Abstract
Human papillomavirus type 16 (HPV-16) has developed numerous ways to modulate host-initiated immune mechanisms. The HPV-16 E6 oncoprotein, for example, can modulate the cellular level, and consequently the activity, of procaspase 8, thus modifying the cellular response to cytokines of the tumor necrosis factor family. E6 from HPV-16, but not E6 from the low-risk types 6b and 11, alters the cellular level of procaspase 8 in a dose-dependent manner. Both the large and small (E6*) isoforms of E6, which originate by way of alternate splicing, can modulate procaspase 8 stability. Intriguingly, although both isoforms bind to procaspase 8, the large isoform accelerates the degradation of procaspase 8 while the small isoform stabilizes it. Binding leads to a change in the ability of procaspase 8 to bind either to itself or to FADD (Fas-associated death domain), with the large version of E6 able to inhibit this binding while the small isoform does not. Consistent with this model, knockdown of the large version of E6 by small interfering RNA leads to increases in the levels of procaspase 8 and its binding to both itself and FADD. Thus, these alternatively spliced isoforms can modulate both the level and the activity of procaspase 8 in opposite directions.
High-risk types of human papillomaviruses (HPV) are causative agents in most cases of cervical carcinoma and are frequently implicated in head and neck cancers, as well. Much of the transforming ability of this virus can be attributed to the activities of its E6 and E7 oncoproteins. The HPV type 16 (HPV-16) E6 oncogene is an early gene expressed during HPV infection that plays an important role in cellular immortalization and transformation. The best-known activity of E6 is its ability to accelerate the degradation of p53 (50); however, not all of its transforming ability can be attributed to this activity (42, 46, 54, 67), and E6 is known to influence additional cellular functions, such as the regulation of transcription and DNA replication (18, 23, 28, 29, 30, 36, 43, 49, 68, 70), epithelial organization and differentiation (6, 9, 11, 64), cell-cell adhesion, polarity and proliferation control (20, 27, 32, 33, 41, 62, 63), the DNA damage response (26, 55), and apoptotic pathways (15, 16, 21, 61, 66, 67).
Activation of the extrinsic apoptotic pathway begins with the binding of a signaling molecule, such as tumor necrosis factor (TNF), FasL, or TRAIL (TNF-related apoptosis inducing ligand), to its receptor. The resulting conformational change in the receptor then allows it to bind to additional adaptor and effector molecules (such as TRADD in the case of TNF and FADD [Fas-associated death domain] for TNF, FasL, and TRAIL) and ultimately results in the binding to and activation of the initiator caspase procaspase 8 (reviewed in references 47, 70, and 71). Activated caspase 8 can then activate caspase 3, which initiates the dissolution of a number of cellular proteins and structures and ultimately leads to apoptosis. Our previous work has shown that the HPV-16 E6 oncoprotein interacts with the extrinsic apoptotic pathway by binding to and altering the functions of upstream signaling molecules. For example, it binds to the death domain of TNF receptor 1 (TNF R1) and thus blocks transmission of the apoptotic signal between TNF R1 and TRADD (16), though it does not appear to accelerate the degradation of the receptor (13). Interestingly, it also binds to FADD, but in contrast to the case with TNF R1, it binds to the death effector domain (DED) rather than to the death domain and does accelerate the degradation of FADD (15). Another intriguing difference between the TNF and Fas-triggered pathways is that while E6 protects cells from apoptosis triggered by Fas in a dose-dependent and monotonic manner (15), we found that while low doses of E6 do indeed protect cells from TNF-mediated apoptosis, high doses sensitize them, indicating additional complexity (13). E6 does not, however, bind with significant affinity to the death domain of TRADD or to Fas (unpublished results), demonstrating the specificity of these interactions. These results led us to wonder whether E6 might interact with procaspase 8, which contains a DED similar to that of FADD.
Procaspase 8 is a key player common to all of the three receptor-mediated pathways mentioned above, those triggered by TNF, FasL, and TRAIL. This makes it an attractive target for viral strategies of immune evasion, and a number of viral proteins have been reported to inhibit apoptosis by inhibiting caspase 8 activation either directly or at steps upstream (reviewed in references 3, 24, and 65). These include the human cytomegalovirus (CMV) UL36 gene product (53), the hepatitis C nonstructural protein 5A (40), the cowpox serpin CrmA (38, 39, 58, 59), the R1 subunit of herpes simplex virus ribonucleotide reductase (31), the herpes simplex virus type 1 latency-associated transcript (26), the human herpesvirus 8 viral FLICE-inhibitory protein (2), the adenovirus 14.7-kDa protein (8, 34), and baculovirus protein p35 (1). We have previously shown that in HCT-116 cells, expression of E6 leads to reduced cellular levels of procaspase 8 (19); however, the mechanism underlying this observation has not yet been described.
Two isoforms of E6 are produced due to alternate splicing: a large version of about 16 kDa (E6large) and a small isoform of approximately half that size (E6small, or E6*). Our published (15) and unpublished results, as well as those of others (17, 22, 45), suggest that the two isoforms might have different biological activities, and we wondered if these different biological activities might contribute to the paradoxical abilities of E6 both to protect cells from and to sensitize cells to TNF. Therefore, to increase our understanding of the role E6 plays in the modulation of apoptotic pathways and to define the relative roles of these two isoforms, we examined the interactions of both versions of E6 with procaspase 8. We found that while both isoforms of E6 can bind to procaspase 8, the small isoform stabilizes procaspase 8 while the large isoform can accelerate its degradation. These results emphasize the complexity of virus-host interactions and also impact the development of molecular models for the interactions of these proteins. As such, they have relevance to the development of novel and effective approaches to the treatment of cervical cancer.
MATERIALS AND METHODS
Reagents.
The following reagents were used in these studies: lyophilized human recombinant TNF-α (R&D Systems, Minneapolis, MN), Fas monoclonal antibodies (clone CH-11; Medical and Biological Laboratories Co., Ltd., Nagoya, Japan), 3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide (MTT) (Sigma, St. Louis, MO), cycloheximide (Sigma), mitomycin C (Sigma), MG132 (Calbiochem, San Diego, CA), and doxycycline (Clontech, Palo Alto, CA).
Cell culture.
U2OS cells, derived from a human osteosarcoma, were obtained from the ATCC (Manassas, VA) and were cultured in McCoy's 5A medium (Invitrogen) supplemented to contain 10% fetal bovine serum (Invitrogen), penicillin (100 U/ml), and streptomycin (100 μg/ml) (Sigma). Production of the stably transfected U2OSE6AS, U2OSE66, U2OSE64A, and U2OSE612 cell lines has been described previously (15), and the U2OSE6L12, U2OSE6S9, U2OSE66siCon, U2OSE66siE6L9, and U2OSE66siE6L10 lines were produced in a similar manner, as were cell lines expressing the HPV-6b and HPV-11 versions of E6. The cell lines U2OSE6tetC (control) and U2OSE6tet24 (E6 expressing) were produced and described previously (13), and the cell lines U2OSE6Ltet12 and U2OSE6Stet37, stably transfected with plasmids that code for the large and small isoforms of E6, respectively, were produced in a similar manner.
Plasmids.
The pHA-E6 S and pHA-E6 AS plasmids were described previously (16) and contain either the sense or the antisense version, respectively, of hemagglutinin epitope-tagged E6 (HA-E6). pHA-E6large produces a transcript (approximately 480 bp) that cannot be spliced to yield the short processed message. The construct was produced using pHA-E6 S as a template and then mutagenizing the donor splice site from AGGT to AGAT (51) using the Quick Change Mutagenesis Kit (Stratagene). In contrast, pHA-E6small/E6* produces only the short transcript and thus codes only for the small version of HA-E6. To produce this plasmid, the reverse transcriptase (RT)-PCR product of 0.3 kb was cloned into pCR2.1 (Invitrogen) and then subcloned into the pE-CMV-1 vector (16). To produce the Flag-tagged versions of the E6large and E6small/E6* proteins from HPV-16, -6b, and -11, as well as the Flag-tagged negative control Flag-LOC5129, the respective E6 genes, as well as LOC5129, were cloned into pFlag-myc CMV-22 (Sigma). pTRE-HA-E6 was obtained by cloning the HindIII bluntend-BamHI fragment from pHA-E6 S into the EcoRI blunt end-BamHI sites of the pTRE2 vector (Clontech).
The sequence for the caspase 8 DED was obtained from cDNA prepared from U2OS cells by PCR amplification using primers 5′-GACTTCAGCAGAAATCTTTATGATATTGGGGAAC-3′ and 5′-GAGATTGTCATTACCCCACACA-3′ and served as the basis for all our additional caspase 8 DED-expressing constructs. The sequence coding for FADD was obtained as described previously (15). The pAD-caspase 8 ded, pBD-caspase 8 ded, pAD-FADD, pBD-FADD, pAD-E6large, pAD-E6small/E6*, pBD-E6large, and pBD-E6small/E6* plasmids needed to perform the mammalian two-hybrid analysis (Stratagene) were obtained by subcloning appropriate fragments in frame with activation and binding domains of the pAD and pBD plasmids. Positive and negative control plasmids for this system were provided in the kit (Stratagene). To express the caspase 8 DED in the Escherichia coli system, in mammalian cells, or in the in vitro transcription/translation system (T7 reticulocyte; Promega), the procaspase 8 DED was cloned in frame with the six-His epitope of pTriEx-4 (Novagen) to produce the plasmid pHis-caspase 8 DED.
To express the glutathione S-transferase (GST)-tagged version of the large (E6large) and small (E6small/E6*) isoforms in E. coli, we cloned E6large and E6small/E6* into pGEX-2T (Amersham/Pharmacia Biotech). In order to inhibit expression of E6large, the small interfering RNA (siRNA) expression vector Silencer 3.1 (Ambion) was used following insertion of the siRNA target (pSilencer 3.1siE6large). This target was specifically designed to inhibit the E6large transcript and was designed using the Ambion design guide and cloned according to the manufacturer's protocol. The scrambled sequence of E6large was also cloned into pSilencer 3.1 and used as a negative control (pSilencer 3.1siCon).
Transfections.
Transfections were carried out using Fugene VI (Roche Molecular Biochemicals) as directed by the manufacturer. For transient transfections, cells were analyzed 40 to 48 h posttransfection. For stable transfections, clones were passaged in selection medium containing G418 (500 μg/ml), hygromycin (100 μg/ml), and puromycin (5 μg/ml). Following 2 to 3 weeks of selection, individual clones were grown and analyzed for protein expression by immunoblotting and/or RT-PCR.
Immunoblotting.
Cells (2.5 × 105 to 1 × 106) were lysed in 50 to 100 μl lysis buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1% Triton X-100, 1 mM EDTA, 5% glycerol, 1 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride, with one tablet of protease inhibitor mixture [Roche Molecular Biochemicals] per 10 ml of buffer added just prior to use) or 100 to 200 μl Laemmli buffer for 10 min on ice. The protein concentrations in cleared lysates were measured using the Bio-Rad Dc Protein Assay (Bio-Rad). Lysates (10 to 40 μg total protein/lane) were then subjected to 8% or 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to Immobilon P membranes (Millipore). Anti-caspase 8 monoclonal antibodies (BD Pharmingen), anti-GST monoclonal antibodies (Santa Cruz Biotechnology), anti-β-actin monoclonal antibodies (Sigma), anti-p53 DO-7 monoclonal antibodies (Novocastra), anti-caspase 8 polyclonal antibodies (MBL International Co., Woburn, MA), anti-Flag monoclonal antibodies (Sigma), or horseradish peroxidase (HRP)-conjugated anti-HA rat polyclonal antibodies were applied at a 1:1,000 dilution (for the anti-β-actin antibodies, a 1:5,000 dilution was used). HRP-coupled secondary anti-mouse or anti-rabbit antibodies (Pierce) (1:2,000 or 1:5,000 dilution, respectively) were used for detection by immunoblotting using the chemiluminescent SuperSignal West Femto or Pico Maximum Sensitivity substrate (Pierce).
Densitometric analysis was then applied to quantitate signals generated by immunoblotting, using a MultiImage Light Cabinet with ChemiImager 4400 v.5.1 software from Alpha Innotech Co. (San Leandro, CA). The film signal was expressed in relative light units. In some cases (Fig. 1A and 2A; also see Fig. 4A), densitometry was done using the Odyssey Infrared Imaging system (LI-COR Biosciences), where the secondary antibody was goat anti-mouse IRDye800CW obtained from LI-COR Biosciences.
FIG. 1.
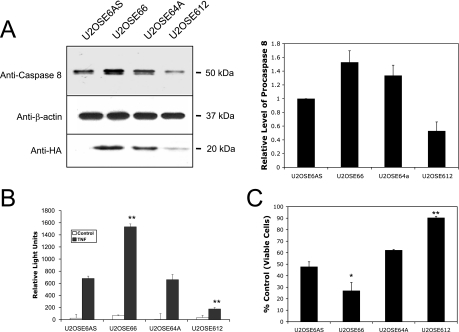
The expression of procaspase 8 in cells is linked to caspase 8 activity and to TNF sensitivity and may be influenced by HPV-16 E6. (A) U2OS cells stably transfected with pHA-E6S express various amounts of procaspase 8. Cells (1 × 106) were lysed in 200 μl Laemmli buffer, and 20 μg (total protein) of the lysate was separated by SDS-PAGE. Procaspase 8 was detected by immunoblotting using anti-caspase 8 antibodies (top), and loading was normalized by immunoblotting using anti-β-actin antibodies (middle). The level of HA-E6 expressed in each of these cell lines was also analyzed by immunoprecipitation (2 × 106 cells per preparation), using anti-HA monoclonal antibodies, and then detected by using anti-HA rat polyclonal antibodies conjugated to HRP (bottom). To obtain the graphical results, immunoblots were performed for three separate experiments and then analyzed using the Odyssey Infrared Imaging System. The error bars represent the standard deviations. (B) Caspase 8 activity measurements correlate well with estimates of protein levels. The indicated cell lines were treated with TNF (5 ng/ml) in the presence of cycloheximide (5 μg/ml) for 6 h and lysed. The level of caspase 8 activity was then measured using IETD-AMC as the substrate in the presence and absence of the caspase 8 inhibitor Ac-IETD-CHO. The activity in wells containing the inhibitor was subtracted from that in wells lacking the inhibitor, following normalization for the amount of protein in each cell lysate. Error bars represent standard deviations. (C) HA-E6 affects the cellular response to TNF. Each of the indicated cell lines was treated with TNF (5 ng/ml) in the presence of cycloheximide (5 μg/ml). After 16 h, cell viability was determined by the MTT assay and is presented as a percentage of viable cells, with cells untreated with TNF serving as the control. *, 0.95 level of confidence; **, 0.99 level of confidence.
FIG. 2.
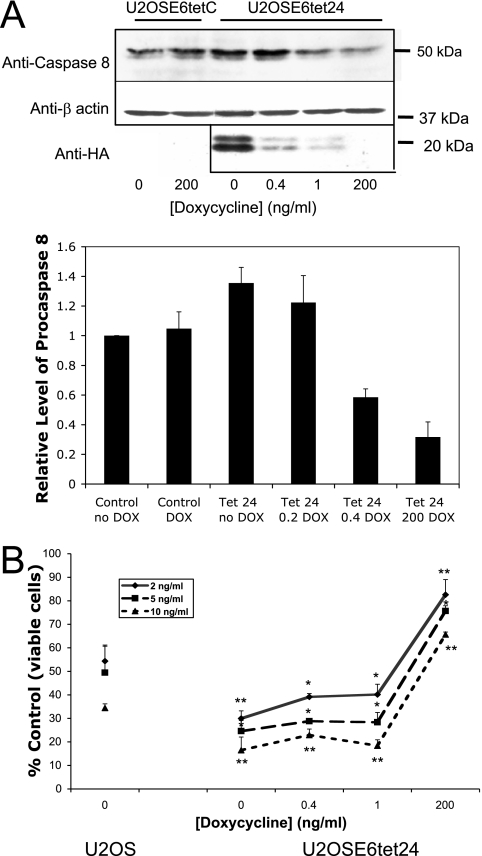
The level of E6 expression affects both the level of endogenous procaspase 8 and the cellular response to TNF. (A) The level of cellular procaspase 8 changes with the amount of E6 expressed. Control cells (U2OSE6tetC) and cells in which the level of E6 could be regulated by the amount of doxycycline (DOX) present in the media (U2OSE6tet24) were cultured in the presence of the indicated concentration of doxycycline and then lysed and analyzed for their levels of procaspase 8 expression by immunoblotting (top). The middle blot analyzes β-actin expression as a loading control. The level of HA-E6 expressed was also analyzed by immunoprecipitation (2 × 106 cells per preparation) using anti-HA monoclonal antibodies and then detected by using anti-HA rat polyclonal antibodies conjugated to HRP (bottom). To obtain the graphical results, immunoblots were performed for three separate experiments and then analyzed using the Odyssey Infrared Imaging System. The error bars represent the standard deviations. (B) The level of E6 expression affects cell viability in the presence of TNF. U2OS cells or U2OSE6tet24 cells, grown in the presence of the indicated levels of doxycycline, were treated with 2, 5, or 10 ng/ml TNF in the presence of cycloheximide (5 μg/ml). Following 16 h of incubation, cell viability was determined by the MTT assay and is presented as a percentage of viable cells (with untreated cells serving as the control). Error bars represent standard deviations. *, 0.95 level of confidence; **, 0.99 level of confidence.
FIG. 4.
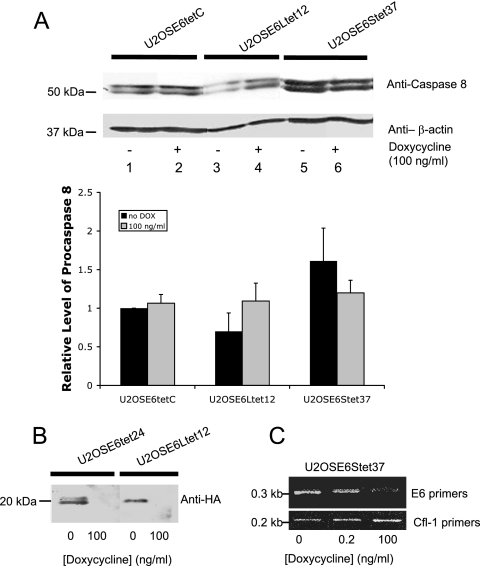
E6large, but not E6small/E6*, reduces the stability of procaspase 8 in a dose-dependent manner. (A) Control cells (U2OSE6tetC) and U2OS cells stably transfected with HA-E6large (U2OSE6Ltet12) or HA-E6small/E6* (U2OSE6Stet37), each under the control of the tet-responsive element, were grown in the presence or absence of 100 ng/ml of doxycycline (1 × 106 per preparation) and then lysed in 100 μl of Laemmli buffer, and 20 μg (total protein) of each cell lysate was separated by SDS-PAGE. Procaspase 8 was detected by immunoblotting using monoclonal anti-caspase 8 antibodies (top). The blot was reblotted with monoclonal antibodies directed against β-actin to normalize for loading (bottom). To obtain the graphical results, immunoblots were performed for three separate experiments and then analyzed using the Odyssey Infrared Imaging System. The error bars represent the standard deviations. (B) The level of E6large in U2OSE6Ltet12 cells is controlled by the presence of doxycycline (DOX) in the media. U2OSE6tet24 and U2OSE6Ltet12 cells were grown in the presence or absence of doxycycline and then lysed, and the amount of HA-E6 was analyzed by immunoprecipitation (2 × 106 cells per preparation) using anti-HA antibodies. (C) The level of E6small/E6* in U2OSE6Stet37 cells is controlled by the presence of doxycycline in the media. U2OSE6Stet37 cells were grown in the presence of 0, 0.2, or 100 ng/ml doxycycline and then lysed, and the amount of E6 message was estimated by RT-PCR using primers specific for E6 (top). Primers for cofilin 1 (bottom) were used for normalization.
Caspase 8 assay.
Cells were plated on black 96-well plates at a density of 2 × 104 per well and incubated overnight. TNF (5 ng/ml) was then added, along with cycloheximide (1 μg/ml) for the times indicated. Caspase activity was then measured using the fluorogenic substrate IETD-AMC for caspase 8 (Alexis Biochemicals) as described previously (5). After 20 h of incubation with fluorogenic substrate, the released fluorochrome was measured with a plate-reading fluorimeter (Flx800; Bio-Tek Instrument Co.) using an excitation of 360 nm and an emission of 460 nm.
Cell viability assay.
Following treatment of cells as described above, cell viability was measured by the MTT assay performed as described previously (15).
RT-PCR.
RT-PCR was used to analyze transcripts in cell lines stably transfected with E6 S, E6large, and E6small/E6*. Total RNA was isolated from each cell line using the TRIZOL reagent (Invitrogen) and was used as a template. cDNA was synthesized using SuperScript II RT (Invitrogen) and an oligo(dT) primer (Amersham Biosciences). The primers for the E6 transcripts, 5′-ATGATATCCTATCCATACGATGTT-3′ and 5′-TGGGTTTCTCTACGTGTTCTTGAT-3′, were then used to amplify the PCR products from the cDNA. The primers for cofilin-1 (5′CCTTCCCAAACTGTTTTGAT-3′ and 5′-CTGGTCCTGCTTCCATGAGTA-3′) were used for normalization of PCRs. The sequences of the Flag 5′ and Flag 3′ primers were obtained from the manufacturer's manual for the use of the pFlag-cmyc plasmid (Sigma).
p53 ELISA.
p53 levels were measured by enzyme-linked immunosorbent assay (ELISA) as described previously (14). The signal was recorded at 405 nm using a microplate reader.
Mammalian two-hybrid assay.
The mammalian two-hybrid binding assay was performed according to the manufacturer's instructions (Stratagene). The indicated combinations of vectors were transfected into U2OS or U2OS-derived cells (7 × 104/well; 24-well plate), along with the pFR-Luc-expressing reporter plasmid (Stratagene). Cells were cotransfected with a plasmid encoding green fluorescent protein (GFP) (pcDNA3 GFP) in order to normalize for transfection efficiency. Forty-eight hours following transfection, luciferase activity was measured using a Luciferase Assay System (Promega) as directed by the manufacturer. The chemiluminescence was then measured using a MicroLuminatPlus LB 96V luminometer from Berthold Technologies, at 0.5 or 2.0 s of integration in chemiluminescent relative light units; 100 μl of cell lysate was also used to measure GFP expression (Flx 800; Bio-Tec Instrument Company) in order to normalize for transfection efficiency.
Expression and purification of recombinant proteins.
To purify the GST, GST-E6large, and GST-E6small/E6* proteins, cleared lysates of transformed, IPTG (isopropyl-β-d-thiogalactopyranoside)-induced E. coli [BL21(DE3); Novagen] cultures (from 100 ml LB broth) were incubated with 1 ml glutathione beads (Sigma) preequilibrated in lysis buffer (50 mM Tris HCl, 0.5 M NaCl, 10 mM EDTA, 10 mM EGTA, 10% glycerol, 1% Triton X-100, 20 μg/ml lysozyme, 1 mM phenylmethylsulfonyl fluoride, 1 tablet of protease inhibitor [Roche] per 10 ml buffer, pH 8.0). Following protein purification according to the manufacturer's recommended protocol (Amersham), beads with their bound GST and GST-E6 proteins were resuspended in phosphate-buffered saline supplemented with 10% glycerol and 2 mM dithiothreitol. The protein concentration for each preparation was estimated using the Bio-Rad Dc Protein Assay. His-procaspase 8 DED expression was also achieved using the T7 TNT Quick Coupled Transcription/Translation System (Promega), according to the manufacturer's protocol.
In vitro pull-down assays, immunoprecipitation, and coimmunoprecipitation.
In vitro pull-down, immunoprecipitation, and coimmunoprecipitation assays were performed as described previously (15). Following pulldown, immunoprecipitation, or coimmunoprecipitation, bound proteins were eluted in 30 μl of 2.5× SDS loading buffer, fractionated by 12% SDS-PAGE, and subjected to immunoblotting. In each case, the membrane used to detect bound proteins was then stripped and reblotted with the appropriate antibodies to measure input.
Centricon fractionation of procaspase 8.
Two 10-cm plates of confluent cells (U2OS and U2OSE6S9) were lysed using 500 μl of 1× lysis buffer as described above (“Caspase 8 assay”). Centrifugation of the cleared lysates using the Microcon YM-30 filter (Millipore) was performed according to the manufacturer's protocol until only 200 μl of retentate remained. The retentate was then transferred and centrifuged (12,500 rpm) through the Microcon YM-100 filter. After 1 minute, 50 μl of filtrate was obtained (filtrate 1). The 50 μl of filtrate obtained during the next 2 min was designated “filtrate 2,” and the 50 μl of filtrate obtained during the final 2 min was designated “filtrate 3.” The filtrate from the YM-30 filter was concentrated to a volume of 50 μl using Microcon YM-3. Each fraction obtained was subjected to 8% PAGE and immunoblotted using antibodies directed against caspase 8.
Statistical analysis.
For each assay, each point was measured in triplicate. Student's one- or two-tailed t test, as appropriate, was used to determine statistical significance.
RESULTS
Expression of procaspase 8 in cells is linked to caspase activity and TNF sensitivity and may be influenced by HPV-16 E6.
Our previously published work (15, 19) led us to hypothesize that E6 might bind to procaspase 8 and accelerate its degradation. To test this prediction, U2OS cells were transfected with a plasmid coding for the epitope-tagged version of HPV-16 E6, and several stable clones were isolated. In U2OSE66, U2OSE64A, and U2OSE612 cells, the HA-E6 sequence was inserted in the sense orientation, and in the U2OSE6AS cells, it was inserted in the antisense orientation and therefore did not encode a functional protein. Unexpectedly, immunoblot analysis using antibodies directed against procaspase 8 demonstrated that in some cases, clones expressing E6 expressed higher (U2OSE66) and in other cases lower (U2OSE612) levels of procaspase 8 than were seen in the control cells (U2OSE6AS) (Fig. 1A). On the bottom of the panel is shown an immunoblot using antibodies directed against HA, confirming that the transfected cells indeed expressed this protein. The results from three such studies were digitally quantitated and are presented graphically to the right of the blot. To correlate the levels of immunoreactive protein with actual caspase 8 activity, cells were treated with TNF for 6 hours and lysed, and their caspase 8 activities were measured by a fluorimetric assay using Ac-IETD-AMC as the substrate (Fig. 1B). The results of the activity assay compare well with those from the immunoblot, with the cells showing the highest level of immunoreactive procaspase 8 (U2OSE66) also displaying the highest level of caspase 8 activity and the cells with the lowest level of immunoreactive procaspase 8 (U2OSE612) displaying the lowest level of caspase 8 activity.
Caspase 8 is an initiator caspase that functions in several apoptotic pathways. To see how the various levels of procaspase 8 observed in these four cell lines affected the cellular response to TNF, each of the cell lines was treated with TNF and then analyzed for cell viability using the MTT assay. The results showed that the levels of procaspase 8 (Fig. 1A and B) correlated well with cellular sensitivity to TNF (Fig. 1C), with cells expressing the highest level of procaspase 8 (U2OSE66) being more sensitive to TNF and the cells with the lowest level of procaspase 8 (U2OSE612) being relatively resistant to TNF. Together, these results suggest that the level and activity of procaspase 8, as well as the cellular response to TNF, may be influenced by E6 expression.
E6 expression modulates the level of procaspase 8 expression and cellular sensitivity to TNF in a dose-dependent manner.
The results discussed above suggest that E6 might be able to modulate the level of procaspase 8 and hence the cellular response to TNF and that the extent and direction of this modulation might depend on the level of E6. To examine this possibility more carefully, we created and characterized a cell line (U2OSE6tet24) in which the level of E6 expression could be regulated by the amount of doxycycline present in the media (15). Control cells (U2OSE6tetC), which carried the tet regulator but not the E6 oncogene, and the E6-expressing U2OSE6tet24 cells were then grown in the presence of various amounts of doxycycline. The resulting levels of procaspase 8 (Fig. 2A, top) and of HA-E6 (Fig. 2A, bottom) were then estimated by immunoblotting. As expected, maximal E6 expression was achieved in the absence of doxycycline, and minimal expression was observed at high levels of doxycycline (200 ng/ml). Intriguingly, the results from the caspase 8 immunoblot (top) showed that high and moderate levels of E6 (associated with 0- and 0.4-ng/ml levels of doxycycline) correspond to high levels of procaspase 8, while low levels of E6 (associated with 200-ng/ml levels of doxycycline) correspond to low levels of procaspase 8. The results from three such studies were digitally quantified and are presented graphically below the blot.
To examine the effects that these various levels of E6, and hence of procaspase 8, might have on the cellular response to TNF, U2OSE6tet24 cells grown in the presence of various amounts of doxycycline were treated with TNF for 16 h, and the resulting cell viability was measured by the MTT assay. The results (Fig. 2B) demonstrate that at high and moderate levels of E6 and procaspase 8 (corresponding to 0, 0.4, and 1 ng/ml doxycycline), cells are in fact sensitized to TNF (compared to U2OS cells), while at low levels of E6 and procaspase 8 (corresponding to 200 ng/ml doxycycline), cells are protected. This is consistent with our previously reported results (15), where we showed that high levels of E6 sensitize cells to TNF while low levels protect them, and with the results from the three independently derived stable clones shown in Fig. 1. Together, these results show that expression of E6 can change the level of procaspase 8 in a dose-dependent manner, with a possible threshold; that high and moderate levels of E6 correspond to high levels of procaspase 8; and that these higher cellular levels of procaspase 8 correspond to a greater sensitivity to TNF.
Procaspase 8 levels decrease in cells expressing HPV-16 E6large and increase in cells expressing HPV-16 E6small/E6*.
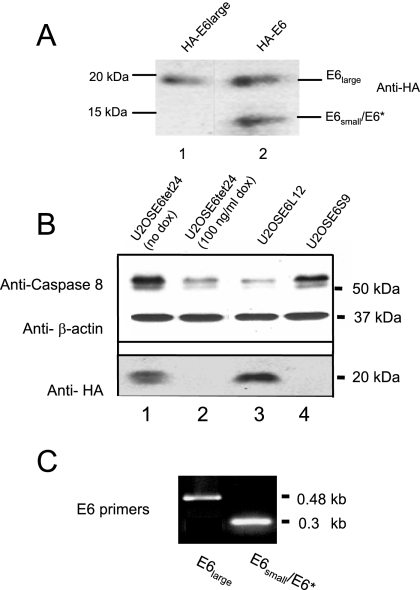
The experiments described above suggest that HPV-16 E6 can both stabilize (i.e., U2OSE66) and destabilize (i.e., U2OSE612) procaspase 8, corresponding, respectively, to either sensitization or resistance to TNF, and indicate that this phenomenon may be dose related. One possible molecular explanation for this could be based on the observation that E6 is produced in both a short form of approximately 7 kDa (E6small/E6*) and a longer form of approximately 16 kDa (E6large) and that these two forms could have different functions. In the experiments described above (Fig. 1 and 2), both the small and large isoforms were present in the E6-expressing cells, so the observations reflect the composite contributions of the two versions. To examine the effect of each isoform individually, plasmids coding for either the small or the large versions only were created (15) and transfected into U2OS cells. Lysates of cells transfected with either the sense version of HA-E6 or a version of HA-E6 coding for only the large isoform were then analyzed for HA-E6 expression by immunoblotting (Fig. 3A). As expected, while only the large isoform was detected in cells transfected with pHA-E6large, both isoforms were detected in cells transfected with pHA-E6. Individual clones stably transfected with either E6large or E6small/E6* were then isolated and analyzed for their levels of procaspase 8. These results (Fig. 3B) showed that cells expressing the large version of E6 (U2OSE6L12) experienced a decrease in their levels of procaspase 8 protein, while cells expressing the small version of E6 (U2OSE6S9) experienced an increase in the amount of procaspase 8. In this experiment, U2OSE6tet24 cells, grown in either the absence or presence of doxycycline (lanes 1 and 2), served as controls and references for cells expressing high (lane 1) or low (lane 2) levels of procaspase 8. RT-PCR analysis for E6 expression confirmed that the selected cells were expressing the expected version(s) of E6 (Fig. 3C). Although we were able to detect both the large and small isoforms in transiently transfected cells (Fig. 3A), our immunoblotting protocol using antibodies directed against HA lacked sufficient sensitivity to detect the small isoform in stably transfected cell lines. This may be due in part to instability of the protein. Another possible reason may be a tendency for E6small/E6* to form complexes with itself, the large version of E6, or other proteins, so that the epitopes to which the antibodies bind are less available.
FIG. 3.
E6large and E6small/E6* influence the cellular level of procaspase 8 in opposite directions. (A) Both E6large and E6small/E6* are expressed in cells. U2OS cells (2 × 106) were transiently transfected with pHAE6large (lane 1) or pHA-E6sense (lane 2). Forty-eight hours posttransfection, the cells were lysed in 100 μl of Laemmli buffer and subjected to separation by SDS-PAGE. Detection of expressed HA-E6 was performed using anti-HA rat polyclonal antibodies conjugated with HRP. (B) Cells expressing E6large display a decreased level of procaspase 8, while cells expressing E6small/E6* display an increased level of procaspase 8. U2OS cells stably transfected with HA-E6large (U2OSE6L12) or HA-E6small/E6* (U2OSE6S9), as well as U2OSE6tet24 cells grown in the absence or presence of 100 ng/ml doxycycline (dox) (1 × 106 per preparation), were lysed in 100 μl of Laemmli buffer, and 20 μg (total protein) of each cell lysate was separated by SDS-PAGE. Procaspase 8 was detected by immunoblotting using monoclonal anti-caspase 8 antibodies (top). The blot was reblotted with monoclonal antibodies directed against β-actin to normalize for loading (second blot). The level of HA-E6 expressed in each of these cell lines was also analyzed by immunoprecipitation (2 × 106 cells per preparation) using anti-HA monoclonal antibodies and then detected by using anti-HA rat polyclonal antibodies conjugated to HRP (third blot). (C) Stable clones transfected with E6large (U2OSE6L12) and E6small/E6* (U2OSE6S9) each expressed the expected version(s) of the E6 message, as determined by RT-PCR.
To examine this observation more carefully and in a dose-dependent manner, plasmids coding for either the large or small isoform of E6 under the control of the Tet-responsive element were constructed and used to create cell lines in which the level of either E6large or E6small/E6* could be individually controlled. Cells were then grown in the presence of 0 (high E6) or 100 (low E6) ng/ml doxycycline and analyzed for the level of procaspase 8 by immunoblotting (Fig. 4A). Control cells unresponsive to doxycycline (U2OStetC) were also tested. The results from cells transfected with the large isoform of E6 (lanes 3 and 4) confirmed that, indeed, the absence of doxycycline (and hence the presence of E6) leads to a decrease in the level of procaspase 8. However, the results from cells transfected with the small isoform of E6 were quite different (lanes 5 and 6). A comparison of the levels of procaspase 8 in the U2OSE6Stet37 cells with that in the control (U2OSE6tetC) cells leads to the conclusion that the level of procaspase 8 is higher in cells expressing the small isoform of E6 and increases as the E6 level is increased by the withdrawal of doxycycline. Results from three such studies were digitally quantitated and are presented graphically below the blot. Figure 4B shows an immunoprecipitation experiment that verifies that the expression of E6large is indeed regulated by the presence of doxycycline, while Fig. 4C shows an RT-PCR experiment that provides this information for the smaller isoform.
E6large destabilizes, and E6small/E6* stabilizes, procaspase 8.
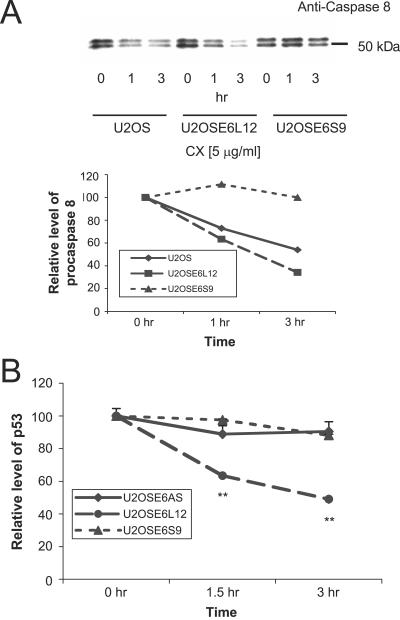
The results described above suggest that the large version of E6 accelerates the degradation of procaspase 8, while the small version of E6 stabilizes the protein. To examine the individual influence of each of the two versions of E6 on endogenous procaspase 8, the rates at which procaspase 8 was degraded in three cell lines—U2OS; U2OSE6L12, expressing the large version of E6; and U2OSE6S9, expressing the small version of E6—were compared (Fig. 5B). In this experiment, protein production was blocked by the addition of cycloheximide, and the level of procaspase 8 was estimated by immunoblotting. The results demonstrate that procaspase 8 is indeed degraded at a higher rate in cells expressing E6large, (U2OSE6L12) and at a lower rate in cells expressing E6small/E6* (U2OSE6S9) than in cells lacking E6 (U2OS).
FIG. 5.
Procaspase 8 is destabilized by E6large and stabilized by E6small/E6*. (A) HPV-16 E6large accelerates, and HPV-16 E6small/E6* inhibits, the degradation of procaspase 8. U2OS, U2OSE6L12, and U2OSE6S9 cells were treated with cycloheximide (CX) (5 μg/ml) and lysed at 0, 1, or 3 h posttreatment. The lysates were then analyzed for their procaspase 8 contents by immunoblotting. The amount of sample loaded in each lane was calculated so that the initial amounts of procaspase 8 protein (zero time point) were approximately equivalent between the three cell lines, with the amounts loaded in the 1- and 3-h lanes equal to that loaded for the 0-h time point for their respective cell lines. The graph shows the densities of the bands on the blot, each normalized to the density at 0 h. (B) The expression of E6large accelerates the degradation of p53. U2OS, U2OSE6L12, and U2OSE6S9 cells were treated with mitomycin C (2 μg/ml) for 16 h in order to increase cellular levels of p53. The cells were treated with cycloheximide (5 μg/ml) and lysed at 0, 1, or 3 h post-cycloheximide treatment. The lysates were then analyzed for their protein contents as described in Materials and Methods and for p53 contents by ELISA. The values (expressed as the level of p53 per mg total protein) were normalized to the level of p53 at 0 h and expressed as a percentage. The error bars indicate standard deviations. **, 0.99 level of confidence.
Perhaps the best-known substrate for degradation mediated by E6 is the p53 tumor suppressor. Since the results described above indicated that the large, but not the small, version of E6 mediated the degradation of procaspase 8, we examined whether the same pattern would be observed in the case of p53 degradation. Cells expressing either the large (U2OSE6L12) or the small (U2OSE6S9) version of E6 were therefore first treated with mitomycin C (to elevate the cellular level of p53) and then with cycloheximide. At the indicated times following cycloheximide treatment, cells were lysed and analyzed for p53 by ELISA. The results (Fig. 5C) show that, as in the case of procaspase 8, E6large, but not E6small/E6*, accelerated p53 degradation.
Both the large and small isoforms of HPV-16 E6 bind to procaspase 8.
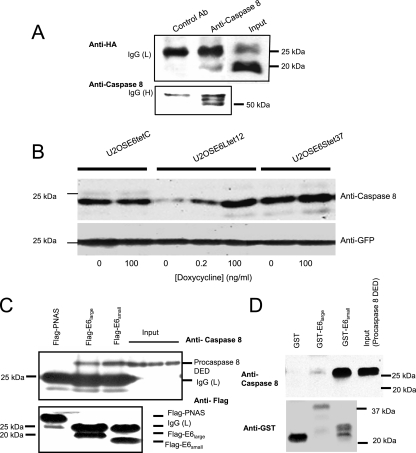
The results described above indicate that E6 can affect the levels and biological activity of procaspase 8 and suggest the possibility that E6 could bind to procaspase 8. To test this, we examined this binding by coimmunoprecipitation (Fig. 6A). In this experiment, beads coupled to either an irrelevant antibody (negative control; lane 1) or anti-procaspase 8 (lane 2) were used to immunoprecipitate bound proteins from lysates of U2OSE6tet24 cells, which were then separated by SDS-PAGE and probed with either anti-HA (top) or anti-procaspase 8 (bottom). The position of HA-E6large is shown in the third lane (input). These results confirm that E6large binds to endogenous procaspase 8. In this experiment, as in many of our immunoblotting and immunoprecipitation experiments, E6small/E6* was not detected, likely due to rapid protein turnover (60, 61) and/or to the unavailability of epitopes.
FIG. 6.
E6 binds to procaspase 8. (A) E6 coimmunoprecipitates with endogenous procaspase 8. U2OSE6tet24 cells (2 × 107) grown in the absence of doxycycline (and hence expressing E6) were lysed with the immunoprecipitation buffer. Five micrograms of irrelevant, isotope-matched control antibodies (left lane) or anti-caspase 8 monoclonal antibodies (middle lane) was used to precipitate a complex which was then bound to Plus A/G agarose. The bound proteins were eluted and subjected to immunoblot analysis. The detection of HA-E6 in the coimmunoprecipitated samples and in the input sample (right lane) was performed using anti-HA rat polyclonal antibodies (top). The same membrane was stripped and reblotted with anti-caspase 8 polyclonal antibodies as a control for precipitation (bottom). IgG (H) and (L), immunoglobulin G heavy and light chains. (B) E6large causes a decrease in the level of exogenously expressed procaspase 8 DED. The indicated cell lines were grown in the indicated concentrations of doxycycline and transiently transfected with pcDNA3-caspase 8 DED and pcDNA3-GFP 48 h prior to lysis. The transfection efficiency was estimated and normalized using anti-GFP antibodies (bottom). (C) Procaspase 8 binds to both E6large and E6small/E6* as analyzed by coimmunoprecipitation. U2OS cells were cotransfected with pFlag-LOC5129 (negative control), pFlag-E6large, or pFlag-E6small/E6* and pTriEX4-procaspase 8 DED. Forty-eight hours posttransfection, the expressed LOC5129-tagged proteins were precipitated using Flag-agarose. The bound proteins were then subjected to separation by SDS-PAGE and immunoblotting with antibodies directed against either caspase 8 (top) or Flag (bottom). The three right lanes show the input levels of procaspase 8 for the incubations shown in the three left lanes. (D) Both HPV-16 E6large and HPV-16 E6small/E6* bind to procaspase 8 DED produced by in vitro transcription/translation, as analyzed by in vitro pull-down assays. Procaspase 8 DED, expressed using the T7 TNT Quick Coupled Transcription/Translation System, was incubated with beads bound to either GST, GST-E6large, or GST-E6small/E6*. The beads were washed, and the bound proteins were eluted with SDS prior to separation by SDS-PAGE and immunoblotting with antibodies directed against procaspase 8 (top) or GST (bottom). An amount of lysate corresponding to the input was also loaded as a control (far right lane).
The coimmunoprecipitation experiment described above (Fig. 6A) examined the binding of E6 to endogenous procaspase 8. To confirm and strengthen these results, we also wished to examine the interaction of E6 with procaspase 8 DED expressed from a transfected plasmid. We first examined whether the stability of this exogenous protein was affected by the large and small isoforms of E6 in the same way as the endogenous protein (Fig. 4A). Procaspase 8 DED was therefore overexpressed in cells expressing doxycycline-regulated amounts of E6large or E6small/E6*, and the resulting level of procaspase 8 DED was estimated by immunoblotting. The transfection efficiency was normalized using GFP. These results (Fig. 6B) demonstrate that for the exogenously supplied version of the procaspase 8 DED, as well as for the endogenous form of the intact protein, the expression of E6large, but not of E6small/E6*, leads to the destabilization of procaspase 8 in a dose-dependent manner.
The binding of both E6large and E6small/E6* was then confirmed by coimmunoprecipitation following transfection of the His-tagged procaspase 8 DED into cells (Fig. 6C). We have found (unpublished observations) that this tagged form of the procaspase 8 DED is much more stable than the nontagged procaspase 8 (which was used in the experiments described above) in the presence of E6large, making it particularly suitable for this binding experiment. In this experiment, lysates were incubated with Flag-tagged PNAS (negative control), Flag-tagged E6large, or Flag-tagged E6small/E6*. After immunoprecipitation, bound proteins were separated by SDS-PAGE and probed using antibodies directed against the procaspase 8 DED (Fig. 6C, top) or anti-Flag (bottom). The presence of procaspase 8 DED in lysates immunoprecipitated with Flag-E6large and Flag-E6small/E6*, but not with Flag-LOC5129, confirmed this interaction (Fig. 6C).
In vitro pull-down analyses using the large or small version of E6 coupled to GST were also performed. The results obtained using the in vitro transcription/translation product of the procaspase 8 DED (Fig. 6D) clearly demonstrate strong binding between E6small/E6* and the DED of procaspase 8. They also show binding between the procaspase 8 DED and E6large. Together, these results clearly demonstrate the binding between both isoforms of E6 and the procaspase 8 DED.
E6large, but not E6small/E6*, inhibits procaspase 8-procaspase 8 and procaspase 8-FADD binding.
The results presented above provide evidence that both the large and small isoforms of HPV-16 E6 can bind to procaspase 8. This binding might then be expected to affect the ability of procaspase 8 to bind to its normal cellular targets, such as procaspase 8 (to form a homodimer) and FADD (as part of the signaling sequence). To determine if this was indeed the case, the influences of the two variants of E6 on procaspase 8-procaspase 8 and procaspase 8-FADD binding were examined.
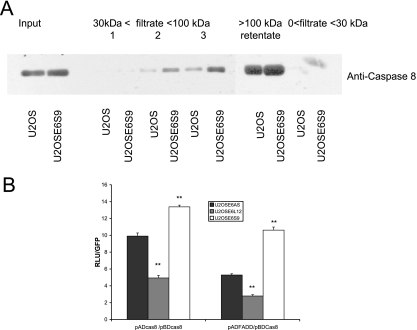
It has been shown that procaspase 8 can exist as a dimer in cells (4, 7). To determine whether E6small/E6* affected dimer formation, lysates from cells expressing E6small/E6* (U2OSE6S9), as well as from the parental cells (U2OS), were prepared, and the proteins were fractionated by filtration. After filtration, the fractions obtained were separated by SDS-PAGE, transferred to a membrane, and immunoblotted using antibodies directed against procaspase 8. The results (Fig. 7A) show that for both U2OS and U2OSE6S9 cells, procaspase 8 appears primarily in the >100-kDa retentate, consistent with its presence as a dimer and providing evidence that E6small/E6* does not interfere with, and may even enhance, dimer formation.
FIG. 7.
Expression of E6 alters the binding between apoptotic signaling-protein partners. (A) E6small/E6* does not interfere with procaspase 8 homodimer formation. Lysates of both the parental U2OS cells and the cells expressing only E6small/E6* (U2OSE6S9) were separated by stepwise centrifugations as described in Materials and Methods. The material in each of the indicated fractions was then separated by SDS-PAGE, transferred to a membrane, and probed using a monoclonal antibody directed against caspase 8. (B) E6large, but not E6small/E6*, inhibits procaspase 8-procaspase 8 and procaspase 8-FADD binding. Cells (7 × 104) stably transfected with the antisense version of HA-E6 (U2OSE6AS), HA-E6large (U2OSE6L12), or HA-E6small/E6* (U2OSE6S9) were plated in wells of a 24-well plate and transfected with the indicated combinations of plasmids, along with a reporter plasmid coding for firefly luciferase under the control of the 5× GAL4 binding site. To control for transfection efficiency, pcDNA3-GFP was cotransfected with the above-mentioned plasmids. Binding was measured by monitoring expression of the luciferase gene, and the results are presented as a ratio of the signal measured for the indicated plasmid combination divided by the signal measured for GFP. The error bars indicate standard deviations. **, 0.99 level of confidence.
To explore this possibility and to further examine the influences of both E6small/E6* and E6large on procaspase 8-procaspase 8 and procaspase 8-FADD interactions, a mammalian two-hybrid system (Stratagene) was utilized. Genes of interest were cloned into the provided plasmids in frame with the activation and binding domains and transfected into cells, along with a luciferase reporter construct. If the genes of interest interacted, the binding could be detected by the expression of the reporter gene. In order to normalize for transfection efficiency, the cells were cotransfected with a plasmid coding for GFP. Initial experiments showed that each cell line used strongly supports the formation and detection of protein-protein interactions; that this assay is capable of detecting caspase 8-caspase 8 binding, as well as FADD-caspase 8 binding; and that all combinations of plasmids designed to function as negative controls resulted in low background signals (data not shown). The plasmids coding for caspase 8 and FADD were then transfected into cells without E6 (U2OSE6AS), cells expressing E6large (U2OSE6L12), and cells expressing E6small/E6* (U2OSE6S9). The results (Fig. 7B) show clearly that both isoforms of E6 have a profound influence on procaspase 8-procaspase 8 interactions, as well as on procaspase 8-FADD interactions. The large isoform decreases both of these interactions by at least 50%, while the small isoform increases the interaction, consistent with the findings from the fractionation experiment. Together, these data support a model in which the large and small variants of E6 both bind to procaspase 8 and exert differential effects both on procaspase 8 stability and on its ability to bind to its normal partner proteins.
siRNA knockdown of E6large leads to increases in procaspase 8 levels, caspase 8-caspase 8 and caspase 8-FADD binding, and sensitivity to TNF.
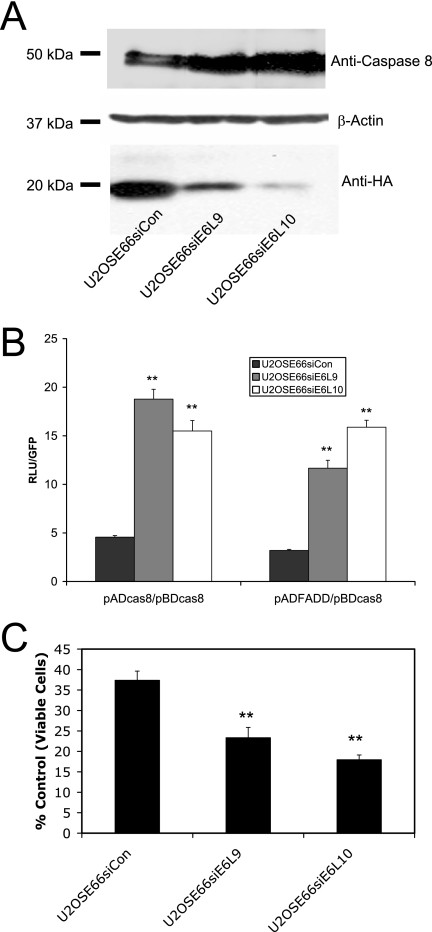
The model suggested by the data described above involves the binding of both the small and large isoforms of E6 to procaspase 8. Binding of the small isoform leads to stabilization of procaspase 8, as well as an increase in caspase 8-caspase 8 and caspase 8-FADD binding. In contrast, binding of the large isoform leads to accelerated degradation of procaspase 8 and a decrease in caspase 8-caspase 8 and caspase 8-FADD binding. One prediction of this model is that if the level of E6large were reduced by siRNA directed against that isoform, the levels of caspase 8 should increase, as well as caspase 8-caspase 8 and caspase 8-FADD interactions. To test this prediction, cells expressing both the small and large isoforms of E6 (U2OSE66) were transfected with constructs coding for either control siRNA or siRNA directed against E6large (siE6large), and stable clones were isolated. The expression levels of procaspase 8 and HA-E6 were then analyzed at both the protein (Fig. 8A) and message (data not shown) levels. Clearly, both siE6large-expressing clones (U2OSE66siE6L9 and U2OSE66siE6L10) expressed procaspase 8 at levels significantly higher than that seen in the control cells, consistent with our prediction (Fig. 8A, top). The third blot, showing the level of HA-E6, demonstrates that significant down-regulation of the large version of E6 was indeed achieved. RT PCR analysis of E6large and E6small/E6* (data not shown) confirmed that in fact, expression of siE6large resulted in down-regulation of the large but not the small isoform of E6.
FIG. 8.
siRNA directed against E6large increases the cellular levels of procaspase 8, as well as procaspase 8-proaspase 8 and procaspase 8-FADD binding. (A) siRNA directed against E6large increases cellular levels of procaspase 8. Cells expressing both isoforms of E6 (U2OSE66) were stably transfected with pSilencer 3.1 siCon or pSilencer 3.1 siE6large. Three resulting cell lines, U2OSE66siCon (siControl), U2OSE66siE6L9 (expressing siE6large), and U2OSE66siE6L10 (expressing siE6large), were lysed and subjected to immunoblot analysis using antibodies directed against procaspase 8 (top) or β-actin (middle) or were immunoprecipitated using anti-HA monoclonal antibodies (bottom). (B) siRNA directed against E6large increases the binding between procaspase 8 and procaspase 8 and between procaspase 8 and FADD. The mammalian two-hybrid analysis was performed as described in Materials and Methods. Error bars represent standard deviations. (C) siRNA directed against E6large affects the cellular response to TNF. Each of the indicated cell lines was treated with TNF (5 ng/ml) in the presence of cycloheximide (5 μg/ml). After 16 h, cell viability was determined by the MTT assay and is presented as a percentage of viable cells, with cells untreated with TNF serving as the control. **, 0.99 level of confidence.
The effect of this loss of E6large on procaspase 8-procaspase 8 and procaspase 8-FADD binding was then analyzed using the mammalian two-hybrid system (Fig. 8B). Consistent with our model, expression of siRNA directed against E6large resulted in increased procaspase 8-procaspase 8 and procaspase 8-FADD interactions. Together, these results strengthen the evidence that the large isoform of E6 accelerates the degradation of procaspase 8, thereby inhibiting its ability to interact with itself (to form homodimers) and with FADD.
The next prediction was that by increasing the level of procaspase 8 and its interactions with itself and with FADD, cells would be further sensitized to TNF. To test this possibility, the U2OSE66siE6L9 and U2OSE66siE6L10 cells, expressing siRNA directed against E6large, were treated with TNF (5 ng/ml; 16 h), and their viability was examined by the MTT assay. The results (Fig. 8C) show that, as predicted, these two cell lines were more sensitive to TNF than were the U2OSE66siCon (control) cells.
The low-risk types HPV-6b and -11 produce only the large isoform of E6, which does not affect the level of procaspase 8.
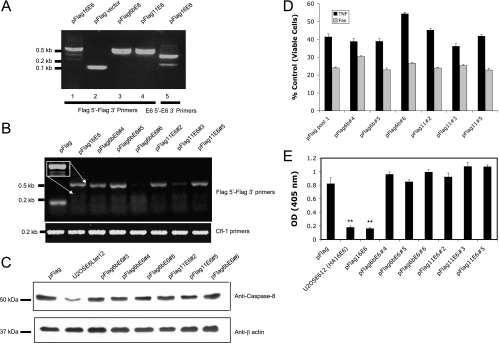
High-risk types of HPV, such as HPV-16 and -18, can cause cervical cancer, while low-risk types, such as HPV-6b and -11, do not. One reason for this difference is thought to be variability in the activities of their respective E6 proteins. For example, it has been reported that the E6 protein from high-risk types accelerates the degradation of p53 while the E6 protein from low-risk types cannot (12, 50), and it seemed likely that other differences in the activities of E6 oncoproteins from these types might also exist. As both the large and small isoforms of HPV-16 E6 appear to have significant and separable biological activities, we began by asking whether either or both of the large and small isoforms of E6 were produced from the HPV-6b and -11 E6 genes. Cells were therefore transfected with plasmids coding for Flag-tagged versions of E6 from HPV-16, -6b, or -11, and stable pools were selected and expanded. The expression of Flag E6 was then analyzed by RT-PCR. The results (Fig. 9A) showed that while both the Flag- and HA-tagged versions of HPV-16 E6 produced transcripts corresponding to both the large and small isoforms, only the transcript corresponding to the large isoform of E6 was produced from HPV-6b or -11 E6.
FIG. 9.
The low-risk types HPV-6b and -11 produce only the large isoform of E6, which does not affect the level of procaspase 8. (A) HPV-6b and -11 produce only the large isoform of E6. Expression of Flag-tagged E6 in stably transfected pools was analyzed by RT-PCR using primers for the vector (Flag 5′ and Flag 3′) (lanes 1 to 4). The presence of the splice variant for E6small/E6* was confirmed using primers 16E6 5′ and 16E63′ (lane 5). (B) Individual clones transfected with E6 from HPV-6b or -11 express the expected sequences at various levels. cDNA was prepared from RNA isolated from cells stably expressing HPV-16 E6, HPV-6b E6, and HPV-11 E6. RT-PCR was performed using primers for the vector (Flag 5′ and Flag 3′) (top). Normalization for loading was done using primers for cofilin (bottom). The inset shows an overexposure of the relevant area for cells transfected with pFlag16E6, demonstrating the presence of both the large and small transcripts. (C) The E6 proteins from the low-risk HPV-6b and -11 types do not decrease the level of endogenously expressed procaspase 8. Cells (2 × 106) from each of the indicated cell lines expressing E6 were lysed in 100 μl Laemmeli buffer. The proteins were then separated by SDS-PAGE (20 μg/lane), and the level of endogenous procaspase 8 in each cell lysate was estimated by immunoblotting. For normalization, anti-β actin antibodies were used to detect the level of the protein. (D) The E6 proteins from HPV-6b and HPV-11 do not affect the cellular response to TNF or Fas. Each of the indicated cell lines was treated with TNF (5 ng/ml) in the presence of cycloheximide (5 μg/ml). After 16 h, cell viability was determined by the MTT assay and is presented as a percentage of viable cells, with cells untreated with TNF serving as the control. Each measurement was made in triplicate, and the error bars indicate the standard deviation of the mean. (E) The E6 proteins from HPV-6b and HPV-11 do not decrease cellular levels of p53. Isolated cell lines stably expressing the indicated versions of E6 (5 × 104 cells per well in a 24-well plate) were untreated or treated with mitomycin C (2 μg/ml) for 16 h. The cells were then lysed with 100 μl of mammalian lysis buffer, and 50 μl of the lysate was subjected to a p53 ELISA (14). The relative level of p53 was estimated by absorbance at 405 nm, and the results are presented as the difference in absorbance between cells treated and untreated with mitomycin C. OD, optical density. Error bars represent standard deviations. **, 0.99 level of confidence.
To examine the impact of E6 oncoproteins from these different types on the level and activity of procaspase 8, we created cell lines expressing HPV-6b E6 and HPV-11 E6 from individual clones. RT PCR using primers for the Flag sequence of the inserted gene was used to examine the presence or absence of the exogenous gene in several individually isolated clones. The results shown in Fig. 9B show that clones 4 and 5 from the HPV-6b E6 transfection, and clones 2 and 5 from the HPV-11 E6 transfection, do indeed express the desired sequence. When these clones were examined for their levels of endogenously expressed procaspase 8 DED, we found that none of the clones expressing E6 from HPV-11 or -6b exhibited a significantly reduced level of this protein (Fig. 9C), and further, that the levels of E6 expression from HPV-16 and HPV-11 were unrelated to the level of procaspase 8. The same pattern of expression was observed when the procaspase 8 DED was expressed from an introduced plasmid (data not shown).
We then investigated whether the E6 proteins from HPV-6b or HPV-11 could protect cells from apoptosis triggered by either TNF or anti-Fas. The results (Fig. 9D) demonstrate that there is no consistent increase in viability following treatment with either TNF or anti-Fas in cells stably expressing E6 from HPV-6b or -11. In the one case where there did appear to be some protection from TNF (HPV-6b, clone 6), the RT-PCR analysis (Fig. 9B) showed an absence of the E6 message, suggesting that the reason for this protection was independent of E6 expression. For comparison, we also examined the ability of the HPV-16, HPV-6b, and HPV-11 versions of E6 to mediate the rapid degradation of p53 (Fig. 9E). As expected, both the HA- and Flag-tagged versions of HPV-16 E6 caused a significant decrease in the cellular level of p53, while neither the HPV-6b nor the HPV-11 version did so.
DISCUSSION
When the HPV-16 E6 protein is expressed in cells, it affects the ability of the cells to respond to apoptotic signals generated by the host in several significant ways. It has long been known, for example, that E6 accelerates the degradation of p53, thus interfering with the cellular response to apoptotic signals generated by genotoxic damage (50). In addition, previous work by our laboratory has shown that E6 also inhibits cell death pathways triggered by receptor-ligand interactions by binding to TNF R1 (16) and to FADD (15) and can thus protect cells from TNF- and Fas-mediated apoptosis. Because procaspase 8 shares substantial structural and functional homology with FADD, it also seemed a likely target for E6. Consistent with this idea, our previous work (19) had demonstrated that in E6-expressing HCT-116 cells, the level of endogenous procaspase 8 was reduced, although the mechanism for this reduction was not defined. Targeting procaspase 8 could provide significant advantages for viral replication, as its central role in extrinsically triggered apoptosis implies that the ability of the virus to control its activity could allow the virus to modulate apoptotic pathways in addition to the TNF and Fas pathways (such as pathways triggered by TRAIL).
In order to define the molecular mechanism underlying the decrease of procaspase 8 in E6-expressing cells, we examined the ability of E6 to bind to and affect the biological activity of procaspase 8. We also looked at the contributions to this binding and modulation made by the two naturally occurring isoforms of E6 separately, as well as in combination. We found that both the small and large versions of E6 can indeed bind to procaspase 8 (Fig. 6) but that the consequences of this binding are very different. When the large isoform of E6 binds, procaspase 8 is destabilized and its rate of degradation is enhanced (Fig. 5), leading to a decrease in the protein level (Fig. 3B and 4A). On the other hand, when the small isoform of E6 binds, procaspase 8 is stabilized and its rate of degradation is lowered (Fig. 5A), leading to an increase in the protein level (Fig. 4A). Several isoforms of E6 are generated by alternative splicing (E6*I to E6*IV), and the E6*I (∼0.20-kb) and E6*II (∼0.3-kb) transcripts code for nearly identical proteins that differ by only the C-terminal 7 amino acids. Our results did not distinguish any possible differences in the activities of these two proteins. In our stable clones, the 0.3-kb transcript appeared to be more abundant. Interestingly, E6 from the low-risk types had no effect on the cellular levels of procaspase 8 (Fig. 9C), suggesting that this particular activity could have a role in tumor formation.
Previous work regarding the effect of HPV-16 E6 on the cellular levels of proteins to which it binds have reported either that E6 has no significant effect (for example, TNF R1 [13], TRIP-Br1 [23], and BRCA1 [69]) or that it reduces their levels by way of accelerated degradation [for example, FADD (15), DLG (46), myc (21), ADA (30), Bak (60), Bax (37), hScrib (41), MUPP1 (32), MAGI-1 (20), MAGI-2 and MAGI-3 (63), O(6)-methylguanine-DNA methyltransferase (55), and tuberin (36)]. To our knowledge, this is the first report of an isoform of E6 stabilizing a protein.
During an actual infection, of course, both the large and small versions of E6 are likely to be available. The consequences of their combined effects can be seen when cells are transfected with the intact E6 gene, which produces both the large and small isoforms. Some of these clones display higher (for example, U2OSE66) and some lower (for example, U2OSE612) levels of procaspase 8 (Fig. 1A), probably reflecting differing relative amounts of the two forms and their opposing activities in the different clones. There are likely to be a number of factors that influence these relative amounts, and our results (Fig. 2) suggest that the dosage of E6 may be one of these factors. For example, high levels of E6 correlate with high cellular levels of procaspase 8, while lower levels of E6 (below a certain threshold) correlate with low levels of procaspase 8 (Fig. 2A). Such a shift could be explained if the absolute and/or relative activities of the two forms varied with increased expression. For example, it may be that at low levels of E6 expression, the activity of E6large predominates and procaspase 8 is destabilized, while at high levels of E6 expression, the activity of E6small/E6* may predominate, leading to procaspase 8 stabilization.
During activation of the apoptotic pathway, the DED of FADD binds to the DED of procaspase 8, and a mutation in caspase 8 at residue L62 can eliminate this binding (35). According to our results, E6large binds to FADD within the first 23 amino acids of its N terminus (15), as well as to the procaspase 8 DED (Fig. 6), and this binding reduces the affinity of procaspase 8 and FADD for each other (Fig. 7B). One can therefore envision the large E6 protein competing for access to the FADD/procaspase 8 binding sites, which leads to an acceleration of proteolysis of FADD and procaspase 8 and results in protection of cells from apoptosis. This is likely the case when E6 levels are low and/or when the large isoform of E6 is the predominant functional species. However, if E6 levels are high and/or if the small isoform of E6 is the predominant functional species, procaspase 8 can be stabilized and the cells therefore sensitized to apoptotic signaling (Fig. 1 and 2) (13).
It has been known for some time that more than one transcript is produced from the E6 genes of oncogenic types of HPV (44, 45, 48, 51, 52). Intriguingly, we found that of the three types tested, HPV-16, -6b, and -11, only the high-risk HPV-16 E6 was able to generate a short isoform, suggesting that the ability to generate E6small/E6* may play a role in oncogenesis. Initially, it was thought that the alternatively spliced versions of the E6 message might function primarily to enhance the translation efficiency of E7 (51), but more recent work suggests that the shorter E6 protein is actually produced (15) and can function by binding to and influencing the biological functions of the large E6 protein. For example, the short version of HPV-18 E6, closely related to the short version of HPV-16 E6, can bind to the large versions of HPV-18 E6, HPV-16 E6, and E6AP; inhibit E6-mediated degradation of p53; and increase p53-mediated transcription and growth arrest in E6-expressing cells (44, 45). Further work by the same laboratory showed that the short isoform appears to be up-regulated during G2/M and that its overexpression can reduce the amount of large E6 present in the insoluble nuclear and membrane fractions (22). In addition, the localization of these variants varies within cells (22). In previous work, our laboratory has shown that the short isoform of HPV-16 E6, though produced, does not bind to FADD or protect cells from apoptosis mediated through the Fas pathway, although the large E6 protein does (15). In contrast, the small variant of E6 does bind to and stabilize procaspase 8. Intriguingly, this action appears to directly oppose that of the large isoform, suggesting that in viruses, as well as in eukaryotic cells, alternatively spliced variants can have opposite biological effects.
It is possible that the relative scarcity of data regarding the roles of E6small/E6* compared to the data for E6large can be partially attributed to the difficulty of detecting this isoform at the protein level. This may be due in part to rapid protein turnover (56, 57). Another reason may be a possible tendency of this isoform to form complexes with itself and/or the large isoform of E6. Although we have been able to demonstrate expression of the small isoform following transient expression (Fig. 3A), in our hands, the sensitivities of immunoblotting and immunoprecipitation approaches have been insufficient to detect the lower levels of expression found in stably transfected cells. For these reasons, we have routinely used RT-PCR analysis to monitor expression of HA-E6small/E6*.
During an actual infection by HPV-16, the level of E6/E7 oncogene expression is repressed, though not entirely eliminated, by E2 (reference 10 and references therein). The resulting low-level expression of E6 may well be effective in protecting cells from immune system mediators, such as TNF, and thus decreasing viral clearance, and may therefore represent the biologically relevant level of this oncoprotein in the context of the virus life cycle. Following the relatively rare event of integration of HPV-16 into the host genome, the E2 gene is frequently disrupted, thus relieving the repression of E6/E7, allowing increased levels of expression of these oncogenes and thereby contributing to their oncogenic potential. A characterization of the levels and ratios of these isoforms at different stages in the life cycle, as well as in the various neoplastic stages of human samples, will be critical for developing a detailed understanding of the various roles of E6.
Together, these results strengthen and extend the emerging sense that HPV-16 has developed multiple approaches, several of which are mediated by E6, that are designed to modify the response of the cells it infects to signals affecting cell death and survival. These mechanisms can be very complex, and a molecular understanding of them is necessary in order to enhance efforts directed toward the best use of existing agents, as well as for the development of novel and effective therapeutic approaches.
Acknowledgments
This work was supported by NCI Grant R01 CA095461 from the National Institutes of Health.
Footnotes
Published ahead of print on 31 January 2007.
REFERENCES
- 1.Beidler, D. R., M. Tewari, P. D. Friesen, G. Poirier, and V. M. Dixit. 1995. The baculovirus p35 protein inhibits Fas- and tumor necrosis factor-induced apoptosis. J. Biol. Chem. 270:16526-16528. [DOI] [PubMed] [Google Scholar]
- 2.Belanger, C., A. Gravel, A. Tomoiu, M. E. Janelle, J. Gosselin, M. J. Tremblay, and L. Flamand. 2001. Human herpesvirus 8 viral FLICE-inhibitory protein inhibits Fas-mediated apoptosis through binding and prevention of procaspase-8 maturation. J. Hum. Virol 4:62-73. [PubMed] [Google Scholar]
- 3.Benedict, C. A. 2003. Viruses and the TNF-related cytokines, an evolving battle. Cytokine Growth Factors Rev. 14:349-357. [DOI] [PubMed] [Google Scholar]
- 4.Boatright, K. M., M. Renatus, F. L. Scott, S. Sperandio, H. Shin, I. M. Pedersen, J. E. Ricci, W. A. Edris, D. P. Sutherlin, D. R. Green, and G. S. Salvesen. 2003. A unified model for apical caspase activation. Mol. Cell 11:529-541. [DOI] [PubMed] [Google Scholar]
- 5.Carrasco, R. A., N. B. Stamm, and B. K. R. Patel. 2003. One-step cellular caspase 3/7 assay. BioTechniques 34:1064-1067. [DOI] [PubMed] [Google Scholar]
- 6.Chen, J. J., C. E. Reid, V. Band, and E. J. Androphy. 1995. Interaction of papillomavirus E6 oncoproteins with a putative calcium-binding protein. Science 269:529-531. [DOI] [PubMed] [Google Scholar]
- 7.Chen, M., A. Orozco, D. M. Spencer, and J. Wang. 2002. Activation of initiator caspases through a stable dimeric intermediate. J. Biol. Chem. 277:50761-50767. [DOI] [PubMed] [Google Scholar]
- 8.Chen, P., J. Tian, I. Kovesdi, and J. T. Bruder. 1998. Interaction of the adenovirus 14.7 kDa protein with FLICE inhibits Fas ligand-induced apoptosis. J. Biol. Chem. 273:5815-5820. [DOI] [PubMed] [Google Scholar]
- 9.Degenhardt, Y. Y., and S. Silverstein. 2001. Interaction of zyxin, a focal adhesion protein, with the E6 protein from human papillomavirus type 6 results in its nuclear translocation. J. Virol. 23:11791-11802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Doorbar, J. 2005. The papillomavirus life cycle. J. Clin. Virol. 32(Suppl. 1):S7-S15. [DOI] [PubMed] [Google Scholar]
- 11.Du, M., X. Fan, E. Hong, and J. J. Chen. 2002. Interaction of oncogenic papillomavirus E6 proteins with fibulin-1. Biochem. Biophys. Res. Commun. 296:962-969. [DOI] [PubMed] [Google Scholar]
- 12.Elbel, M., S. Carl, S. Spaderna, and T. Iftner. 1997. A comparative analysis of the interactions of the E6 proteins from cutaneous and genital papillomaviruses with p53 and E6AP in correlation to their transforming potential. Virology 239:132-149. [DOI] [PubMed] [Google Scholar]
- 13.Filippova, M., T. A. Brown-Bryan, C. A. Casiano, and P. J. Duerksen-Hughes. 2005. The human papillomavirus 16 E6 protein can render cells either sensitive or resistant to TNF: effect of dose. Cell Death Differ. 12:1622-1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Filippova, M., and P. J. Duerksen-Hughes. 2003. Inorganic and dimethylated arsenic species induce cellular p53. Chem. Res. Toxicol. 16:423-431. [DOI] [PubMed] [Google Scholar]
- 15.Filippova, M., L. Parkhurst, and P. J. Duerksen-Hughes. 2004. The human papillomavirus 16 E6 protein binds to Fas-associated death domain and protects cells from Fas-triggered apoptosis. J. Biol. Chem. 279:25729-25744. [DOI] [PubMed] [Google Scholar]
- 16.Filippova, M., H. Song, J. L. Connolly, T. S. Dermody, and P. J. Duerksen-Hughes. 2002. The human papillomavirus 16 E6 protein binds to tumor necrosis factor (TNF) R1 and protects cells from TNF-induced apoptosis. J. Biol. Chem. 277:21730-21739. [DOI] [PubMed] [Google Scholar]
- 17.Finzer, P., A. Aguilar-Lemarroy, and F. Rosl. 2002. The role of human papillomavirus oncoproteins E6 and E7 in apoptosis. Cancer Lett. 188:15-24. [DOI] [PubMed] [Google Scholar]
- 18.Gao, Q., S. Srinivasan, S. N. Boyer, D. E. Wazer, and V. Band. 1999. The E6 oncoproteins of high-risk papillomaviruses bind to a novel putative GAP protein, E6TP1, and taraget it for degradation. Mol. Cell Biol. 19:733-744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Garnett, T. O., M. Filippova, and P. J. Duerksen-Hughes. 2006. Accelerated degradation of FADD and procaspase 8 in cells expressing human papilloma virus 16 E6 impairs TRAIL-mediated apoptosis. Cell Death Differ. 13:1915-1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Glausinger, B. A., S. S. Lee, M. Thomas, L. Banks, and R. T. Javier. 2000. Interactions of the PDZ-protein MAGI-1 with adenovirus E4 ORF1 and high-risk papillomavirus E6 oncoproteins. Oncogene 19:5270-5280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gross-Mesilaty, S., E. Reinstein, B. Bercovich, K. E. Tobias, A. L. Schwartz, C. Kahana, and A. Ciechanover. 1998. Basal and human papillomavirus E6 oncoprotein-induced degradation of Myc proteins by the ubiquitin pathway. Proc. Natl. Acad. Sci. USA 95:8058-8063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guccione, E., D. Pim, and L. Banks. 2004. HPV-18 E6*I modulates HPV-18 full-length E6 functions in a cell cycle dependent manner. Int. J. Cancer 110:928-933. [DOI] [PubMed] [Google Scholar]
- 23.Gupta, S., P. P. Takhar, R. Degenkolbe, C. H. Koh, H. Zimmermann, C. M. Yang, K. Guan Sim, S. I. Hsu, and H. U. Bernard. 2003. The human papillomavirus type 11 and 16 E6 proteins modulate the cell-cycle regulator and transcription cofactor TRIP-Br1. Virology 317:155-164. [DOI] [PubMed] [Google Scholar]
- 24.Hay, S., and G. Kannourakis. 2002. A time to kill: viral manipulation of the cell death program. J. Gen. Virol. 83:1547-1564. [DOI] [PubMed] [Google Scholar]
- 25.Henderson, G., W. Peng, L. Jin, G. C. Perng, A. B. Nesburn, S. L. Wechsler, and C. Jones. 2002. Regulation of caspase 8- and caspase 9-induced apoptosis by the herpes simplex virus type 1 latency-associated transcript. J. Neurovirol. 8(Suppl. 2):103-111. [DOI] [PubMed] [Google Scholar]
- 26.Iftner, T., M. Ebel, B. Schopp, T. Hiler, J. I. Loizou, K. W. Caldecott, and F. Stubrenrauch. 2002. Interference of papillomavirus E6 protein with single-strand break repair by interaction with XRCC1. EMBO J. 21:47141-47148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kiyono, T., A. Hiraiwa, M. Fujita, Y. Hayashi, T. Akiyama, and M. Ishibashi. 1997. Binding of high-risk human papillomavirus E6 oncoproteins to the human homologue of the Drosophila discs large tumor suppressor protein. Proc. Natl. Acad. Sci. USA 94:11612-11616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kuhne, C., and L. Banks. 1998. E3-ubiquitin ligase/E6-AP links multicopy maintenance protein 7 to the ubiquitination pathway by a novel motif, the L2G box. J. Biol. Chem. 273:34302-34309. [DOI] [PubMed] [Google Scholar]
- 29.Kukimoto, I., S. Aihara, K. Yoshiike, and T. Kanda. 1998. Human papillomavirus oncoprotein E6 binds to the C-terminal region of human minichromosome maintenance 7 protein. Biochem. Biophys. Res. Commun. 249:258-262. [DOI] [PubMed] [Google Scholar]
- 30.Kumar, A., Y. Zhao, G. Meng, M. Zeng, S. Srinivasan, L. M. Delmolino, Q. Gao, G. Dimri, G. F. Weber, D. E. Wazer, H. Band, and V. Band. 2002. Human papillomavirus oncoprotein E6 inactivates the transcriptional coactivator human ADA3. Mol. Cell. Biol. 22:5801-5812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Langelier, Y., S. Bergeron, S. Chabaud, J. Lippens, C. Guilbault, A. M.-J. Sasseville, S. Denis, D. D. Mosser, and B. Massie. 2002. The R1 subunit of herpes simplex virus ribonucleotide reductase protects cells against apoptosis at, or upstream of, caspase-8 activation. J. Gen. Virol. 83:2779-2789. [DOI] [PubMed] [Google Scholar]
- 32.Lee, S. S., B. A. Glausinger, F. Mantovani, L. Banks, and R. T. Javier. 2000. Multi-PDZ domain protein MUPP1 is a cellular target for both adenovirus E4-ORF1 and high-risk papillomavirus type 18 E6 oncoproteins. J. Virol. 74:9680-9693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee, S. S., R. S. Weiss, and R. T. Javier. 1997. Binding of human virus oncoproteins to hDlg/SAP97, a mammalian homolog of the Drosophila disks large tumor suppressor protein. Proc. Natl. Acad. Sci. USA 94:6670-6675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li, Y., J. Kang, and M. S. Horwitz. 1997. Interaction of an adenovirus 14.7-kilodalton protein inhibitor of tumor necrosis factor alpha cytolysis with a new member of the GTPase superfamily of signal transducers. J. Virol. 71:1576-1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu, B., D. Peng, Y. Lu, W. Jin, and Z. Fan. 2002. A novel single amino acid deletion caspase-8 mutant in cancer cells that lost proapoptotic activity. J Biol. Chem. 277:30159-30164. [DOI] [PubMed] [Google Scholar]
- 36.Lu, Z., X. Hu, Y. Li, L. Zheng, Y. Zhou, H. Jiang, T. Ning, Z. Basang, C. Zhang, and Y. Ke. 2004. Human papillomavirus 16 E6 oncoprotein interferences with insulin signaling pathway by binding to tuberin. J. Biol. Chem. 279:35664-35670. [DOI] [PubMed] [Google Scholar]
- 37.Magal, S. S., A. Jackman, S. Ish-Shalom, L. E. Botzer, P. Gonen, R. Schlegel, and L. Sherman. 2005. Downregulation of Bax mRNA expression and protein stability by the E6 protein of human papillomavirus 16. J. Gen. Virol. 86:611-621. [DOI] [PubMed] [Google Scholar]
- 38.Marsters, S. A., R. M. Pitti, C. J. Donahue, S. Ruppert, K. D. Bauer, and A. Askenazi. 1996. Activation of apoptosis by Apo-2 ligand is independent of FADD but blocked by CrmA. Curr. Biol. 6:750-752. [DOI] [PubMed] [Google Scholar]
- 39.Miura, M., R. M. Friedlander, and J. Yuan. 1995. Tumor necrosis factor-induced apoptosis is mediated by a CrmA-sensitive cell death pathway. Proc. Natl. Acad. Sci. USA 92:8318-8322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Miyasaka, Y., N. Enomoto, M. Kurosaki, N. Sakamoto, N. Kanazawa, T. Kohashi, E. Ueda, S. Maekawa, H. Watanabe, N. Izumi, C. Sato, and M. Watanabe. 2003. Hepatitis C virus nonstructural protein 5A inhibits tumor necrosis factor-alpha-mediated apoptosis in Huh7 cells. J. Infect. Dis. 188:1537-1544. [DOI] [PubMed] [Google Scholar]
- 41.Nakagawa, S., and J. M. Huibregtse. 2000. Human scribble (Vartul) is targeted for ubiquitin-mediated degradation by the high-risk papillomavirus E6 proteins and the E6AP ubiquitin-protein ligase. Mol. Cell. Biol. 20:8244-8253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pan, H., and A. E. Griep. 1995. Temporally distinct patterns of p53-dependent and p53-independent apoptosis during mouse lens development. Genes Dev. 9:2157-2169. [DOI] [PubMed] [Google Scholar]
- 43.Patel, D., S. M. Huang, L. A. Baglia, and D. J. McCance. 1999. The E6 protein of human papillomavirus type 16 binds to and inhibits co-activation by CBP and p300. EMBO J. 18:5061-5072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pim, D., and L. Banks. 1999. HPV-18 E6*I protein modulates the E6-directed degradation of p53 by binding to full-length HPV-18 E6. Oncogene 18:7403-7408. [DOI] [PubMed] [Google Scholar]
- 45.Pim, D., P. Massimi, and L. Banks. 1997. Alternatively spliced HPV-18 E6* protein inhibits E6 mediated degradation of p53 and suppresses transformed cell growth. Oncogene 15:257-264. [DOI] [PubMed] [Google Scholar]
- 46.Pim, D., M. Thomas, R. Javier, D. Gardiol, and L. Banks. 2000. HPV E6 targeted degradation of the discs large protein: evidence for the involvement of a novel ubiquitin ligase. Oncogene 19:719-725. [DOI] [PubMed] [Google Scholar]
- 47.Riedl, S. J., and Y. Shi. 2004. Molecular mechanisms of caspase regulation during apoptosis. Nat. Rev. Mol. Cell. Biol. 5:897-907. [DOI] [PubMed] [Google Scholar]
- 48.Roggenbuck, B., P. M. Larsen, S. J. Fey, D. Bartsch, L. Gissmann, and E. Schwarz. 1991. Human papillomavirus type 18 E6*, E6, and E7 protein synthesis in cell-free translation systems and comparison of E6 and E7 in vitro translation products to proteins immunoprecipitated from human epithelial cells. J. Virol. 65:5068-5072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ronco, L. V., A. Y. Karpova, M. Vidal, and P. M. Howley. 1998. Human papillomavirus 16 E6 oncoprotein binds to interferon regulatory factor-3 and inhibits its transcriptional activity. Genes Dev. 12:2061-2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Scheffner, M., B. A. Werness, J. M. Huibregtse, A. J. Levine, and P. M. Howley. 1990. The E6 oncoprotein encoded by human papillomavirus types 16 and 18 promotes the degradation of p53. Cell 63:1129-1136. [DOI] [PubMed] [Google Scholar]
- 51.Sedman, S. A., M. S. Barbosa, W. C. Vass, N. L. Hubbert, J. A. Haas, D. R. Lowy, and J. T. Schiller. 1991. The full-length E6 protein of human papillomavirus type 16 has transforming and trans-activating activities and cooperates with E7 to immortalize keratinocytes in culture. J. Virol. 65:4860-4866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shally, M., N. Alloul, A. Jackman, M. Muller, L. Gissmann, and L. Sherman. 1996. The E6 variant proteins E6I-E6IV of human papillomavirus 16: expression in cell free systems and bacteria and study of their interaction with p53. Virus Res. 42:81-96. [DOI] [PubMed] [Google Scholar]
- 53.Skaletskaya, A., L. M. Bartle, T. Chittenden, L. McCormick, E. S. Maocarski, and V. S. Goldmacher. 2001. A cytomegalovirus encoded inhibitor of apoptosis that suppresses caspase-8 activation. Proc. Natl. Acad. Sci. USA 98:7829-7834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Song, S., G. A. Gulliver, and P. F. Lambert. 1998. Human papillomavirus type 16 E6 and E7 oncogenes abrogate radiation-induced DNA damage responses in vivo through p53-dependent and p53-independent pathways. Proc. Natl. Acad. Sci USA 95:2290-2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Srivenugopal, K. S., and F. Ali-Osman. 2002. The DNA repair protein, O(6)-methylguanine-DNA methyltransferase is a proteolytic target for the E6 human papillomavirus oncoprotein. Oncogene 21:5940-5945. [DOI] [PubMed] [Google Scholar]
- 56.Stacey, S. N., D. Jordan, P. J. Snijders, M. Mackett, J. M. Walboomers, and J. R. Arrand. 1995. Translation of the human papillomavirus type 16 E7 oncoprotein from bicistronic mRNA is independent of splicing events within the E6 open reading frame. J. Virol. 69:7023-7031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Stacey, S. N., D. Jordan, A. J. Williamson, M. Brown, J. H. Coote, and J. R. Arrand. 2000. Leaky scanning is the predominant mechanism for translation of human papillomavirus type 16 E7 oncoprotein from E6/E7 bicistronic mRNA. J. Virol. 74:7284-7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Talley, A. K., S. Dewhurst, S. W. Perry, S. C. Dollard, S. Gummuluru, S. M. Fine, D. New, L. G. Epstein, H. E. Gendelman, and H. A. Gelbard. 1995. Tumor necrosis factor alpha-induced apoptosis in human neuronal cells: protection by the antioxidant N-acetylcysteine and the genes bcl-2 and crmA. Mol. Cell. Biol. 15:2359-2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tewari, M., and V. M. Dixit. 1995. Fas- and tumor necrosis factor-induced apoptosis is inhibited by the poxvirus crmA gene product. J. Biol. Chem. 270:3255-3260. [DOI] [PubMed] [Google Scholar]
- 60.Thomas, M., and L. Banks. 1999. Human papillomavirus (HPV) E6 interactions with Bak are conserved amongst E6 proteins from high and low risk HPV types. J. Gen. Virol. 80:1513-1517. [DOI] [PubMed] [Google Scholar]
- 61.Thomas, M., and L. Banks. 1998. Inhibition of Bak-induced apoptosis by HPV-18 E6. Oncogene 17:2943-2954. [DOI] [PubMed] [Google Scholar]
- 62.Thomas, M., B. A. Glausinger, D. Pim, R. T. Javier, and L. Banks. 2001. HPV E6 and MAGUK protein interactions: determination of the molecular basis for specific protein recognition and degradation. Oncogene 20:5431-5439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Thomas, M., R. Laura, K. Hepner, E. Guccione, C. Sawyers, L. Lasky, and L. Banks. 2002. Oncogenic human papiillomavirus E6 proteins target the MAGI-2 and MAGI-3 proteins for degradation. Oncogene 21:5088-5096. [DOI] [PubMed] [Google Scholar]
- 64.Tong, X., and P. M. Howley. 1997. The bovine papillomavirus E6 oncoprotein interacts with paxillin and disrupts the actin cytoskeleton. Proc. Natl. Acad. Sci. USA 94:4412-4417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tortorella, D., B. E. Gewurz, M. H. Furman, D. J. Schust, and H. L. Ploegh. 2000. Viral subversion of the immune system. Annu. Rev. Immunol. 18:861-926. [DOI] [PubMed] [Google Scholar]
- 66.Tross-Mesilaty, S., E. Reinstein, B. Bercovich, K. E. Tobias, A. L. Schwartz, C. Kahana, and A. Ciechanover. 1998. Basal and human papillomavirus E6 oncoprotein-induced degradation of Myc proteins by the ubiquitin pathway. Proc. Natl. Acad. Sci. USA 95:8058-8063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Veldman, T., X. Liu, H. Yuan, and R. Schlegel. 2003. Human papillomavirus E6 and Myc proteins associate in vivo and bind to and cooperatively activate the telomerase reverse transcriptase promoter. Proc. Natl. Acad. Sci. USA 100:8211-8216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zeng, M., A. Kumar, G. Meng, Q. Gao, G. Dimri, D. E. Wazer, H. Band, and V. Band. 2002. Human papillomavirus 16 E6 oncoprotein inhibits retinoic X receptor-mediated transactivation by targeting human ADA3 coactivator. J. Biol. Chem. 277:45611-45618. [DOI] [PubMed] [Google Scholar]
- 69.Zhang, Y., S. Fan, Q. Meng, Y. Ma, P. Katiyar, R. Schlegel, and E. M. Rosen. 2005. BRCA1 interaction with human papillomavirus oncoproteins. J. Biol. Chem. 280:33165-33177. [DOI] [PubMed] [Google Scholar]
- 70.Zimmermann, H., R. Degenkolbe, H. Bernard, and M. O'Connor. 1999. The human papillomavirus type 16 E6 oncoprotein can down-regulate p53 activity by targeting the transcriptional coactivator CBP/p300. J. Virol. 73:6209-6219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zimmermann, K. C., C. Bonzon, and D. R. Green. 2001. The machinery of programmed cell death. Pharmacol. Therap. 92:57-70. [DOI] [PubMed] [Google Scholar]