis.
At 24 h posttransfection, HUH-7 cells were washed with PBS and fixed with 4% paraformaldehyde in Dulbecco's modified Eagle medium for 30 min. The fixative was removed, and free aldehydes were quenched with 100 mM glycine in PBS. Afterwards, the samples were washed once with PBS and permeabilized with PBS containing 0.1% Triton X-100. Cells were incubated in blocking solution (2% bovine serum albumin, 0.2% Tween 20, 5% glycerol, and 0.05% sodium azide in PBS) and stained with primary and secondary antibodies as indicated below (see figure legends). Microscopic analysis was performed with an Axiomat fluorescence microscope (Zeiss).
In the case of native IF analysis, at 24 h posttransfection, HUH-7 cells were washed with cold PBS and incubated with the primary antibody at 4°C for 1 h. The cells were rinsed twice with PBS, fixed in 4% paraformaldehyde for 10 min at room temperature, and incubated in PBS containing 100 mM glycine for 10 min. After the incubation in blocking solution, cells were stained with secondary antibodies as indicated below (see figure legends).
IEM analysis.
For microscopic analysis, we used the fractions of the Nycodenz gradient containing the filamentous membranes. A drop of this suspension was deposited on Formvar-carbon-coated nickel grids for 1 h. The excess fluid was blotted away with Whatman filter paper, and the grids were incubated with blocking buffer (see “Indirect IF analysis” above) for 10 min. Indirect immunostaining was performed by incubating the grids with the primary antibodies (see figure legends) for 60 min. After washing the grids six times for 2 min in PBS, bound immunoglobulins (IgG) were detected with a secondary antibody coupled to colloidal gold particles. After fixation with 0.25% glutaraldehyde in 1× PBS, samples were negatively stained with 2% phosphotungstic acid solution and examined with a Zeiss 109 electron microscope.
RESULTS AND DISCUSSION
Intracellular accumulation of GP in VP40-enriched MVBs does not require the cytoplasmic domain of GP.
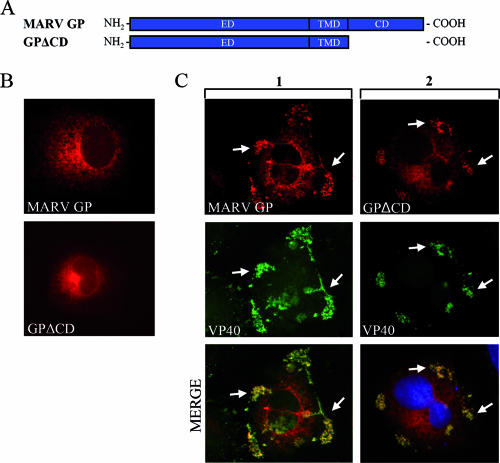
For many viruses, it is suggested that the spike glycoproteins interact with matrix proteins via their cytoplasmic tails (9, 13, 17, 41). Therefore, the role of the cytoplasmic domain in the accumulation of GP in the VP40-positive MVBs was evaluated. To this end, a mutant of GP was constructed in which the cytoplasmic domain was completely removed (GPΔCD) (Fig. 1A). IF analysis revealed that both wild-type GP and GPΔCD were predominantly concentrated in the perinuclear region, which is consistent with their accumulation in the ER or Golgi apparatus (3, 21) (Fig. 1B). Coexpression with VP40 resulted in partial redistribution of wild-type GP and GPΔCD to peripheral VP40-positive clusters, indicating that accumulation of GP in the VP40-enriched MVBs is independent of its cytoplasmic tail (Fig. 1C).
FIG. 1.
The cytoplasmic domain of MARV GP does not influence its accumulation in MVBs. (A) Schematic diagram of MARV GP with deletion of the cytoplasmic tail. ED, ectodomain; TMD, transmembrane domain; CD, cytoplasmic domain. (B) HUH-7 cells were transfected with MARV GP or GPΔCD expression plasmids under the control of a beta-actin promoter. At 24 h posttransfection, cells were fixed with 4% paraformaldehyde and stained with a rabbit anti-GP IgG and a donkey anti-rabbit IgG conjugated with rhodamine. (C) HUH-7 cells were transfected with plasmids encoding VP40 and GP or VP40 and GPΔCD. Cells were fixed at 24 h posttransfection and immunostained with a rabbit anti-GP and a mouse anti-VP40 antibody. Bound antibodies were detected using secondary donkey anti-rabbit IgG coupled with rhodamine and a donkey anti-mouse IgG conjugated with FITC. The arrows indicate colocalization of MARV VP40 and MARV wild-type GP or GPΔCD.
The transmembrane domain of GP mediates its accumulation in VP40-positive MVBs.
To further elucidate which domain(s) of MARV GP may be essential for accumulation in VP40-enriched MVBs, we mutually exchanged the ectodomain, the transmembrane domain, or cytoplasmic domain of GP with the respective domains of the LASV GPC. Schematic diagrams of the constructed mutants are shown in Fig. 2B. LASV GPC was selected as a reporter protein because it shows no accumulation in VP40-enriched MVBs (Fig. 2A). LASV GPC, like MARV GP, is a type I transmembrane protein which is synthesized as a precursor and cleaved into two subunits, GP1 and GP2, by the cellular protease SKI-1/S1P in the ER or an early Golgi compartment (11, 22).
Expression of chimeric and wild-type glycoproteins was performed in HUH-7 cells, and their respective intracellular distributions were analyzed by IF microscopy. All constructed mutants showed predominantly a perinuclear staining pattern that was similar to that of the wild-type LASV GPC and MARV GP (Fig. 2C). Staining of the cell surface for the presence of the chimeric proteins showed that, with the exception of LLM, all chimeric glycoproteins were able to reach the plasma membrane (Fig. 2D). Since LLM was also the only construct which was not cleaved (Fig. 3B, right panel), it is presumed that severe misfolding prevented its exit from the ER.
FIG. 3.
MARV GP transmembrane domain mediates incorporation of GP in filamentous VP40-induced VLPs. HEK 293 cells were transfected with plasmids encoding VP40 and MARV GP, MARV GPΔCD, LASV GPC, or chimeric glycoproteins as indicated next to the figures. At 48 h posttransfection, particulate material of cellular supernatant was harvested and pelleted through a 20% sucrose cushion following a separation of vesicular and filamentous particles by centrifugation through a Nycodenz gradient. The filamentous particles were subjected to IEM analysis. Cells were lysed and subjected to WB analysis. (A, C, E, left panel) Chimeric and wild-type glycoproteins were detected by rabbit or mouse anti-LASV GPC or a rabbit anti-MARV GP IgG and a goat anti-rabbit or anti-mouse IgG coupled with colloidal gold (10- or 12-nm-diameter gold beads). Bar, 100 nm. (D, E, right panel) To evaluate the incorporation of GP into VLPs, the density of gold labeling was quantified (counted VLPs: n ≥ 5). The data represent the mean values and standard deviations. Asterisks indicate statistically significant differences (*, P < 0.05; **, P < 0.01; ***, P < 0.001) in comparison to MARV GP/VP40 (D, left panel) or LMM/VP40 (D, right panel). (B, E, middle panel) Lysed cells were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis and WB analyses. Proteins were detected using a mouse anti-LASV GPC or a rabbit/goat anti-MARV GP IgG followed by incubation with anti-goat, anti-mouse, or anti-rabbit-HRP antibody.
Subsequently, the chimeric glycoproteins were, respectively, coexpressed with MARV VP40 and subjected to IF analysis. Among the chimeric glycoproteins that contained the ectodomain of MARV GP, only MML colocalized with the VP40-enriched large peripheral MVBs (Fig. 2E, column 1). When the MARV GP transmembrane domain alone (MLM) or in combination with the cytoplasmic domain (MLL) was substituted, resulting mutants showed a perinuclear staining pattern and no accumulation in MVBs (Fig. 2E, columns 2 and 3). These results suggested that the transmembrane domain of MARV GP was required for the accumulation of GP in the VP40-positive MVBs.
Coexpression of wild-type LASV GPC with VP40 did not induce colocalization of GPC in peripheral VP40-enriched MVBs (Fig. 2A). In contrast, the presence of the transmembrane domain of MARV GP in the background of LASV GPC (LML) resulted in accumulation of the chimeric protein in VP40-enriched MVBs. Likewise, combined transfer of the transmembrane and cytoplasmic domain of MARV GP into LASV GPC (LMM) led to accumulation of the resulting chimeric protein in MVBs (Fig. 2E, columns 4 and 6). In contrast, the mutant LLM that contained only the cytoplasmic domain of MARV GP localized in the perinuclear region, and colocalization with VP40 in MVBs was not observed (Fig. 2E, column 5). This is consistent with a block in transport that was also observed upon expression of the chimeric protein alone (Fig. 2D, right panel). These results indicated that the transmembrane domain of MARV GP is necessary and sufficient to mediate accumulation in VP40-positive MVBs. Moreover, these results confirm that the cytoplasmic domain is not essential for accumulation of GP in VP40-positive MVBs (Fig. 1).
The mechanism whereby the transmembrane domain mediates accumulation of MARV GP in MVBs remains unknown. Amino acids with polar side chains that have been associated with endosomal targeting (29) are present in the membrane-spanning domain of both MARV and LASV glycoproteins and therefore cannot explain why the transmembrane domain of MARV GP mediated recruitment in MVBs and the transmembrane domain of LASV GPC did not. The hydrophobicity values of the two transmembrane domains are also similar. Using the program ProtParam (15), we determined the grand average of the hydropathicity index of the two transmembrane domains and found values of 1.771 for LASV GPC and 1.703 for the transmembrane domain of MARV GP. In contrast, the length of both transmembrane domains (30 versus 24 amino acids) is different and may account for the different phenotype of chimeric proteins containing the LASV GPC or MARV GP transmembrane domain. For several membrane-anchored glycoproteins, the transmembrane domain has been reported to determine their intracellular localization (6, 26). Whether this holds true also for the transmembrane domains of MARV GP and LASV GPC needs to be investigated.
Incorporation of MARV GP in VLPs is dependent on its transmembrane domain.
Colocalization of VP40 and GP in MVBs was presumed to be important for formation of the MARV envelope (21). Therefore, we were interested in determining whether chimeric glycoproteins that did not accumulate in MVBs upon coexpression with VP40 could be rescued into VP40-induced VLPs. To this end, the constructed chimeric glycoproteins were coexpressed with VP40, and released filamentous VLPs were tested for the presence of the respective glycoprotein. First, to investigate whether LASV GPC could be incorporated into VP40-induced VLPs, VP40 and LASV GPC were coexpressed in HEK 293 cells, and released VLPs were centrifuged through a sucrose cushion and subsequently separated by centrifugation through a Nycodenz step gradient at 48 h posttransfection. Transfected cells were lysed, and the resultant protein lysates were subjected to immunoblot analysis. Final pellets from centrifugation were analyzed by IEM (Fig. 3A). HEK 293 cells were used for these experiments, since VLPs arereleased from these cells with a higher efficiency than from HUH-7 cells. As shown in Fig. 3B, lane 7, LASV GPC was detected intracellularly in its cleaved form but was not integrated into released filamentous VLPs (Fig. 3A). Next, MARV VP40 was coexpressed with the constructed chimeric glycoproteins (Fig. 2B), and released VLPs were analyzed as described above. Incorporation of chimeric glycoproteins at the surface of VLPs was evaluated by the appearance of gold particles representing antibodies bound to the respective chimera. Quantification of GP determined by the number of gold particles decorating secondary antibodies showed that, while MARV GP was readily incorporated into VLPs (68.6 ± 11.5 gold particles per μm) (Fig. 3A, left panel, and D), replacement of the transmembrane domain with that of LASV GPC (MLM) resulted in a dramatic loss of incorporation into VLPs (5.1 ± 2.2 gold particles per μm) (Fig. 3C, panel 3, and D). Likewise, substitution of both the transmembrane and cytoplasmic domains with the corresponding domains from LASV GPC (MLL) completely blocked glycoprotein incorporation into VLPs (1.9 ± 2 gold particles per μm) (Fig. 3C, panel 2, and D). In contrast, chimera MML was incorporated into VLPs, although to a lesser extent than wild-type GP (41 ± 11.6 gold particles per μm) (Fig. 3C, panel 1, and D). These observations suggested that the transmembrane domain of MARV GP is essential for incorporation into VP40-induced VLPs. This view was supported by analysis of chimeric proteins LML and LMM. In contrast to wild-type LASV GPC (Fig. 3A, right panel), LML and LMM were both incorporated into VLPs (Fig. 3C, panel 5 and 6). While the incorporation of LML was 20.2 ± 4.1 gold particles per μm, the presence of the MARV cytoplasmic domain clearly increased the incorporation efficiency to 46.3 ± 14.3 gold particles per μm. Therefore, we concluded that the MARV GP transmembrane domain was necessary and sufficient to mediate incorporation into VLPs. Nevertheless, incorporation efficiency could be enhanced in the presence of the cytoplasmic domain. Chimeric glycoproteins that did not accumulate in VP40-enriched MVBs were not incorporated into VLPs, although they were expressed at the same level as wild-type MARV GP (Fig. 3B, left panel) and were able to reach the plasma membrane (Fig. 2D) (MLL and MLM). Furthermore, all chimeric glycoproteins that were able to accumulate in VP40-enriched MVBs could be incorporated into VLPs (MML, LMM, and LML).
Finally, the tailless mutant of MARV GP, GPΔCD, was tested for its ability to incorporate into VLPs. GPΔCD integrated into VLPs as efficiently as wild-type GP (Fig. 3E). This showed that the cytoplasmic domain is not required for the accumulation of GP in VP40-enriched MVBs nor for its incorporation into VLPs.
Although in the order Mononegavirales efficient incorporation of transmembrane glycoproteins into progeny viral particles is often mediated by the glycoprotein's cytoplasmic domain (7, 19, 24, 25, 33, 41), our results are not unprecedented. They are in line with data from Naim (27) showing that the specific sequence of the influenza virus HA transmembrane domain was essential for its incorporation into virions and the cytoplasmic tail only modulated the incorporation efficiency.
In summary, our data provide evidence for a critical role of the transmembrane domain of MARV GP in both its accumulation in peripheral VP40-enriched MVBs and in the incorporation of GP into VP40-induced VLPs. Additionally, our study suggests that trapping of GP in the VP40-enriched late endosomal compartment is critical for the formation of the viral envelope. Since VLPs derived from filoviral VP40 have been shown to be very efficient in raising the immune response in experimental animals (35, 38), our findings are also of importance for the development of VP40-based virosomes that could be used as vaccine candidates.
Acknowledgments
We thank Angelika Lander for expert technical support and Beate Berghoefer for providing the plasmid pCAGGS-MARV-GPΔCD.
This work was supported by the Deutsche Forschungsgemeinschaft, Sonderforschungsbereich 535, TPA13, and Schwerpunktprogramm SPP 1175/BE 1325/5-1.
Footnotes
Published ahead of print on 31 January 2007.
REFERENCES
- 1.Ali, A., and D. P. Nayak. 2000. Assembly of Sendai virus: M protein interacts with F and HN proteins and with the cytoplasmic tail and transmembrane domain of F protein. Virology 276:289-303. [DOI] [PubMed] [Google Scholar]
- 2.Bamberg, S., L. Kolesnikova, P. Moller, H. D. Klenk, and S. Becker. 2005. VP24 of Marburg virus influences formation of infectious particles. J. Virol. 79:13421-13433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Becker, S., H. D. Klenk, and E. Mühlberger. 1996. Intracellular transport and processing of the Marburg virus surface protein in vertebrate and insect cells. Virology 225:145-155. [DOI] [PubMed] [Google Scholar]
- 4.Becker, S., C. Rinne, U. Hofsass, H. D. Klenk, and E. Mühlberger. 1998. Interactions of Marburg virus nucleocapsid proteins. Virology 249:406-417. [DOI] [PubMed] [Google Scholar]
- 5.Bilsel, P., M. R. Castrucci, and Y. Kawaoka. 1993. Mutations in the cytoplasmic tail of influenza A virus neuraminidase affect incorporation into virions. J. Virol. 67:6762-6767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bulbarelli, A., T. Sprocati, M. Barberi, E. Pedrazzini, and N. Borgese. 2002. Trafficking of tail-anchored proteins: transport from the endoplasmic reticulum to the plasma membrane and sorting between surface domains in polarised epithelial cells. J. Cell Sci. 115:1689-1702. [DOI] [PubMed] [Google Scholar]
- 7.Cathomen, T., H. Y. Naim, and R. Cattaneo. 1998. Measles viruses with altered envelope protein cytoplasmic tails gain cell fusion competence. J. Virol. 72:1224-1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cosson, P. 1996. Direct interaction between the envelope and matrix proteins of HIV-1. EMBO J. 15:5783-5788. [PMC free article] [PubMed] [Google Scholar]
- 9.Dubay, J. W., S. J. Roberts, B. H. Hahn, and E. Hunter. 1992. Truncation of the human immunodeficiency virus type 1 transmembrane glycoprotein cytoplasmic domain blocks virus infectivity. J. Virol. 66:6616-6625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eichler, R., O. Lenz, T. Strecker, and W. Garten. 2003. Signal peptide of Lassa virus glycoprotein GP-C exhibits an unusual length. FEBS Lett. 538:203-206. [DOI] [PubMed] [Google Scholar]
- 11.Eschli, B., K. Quirin, A. Wepf, J. Weber, R. Zinkernagel, and H. Hengartner. 2006. Identification of an N-terminal trimeric coiled-coil core within arenavirus glycoprotein 2 permits assignment to class I viral fusion proteins. J. Virol. 80:5897-5907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Feldmann, H., C. Will, M. Schikore, W. Slenczka, and H. D. Klenk. 1991. Glycosylation and oligomerization of the spike protein of Marburg virus. Virology 182:353-356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Freed, E. O., and M. A. Martin. 1996. Domains of the human immunodeficiency virus type 1 matrix and gp41 cytoplasmic tail required for envelope incorporation into virions. J. Virol. 70:341-351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Funke, C., S. Becker, H. Dartsch, H. D. Klenk, and E. Muhlberger. 1995. Acylation of the Marburg virus glycoprotein. Virology 208:289-297. [DOI] [PubMed] [Google Scholar]
- 15.Gasteiger, E., C. Hoogland, A. Gattiker, S. Duvaud, M. R. Wilkins, R. D. Appel, and A. Bairoch. 2005. Protein identification and analysis tools on the ExPASy server, p. 571-607. In J. M. Walker (ed.), The proteomics protocols handbook. Humana Press, Totowa, NJ.
- 16.Geyer, H., C. Will, H. Feldmann, H. D. Klenk, and R. Geyer. 1992. Carbohydrate structure of Marburg virus glycoprotein. Glycobiology 2:299-312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gray, K. D., and M. J. Roth. 1993. Mutational analysis of the envelope gene of Moloney murine leukemia virus. J. Virol. 67:3489-3496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Higuchi, R., B. Krummel, and R. K. Saiki. 1988. A general method of in vitro preparation and specific mutagenesis of DNA fragments: study of protein and DNA interactions. Nucleic Acids Res. 16:7351-7367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jin, H., G. P. Leser, J. Zhang, and R. A. Lamb. 1997. Influenza virus hemagglutinin and neuraminidase cytoplasmic tails control particle shape. EMBO J. 16:1236-1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kolesnikova, L., S. Bamberg, B. Berghofer, and S. Becker. 2004. The matrix protein of Marburg virus is transported to the plasma membrane along cellular membranes: exploiting the retrograde late endosomal pathway. J. Virol. 78:2382-2393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kolesnikova, L., B. Berghofer, S. Bamberg, and S. Becker. 2004. Multivesicular bodies as a platform for formation of the Marburg virus envelope. J. Virol. 78:12277-12287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lenz, O., J. ter Meulen, H. D. Klenk, N. G. Seidah, and W. Garten. 2001. The Lassa virus glycoprotein precursor GP-C is proteolytically processed by subtilase SKI-1/S1P. Proc. Natl. Acad. Sci. USA 98:12701-12705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Martini, G. A., H. G. Knauff, H. A. Schmidt, G. Mayer, and G. Baltzer. 1968. On the hitherto unknown, in monkeys originating infectious disease: Marburg virus disease. Dtsch. Med. Wochenschr. 93:559-571. [DOI] [PubMed] [Google Scholar]
- 24.Mebatsion, T., F. Weiland, and K. K. Conzelmann. 1999. Matrix protein of rabies virus is responsible for the assembly and budding of bullet-shaped particles and interacts with the transmembrane spike glycoprotein G. J. Virol. 73:242-250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mitnaul, L. J., M. R. Castrucci, K. G. Murti, and Y. Kawaoka. 1996. The cytoplasmic tail of influenza A virus neuraminidase (NA) affects NA incorporation into virions, virion morphology, and virulence in mice but is not essential for virus replication. J. Virol. 70:873-879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Munro, S. 1995. An investigation of the role of transmembrane domains in Golgi protein retention. EMBO J. 14:4695-4704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Naim, R. 1993. Basis for selective incorporation of glycoproteins into the influenza virus envelope. J. Virol. 67:4831-4841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Owen, K. E., and R. J. Kuhn. 1997. Alphavirus budding is dependent on the interaction between the nucleocapsid and hydrophobic amino acids on the cytoplasmic domain of the E2 envelope glycoprotein. Virology 230:187-196. [DOI] [PubMed] [Google Scholar]
- 29.Reggiori, F., and H. R. Pelham. 2002. A transmembrane ubiquitin ligase required to sort membrane proteins into multivesicular bodies. Nat. Cell Biol. 4:117-123. [DOI] [PubMed] [Google Scholar]
- 30.Robison, C. S., and M. A. Whitt. 2000. The membrane-proximal stem region of vesicular stomatitis virus G protein confers efficient virus assembly. J. Virol. 74:2239-2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sänger, C., E. Mühlberger, B. Lötfering, H.-D. Klenk, and S. Becker. 2002. The Marburg virus surface protein is phosphorylated at its ectodomain. Virology 295:20-29. [DOI] [PubMed] [Google Scholar]
- 32.Sanger, C., E. Muhlberger, E. Ryabchikova, L. Kolesnikova, H. D. Klenk, and S. Becker. 2001. Sorting of Marburg virus surface protein and virus release take place at opposite surfaces of infected polarized epithelial cells. J. Virol. 75:1274-1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schnell, M. J., L. Buonocore, E. Boritz, H. P. Ghosh, R. Chernish, and J. K. Rose. 1998. Requirement for a non-specific glycoprotein cytoplasmic domain sequence to drive efficient budding of vesicular stomatitis virus. EMBO J. 17:1289-1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Swenson, D. L., K. L. Warfield, K. Kuehl, T. Larsen, M. C. Hevey, A. Schmaljohn, S. Bavari, and M. J. Aman. 2004. Generation of Marburg virus-like particles by co-expression of glycoprotein and matrix protein. FEMS Immunol. Med. Microbiol. 40:27-31. [DOI] [PubMed] [Google Scholar]
- 35.Swenson, D. L., K. L. Warfield, D. L. Negley, A. Schmaljohn, M. J. Aman, and S. Bavari. 2005. Virus-like particles exhibit potential as a pan-filovirus vaccine for both Ebola and Marburg viral infections. Vaccine 23:3033-3042. [DOI] [PubMed] [Google Scholar]
- 36.Takimoto, T., T. Bousse, E. C. Coronel, R. A. Scroggs, and A. Portner. 1998. Cytoplasmic domain of Sendai virus HN protein contains a specific sequence required for its incorporation into virions. J. Virol. 72:9747-9754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Volchkov, V. E., V. A. Volchkova, U. Stroher, S. Becker, O. Dolnik, M. Cieplik, W. Garten, H. D. Klenk, and H. Feldmann. 2000. Proteolytic processing of Marburg virus glycoprotein. Virology 268:1-6. [DOI] [PubMed] [Google Scholar]
- 38.Warfield, K. L., D. L. Swenson, D. L. Negley, A. L. Schmaljohn, M. J. Aman, and S. Bavari. 2004. Marburg virus-like particles protect guinea pigs from lethal Marburg virus infection. Vaccine 22:3495-3502. [DOI] [PubMed] [Google Scholar]
- 39.World Health Organization. 2005. Marburg haemorrhagic fever in Angola-update 26: MOH declares outbreak over. World Health Organization, Geneva, Switzerland. http://www.who.int/csr/don/2005_11_07a/en/.
- 40.World Health Organization. 2005. Marburg haemorrhagic fever, Angola. Wkly. Epidemiol. Rec. 80:158-159. [PubMed] [Google Scholar]
- 41.Yu, X., X. Yuan, M. F. McLane, T. H. Lee, and M. Essex. 1993. Mutations in the cytoplasmic domain of human immunodeficiency virus type 1 transmembrane protein impair the incorporation of Env proteins into mature virions. J. Virol. 67:213-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhao, H., B. Lindqvist, H. Garoff, C. H. von Bonsdorff, and P. Liljestrom. 1994. A tyrosine-based motif in the cytoplasmic domain of the alphavirus envelope protein is essential for budding. EMBO J. 13:4204-4211. [DOI] [PMC free article] [PubMed] [Google Scholar]