Abstract
Infections with RNA viruses are sensed by the innate immune system through membrane-bound Toll-like receptors or the cytoplasmic RNA helicases RIG-I and MDA-5. It is believed that MDA-5 is crucial for sensing infections by picornaviruses, but there have been no studies on the role of this protein during infection with poliovirus, the prototypic picornavirus. Beginning at 4 h postinfection, MDA-5 protein is degraded in poliovirus-infected cells. Levels of MDA-5 declined beginning at 6 h after infection with rhinovirus type 1a or encephalomyocarditis virus, but the protein was stable in cells infected with rhinovirus type 16 or echovirus type 1. Cleavage of MDA-5 is not carried out by either poliovirus proteinase 2Apro or 3Cpro. Instead, degradation of MDA-5 in poliovirus-infected cells occurs in a proteasome- and caspase-dependent manner. Degradation of MDA-5 during poliovirus infection correlates with cleavage of poly(ADP) ribose polymerase (PARP), a hallmark of apoptosis. Induction of apoptosis by puromycin leads to cleavage of both PARP and MDA-5. The MDA-5 cleavage product observed in cells treated with puromycin is ∼90 kDa, similar in size to the putative cleavage product observed in poliovirus-infected cells. Poliovirus-induced cleavage of MDA-5 may be a mechanism to antagonize production of type I interferon in response to viral infection.
The first line of the host immune response against viruses comprises the innate immune system, which responds within minutes after viruses infect a cell by mounting an antiviral response (16). Sensing of RNA viruses occurs through recognition of viral components such as double-stranded RNA (dsRNA), which is produced during viral RNA replication and may also be present within single-stranded RNA genomes. Detection of dsRNA triggers the innate immune system to produce the antiviral cytokines alpha interferon (IFN-α) and IFN-β.
Sensing of dsRNA by the innate immune system is accomplished either by Toll-like receptors or by cytoplasmic sensors such as PKR, RIG-I, and MDA-5 (1). Both RIG-I and MDA-5 proteins contain an RNA helicase domain, which binds dsRNA, and caspase recruitment domains that are involved in signaling (19, 20, 33). Binding of dsRNA to RIG-I and MDA-5 leads to interaction with a caspase recruitment domain-containing adaptor protein called IPS-1, MAVS, VISA, or Cardif (22, 28, 30, 32). This outer mitochondrial membrane protein mediates the recruitment and activation of protein kinases that phosphorylate the transcription factor IFN-regulatory factor 3, leading to the synthesis of type I IFN.
Although RIG-I and MDA-5 are similar proteins that induce type I IFN synthesis through the same pathway, they appear to specialize in recognition of different viruses. Analyses of mice lacking the gene encoding either RIG-I or MDA-5 have revealed that RIG-I is essential for detecting infection by influenza viruses, paramyxoviruses, and Japanese encephalitis virus (21). In contrast, MDA-5 is critical for detecting infection with encephalomyocarditis virus (EMCV) (11, 21). Mice lacking the mda-5 gene are deficient in the production of type I IFN in response to EMCV infection and are more susceptible to infection with this virus. It has therefore been concluded that mda-5 is critical for sensing infection with picornaviruses.
The importance of the RIG-I/MDA-5 sensing pathway is underscored by the existence of viral proteins that antagonize its function. The V proteins of paramyxoviruses bind MDA-5, blocking activation of the IFN-β promoter (2). RIG-I-mediated activation of IFN-regulatory factor 3 is suppressed in cells infected with hepatitis A virus, although the mechanism is not known (9). The NS3-4a proteinase of hepatitis C virus cleaves IPS-1, inhibiting type I IFN responses (28).
Despite the rapid advances in our understanding of the role of RIG-I and MDA-5 in sensing viral dsRNA, there have been no studies on the function of these proteins during infection with poliovirus, the prototypic picornavirus. In this study, we found that MDA-5 protein is degraded in poliovirus-infected cells. MDA-5 is not directly cleaved by either viral proteinase 2Apro or 3Cpro. Instead, degradation of MDA-5 in poliovirus-infected cells occurs in a proteasome- and caspase-dependent manner, and correlates with cleavage of poly(ADP) ribose polymerase (PARP), a hallmark of apoptosis. Poliovirus-induced cleavage of MDA-5 may be a mechanism to antagonize production of type I IFN in response to viral infection.
MATERIALS AND METHODS
Cells and viruses.
S3 HeLa and CHP100L cells were grown in Dulbecco's modified Eagle medium (Invitrogen, Carlsbad, CA), 10% bovine calf serum (HyClone, Logan, UT), and 1% penicillin-streptomycin (Invitrogen). For plaque assays HeLa cells were grown in Dulbecco's modified Eagle medium (Specialty Media, Philipsburg, NJ), 0.2% NaHCO3, 5% bovine calf serum, 1% penicillin-streptomycin, and 0.9% Bacto-agar (Difco, Franklin Lakes, NJ).
Replication-defective human adenoviruses with or without the coding region for mda-5 were produced as described previously (15). Virus titer was determined by fluorescent-focus assay in HEK 293 cells. For expression of mda-5, viruses were adsorbed to cells at a multiplicity of infection (MOI) of 50 PFU per cell for 2 h. After the addition of culture medium, cells were incubated for 16 h before infection with poliovirus.
Stocks of poliovirus strain P1/Mahoney, rhinovirus type 16, and EMCV were produced by transfecting HeLa cells with RNA transcripts derived by in vitro transcription of plasmids harboring complete DNA copies of the viral genomes (8, 26, 29). Stocks of echovirus type 1 and rhinovirus type 1a were obtained from the American Type Culture Collection, Manassas, VA, and were propagated in HeLa cells. Poliovirus mutant Se1-3C-02, which contains the single amino acid change V54A in 3Cpro (7), was obtained from B. Semler, University of California, Irvine. A poliovirus mutant with a single amino acid change, Y88L (35), in 2Apro was constructed by site-directed mutagenesis of a full-length DNA copy of the genome of poliovirus strain P1/Mahoney. Both amino acid changes, Y88L in 2Apro and V54A in 3Cpro, were introduced into the poliovirus genome by site-directed mutagenesis.
Reagents.
Rabbit antibody against MDA-5 was produced by immunizing rabbits with a purified protein encompassing the first 335 amino acids of the protein linked to glutathione S-transferase (27). Mouse monoclonal anti-EF1α was purchased from Upstate USA Inc., Chicago, IL. Rabbit polyclonal anti-PARP was purchased from Cell Signaling Technology, Danvers, MA. Mouse monoclonal anti-poly(A)-binding protein (anti-PABP) was purchased from Abcam, Cambridge, MA. The proteasome inhibitors MG132 and epoxomicin were purchased from Calbiochem, San Diego, CA. The general caspase inhibitor benzyloxycarbonyl-Val-Ala-Asp-(OMe) fluoromethylketone (Z-VAD-FMK) was purchased from R&D Systems, Minneapolis, MN. Poly(IC) was purchased from Amersham BioSciences, Piscataway, NJ. Purified poliovirus proteinase 3CDpro was a gift of Bert Semler, University of California, Irvine. Poliovirus 3CDpro is a precursor to 3Cpro that is believed to be the proteinase responsible for most cleavages at QG pairs in the viral polyprotein (34). Purified coxsackievirus B3 2Apro was a gift of Richard Lloyd, Baylor College of Medicine (17).
Virus replication in cultured cells.
Monolayers of adherent cells were infected with poliovirus at a multiplicity of 10 PFU per cell. At the specified time after infection, cells and medium were collected, subjected to three freeze-thaw cycles to release intracellular virus, and clarified by centrifugation. The virus titer was determined by plaque assay on monolayers of HeLa cells.
In vitro translation.
Coupled transcription-translation experiments were performed using the TNT quick-coupled transcription-translation system (Promega Corp, Madison, WI) in the presence of [35S]methionine (Amersham BioSciences, Piscataway, NJ). MDA-5 DNA was cloned into the vector pcDNA3.1, which drives transcription with the T7 promoter. One microgram of DNA was added to each 50 μl reaction mixture, and the manufacturer's protocol was followed. The in vitro-translated MDA-5 was added to cleavage buffer containing 100 mM NaCl2, 5 mM MgCl2, and 10 mM HEPES-KOH (pH 7.4) in the presence or absence of 3CDpro or 2Apro. The cleavage reaction mixtures were incubated at 37°C for 4 h. The products were separated by 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and the gel was dried, exposed to a phosphorimager plate, and developed after 24 h.
IFN-β RNA expression analysis.
At different times after viral or mock infection, HeLa cells were lysed with RLT buffer (QIAGEN, Valencia, CA) and frozen at −80°C overnight. RNA was then extracted from thawed samples with the RNeasy kit (QIAGEN) according to the protocol provided by the manufacturer. An on-column DNase treatment step was carried out using an RNase-free DNase set (QIAGEN). RNA concentrations were measured using the ND-1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE). Two micrograms of RNA from each time point were converted to cDNA by using Superscript III RNA polymerase, random primer, and First-Strand buffer from Invitrogen according to the protocol provided by the manufacturer. Deoxynucleoside triphosphates were purchased from Stratagene (La Jolla, CA), while RNase inhibitor (rRNasin) was purchased from Promega, Madison, WI. Real-time PCR analyses were carried out on the Prism 7500 real-time PCR system (Applied Biosystems, Foster City, CA). Triplicate reactions with a final volume of 25 μl were performed in 96-well Micro Optical plates (Applied Biosystems). For each sample, 20 ng of cDNA was added to a PCR mix made with the 5× Fast-Start SYBR green master mix containing Rox (Roche Diagnostics, Indianapolis, IN) and a final concentration of 400 nM β-actin or IFN-β primers. The primers were designed using the Universal Probe Library software provided at the Roche Diagnostics website, and their sequences are as follows: IFN-β forward primer, CGA CAC TGT TCG TGT TGT CA; IFN-β reverse primer, GAA GCA CAA CAG GAG AGC AA; β-actin forward primer, AGG GAA ATC GTG CGT GAC AT; β-actin reverse primer, CAT CTG CTG GAA GGT GGA CA. Cycling conditions were 95°C for 10 minutes, followed by 45 cycles of 95°C for 31 seconds, 50°C for 31 seconds, and 72°C for 31 seconds. The ΔΔCT method of relative quantitation (as described in the Applied Biosystems user manual) was used to calculate fold change, with β-actin serving as the endogenous control for normalization.
Western blot analysis.
Cells were harvested into the culture medium with a plastic scraper, collected by centrifugation, washed with phosphate-buffered saline (PBS) (136.9 mM NaCl, 2.68 mM KCl, 8.1 mM Na2HPO4, 1.47 mM KH2PO4), and lysed in radioimmunoprecipitation buffer (PBS containing 1% NP-40, 0.5% sodium deoxycholate, and 0.1% SDS). Aliquots containing 50 μg total protein were fractionated by 10% SDS-PAGE. Proteins were transferred electrophoretically to a nitrocellulose membrane, which was then incubated with anti-MDA-5 or anti-EF1α antibody in PBS containing 5% nonfat milk for 2 h at room temperature. The membrane was washed in PBS containing 0.1% Tween, followed by addition of the appropriate secondary antibody. Proteins were visualized using the ECL chemiluminescence system (Amersham BioSciences, Piscataway, NJ).
RESULTS
Cleavage of MDA-5 during poliovirus infection.
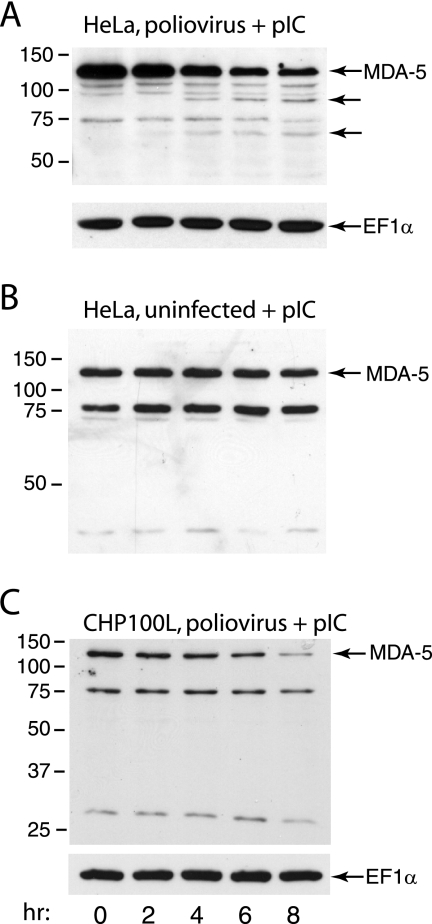
It has been suggested that picornavirus infections are detected by MDA-5, not RIG-I (21). However, sensing of picornavirus infections has been studied only with members of the cardiovirus genus, EMCV, mengovirus, and Theiler's virus (21). The poliovirus genome encodes two proteinases that could be involved in cleavage of MDA-5 during poliovirus infection as a mechanism for attenuating the IFN response. We therefore determined whether MDA-5 was degraded during poliovirus infection. HeLa cells were treated with poly(IC) for 16 h to induce the synthesis of MDA-5 and then infected with poliovirus (MOI of 10). At different times after infection, MDA-5 protein was examined by Western blot analysis. Beginning at 4 h postinfection, levels of MDA-5 protein declined, and two proteins of ∼90 and ∼70 kDa which might be cleavage products appeared (Fig. 1A). No degradation of MDA-5 protein was detected in uninfected cells up to 8 h after removal of poly(IC) (Fig. 1B). These results show that degradation of MDA-5 does not occur in uninfected cells but is induced by virus infection. Degradation of MDA-5 protein was also observed during poliovirus infection of the neuroepithelioma cell line CHP100L (Fig. 1C), indicating that cleavage is not restricted to HeLa cells. However, putative MDA-5 cleavage products were not observed in CHP100L cells.
FIG. 1.
Degradation of MDA-5 during poliovirus infection. Monolayers of HeLa cells (A, B) or CHP100L cells (C) were treated with poly(IC) (20 μg/ml) for 16 h and then infected with poliovirus (MOI, 10) (A, C) or mock infected (B). At the indicated times after infection, cell extracts were prepared and fractionated by SDS-PAGE, and MDA-5 was detected by Western blot analysis. Unlabeled arrows indicate putative cleavage products of MDA-5. The identities of the ∼75-kDa protein and smaller proteins detected by the antibody are not known; the intensities of these proteins vary depending on the batch of anti-MDA-5 antibody used. Separate bottom panels show detection of EF1α to ensure that equal amounts of protein were applied to each lane.
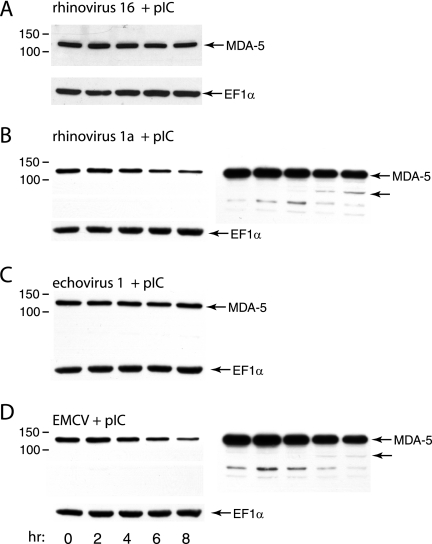
To determine if MDA-5 cleavage is restricted to cells infected with poliovirus, MDA-5 was examined at different times after infection with other picornaviruses. No evidence of MDA-5 cleavage was observed in cells infected with rhinovirus type 16 or echovirus type 1 (Fig. 2A and C). In cells infected with rhinovirus type 1a or EMCV, levels of MDA-5 declined beginning at 6 h postinfection, and a putative cleavage product of ∼90 kDa was observed (Fig. 2B and D).
FIG. 2.
Effect of picornavirus infection on MDA-5. Monolayers of HeLa cells were treated with poly(IC) (20 μg/ml) for 16 h and then infected with rhinovirus type 16 (A) or type 1a (B), echovirus type 1 (C), or EMCV (D) (MOI, 10). At the indicated times after infection, cell extracts were prepared and fractionated by SDS-PAGE, and MDA-5 was detected by Western blot analysis. The panels at the right show longer exposures of the Western blot to enable detection of putative MDA-5 cleavage products (unlabeled arrows). Separate bottom panels show detection of EF1α to ensure that equal amounts of protein were applied to each lane.
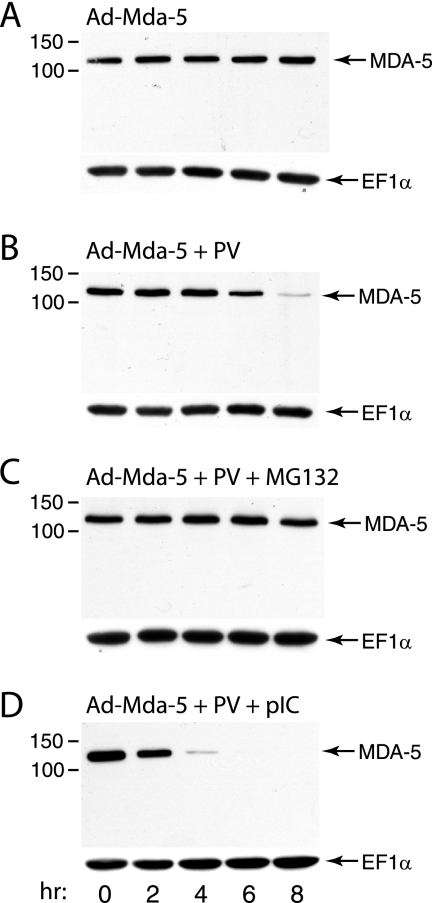
In the absence of poly(IC) treatment, levels of MDA-5 in HeLa cells are too low to allow reproducible detection by Western blot analysis during poliovirus infection. However, treatment of cells with poly(IC) induces a variety of IFN-stimulated genes, some of which could be responsible for cleavage of MDA-5. It was therefore necessary to determine whether poliovirus-induced degradation of MDA-5 occurs in the absence of poly(IC) treatment. To address this question, hemagglutinin (HA)-tagged MDA-5 was synthesized in HeLa cells from an adenovirus vector. Cells were then infected with poliovirus, and MDA-5 was examined by Western blot analysis using anti-HA antibody. Degradation of HA-MDA-5 was observed at 6 h postinfection (Fig. 3B) but not in uninfected cells (Fig. 3A). When cells were treated with poly(IC), degradation of HA-MDA-5 was observed at 4 h after infection with poliovirus (Fig. 3D). No putative cleavage products of HA-MDA-5 were observed in these experiments.
FIG. 3.
Poliovirus (PV)-induced cleavage of HA-MDA-5 produced from an adenovirus (Ad) vector. Monolayers of HeLa cells were infected with Ad.mda-5 (MOI, 50) and 16 h later were either mock infected (A) or infected with poliovirus (MOI, 10) (B, C, D). After adsorption of Ad.mda-5, cells were treated with MG132 (20 μM) (C) or poly(IC) (20 μg/ml) (D). At the indicated times after poliovirus infection, cell extracts were prepared and fractionated by SDS-PAGE, and MDA-5 was detected by Western blot analysis using anti-HA antibody. Separate bottom panels show detection of EF1α to ensure that equal amounts of protein were applied to each lane.
Role of viral proteinases in poliovirus-induced MDA-5 cleavage.
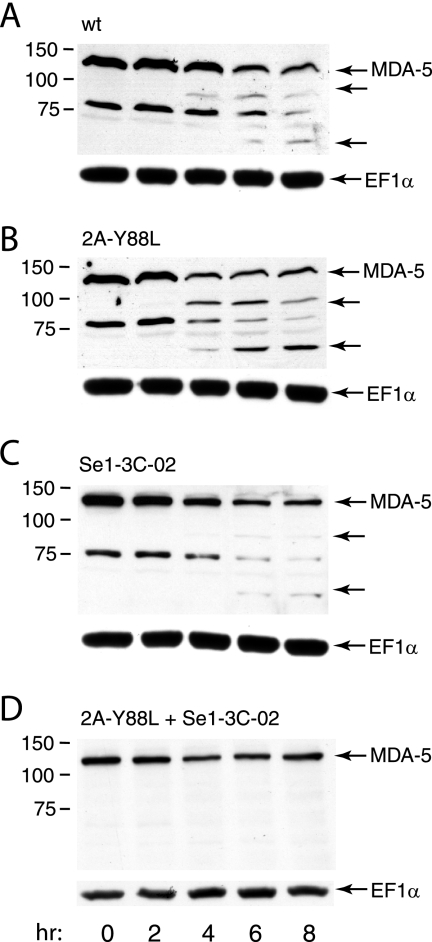
The poliovirus genome encodes two viral proteinases, 2Apro and 3Cpro, that cleave the viral polyprotein to produce the functional viral proteins. These proteinases are also known to degrade cellular proteins. Poliovirus 2Apro cleaves the translation initiation protein eIF4G, leading to inhibition of 5′-end-dependent translation (24). Cellular transcription proteins are cleaved by poliovirus 3Cpro, causing inhibition of host cell RNA synthesis (31). Polioviruses with single amino acid changes in either 2Apro or 3Cpro which abrogate cleavage of cellular proteins have been described. These poliovirus mutants were used to determine whether either viral proteinase plays a role in poliovirus-induced cleavage of MDA-5. Poliovirus mutant 2Apro Y88L contains a single amino acid change in 2Apro that abolishes cleavage in trans, e.g., prevents cleavage of eIF4G, but does not affect cis cleavage of the viral polyprotein (36). MDA-5 cleavage was observed in cells infected with 2Apro Y88L (Fig. 4B), suggesting that 2Apro does not directly cleave MDA-5. The kinetics of MDA-5 degradation in cells infected with the mutant virus are slightly accelerated compared to that in cells infected with the wild type: putative MDA-5 cleavage products are observed at 2 h postinfection in cells infected with 2Apro Y88L (Fig. 4B) but not in cells infected with wild-type virus (Fig. 4A).
FIG. 4.
Effect of amino acid changes in viral proteinases 2Apro and 3Cpro on poliovirus-induced degradation of MDA-5. Monolayers of HeLa cells were treated with poly(IC) (20 μg/ml) for 16 h and then infected with wild-type (wt) poliovirus (A), the single mutant 2AproY88L (B) or Se1-3C-02 (C), or the double mutant 2AproY88L+Se1-3C-02 (D) (MOI, 10). At the indicated times after infection, cell extracts were prepared and fractionated by SDS-PAGE, and MDA-5 was detected by Western blot analysis. Unlabeled arrows indicate putative cleavage products of MDA-5. The identity of the ∼75-kDa species in panels A to C is not known; the intensity of this protein varies depending on the batch of anti-MDA-5 antibody used. This protein is not present in panel D. Separate bottom panels show detection of EF1α to ensure that equal amounts of protein were applied to each lane.
The effect on MDA-5 cleavage of a mutant poliovirus with a single amino acid change in 3Cpro was also determined. Poliovirus mutant Se1-3C-02 contains a single amino acid change in 3Cpro that has been reported to block cleavage of host cell transcription proteins but not processing of the viral polyprotein (6, 7). MDA-5 cleavage was observed in cells infected with Se1-3C-02, suggesting that 3Cpro does not directly degrade MDA-5 (Fig. 4C).
In contrast to these findings, degradation of MDA-5 and the putative cleavage products were not observed in cells infected with a poliovirus mutant containing single amino acid changes in both 2Apro and 3Cpro (Fig. 4D).
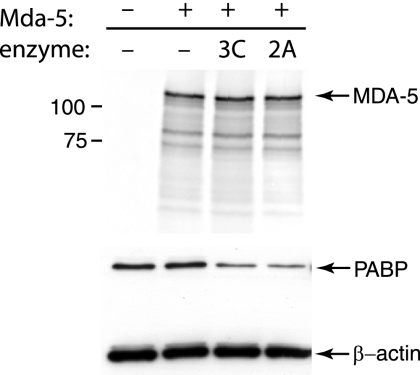
It is possible that the single amino acid changes in poliovirus mutants 2Apro Y88L and Se1-3C-02 block cleavage of other cell proteins but not MDA-5. To address this possibility, we determined whether MDA-5 can be directly cleaved in vitro by either viral proteinase. MDA-5 was produced by in vitro translation in a reticulocyte lysate, and purified poliovirus 3CDpro or coxsackievirus B3 2Apro was then added to the reticulocyte extract. After incubation, the [35S]methionine-labeled MDA-5 protein was detected by phosphorimaging. Coxsackievirus B3 2Apro was used because it has not been possible to purify poliovirus 2Apro (R. Lloyd, personal communication). No degradation of MDA-5 was observed after incubation with either enzyme (Fig. 5). Proteinase activity was confirmed by demonstrating cleavage of PABP, a known substrate of both enzymes (Fig. 5).
FIG. 5.
Effect of 2Apro and 3CDpro on MDA-5 in vitro. MDA-5 was produced by in vitro translation in a reticulocyte lysate in the presence of [35S]methionine. The lysate was subsequently incubated with purified 2Apro or 3CDpro and fractionated by SDS-PAGE, and [35S]methionine labeled proteins were detected by phosphorimaging. PABP (to confirm enzyme activity) and β-actin (loading control) were detected by Western blot analysis of a separate gel. The plasmid encoding MDA-5 was omitted from the reaction in lane 1.
Effect of proteasome and caspase inhibitors on poliovirus-induced MDA-5 cleavage.
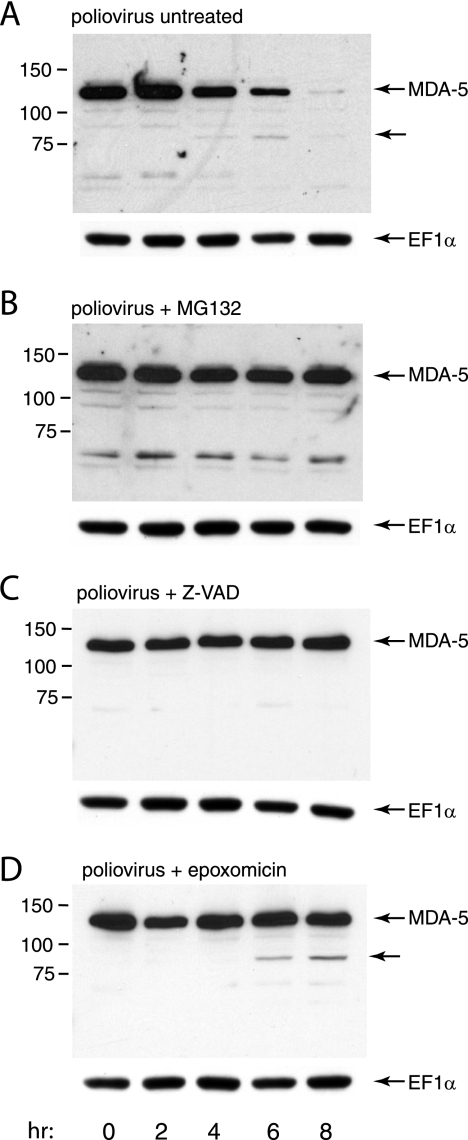
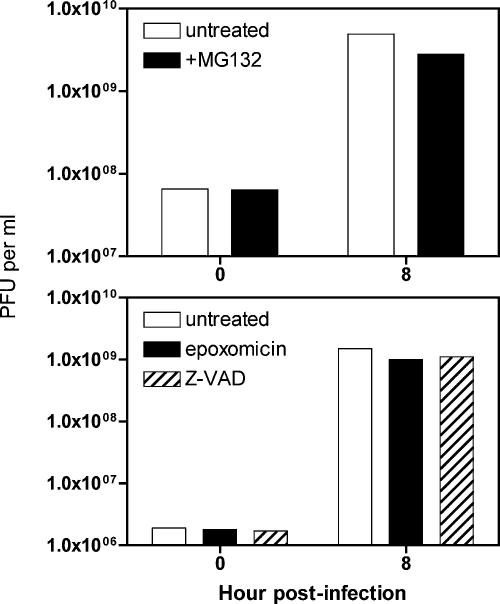
To determine whether cellular proteinases are involved in poliovirus-induced cleavage of MDA-5, the effect of inhibitors of the proteasome (MG132 and epoxomicin) and caspases (Z-VAD) on MDA-5 degradation was determined. After induction of MDA-5 synthesis by poly(IC) treatment of HeLa cells, cells were infected with poliovirus (MOI of 10), and culture medium was added with or without inhibitor. The levels of MDA-5 at different times after infection were determined by Western blot analysis. The results show that poliovirus-induced degradation of MDA-5 is inhibited by MG132, epoxomicin, and Z-VAD (Fig. 6). MG132 and Z-VAD completely blocked degradation of MDA-5, while epoxomicin did not fully prevent cleavage at the concentration used. Poliovirus-induced cleavage of HA-MDA-5 produced from an adenovirus vector was also blocked by treatment of cells with MG132 (Fig. 3C). None of these proteasome or caspase inhibitors significantly reduced poliovirus yields, indicating that they do not inhibit the viral proteinases 2Apro and 3Cpro (Fig. 7). These results indicate that poliovirus-induced degradation of MDA-5 occurs by a process that requires both caspases and the cellular proteasome.
FIG. 6.
Effect of proteinase inhibitors on poliovirus-induced MDA-5 cleavage. Monolayers of HeLa cells were treated with poly(IC) (20 μg/ml) for 16 h and then infected with poliovirus (MOI, 10) in the absence of inhibitor (A) or in the presence of MG132 (20 μM) (B), Z-VAD-FMK (100 μM) (C), or epoxomicin (10 μM) (D). At the indicated times after infection, cell extracts were prepared and fractionated by SDS-PAGE, and MDA-5 was detected by Western blot analysis. Unlabeled arrows indicate a putative cleavage product of MDA-5. The identity of the protein migrating faster than the 75-kDa marker in panel B is not known, but this protein was not consistently observed. Separate bottom panels show detection of EF1α to ensure that equal amounts of protein were applied to each lane.
FIG. 7.
Effect of proteinase inhibitors on poliovirus yields in HeLa cells. Monolayers of HeLa cells were infected with poliovirus (MOI, 10) in the absence or presence of MG132 (20 μM), Z-VAD-FMK (100 μM), or epoxomicin (10 μM). At the indicated times after infection, virus titers were determined by plaque assay on HeLa cell monolayers.
Correlation between cleavage of PARP and cleavage of MDA-5.
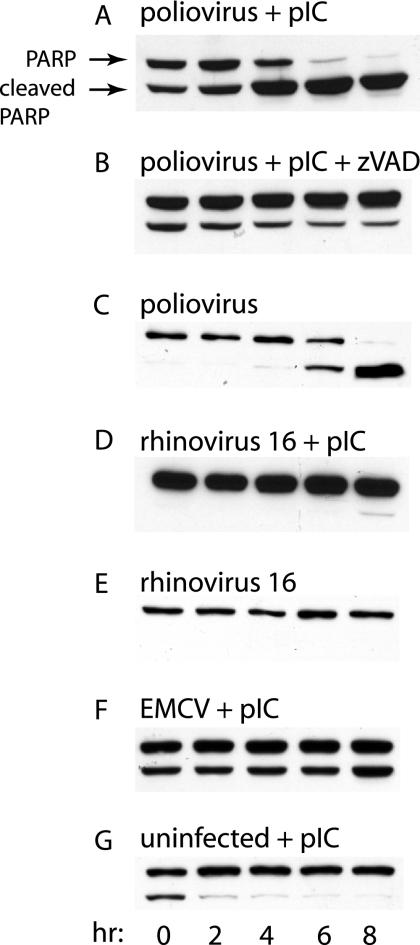
The mouse ortholog of human MDA-5 has been shown to undergo caspase-dependent cleavage in cells that are induced to enter apoptosis (23). Based on our observations, a plausible hypothesis is that poliovirus infection induces apoptosis, which in turn leads to MDA-5 cleavage. To address this hypothesis, we determined whether poliovirus infection leads to caspase-dependent cleavage of PARP, a hallmark of apoptosis. In cells infected with poliovirus, significant PARP cleavage was observed at 6 h postinfection (Fig. 8C), a time at which cleavage of HA-MDA-5 is also observed (Fig. 3B). When cells were pretreated with poly(IC), PARP cleavage was observed at 4 h after poliovirus infection (Fig. 8A), as is cleavage of MDA-5 under these conditions (Fig. 1A and 3D). As expected, poliovirus-induced cleavage of PARP was inhibited by treatment of cells with Z-VAD (Fig. 8B). A small amount of PARP cleavage was observed at 8 h after infection with rhinovirus type 16 (Fig. 8D), consistent with absence of MDA-5 cleavage in cells infected with this picornavirus (Fig. 2A). Treatment of HeLa cells with poly(IC) alone induced only transient cleavage of MDA-5 (Fig. 8G), consistent with the fact that MDA-5 cleavage is not observed after poly(IC) treatment alone (Fig. 1B).
FIG. 8.
Cleavage of PARP during picornavirus infection. Monolayers of HeLa cells were treated with poly(IC) (20 μg/ml) (A, B, D, F, G) for 16 h and then mock infected (G) or infected with poliovirus (A, B, C), rhinovirus type 16 (D, E), or EMCV (F) (MOI, 10). In panel B, Z-VAD-FMK (100 μM) was included during infection. At the indicated times after infection, cell extracts were prepared and fractionated by SDS-PAGE, and PARP was detected by Western blot analysis.
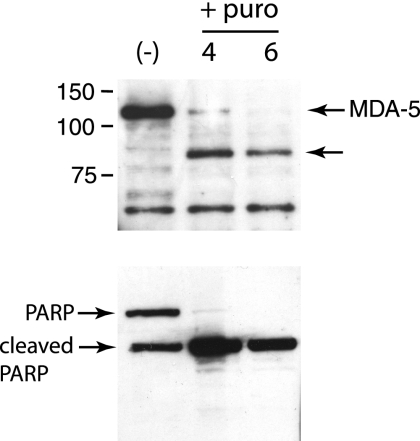
To confirm that induction of apoptosis leads to MDA-5 cleavage, HeLa cells were treated with puromycin and MDA-5 was examined by Western blot analysis. Puromycin treatment for 4 and 6 h induced apoptosis as expected, as shown by cleavage of PARP (Fig. 9). In the same cells, a ∼90-kDa cleavage product of MDA-5 was observed (Fig. 9).
FIG. 9.
Cleavage of MDA-5 and PARP in cells induced to undergo apoptosis by treatment with puromycin. Monolayers of HeLa cells were treated with poly(IC) (20 μg/ml) for 16 h, and then puromycin (puro) (10 μM) was added to induce apoptosis. At the indicated times after addition of puromycin to the culture medium, cell extracts were prepared and fractionated by SDS-PAGE, and MDA-5 or PARP were detected by Western blot analysis. The unlabeled arrow indicates a putative ∼90-kDa cleavage product of MDA-5. The identity of the protein migrating faster than the 75-kDa marker is not known.
Induction of IFN-β synthesis during poliovirus infection.
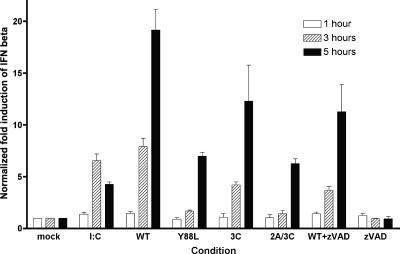
Poliovirus-induced cleavage of MDA-5 might be a mechanism to enhance viral replication by dampening the innate response to infection. To address this possibility, induction of IFN-β RNA during poliovirus infection of HeLa cells was examined by quantitative real-time PCR (Fig. 10). As expected, treatment of HeLa cells with poly(IC) induced IFN-β synthesis. Induction of IFN-β RNA synthesis was observed at 3 and 5 h postinfection with poliovirus. Infection with mutant polioviruses harboring single amino acid changes in 2Apro or 3Cpro, or infection with a double mutant with changes in both viral proteinases, led to reduced induction of IFN-β RNA synthesis. After treatment of cells with Z-VAD-FMK, which blocks poliovirus-induced cleavage of MDA-5, less induction of IFN-β was observed compared with that in untreated, poliovirus-infected cells.
FIG. 10.
Induction of IFN-β RNA in poliovirus-infected cells. Monolayers of HeLa cells were either mock treated, treated with poly(IC) (20 μg/ml) or Z-VAD-FMK (100 μM) for 16 h, or infected with wild-type (WT) or mutant polioviruses (MOI, 10). At 1, 3, and 5 h after infection, RNA was extracted from cells and analyzed for IFN-β expression by quantitative real-time PCR, using SYBR green to detect dsDNA. RNA levels were normalized to β-actin.
DISCUSSION
Cleavage of MDA-5 was observed in cells infected with poliovirus, rhinovirus type 1a, and EMCV but not in cells infected with echovirus type 1 or rhinovirus type 16. Putative MDA-5 cleavage products of ∼90 and ∼70 kDa were observed in cells infected with poliovirus. Poliovirus-induced degradation of HA-MDA-5 produced from an adenovirus vector was also observed, although cleavage products were not detected.
Likely candidates for cleavage of MDA-5 during poliovirus infection are the two viral proteinases, 2Apro or 3Cpro. These proteinases not only process the viral polyprotein but have been shown to cleave a variety of cell proteins, including eIF4G (24), PABP (25), the catalytic subunit of DNA-dependent protein kinase (13) (cleaved by 2Apro), Nup62 and Nup153 (14), and proteins involved in RNA polymerase transcription such as TATA-binding protein, CREB, Oct-1, p53, IIICα, and SL-1 (31) (cleaved by 3Cpro). To determine whether the viral proteinases participate in MDA-5 cleavage, we used mutant polioviruses with single amino acid changes in either enzyme that have been previously shown to block cleavage of host cell proteins. Degradation of MDA-5 proceeded in cells infected with these mutant viruses, suggesting that the viral proteinases do not directly cleave MDA-5. To address the possibility that the mutant proteinases are still able to effect MDA-5 cleavage, we determined whether the purified viral proteinases can directly cleave MDA-5 protein in vitro. The results indicate that neither poliovirus 3CDpro nor coxsackievirus B3 2Apro cleaves MDA-5 in a rabbit reticulocyte lysate. It was necessary to use the coxsackievirus 2Apro for this experiment because it has not been possible to purify the poliovirus enzyme. The 2Apro enzymes of poliovirus and coxsackievirus have similar substrate specificities and cleave similar sites within the viral polyprotein as well as cellular proteins such as eIF4G and PABP (17). It would therefore be expected that the poliovirus and coxsackievirus 2Apro proteinases would both have the ability to cleave MDA-5.
The results presented here, together with previous findings, are consistent with the hypothesis that poliovirus infection activates the apoptotic pathway, which then leads to MDA-5 cleavage. The mouse ortholog of human MDA-5 undergoes caspase-dependent cleavage in cells that are induced to enter apoptosis (23). It was shown previously that poliovirus infection induces apoptosis, causing mitochondrial damage, release of cytochrome c, and activation of caspase-9 and caspase-3 (4). We have shown that poliovirus-induced cleavage of MDA-5 is blocked by the caspase inhibitor Z-VAD-FMK. Our results also indicate that cleavage of PARP occurs in poliovirus-infected cells, an event that correlates with MDA-5 cleavage. Pretreatment of cells with poly(IC) leads to accelerated cleavage of both PARP and MDA-5 in poliovirus-infected cells. Treatment of uninfected HeLa cells with puromycin, a known inducer of apoptosis, leads to cleavage of both PARP and MDA-5. The putative MDA-5 cleavage product observed in cells treated with puromycin is ∼90 kDa, similar in size to the putative cleavage product observed in poliovirus-infected cells. Infection of HeLa cells with rhinovirus type 16 does not lead to apoptosis, as judged by the very low level of cleaved PARP observed by 8 h postinfection, and MDA-5 cleavage is not observed.
The main triggers of apoptosis in poliovirus-infected cells appear to be the two viral proteinases, as production of either poliovirus 2Apro or 3Cpro in cells is sufficient to induce apoptosis (3, 12). It would therefore be expected that amino acid changes in either viral proteinase would not be sufficient to block virus-induced apoptosis. Our observation that MDA-5 cleavage is not reduced in cells infected with polioviruses containing a single amino acid change in either viral proteinase is in accord with this expectation. Furthermore, MDA-5 cleavage does not occur in cells infected with a poliovirus mutant containing single amino acid changes in both viral proteinases 2Apro and 3Cpro. These findings provide additional evidence that induction of apoptosis by 2Apro and 3Cpro is central to induction of MDA-5 degradation.
Sites of caspase cleavage in murine MDA-5 have been identified at amino acids 208, 216, 251, and 260 to 280 (23), and similar sequences are present in the human MDA-5 protein. Cleavage at some of these sites could produce the less-than-full-length MDA-5 proteins observed in poliovirus-infected cells. For example, cleavage at amino acid 216 of MDA-5 could produce the ∼90-kDa protein, and cleavage at a putative caspase cleavage site at amino acid 673 could produce the ∼70-kDa protein (Fig. 1A). It is not clear why cleavage products were not detected when HA-MDA-5 was degraded during poliovirus infection (Fig. 3). It will be necessary to determine amino acid sequences from the ∼90- and ∼70-kDa proteins to determine whether these are bona fide cleavage products.
The inhibition of poliovirus-induced cleavage of MDA-5 by MG132 and epoxomicin indicates that the proteasome is also involved in MDA-5 degradation. The proteasome has been implicated in the regulation of apoptosis by modulating the levels of pro- and antiapoptotic molecules (10). MDA-5 is a proapoptotic protein (1, 18), and therefore its cleavage by the proteasome is not unexpected. Recent studies investigating potential signal transduction pathways involved in regulating mda-5-induced apoptosis in mammalian cells demonstrate the importance of Raf/Ras/MEK/EFK signaling pathways in apoptosis induction by mda-5 (1). The results of these studies demonstrate that rodent and human cells containing an activated Raf/Ras/MEK/ERK are resistant to mda-5-induced killing. Accordingly, this resistance is antagonized by inhibiting this important signal transduction cascade either by directly inhibiting ras activity using an antisense strategy or by targeting ras-downstream factors, such as MEK1/2, with the pharmacological inhibitor PD98059. The role of this pathway in poliovirus-induced MDA-5 cleavage remains to be defined.
The observation that inhibitors of either the proteasome or caspases can block cleavage of MDA-5 during poliovirus infection shows that both cellular proteases are required for degradation. The simplest explanation for this finding is that caspases and the proteasome act in a linear manner on MDA-5. For example, cleavage of MDA-5 by caspases might be required before the protein can be a substrate for the proteasome. Because inhibitors of either protease block MDA-5 degradation, it is not possible to determine whether caspases or the proteasome are first in this pathway.
Cleavage of MDA-5 during poliovirus infection might be a mechanism to evade the innate immune response by attenuating the production of IFN, thereby allowing higher levels of viral replication. If this hypothesis were correct, we would expect to observe higher poliovirus yields when cleavage of MDA-5 is blocked by MG132 or epoxomicin. The presence of MG132 or epoxomicin had no effect on poliovirus yields during infection at a high MOI (Fig. 8 and unpublished data). Furthermore, induction of IFN-β RNA synthesis during poliovirus infection was not stimulated by conditions which block MDA-5 cleavage, such as infection with a viral mutant with amino acid changes in both proteinases, or the presence of Z-VAD-FMK. Taken together, these observations suggest that virus-induced cleavage of MDA-5 has no beneficial effect on poliovirus replication. To provide a definitive answer to the question of whether poliovirus-induced cleavage of MDA-5 leads to enhanced viral replication, it will be necessary to produce a variant of MDA-5 that is resistant to virus-induced cleavage and to determine whether the production of the altered protein in cells reduces poliovirus yields.
MDA-5, not RIG-I, is believed to be crucial for sensing picornavirus infection. This conclusion derives from studies in which mice lacking the genes encoding MDA-5 or RIG-I were infected with EMCV (11, 21). Mice lacking the mda-5 gene failed to produce IFN when infected with EMCV, and the animals were more susceptible to infection. However, absence of the gene encoding RIG-I had no effect on IFN production after EMCV infection of mice. While these results are compelling, it seems premature to conclude that all picornaviruses are sensed by MDA-5, because only cardioviruses have been studied. Other unanswered questions include the effect of absence of MDA-5 or RIG-I on picornavirus yields in different cell types and whether the effects of these sensor molecules on viral replication differ in cultured cells and on IFN production in animals. For example, overproduction of RIG-I in cultured cells reduces yields of vesicular stomatitis virus and EMCV (33), but mice lacking the gene for RIG-I have no impairment in IFN production in response to EMCV infection (21). Additional experiments with cells and with mice lacking RIG-I and mda-5 are required to assess the physiological role of these proteins in poliovirus infection.
Forced expression of mda-5, but not RIG-I or Toll-like receptor 3, leads to enhanced IFN-β promoter activity in measles virus-infected A549 non-small-cell lung carcinoma cells (5). In this context IFN-induced mda-5 is involved in measles virus-induced expression of antiviral cytokines. These findings provide yet another example of the potential subtle and unique roles of MDA-5 and RIG-I in sensing and responding to specific viral infections. Further studies on MDA-5 and RIG-I in cells infected with different viruses is clearly important in understanding and potentially developing strategies for selectively and effectively intervening in viral pathogenesis.
Acknowledgments
This work was supported in part by Public Health Service grant AI50754 from the National Institute of Allergy and Infectious Diseases to V.R.R. and by grants from the Public Health Service (grant GM68448), the Samuel Waxman Cancer Research Foundation, and the Chernow Endowment to P.B.F., who is the Michael and Stella Chernow Urological Cancer Research Scientist and an SWCRF Investigator.
We thank Richard Llyod and Bert Semler for the gifts of 2Apro and 3CDpro, respectively.
Footnotes
Published ahead of print on 31 January 2007.
REFERENCES
- 1.Akira, S., S. Uematsu, and O. Takeuchi. 2006. Pathogen recognition and innate immunity. Cell 124:783-801. [DOI] [PubMed] [Google Scholar]
- 2.Andrejeva, J., K. S. Childs, D. F. Young, T. S. Carlos, N. Stock, S. Goodbourn, and R. E. Randall. 2004. The V proteins of paramyxoviruses bind the IFN-inducible RNA helicase, mda-5, and inhibit its activation of the IFN-beta promoter. Proc. Natl. Acad. Sci. USA 101:17264-17269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barco, A., E. Feduchi, and L. Carrasco. 2000. Poliovirus protease 3C(pro) kills cells by apoptosis. Virology 266:352-360. [DOI] [PubMed] [Google Scholar]
- 4.Belov, G. A., L. I. Romanova, E. A. Tolskaya, M. S. Kolesnikova, Y. A. Lazebnik, and V. I. Agol. 2003. The major apoptotic pathway activated and suppressed by poliovirus. J. Virol. 77:45-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berghall, H., J. Siren, D. Sarkar, I. Julkunen, P. B. Fisher, R. Vainionpaa, and S. Matikainen. 2006. The interferon-inducible RNA helicase, mda-5, is involved in measles virus-induced expression of antiviral cytokines. Microbes Infect. 8:2138-2144. [DOI] [PubMed] [Google Scholar]
- 6.Clark, M. E., T. Hämmerle, E. Wimmer, and A. Dasgupta. 1991. Poliovirus proteinase 3C converts an active form of transcription factor IIIC to an inactive form: a mechanism for inhibition of host cell polymerase III transcription by poliovirus. EMBO J. 10:2941-2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dewalt, P. G., and B. L. Semler. 1987. Site-directed mutagenesis of proteinase 3C results in a poliovirus deficient in synthesis of viral RNA polymerase. J. Virol. 61:2162-2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Duke, G. M., and A. C. Palmenberg. 1989. Cloning and synthesis of infectious cardiovirus RNAs containing short, discrete poly(C) tracts. J. Virol. 63:1822-1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fensterl, V., D. Grotheer, I. Berk, S. Schlemminger, A. Vallbracht, and A. Dotzauer. 2005. Hepatitis A virus suppresses RIG-I-mediated IRF-3 activation to block induction of beta interferon. J. Virol. 79:10968-10977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Friedman, J., and D. Xue. 2004. To live or die by the sword: the regulation of apoptosis by the proteasome. Dev. Cell 6:460-461. [DOI] [PubMed] [Google Scholar]
- 11.Gitlin, L., W. Barchet, S. Gilfillan, M. Cella, B. Beutler, R. A. Flavell, M. S. Diamond, and M. Colonna. 2006. Essential role of mda-5 in type I IFN responses to polyriboinosinic:polyribocytidylic acid and encephalomyocarditis picornavirus. Proc. Natl. Acad. Sci. USA 103:8459-8464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goldstaub, D., A. Gradi, Z. Bercovitch, Z. Grosmann, Y. Nophar, S. Luria, N. Sonenberg, and C. Kahana. 2000. Poliovirus 2A protease induces apoptotic cell death. Mol. Cell. Biol. 20:1271-1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Graham, K. L., K. E. Gustin, C. Rivera, N. M. Kuyumcu-Martinez, S. S. Choe, R. E. Lloyd, P. Sarnow, and P. J. Utz. 2004. Proteolytic cleavage of the catalytic subunit of DNA-dependent protein kinase during poliovirus infection. J. Virol. 78:6313-6321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gustin, K. E., and P. Sarnow. 2001. Effects of poliovirus infection on nucleo-cytoplasmic trafficking and nuclear pore complex composition. EMBO J. 20:240-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Holmes, M., E. Rosenberg, and K. Valerie. 2003. Adenovirus expressing p53. Methods Mol. Biol 234:1-16. [DOI] [PubMed] [Google Scholar]
- 16.Janeway, C. A., Jr., and R. Medzhitov. 2002. Innate immune recognition. Annu. Rev. Immunol. 20:197-216. [DOI] [PubMed] [Google Scholar]
- 17.Joachims, M., P. C. Van Breugel, and R. E. Lloyd. 1999. Cleavage of poly(A)-binding protein by enterovirus proteases concurrent with inhibition of translation in vitro. J. Virol. 73:718-727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kang, D. C., R. V. Gopalkrishnan, L. Lin, A. Randolph, K. Valerie, S. Pestka, and P. B. Fisher. 2004. Expression analysis and genomic characterization of human melanoma differentiation associated gene-5, mda-5: a novel type I interferon-responsive apoptosis-inducing gene. Oncogene 23:1789-1800. [DOI] [PubMed] [Google Scholar]
- 19.Kang, D. C., R. V. Gopalkrishnan, Q. Wu, E. Jankowsky, A. M. Pyle, and P. B. Fisher. 2002. mda-5: An interferon-inducible putative RNA helicase with double-stranded RNA-dependent ATPase activity and melanoma growth-suppressive properties. Proc. Natl. Acad. Sci. USA 99:637-642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kato, H., S. Sato, M. Yoneyama, M. Yamamoto, S. Uematsu, K. Matsui, T. Tsujimura, K. Takeda, T. Fujita, O. Takeuchi, and S. Akira. 2005. Cell type-specific involvement of RIG-I in antiviral response. Immunity 23:19-28. [DOI] [PubMed] [Google Scholar]
- 21.Kato, H., O. Takeuchi, S. Sato, M. Yoneyama, M. Yamamoto, K. Matsui, S. Uematsu, A. Jung, T. Kawai, K. J. Ishii, O. Yamaguchi, K. Otsu, T. Tsujimura, C. S. Koh, C. Reis e Sousa, Y. Matsuura, T. Fujita, and S. Akira. 2006. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature 441:101-105. [DOI] [PubMed] [Google Scholar]
- 22.Kawai, T., K. Takahashi, S. Sato, C. Coban, H. Kumar, H. Kato, K. J. Ishii, O. Takeuchi, and S. Akira. 2005. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat. Immunol. 6:981-988. [DOI] [PubMed] [Google Scholar]
- 23.Kovacsovics, M., F. Martinon, O. Micheau, J. L. Bodmer, K. Hofmann, and J. Tschopp. 2002. Overexpression of Helicard, a CARD-containing helicase cleaved during apoptosis, accelerates DNA degradation. Curr. Biol. 12:838-843. [DOI] [PubMed] [Google Scholar]
- 24.Krausslich, H. G., M. J. H. Nicklin, H. Toyoda, D. Etchison, and E. Wimmer. 1987. Poliovirus proteinase 2A induces cleavage of eukaryotic initiation factor 4F polypeptide p220. J. Virol. 61:2711-2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kuyumcu-Martinez, N. M., M. E. Van Eden, P. Younan, and R. E. Lloyd. 2004. Cleavage of poly(A)-binding protein by poliovirus 3C protease inhibits host cell translation: a novel mechanism for host translation shutoff. Mol. Cell. Biol. 24:1779-1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee, W. M., W. Wang, and R. R. Rueckert. 1995. Complete sequence of the RNA genome of human rhinovirus 16, a clinically useful common cold virus belonging to the ICAM-1 receptor group. Virus Genes 9:177-181. [DOI] [PubMed] [Google Scholar]
- 27.Lin, L., Z. Su, I. V. Lebedeva, P. Gupta, H. Boukerche, T. Rai, G. N. Barber, P. Dent, D. Sarkar, and P. B. Fisher. 2006. Activation of Ras/Raf protects cells from melanoma differentiation-associated gene-5-induced apoptosis. Cell Death Differ. 13:1982-1993. [DOI] [PubMed] [Google Scholar]
- 28.Meylan, E., J. Curran, K. Hofmann, D. Moradpour, M. Binder, R. Bartenschlager, and J. Tschopp. 2005. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature 437:1167-1172. [DOI] [PubMed] [Google Scholar]
- 29.Racaniello, V. R., and D. Baltimore. 1981. Cloned poliovirus complementary DNA is infectious in mammalian cells. Science 214:916-919. [DOI] [PubMed] [Google Scholar]
- 30.Seth, R. B., L. Sun, C. K. Ea, and Z. J. Chen. 2005. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell 122:669-682. [DOI] [PubMed] [Google Scholar]
- 31.Weidman, M. K., R. Sharma, S. Raychaudhuri, P. Kundu, W. Tsai, and A. Dasgupta. 2003. The interaction of cytoplasmic RNA viruses with the nucleus. Virus Res. 95:75-85. [DOI] [PubMed] [Google Scholar]
- 32.Xu, L. G., Y. Y. Wang, K. J. Han, L. Y. Li, Z. Zhai, and H. B. Shu. 2005. VISA is an adapter protein required for virus-triggered IFN-beta signaling. Mol. Cell 19:727-740. [DOI] [PubMed] [Google Scholar]
- 33.Yoneyama, M., M. Kikuchi, T. Natsukawa, N. Shinobu, T. Imaizumi, M. Miyagishi, K. Taira, S. Akira, and T. Fujita. 2004. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat. Immunol. 5:730-737. [DOI] [PubMed] [Google Scholar]
- 34.Ypma-Wong, M. F., P. G. Dewalt, V. H. Johnson, J. G. Lamb, and B. L. Semler. 1988. Protein 3CD is the major poliovirus proteinase responsible for cleavage of the P1 capsid precursor. Virology 166:265-270. [DOI] [PubMed] [Google Scholar]
- 35.Yu, S. F., P. Benton, M. Bovee, J. Sessions, and R. E. Lloyd. 1995. Defective RNA replication by poliovirus mutants deficient in 2A protease cleavage activity. J. Virol. 69:247-252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yu, S. F., and R. E. Lloyd. 1991. Identification of essential amino acid residues in the functional activity of poliovirus 2A protease. Virology 182:615-625. [DOI] [PubMed] [Google Scholar]