Abstract
Rabies virus P protein inhibits alpha interferon (IFN-α)- and IFN-γ-stimulated Jak-STAT signaling by retaining phosphorylated STAT1 in the cytoplasm. Here, we show that P also blocks an intranuclear step that is the STAT1 binding to the DNA promoter of IFN-responsive genes. As P is a nucleocytoplasmic shuttling protein, we first investigated the effect of the cellular distribution of P on the localization of STAT1 and consequently on IFN signaling. We show that the localization of STAT1 is correlated with the localization of P: in cells expressing a nuclear form of P (the short P3 isoform or the complete P in the presence of the export inhibitor leptomycin B), STAT1 is nuclear, whereas in cells expressing a cytoplasmic form of P, STAT1 is cytoplasmic. However, the expression of nuclear forms of P inhibits the signaling of both IFN-γ and IFN-α, demonstrating that the retention of STAT1 in the cytoplasm is not the only mechanism involved in the inhibition of IFN signaling. Electrophoretic mobility shift analysis indicates that P expression in the cell extracts of infected cells or in stable cell lines prevents IFN-induced DNA binding of STAT1. The loss of the DNA binding of STAT1 and ISGF3 was also observed when purified recombinant P or P3 was added to the extracts of IFN-γ- or IFN-α-treated cells, indicating that P directly affects the DNA binding activity of STAT1. Then products of the rabies virus P gene are able to counteract IFN signaling by creating both cytoplasmic and nuclear blocks for STAT1.
The interferon (IFN) response is one of the host response system's primary defense mechanisms against viral infection. Type I IFN (alpha/beta interferon [IFN-α/β]) is produced by most cells as a direct response to viral infection, while type II IFN (IFN-γ) is synthesized almost exclusively by activated NK cells and activated T cells in response to virus-infected cells. Both type I and II IFNs achieve antiviral effects by binding to their respective receptors (IFNAR for IFN-α/β or IFNGR for IFN-γ), resulting in the activation of a distinct but related “Janus” tyrosine kinase/signal transducer and activator of transcription (Jak/STAT) pathway (12). Briefly, the interaction of IFN-α/β with IFNAR leads to the activation of the Jak protein tyrosine kinases (Tyk 2 and Jak1) that phosphorylate STAT1 and STAT2. The phosphorylated STATs heterodimerize and bind to a DNA binding protein, IFN regulatory factor 9 (IRF9), to form a complex, IFN-stimulated growth factor 3 (ISGF3). ISGF3 translocates into the nucleus and binds to an IFN-stimulated response element (ISRE) to induce IFN-stimulated genes (ISGs). The binding of IFN-γ to its receptor, IFNGR, results in the phosphorylation of STAT1 by Jak1 and Jak2. STAT1 homodimers form, migrate to the nucleus, and bind to a DNA element termed GAS (for gamma-activated sequence) to induce specifically the transcription of IFN-γ target genes (12).
All the IFN-induced biological responses are believed to be mediated by ISG products that have been shown to display intrinsic antiviral activities (8, 24).
Viruses that require cellular machinery for their replication have evolved different strategies to counteract IFN action, particularly by altering IFN induction, IFN signaling, and IFN-induced mediators (1, 9, 15). Several viral proteins acting as IFN antagonists have been identified in Mononegavirales, such as members of the Paramyxoviridae families (10, 16). Very recently, interference with IFN production and signaling was described for rabies virus of the Lyssavirus genus that belongs to the Rhabdoviridae family (4, 5, 6, 28).
Rabies virus has a linear, nonsegmented, single-strand RNA genome of negative polarity. The ribonucleoprotein contains the RNA genome tightly encapsidated by the viral nucleoprotein (N) and the RNA polymerase complex, which consists of the large protein (L) and its cofactor, the phosphoprotein (P). Both L and P are involved in transcription and replication. A positive-stranded leader RNA and five mRNAs are synthesized during transcription. The replication process yields nucleocapsids containing full-length antisense genome RNA, which in turn serves as a template for the synthesis of sense genome RNA.
The rabies virus P protein is a noncatalytic cofactor and a regulatory protein that plays a role in viral transcription and replication: it stabilizes the RNA polymerase L to the N-RNA template and binds to the soluble N, preventing its aggregation and keeping it in a suitable form for specific encapsidation of viral RNA. P protein has other specific functions in the host cells (6). Interestingly, rabies virus P protein interacts directly with two proteins, STAT1 and promyelocytic leukemia protein (PML) (3, 28), playing an important role in the IFN-induced antiviral response. In addition, P protein impairs IRF-3 phosphorylation, leading to the inhibition of IFN production (4). This multifunctionality of P may be linked to the high polymorphism of protein expression. It is phosphorylated by two kinases, rabies virus protein kinase and protein kinase C, leading to the formation of different phosphorylated forms of the P protein (14). In addition, the P gene encodes not only P but also additional shorter P products (P2, P3, P4, and P5) whose translation is initiated from downstream and in-frame AUG codons by a leaky scanning mechanism (7). These small versions of P have different intracellular distributions. The nuclear localizations of P3, P4, and P5 are due to the presence of a nuclear localization signal (NLS) located in the C-terminal part of the protein, whereas the cytoplasmic distributions of P and P2 are the result of a CRM1 nuclear export signal (NES) located in the N-terminal part of the protein (20).
We and others have previously shown that rabies virus P protein inhibits signaling by blocking the nuclear accumulation of STAT1 (5, 28). By analyzing the molecular mechanisms leading to the inhibition of IFN signaling by rabies virus P protein, we have shown that P protein and the nuclear P3 isoform inhibit an additional step that occurs in the nucleus: the binding of STAT1 or ISGF3 to the DNA promoters (i.e., to GAS and ISRE) of IFN-γ- or IFN-α-responsive genes, respectively.
MATERIALS AND METHODS
Cells and viruses.
All experiments were performed with human glioblastoma astrocytoma cells (U373-MG). Cells were grown in Dulbecco's modified Eagle medium supplemented with 10% fetal calf serum.
The CVS strain of rabies virus was grown in BSR cells cloned from BHK21 (baby hamster kidney) cells.
Stably transfected U373-MG cells.
Stable P-expressing cell lines were produced by transfecting U373-MG cells with plasmid pCDNA3.1-Hygro (Invitrogen) (encoding the wild-type P protein described below) by the calcium phosphate coprecipitation procedure. After 48 h, the transfection medium was replaced by Dulbecco's modified Eagle's medium containing 500 μg/ml hygromycin B (Invitrogen). Surviving cells were transferred and expanded in the presence of hygromycin B. Control U373-MG cells were generated the same way with pCDNA3.1-Hygro.
Interferons, antibodies, and leptomycin B (LMB) treatment.
Human IFN-α (hIFN-α-100) with a specific activity of 5 × 106 U/ml was from Strathmann Biotec, and hIFN-γ with a specific activity of 2 × 107 U/mg was from Roussel Uclaf (Romainville, France).
The mouse polyclonal anti-P antibody has been described previously (22). Rabbit anti-STAT1 (catalog no. sc-346), anti-STAT1 phosphotyrosine 701 (catalog no. sc-7988), anti-PML (catalog no. H-238), anti-PKR (catalog no. sc-707), and anti-IRF1 (catalog no. sc-497) antibodies were obtained from Santa Cruz Biotechnology, Inc. Rabbit anti-STAT2 (catalog no. 06-502) and anti-STAT2 phosphotyrosine 689 (catalog no. 07-224) antibodies were obtained from Upstate Biotechnology.
Monoclonal anti-tubulin antibody from Amersham (N356) was used. LMB (Sigma) was added to culture medium to a final concentration of 20 nM for 1.5 h before IFN treatment.
Plasmid constructions.
The constructs p-P-GFP, p-PΔN52-GFP, and pPΔN44-GFP have been described previously (20). The plasmids pLex-P and pLex-PΔN52 have been described previously by Raux et al. (23), and the plasmids pET22-P-his and pET22-PΔN52 (P3-his) have been described previously by Gigant et al. (11).
The plasmid pLex-PΔN44 differed from pLex-P by a deletion of 162 bp at the 5′ end terminus of the P gene. The deletion was introduced by PCR amplification of the wild-type P gene using the forward oligonucleotide GCCGAATTCGAAGTGGACAACCTCCT with an EcoRI site (underlined) and the backward oligonucleotide GCCGTCGACTTATATTCCTGAAGATCG (complementary to the 3′ end of the P mRNA) with a SalI site (underlined). The amplified double-stranded cDNA was digested by EcoRI and SalI and inserted in frame with LexA-BD into the corresponding cloning sites of pLex10 as described previously (28).
The construct pCDNA3.1-P was obtained by inserting the P gene into pCDNA3.1-Hygro (Invitrogen). The P gene was amplified by PCR using a forward oligonucleotide (GCCGCTAGCATGAGCAAGATCTTTGTT) containing an NheI site (underlined) and a backward oligonucleotide (GCCTCTAGATTAGCAGGATGTATAGCG) which was complementary to the 3′ end of the P mRNA. The XbaI site of the backward oligonucleotide is underlined. The amplified double-stranded cDNA was digested by NheI and XbaI and inserted into the corresponding cloning sites of pCDNA3.1-Hygro.
Cell infection and transient transfections.
Monolayers of U373-MG cells were grown to 80% confluence in 6-cm dishes and infected with 5 PFU/cell of rabies virus (CVS strain). Cells were used for experiments at 24 h postinfection.
Monolayers of U373-MG cells were grown in 12-well plates or on a sterile glass coverslip in 6-well plates (from 50 to 80% confluence) and were transfected by the calcium phosphate coprecipitation procedure with 2.5 μg or 5 μg of plasmid DNA.
Luciferase assays.
Cells in 12-well plates were transfected with 2.5 μg of plasmid encoding P-green fluorescent protein (GFP), P3-GFP, or PΔN44-GFP; 0.75 μg of pRL-TK; and 2.5 μg of pISREluc (or pGASluc). At 48 h posttransfection, cells were untreated or treated with 2,000 U/ml of human recombinant IFN-α (hIFN-α) or hIFN-γ. Cells were harvested at 6 h after IFN treatment and assayed for firefly and Renilla luciferase activities as described by the manufacturer (dual-luciferase reporter assay system; Promega). Relative expression levels were calculated by dividing the values for firefly luciferase by those for Renilla luciferase. In some cases, P-expressing cells were transfected with pRL-TK and pISREluc (or pGASluc) and treated as described above.
P expression and purification.
Recombinant His-tagged P and P3 proteins were produced in Escherichia coli and purified as described previously by Gigant et al. (11).
EMSA.
Uninfected or infected cells were not treated or treated with 2,000 U/ml of hIFN-γ for 30 min. Cells were harvested, and total cell extracts were prepared. Briefly, 3 × 107 cells were washed with cold phosphate-buffered saline and lysed in 800 μl of cold freshly prepared lysis buffer (0.5% NP-40, 50 mM Tris [pH 8.0], 10% glycerol, 0.1 mM EDTA, 200 mM NaCl, 1 mM dithiothreitol [DTT]) with a mixture of proteases inhibitors (Complete protease inhibitor cocktail; Boehringer Mannheim). Proteins were examined by electrophoretic mobility shift assays (EMSA) as described elsewhere (21) with a 32P-labeled GAS probe. The probe was generated with the duplex oligonucleotide 5′-TACAACAGCCTGATTTCCCCGAAATGACGC-3′ (the respective antisense oligonucleotide is not shown; the GAS-like site is underlined). The presence of specific gamma-activated factor (GAF) complexes was confirmed with specific anti-STAT1 antibody.
EMSAs were also performed with cell extracts from IFN-γ- or IFN-α-treated cells in the presence of recombinant His-tagged P, His-tagged P3, or His-tagged Gp17 protein (protein of the phage Spp1 provided by I. Petitpas, LVMS, CNRS, Gif sur Yvette, France). In the case of IFN-γ treatment, the GAS probe was used as described above. In the case of IFN-α treatment, EMSA was performed with nuclear cell extracts and an ISRE probe. Briefly, 5 × 105 cells were lysed in 125 μl of cold freshly prepared buffer A (0.5% NP-40, 10 mM HEPES [pH 7.9], 10 mM KCl, 0.1 mM EDTA, 1 mM DTT, 1 mM phenylmethylsulfonyl fluoride). Nuclear extracts were incubated in 50 μl of cold freshly prepared buffer N (20 mM HEPES [pH 7.9], 400 mM NaCl, 0.1 mM EDTA, 1 mM DTT, 1 mM phenylmethylsulfonyl fluoride) completed with protease inhibitors. Proteins were examined by EMSA with a 32P-labeled ISRE probe. The probe was generated with the duplex oligonucleotide 5′-AAAGGGAAAGTGAAACTAGAAAGTGAAAGA-3′ (ISRE element). The binding of ISGF3 to the probe was confirmed with specific anti-STAT2 antibody.
Cells extracts and immunoblotting.
In some experiments, cells were lysed in hot Laemmli sample buffer for 5 min and proteins were analyzed by 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (PAGE) and transferred onto a nitrocellulose membrane as described previously (28).
Immunofluorescence staining and confocal microscopy.
Cells were fixed and permeabilized for 5 min with methanol at −20°C. They were then prepared for double-immunofluorescence staining and analyzed by confocal microscopy. The intracellular distribution of STAT1 or phosphorylated STAT1 (pSTAT1) was analyzed by using rabbit anti-STAT1 or anti-pSTAT1 antibodies at a dilution of 1/100 or 1/50, respectively, and the corresponding anti-rabbit immunoglobulin G (IgG) antibody conjugated to Alexa Fluor 568 (Molecular Probes). The viral P protein was stained by using mouse polyclonal anti-P antibody at a dilution of 1/1,000 and the corresponding anti-mouse IgG antibody conjugated to Alexa Fluor 488 (Molecular Probes). The cells were mounted in mounting medium containing 4,6-diamidino-2-phenylindole (DAPI) to stain nuclei.
Confocal laser microscopy was performed with a Leica SP2 microscope (63× oil immersion objective) using ultraviolet excitation at 351 nm (DAPI), blue laser excitation at 488 nm (Alexa Fluor 488), and green laser excitation at 545 nm (Alexa Fluor 568) in sequential recording mode.
RESULTS
The localization of STAT1 depends on the localization of P: STAT1 colocalizes with P in the cytoplasm and with P3 in the nucleus.
In order to study the mechanism involved in the cytoplasmic retention of STAT1 in the presence of P, we first investigated whether the localization of STAT1 is correlated with the localization of P. We took advantage of our previous data (20) demonstrating that P protein is a nucleocytoplasmic shuttling protein that contains an NLS in the C-terminal domain and a CRM1-dependent NES in the N-terminal domain (Fig. 1A). These signals determine the localization of the N-terminally truncated P proteins (P2, P3, P4, and P5) synthesized from the P mRNA: P and P2 are excluded from the nucleus due to the NES, and P3 to P5 are nuclear because they have only the NLS (20).
FIG. 1.
Rabies virus P protein is a nucleocytoplasmic protein. (A) P contains a CRM1-dependent NES between the residues 48 and 59 (underlined) and a conformational NLS in the carboxy-terminal part of P containing a short lysine-rich stretch located in close proximity to arginine 260 (211KKYK214-R260) (20). P3 (residues 53 to 297) is an original product translated from the P gene and is present in infected cells (7); the first methionine (bold) of P3 is located inside the NES. The protein PΔN44 contains nine more residues than does P3 and also contains the NES. (B) Interaction of P3 and PΔN44 with STAT1 by a two-hybrid system. L40 yeast cells expressing the indicated bait and prey pairs were streaked onto plates lacking tryptophan and leucine. The induction of the lacZ reporter gene was assayed by the appearance of blue colonies as previously described by Vidy et al. (28).
In order to analyze the effect of P localization on IFN-induced STAT1 nuclear accumulation, we used first LMB to inhibit the CRM1-dependent nuclear export of P (Fig. 2A and 3A) and second deleted P mutants (Fig. 2B). Indirect immunofluorescence was performed to analyze the subcellular distribution of STAT1 after stimulation with IFN.
FIG. 2.
Localization of pSTAT1 is correlated with the localization of P. (A) U373-MG control (upper panel) or P-expressing cell lines were incubated in the absence (middle panel) or presence of LMB (+LMB) at a final concentration of 20 nM for 1.5 h (lower panel). Cells were unstimulated or stimulated with 2,000 U/ml of hIFN-α or IFN-γ for 30 min as indicated, and they were fixed, permeabilized, and then stained with anti-P and anti-pSTAT1 antibodies and DAPI. (B) U373-MG cells were transfected with plasmids expressing P-GFP, P3-GFP (PΔN52-GFP), or PΔN44-GFP. Forty-eight hours after transfection, cells were treated with 2,000 U/ml of hIFN-α or hIFN-γ for 30 min as indicated. Cells were then stained with anti-P and anti-pSTAT1 antibodies and DAPI.
FIG. 3.
Localization of total STAT1 is not sensitive to LMB and depends on the P localization. (A) U373-MG cells transfected with plasmids expressing P-GFP were incubated in the absence (upper panel) or presence of LMB (+LMB) (lower panel) at a final concentration of 20 nM for 1.5 h and then treated with 2,000 U/ml of hIFN-α or hIFN-γ for 30 min as indicated. Cells were then stained with anti-P and anti-STAT1 antibodies and DAPI. The scale bars correspond to 40 μM. (B) U373-MG cells were incubated in the absence or presence of LMB (+LMB) at a final concentration of 20 nM for 1.5 h and then untreated (−) (left panel) or treated (+) (right panel) with 2,000 U/ml of hIFN-α for 30 min. Cells were then stained with anti-STAT1 antibodies and DAPI.
Control or P-expressing U373-MG cell lines were stained with anti-P antibody and anti-pSTAT1 (Fig. 2A). In control cells, pSTAT1 was rapidly redistributed into the nucleus following IFN-α or IFN-γ stimulation (Fig. 2A, upper panel). As expected, P displayed cytoplasmic localization and its expression prevented the nuclear accumulation of STAT1 in response to IFN-α or IFN-γ, resulting in the cytoplasmic localization of pSTAT1 (Fig. 2A, middle panel) (28). Accordingly to our previous results, similar cytoplasmic localization of total STAT1 in response to IFN was observed in the presence of rabies virus P (Fig. 3A) (28). Although CRM1-dependent NES elements have been identified on STAT1, it has been reported that the addition of LMB for 1 or 2 h before IFN treatment (30 min) influenced neither STAT1 cytoplasmic localization in the resting state nor its nuclear accumulation upon activation, indicating the existence of additional export mechanism (2). By using this condition, we observed the same insensitivity of STAT1 to the drug (Fig. 3B). In contrast, the localization of P was sensitive to LMB treatment, resulting in the nuclear retention of P as previously described (Fig. 2A, lower panel, and 3A) (28). In addition, the nuclear P localization appeared to be correlated with the nuclear accumulation of pSTAT1 (Fig. 2A, lower panel) or total STAT1 (Fig. 3A) upon IFN-α and IFN-γ treatment.
U373-MG cells were also transfected with plasmids encoding complete P and truncated P proteins in fusion with GFP. As previously shown, the amino-terminally truncated PΔN52-GFP, also named P3-GFP, was nuclear because it contains only the NLS (Fig. 1A and 2B); in contrast, PΔN44-GFP that contains more residues than P3-GFP does to reconstitute the NES was cytoplasmic (Fig. 1A and 2B). Both mutants contain the STAT1 binding domain that is located in the C-terminal domain of P and interacted with STAT1 (Fig. 1B) (28). In cells expressing P3-GFP, pSTAT1 displayed a nuclear localization (Fig. 2B, medium panel), whereas in cells expressing PΔN44-GFP, pSTAT1 was cytoplasmic (Fig. 2B, lower panel). These results indicate that the localization of STAT1 in response to IFN is correlated with the localization of P.
The inhibition of IFN signaling is not correlated to the retention of STAT1 in the cytoplasm.
We analyzed the effect of the localization of P or STAT1 on the IFN transcriptional responses. IFN-α/β and IFN-γ luciferase reporter gene assays were conducted with transiently and stably transfected U373-MG cells (Fig. 4).
FIG. 4.
Expression of the nuclear forms of P inhibits IFN-α and IFN-γ signaling. (A) U373-MG cells were transfected with an ISRE-firefly luciferase reporter plasmid (pISRE-f. luc) and a Renilla luciferase expression vector (pTK-r.luc) and either an empty vector or a plasmid expressing P-GFP, P3-GFP, or PΔΝ44-GFP as indicated. At 48 hours after transfection, cells were untreated (−) or treated (+) with 2,000 U/ml of hIFN-α for 6 h prior to lysis and luciferase assays. (B) U373-MG cell lines expressing P or a control plasmid were transfected with pISRE-f.luc and pTK-r.luc. At 48 hours after transfection, cells were incubated in the absence or presence of LMB at a final concentration of 20 nM for 1 h. Cells were then unstimulated (−) or stimulated (+) with 2,000 U/ml of hIFN-α for 6 h prior to lysis and luciferase assays. (C) Same as described for panel B, but an IFN-γ-responsive GAS luciferase reporter was used instead of ISRE luciferase (pGAS-luc) and hIFN-γ was used instead of hIFN-α. All bars represent average values of firefly luciferase from triplicate samples, normalized to the expression of Renilla luciferase and expressed as percentages of IFN-stimulated controls; error bars indicate standard deviations. (D) U373-MG control or P-expressing cell lines were untreated (−) or treated with 2,000 U/ml of hIFN-γ (+γ) or hIFN-α (+α) for 24 h. The expression of PML, PKR, and IRF1 was studied by Western blot analysis with specific antibodies.
As expected, cells receiving IFN-α treatment resulted in the induction of the luciferase reporter gene activity compared to that for untreated cells (Fig. 4A). Expression of the cytoplasmic P protein in transfected cells (Fig. 4A) inhibited IFN-α-responsive transcription, as did the cytoplasmic PΔN44-GFP protein (Fig. 4A). IFN-α signaling inhibition was also observed in the presence of the nuclear P3 protein (Fig. 4A). Similar results were obtained after IFN-γ treatment (data not shown). These data indicate that the IFN evasion activity does not depend on the localization of P and suggest that the nuclear P3 product interferes with an intranuclear step of IFN signaling. To confirm these data, we studied the effect of P on IFN-α and IFN-γ responses in cell lines stably expressing P. Experiments that induced the nuclear localization of P were performed in the absence or presence of LMB, as shown in Fig. 2A. Similar inhibition of IFN signaling by P protein was observed in the absence or presence of LMB (Fig. 4B and C), demonstrating that the retention of STAT1 in the cytoplasm is not the only mechanism involved in this inhibition. In addition, P protein expressed in a stable cell line was able to impair the synthesis of some ISG products, such as PML, PKR, and IRF1, induced by IFN-α or IFN-γ (Fig. 4D).
The inhibition of IFN signaling is due to a reduction of binding of pSTAT1 and ISGF3 on DNA promoters.
We further investigated the effect of P on the downstream intranuclear step of IFN-γ and IFN-α signaling. After its nuclear translocation, the pSTAT1 homodimer termed GAF binds to a DNA element termed GAS to induce specifically the transcription of ISGs.
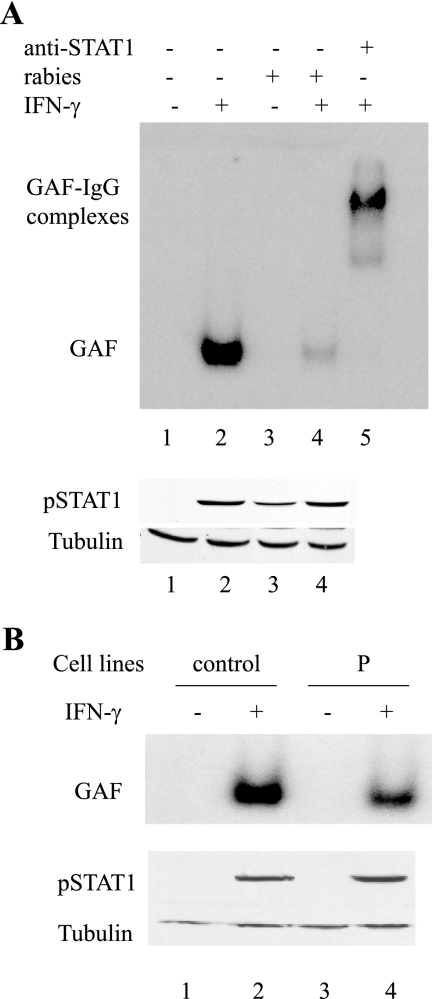
Therefore, we analyzed the binding activity of the pSTAT1 homodimer to the GAS motif by EMSA with infected U373-MG cell extracts (Fig. 5A, upper panel). Cells were uninfected or infected and then untreated or treated with IFN-γ for 30 min, and cell extracts were analyzed by EMSA with a [γ-32P]ATP-labeled GAS DNA probe. As expected, upon induction with IFN-γ, a band corresponding to the slower-migrating product predicted to be a GAF complex was apparent in uninfected cell extracts (Fig. 5A, lane 2). This band was absent in nontreated IFN-γ cells that were either uninfected or infected (Fig. 5A, lanes 1 and 3). Interestingly, a significant reduction in GAF complexes in response to IFN-γ was observed in infected cells (Fig. 5A, lane 4). Western blot analysis was performed on the same cell extracts to detect pSTAT1 (Fig. 5A, lower panel). The results confirmed that levels of pSTAT1 were similar in uninfected and infected cells upon IFN-γ treatment (lanes 2 and 4) and indicated that rabies virus infection inhibits the binding of STAT1 to DNA. The incubation of cell extracts with the anti-STAT1 antibody prior to incubation with the probe revealed the presence of a supershifted band, supporting the possibility that the GAF complex was composed of the STAT1 homodimer (Fig. 5A, lane 5).
FIG. 5.
Effect of P expressed in infected cells and in P-expressing cell lines on the formation of GAF complexes. (A) U373-MG cells were not infected (−) (lanes 1 and 2) or infected (+) (lanes 3 and 4). Twenty-four hours after infection, cells were not treated (−) (lanes 1 and 3) or treated (+) (lanes 2 and 4) with 2,000 U/ml of hIFN-γ for 30 min. Total cell lysates were analyzed by EMSA using a GAS γ-32P-labeled probe and native PAGE (upper panel). The same cell extracts were analyzed by immunoblotting with anti-pSTAT1 and anti-tubulin antibodies (lower panel). In addition, extracts of noninfected and IFN-γ-treated cells were incubated with anti-STAT1 antibodies prior to incubation with GAS γ-32P-labeled probe (lane 5). The GAF complex and the supershifted GAF complex-IgG are indicated. (B) Control and P-expressing cell lines were not treated (−) (lanes 1 and 3) or treated (+) (lanes 2 and 4) with 2,000 U/ml of hIFN-γ for 30 min. Total cell lysates were analyzed by EMSA using a GAS γ-32P-labeled probe and native PAGE. The same cell extracts were analyzed by immunoblotting with anti-pSTAT1 and anti-tubulin antibodies (lower panel).
In order to verify that this inhibition was mediated by P protein, EMSA analysis was also performed in P-expressing U373-MG cells (Fig. 5B). As shown in the context of viral infection (Fig. 5A) (28), P expression did not induce STAT1 degradation and did not interfere with STAT1 phosphorylation (Fig. 5B, lower panel). A reduction of the formation of GAF complexes was observed, demonstrating that P inhibits the binding of the pSTAT1 to the GAS DNA promoter (Fig. 5B, upper panel).
To further confirm the direct role of P or P3 in the inhibition of the DNA binding of STAT1, in vitro assays were performed by using purified proteins. Recombinant His-tagged P and P3 proteins were produced in Escherichia coli and purified as described previously by Gigant et al. (11). Extracts of IFN-γ-treated cells containing pSTAT1 were mixed with increasing concentrations of His-tagged P and His-tagged P3 and analyzed by EMSA (Fig. 6A and B). A concentration of 1 μM of P or P3 inhibits the formation of GAF complexes (Fig. 6A and B, lanes 2 and 3), whereas the same concentration of an unrelated His-tagged protein (gp17 protein of the phage Spp1) had no effect on the formation of GAF complexes (Fig. 6C). The capacity of P or P3 to impair STAT1-GAS binding increased in a P or P3 concentration-dependent manner (Fig. 6A and B). These results demonstrated the ability of purified P or P3 to inhibit the binding of the pSTAT1 dimers to the GAS probe, probably by directly interacting with STAT1. EMSA analysis was also performed to analyze the effect of P on the complex formation of ISGF3 with DNA. Extracts from IFN-α-treated cells were mixed with 1 μM of His-tagged P or His-tagged P3 or gp17. Similar P- and P3-dependent inhibition of the complex formation with the ISRE probe was observed (Fig. 7), demonstrating that P is also able to block the binding of ISGF3 to ISRE in response to IFN-α. The fact that STAT1 concentration in the extract was very low (less than 100 nM) and that the labeled probe was present under nonsaturing conditions led us to estimate the dissociation constant (KD) between P and STAT1 that corresponds to the P concentration responsible for 50% of the STAT1 DNA binding inhibition; the apparent KD value is in the 100 nM range.
FIG. 6.
Effect of recombinant P and P3 proteins on the formation of GAF complexes. Total cell extracts of hIFN-γ-treated U373-MG cells were mixed with increasing concentrations of His-tagged P (A) or His-tagged P3 (B) protein or unrelated His-tagged Gp17 protein (C) and submitted to EMSA analysis. (D) The same cell extracts were analyzed by immunoblotting with anti-pSTAT1 and anti-tubulin antibodies.
FIG. 7.
Effect of recombinant P and P3 proteins on the formation of ISGF3 complexes to the ISRE promoter. (A) U373-MG cells were not treated (−) (lane 1) or treated (+) (lanes 2 to 7) with 2,000 U/ml of hIFN-α for 30 min. Nuclear cell extracts were mixed with 1 μM of His-tagged P (lane 5), His-tagged P3 (lane 6), or His-tagged Gp17 (lane 7) proteins and submitted to EMSA analysis using a ISRE γ-32P-labeled probe and native PAGE gel. In addition, extracts of IFN-α-treated cells were incubated with anti-STAT2 antibodies prior to incubation with γ-32P-labeled probe (lane 3, *). (B) The same cell extracts were analyzed by immunoblotting with anti-pSTAT2 and anti-tubulin antibodies.
DISCUSSION
We have previously shown that rabies virus P protein interacts with STAT1 and inhibits IFN signaling pathways (28). As previously shown by Brzózka et al. (4), the interaction of P with pSTAT1 is much stronger than that with non-pSTAT1 (5). P does not target STAT1 for degradation or interfere with STAT1 phosphorylation, but it retains STAT1 in the cytoplasm (5, 28). By analyzing the molecular mechanism involved in the cytoplasmic retention of STAT1, we show in this study that P is also able to block an intranuclear step of type I and type II IFN signaling: the binding of STAT1 and ISGF3 to the DNA promoters.
Previous data have shown that (i) P is a nucleocytoplamic protein that shuttles between the cytoplasm and the nucleus (20), (ii) the N-terminally truncated P3 is nuclear (20), and (iii) the STAT1 binding site is located in the carboxyl-terminal domain of P (28). We confirm here that P3 shares the STAT1 binding site with P.
We first show that following IFN activation, the localization of STAT1 is correlated with the localization of P. In cells stably or transiently expressing a nuclear form of P (P in the presence of LMB or P3), STAT1 is nuclear, whereas in cells expressing a cytoplasmic form of P (PΔN44 or P), STAT1 is cytoplasmic. It should be noted that in the absence of IFN treatment, STAT1 does not relocalize to the nucleus in the presence of P3, indicating that P or P3 interacts more efficiently with the phosphorylated form of STAT1 as previously shown by Brzózka et al. (4). Surprisingly, the nuclear forms of P are able to inhibit IFN signaling as tested by luciferase activity, demonstrating that this inhibition is not due to the retention of STAT1 in the cytoplasm. Therefore, we examined the following nuclear step that is the DNA binding activity of STAT1. We show by EMSA of cell extracts from infected cells or cells stably expressing P that the capacity of IFN-γ to induce DNA binding of STAT1 was inhibited. Interestingly, the addition of purified recombinant P or P3 to extracts from IFN-γ- or IFN-α-treated cells prevents the binding of pSTAT1 to the GAS or of ISGF3 to the ISRE, demonstrating that P interacts directly with STAT1, leading to the inhibition of type I and type II IFN responses.
It is unclear at present how P protein inhibits the binding activity of pSTAT1 to the DNA. As described previously, rabies virus P protein interacts with the coiled-coil or DNA binding domains of STAT1 (28); therefore, the direct interaction of P with the DNA binding domain of STAT1 could interfere with the DNA binding activity of STAT1.
The fact that viral P and P3 proteins share the STAT1 binding domain and localize to different compartments of the cell provides the virus a dual strategy for blocking both cytoplasmic and nuclear forms of STAT1. This is also the case with Nipah virus V and W proteins that inhibit STAT1 activation from the cytoplasm and the nucleus, respectively (25, 26, 27).
P has been also shown to impair nuclear accumulation of STAT1 (5, 28), suggesting that P may inhibit IFN signaling at two different and independent steps. However, we cannot exclude the possibility that both steps are related and the inhibition of nuclear accumulation of STAT1 is due to a reduction of the DNA binding activity. Indeed, it has been proposed that DNA binding controls the nuclear accumulation of STAT1: DNA binding protects STAT1 from dephosphorylation, and the DNA-bound STAT1 is thus retained in the nucleus (17, 18, 19). In this model, the loss of DNA binding is associated with the cytoplasmic accumulation of STAT1. In our case, the loss of DNA binding is necessary but not sufficient to explain the different localization of STAT1 in the presence of P or P3; in addition, the presence of a strong export signal in the N-terminal part of P may be involved in the nuclear export of STAT1, as suggested by the results obtained with the PΔN44 mutant.
Viral inhibition of the Jak-STAT pathway has been shown in other negative-strand RNA viruses, and among members of the Paramyxoviridae family, there is a great diversity in the evasion STAT signaling. Viral proteins can target STAT1 and STAT2 for degradation and inhibit phosphorylation and dimerization or nuclear accumulation of STAT1 (6, 15, 16). Very few cases of inhibition of the DNA binding activity of STAT1 have been reported, and this inhibition is not direct but described as a consequence of the impairment of one of the upstream steps. To our knowledge, only one report has shown that Sendai C protein directly inhibits the binding of the STAT1 homodimer on DNA (13).
It is interesting that rabies virus P protein, in addition to inhibiting IFN type I synthesis, acts at three different levels of the IFN signaling: it inhibits the nuclear accumulation of STAT1, the binding of STAT1 to the DNA, and the function of ISG products such as PML. Quite frequently, viruses use more than one strategy to evade the IFN system at one or more levels, and this may reflect how difficult it is to completely shut down this host antiviral response. In that sense, rabies virus P can be termed a multifunctional IFN antagonist. This gives a rabies virus with limited coding capacity the ability to inhibit multiple arms of the host's innate immune response.
Acknowledgments
We are grateful to Yves Gaudin for helpful discussions and careful reading of the manuscript. We acknowledge Spencer Brown and Susanne Bolt for help and assistance with confocal microscopy.
The microscopy was performed on the “Plate-forme Imagerie et Biologie Cellulaire” of the CNRS campus supported by the Institut Fédératif de Recherche 87 “La plante et son environnement” and the program ASTRE of the Conseil général de l'Essonne. The work in the team of M. Chelbi-Alix is supported by the “Association pour la Recherche sur le Cancer.”
Footnotes
Published ahead of print on 7 February 2007.
REFERENCES
- 1.Basler, C. F., and A. Garcia-Sastre. 2002. Viruses and the type I interferon antiviral system: induction and evasion. Int. Rev. Immunol. 21:305-337. [DOI] [PubMed] [Google Scholar]
- 2.Begitt, A., T. Meyer, M. van Rossum, and U. Vinkemeier. 2000. Nucleocytoplasmic translocation of Stat1 is regulated by a leucine-rich export signal in the coiled-coil domain. Proc. Natl. Acad. Sci. USA 97:10418-10423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blondel, D., T. Regad, N. Poisson, B. Pavie, H. Harper, P. P. Pandolfi, H. De Thé, and M. K. Chelbi-Alix. 2002. Rabies virus P and small P products interact directly with PML and reorganize PML nuclear bodies. Oncogene 21:7957-7970. [DOI] [PubMed] [Google Scholar]
- 4.Brzózka, K., S. Finke, and K. K. Conzelmann. 2005. Identification of the rabies virus alpha/beta interferon antagonist: phosphoprotein P interferes with phosphorylation of interferon regulatory factor 3. J. Virol. 79:7673-7681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brzózka, K., S. Finke, and K. K. Conzelmann. 2006. Inhibition of interferon signalling by rabies virus phosphoprotein P: activation-dependent binding of STAT1 and STAT2. J. Virol. 80:2675-2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chelbi-Alix, M. K., A. Vidy, J. El. Bougrini, and D. Blondel. 2006. Rabies viral mechanisms to escape the IFN system: the viral protein P interferes with IRF-3, Stat1, and PML nuclear bodies. J. Interferon Cytokine Res. 26:271-280. [DOI] [PubMed] [Google Scholar]
- 7.Chenik, M., K. Chebli, and D. Blondel. 1995. Translation initiation at alternate in-frame AUG codons in the rabies virus phosphoprotein mRNA is mediated by a ribosomal leaky scanning mechanism. J. Virol. 69:707-712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Darnell, J. E., Jr., I. M. Kerr, and G. R. Stark. 1994. Jak-STAT pathways and transcriptional activation in response to IFNs and extracellular signalling proteins. Science 264:1415-1421. [DOI] [PubMed] [Google Scholar]
- 9.García-Sastre, A. 2002. Mechanisms of inhibition of the host interferon alpha/beta-mediated antiviral responses by viruses. Microbes Infect. 4:647-655. [DOI] [PubMed] [Google Scholar]
- 10.Garcin, D., J. Curran, and D. Kolakofsky. 2000. Sendai virus C proteins must interact directly with cellular components to interfere with interferon action. J. Virol. 74:8823-8830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gigant, B., F. Iseni, Y. Gaudin, M. Knossow, and D. Blondel. 2000. Neither phosphorylation nor the amino-terminal part of rabies virus phosphoprotein is required for its oligomerization. J. Gen. Virol. 81:1757-1761. [DOI] [PubMed] [Google Scholar]
- 12.Goodbourn, S., L. Didcock, and R. E. Randall. 2000. Interferons: cell signaling, immune modulation, antiviral response and virus countermeasures. J. Gen. Virol. 81:2341-2364. [DOI] [PubMed] [Google Scholar]
- 13.Gotoh, B., T. Komatsu, K. Takeuchi, and J. Yokoo. 2003. The C-terminal half-fragment of the Sendai virus C protein prevents the gamma-activated factor from binding to a gamma-activated sequence site. Virology 316:29-40. [DOI] [PubMed] [Google Scholar]
- 14.Gupta, A. K., D. Blondel, S. Ghoudhary, and A. K. Banerjee. 2000. The phosphoprotein of rabies virus is phosphorylated by a unique cellular protein kinase and specific isomers of protein kinase C. J. Virol. 74:91-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haller, O., G. Kochs, and F. Weber. 2006. The interferon response circuit: induction and suppression by pathogenic viruses. Virology 344:119-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Horvath, C. M. 2004. Silencing STATs: lessons from paramyxovirus interferon evasion. Cytokine Growth Factor Rev. 15:117-127. (Erratum, 15:477.) [DOI] [PubMed] [Google Scholar]
- 17.Meyer, T., A. Begitt, L. Lodige, M. Van Rossum, and U. Vinkemeier. 2002. Constitutive and IFN-gamma-induced nuclear import of STAT1 proceed through independent pathways. EMBO J. 21:344-354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meyer, T., A. Marg, P. Lemke, B. Wiesner, and U. Vinkemeier. 2003. DNA binding controls inactivation and nuclear accumulation of the transcription factor Stat1. Genes Dev. 17:1992-2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meyer, T., L. Hendry, A. Begitt, S. John, and U. Vinkemeier. 2004. A single residue modulates tyrosine dephosphorylation, oligomerization, and nuclear accumulation of STAT transcription factors. J. Biol. Chem. 279:18998-19007. [DOI] [PubMed] [Google Scholar]
- 20.Pasdeloup, D., N. Poisson, H. Raux, Y. Gaudin, R. W. Ruigrok, and D. Blondel. 2005. Nucleocytoplasmic shuttling of the rabies virus P protein requires a nuclear localization signal and a CRM1-dependent nuclear export signal. Virology 334:284-931. [DOI] [PubMed] [Google Scholar]
- 21.Pelicano, L., F. Li, C., Schindler, and M. K. Chelbi-Alix. 1997. Retinoic acid enhances the expression of interferon-induced proteins: evidence for multiple mechanisms of action. Oncogene 15:2349-2359. [DOI] [PubMed] [Google Scholar]
- 22.Raux, H., F. Iseni, F. Lafay, and D. Blondel. 1997. Mapping of monoclonal antibody epitopes of the rabies virus P protein. J. Gen. Virol. 78:119-124. [DOI] [PubMed] [Google Scholar]
- 23.Raux, H., A. Flamand, and D. Blondel. 2000. Interaction of the rabies virus P protein with the LC8 dynein light chain. J. Virol. 74:10212-10216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Regad, T., and M. K. Chelbi-Alix. 2001. Role and fate of PML nuclear bodies in response to interferon and viral infections. Oncogene 20:7274-7286. [DOI] [PubMed] [Google Scholar]
- 25.Rodriguez, J. J., C. D. Cruz, and C. M. Horvath. 2004. Identification of the nuclear export signal and STAT-binding domains of the Nipah virus V protein reveals mechanisms underlying interferon evasion. J. Virol. 78:5358-5367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shaw, M. L., A. Garcia-Sastre, P. Palese, and C. F. Basler. 2004. Nipah virus V and W proteins have a common STAT1-binding domain yet inhibit STAT1 activation from the cytoplasmic and nuclear compartments, respectively. J. Virol. 78:5633-5641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shaw, M. L., W. B. Cardenas, D. Zamarin, P. Palese, and C. F. Basler. 2005. Nuclear localization of the Nipah virus W protein allows for inhibition of both virus- and Toll-like receptor 3-triggered signaling pathways. J. Virol. 79:6078-6088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vidy, A., M. Chelbi-Alix, and D. Blondel. 2005. Rabies virus P protein interacts with STAT1 and inhibits interferon signal transduction pathways. J. Virol. 79:14411-14420. [DOI] [PMC free article] [PubMed] [Google Scholar]