Abstract
Infection with adenovirus vectors (AdV) results in rapid activation of innate immunity, which serves the dual purpose of stimulating inflammatory antiviral host defenses and the adaptive immune system. Viral recognition by macrophages, dendritic cells, and other cell types requires an ability to sense the presence of a foreign molecular pattern by “pattern recognition receptors.” The nature of the adenoviral sensor, the target ligand of the sensor, and the downstream antiviral signaling response triggered by virus infection have not been defined for this nonenveloped double-stranded DNA (dsDNA) virus. We have identified four critical links involved in AdV recognition by murine antigen-presenting cells (APC) and primary lung fibroblasts: (i) viral recognition occurs chiefly via a Toll-like receptor (TLR)-independent nucleic acid-sensing mechanism recognizing the viral dsDNA genome, (ii) the intact viral particle and capsid proteins are required for efficient intracellular delivery of the viral genome, (iii) delivery of the viral genome triggers interferon regulatory factor 3 (IRF3) phosphorylation, and (iv) IRF3 activation is the required dominant antiviral signaling pathway used by APC, whereas the “primary” involvement of NF-κB, mitogen-activated protein kinase, or Akt pathways is less prominent. In this study we provide the first direct evidence that infection by a dsDNA virus stimulates an IRF3-mediated interferon and proinflammatory response through a TLR-independent DNA-sensing mechanism.
Adenovirus (Ad) is a nonenveloped, double-stranded, linear DNA virus with a genome of approximately 35 kilobases and a particle size of 70 to 100 nm. The majority of Ad serotypes infect cells through a well-characterized, multistep process that includes high-affinity binding of the capsid protein fiber to the coxsackie-Ad receptor cell surface protein (1) and endocytic internalization stimulated by an RGD motif present in penton base binding to membrane integrins (61). Following internalization and subsequent endosome acidification, Ad escapes the endosomal compartment by directed membrane disruption (62) and transmigrates to the nuclear envelope by way of dynein/microtubule-mediated transport (31). Viral DNA passes through the nuclear pore complex (7) and establishes a noncovalent association with the nuclear matrix as a prerequisite to transcription by the host transcriptional machinery (51).
The entire process of virus entry is extremely efficient, which in combination with a well-defined and easily manipulated viral genome prompted development of Ad vectors (AdV) for gene transfer applications. First-generation AdV are deficient for the E1 and E3 regions, rendering them replication deficient and able to carry up to 7 kb of foreign genomic material. Systemic administration of AdV results in high levels of transgene expression but also results in a robust inflammatory immune response (41, 66), which is followed by an equally vigorous adaptive immune response (65). The combination of the innate inflammatory response with the adaptive immune response eradicates infecting virus as well as cells harboring virus. Thus, host innate immunity to AdV represents a major barrier to the effective use of these agents for a variety of therapeutic applications.
Macrophages and dendritic cells (DC) are among the frontline sentinel cells that initiate a response to pathogenic stimuli, including Ad. Murine bone marrow-derived macrophages (BMMO) or DC (BMDC) are relatively nonpermissive to AdV transduction (44, 52), consistent with low levels of coxsackie-Ad receptor in murine antigen-presenting cells (APC) (59). However, they undergo activation and maturation when exposed to AdV (38, 59, 60). Studies indicate that penton base-RGD binding to membrane integrins contributes to virus binding and internalization in these cell types (15, 59). While these studies demonstrate that APC respond to AdV, the molecular mechanisms by which these cells sense infection by Ad or AdV are not understood.
To date, two types of innate immune recognition receptors that link virus detection with inflammation and activation of adaptive immunity have been characterized. The first type, the membrane-associated Toll-like receptors (TLRs), are expressed primarily by macrophages and DC. There are 13 murine TLRs, and they have been extensively studied (25). Activation occurs on the surface of the cell membrane or within the endosomal compartment and requires either a TRIF or MyD88 adaptor molecule. TLR3, TLR7/8, and TLR9 recognize double-stranded RNA (dsRNA), single-stranded RNA, and double-stranded DNA (dsDNA) with unmethylated CpG motifs, respectively (24). TLR4 mediates the recognition of mouse mammary tumor virus (46) and the fusion protein of respiratory syncytial virus (30), and TLR2 mediates the cytokine responses to human cytomegalovirus (3), herpes simplex virus type 1 (HSV-1) envelope proteins (29), and measles virus hemagglutinin (2).
Engagement of a TLR family member initiates a network of signaling cascades, invariably involving the intracellular adaptor proteins MyD88 and/or TRIF (24). The adaptor proteins facilitate activation of downstream signaling cascades, which in turn lead to activation of transcription factors, including NF-κB, AP1, and interferon (IFN) regulatory factor (IRF) family members (24, 58). TLR4 is the only family member known to activate through both TRIF and MyD88 adaptor molecules; all others use either MyD88 or TRIF (25). Lipopolysaccharide (LPS) stimulation of TLR4 results in activation of gene families which are TRIF dependent, gene families which are MyD88 dependent, and genes that are activated by both pathways (11). For signaling responses that are stimulated by both TRIF and MyD88 adaptors, Akira's group (23, 26, 64) and others (4) have shown that in single-knockout (KO) cells, signaling for these responses occurs with altered kinetics compared to wild-type (WT) cells. Importantly no cytokines or IFNs are controlled by the pathway that utilizes both MyD88 and TRIF adaptors (11).
The second type of viral innate immune sensor belongs to the RNA helicase family and includes RIG-1 and MDA-5, which serve as cytosolic dsRNA detectors. RIG-1 is critical for vesicular stomatitis virus, Newcastle disease virus, Sendai virus, and influenza virus recognition (21, 22), while MDA-5 is the principal sensor for transfected poly(I·C) and encephalomyocarditis virus (6, 22) infection. Recognition by the RNA helicases results in the recruitment of the adaptor protein IPS1 (also known as MAVS, VISA, or Cardif), which stimulates the TANK-related kinases TBK1 and IKKɛ, leading to the phosphorylation of IRF3 and IRF7, which are transcription factors essential for the expression of type I IFNs (28).
Recent studies indicate the existence of a novel antimicrobial recognition mechanism. Exposure of cells to intracellular bacteria or transfected DNA results in induction of a type I IFN response that is independent of the TLR or RNA helicase pathway, which implies the existence of a cytosolic DNA-sensing mechanism that activates the IRF3 transcriptional regulator (17, 55).
In this report, we provide the first demonstration that a dsDNA virus activates APC and fibroblasts through TLR-independent dsDNA-mediated activation of IRF3 and that IRF3 functions as a master regulator of the antiviral immune response in these cells. BMDC and BMMO respond to AdV infection by producing large amounts of type I IFNs and modest levels of interleukin-6 (IL-6) and tumor necrosis factor alpha (TNF-α). AdV activation is dependent on IRF3; both APC and primary lung fibroblasts from IRF3-deficient animals are largely nonresponsive to AdV infection. Activation of this pathway does not depend on MyD88 or TRIF adaptor molecules. AdV recognition by APC occurs through two basic events: the intact viral capsid interacts with the target cell and facilitates vector internalization; then, following internalization, breakdown of the capsid reveals viral DNA to an intracellular sensor that triggers a cascade leading to IRF3 activation.
MATERIALS AND METHODS
Animals.
Eight- to 12-week-old male and female C57BL6 mice were obtained from Jackson Laboratories. C.B17/lcr-SCID and C.B17/lcr controls were purchased from Charles River Laboratories. MyD88-KO and TLR9-KO mice were generously provided by Shizuo Akira (Osaka University, Osaka, Japan). TRIFLps2/Lps2 (TRIF−/−) mice were kindly provided by Bruce Beutler, and IRF3-KO mice were kindly provided by Tadatsugu Taniguchi.
Viruses and AdV DNA.
Ad5CiG and Ad5CiG-ΔRGD (52) and ts1-Ad2 (8) were previously described. Viruses were purified through two rounds of CsCl gradient ultracentrifugation and stored at −70°C in storage buffer (10 mM Tris, 2 mM MgCl2, 40% sucrose, pH 7.5). AdV DNA was purified by standard procedures (52). All viral preparations contained less than 0.1 endotoxin unit per ml, and DNA preparations contained less than 0.001 U/mg DNA as determined by the Limulus amebocyte lysate assay (BioWhittaker). Psoralen-UV inactivation was performed by mixing 0.01 volume of 33-mg/ml 8-methoxy-psoralen (Sigma) with the purified viral vector and exposing it to a 365-nm UV light source 4 cm from the light filter for 60 min on ice. Residual psoralen was removed by dialysis. For heat inactivation, Ad5CiG was incubated at 56°C for 1 h. AdV empty capsids (eAdV) were purified by placing the upper band from an AdV-maxiprep CsCl step gradient onto a continuous gradient (1.2 to 1.45 gm/ml) and centrifugation at 100,000 × g for 8 h. The eAdV band from this gradient was applied to a second continuous CsCl gradient for 12 h, and the eAdV band was then isolated and dialyzed into storage buffer. The ratio of contaminating intact AdV to eAdV determined by real-time PCR with hexon primers was 1:40. AdV and eAdV amounts were matched by protein content and by Western blotting with rabbit polyclonal anti-Ad5 serum. HSV-1 (strain KOS) was kindly provided by F. Homa (University of Pittsburgh).
BMMO and BMDC.
Bone marrow cells were extracted from the femurs and tibiae of mice, and red cells were removed with lysis solution (0.15 M NH4Cl, 1 mM KHCO3, 0.1 mM EDTA). For BMMO, bone marrow cells were cultured in Dulbecco modified Eagle medium (DMEM) supplemented with 20% fetal bovine serum (FBS) and 25% supernatant derived from confluent L929 cell cultures. At day 7, immature macrophages were collected. This procedure yields a pure population of macrophage colony-stimulating factor-dependent, adherent macrophages. For BMDC, bone marrow cells were cultured in DMEM containing 10% FBS and 30% supernatant derived from J558L cells (16). On day 2, two-thirds of the supernatant was replaced to remove granulocytes. On day 7, nonadherent cells were collected. Typically more than 80% of the BMDC population was CD11c+ CD11b+ GR1− as determined by fluorescence-activated cell sorter analysis of specific surface markers.
Fluorescence-activated cell sorter analysis of cell surface phenotype.
The following antibodies against murine molecules were used: fluorescein isothiocyanate-conjugated anti-CD11c and anti-CD11b and phycoerythrin-conjugated anti-CD11c, anti-CD86, anti-CD80, anti-CD40, and anti-major histocompatibility complex class II. Cells were incubated for 15 min with rat monoclonal antibody to CD16/32 to block Fc receptors and then with the primary antibodies for 30 min on ice. Cells were fixed in 1% paraformaldehyde and analyzed using a Beckman Coulter XL flow cytometer. All antibodies were from BD Biosciences.
Cell treatments.
The dsRNA mimetic poly(I·C) was from Amersham Biosciences. Ultrapure LPS from Escherichia coli and nuclease-resistant phosphorothioate-modified CpG ODN 1826 were purchased from InvivoGen.
Bone marrow-derived cells differentiated for 7 days were replated into 60-mm plates at a density of 5 × 106 cells/plate in 3 ml DMEM with 10% FBS. At day 9, the medium volume was reduced to 1.5 ml and cells were treated with CpG ODN 1826 (2 μM), LPS (100 ng/ml), or Ad5CiG at a ratio of 25,000 particles per cell unless otherwise indicated. Herpesvirus infections were carried out at 5 PFU/cell. For transfection experiments, medium was replaced at day 8 with OPTI-MEM I (Invitrogen). At day 9, 10 μg of Ad DNA or poly(I·C) and 10 μl of Lipofectamine 2000 (Invitrogen) were incubated in 100 μl of OPTI-MEM I for 15 min before being used for stimulation.
For inhibition studies, primary macrophages were pretreated for 60 min with drugs prior to exposure to various stimuli. Actin or tubulin cytoskeleton polymerization was inhibited with 10 μM cytochalasin D or 20 μM nocodazole, respectively. Cycloheximide (CHX) (50 μg/ml) and puromycin (25 μg/ml) were used for de novo protein synthesis inhibition. Cell viability was above 85% for all treatments as confirmed by trypan blue exclusion assay. All inhibitors were from Sigma-Aldrich.
Western blots.
Whole-cell extracts were prepared by washing cells twice with ice-cold phosphate-buffered saline and harvesting them in lysis buffer (50 mM Tris [pH 7.5], 150 mM NaCl, 1 mM EDTA, 1% NP-40) with addition of phosphatase inhibitor cocktails 1 and 2 (Sigma-Aldrich) and protease inhibitors (30 mM sodium fluoride, 100 μM sodium orthovanadate, 1 mM phenylmethylsulfonyl fluoride, 10 μg/ml aprotinin, 10 μg/ml leupeptin, 1 μg/ml pepstatin, 1 μg/ml microcystin-LR, and 1 mM benzamidine). The lysates were cleared by centrifugation at 13,000 × g for 15 min at 4°C, and protein quantification was performed with the DC protein assay kit (Bio-Rad Laboratories).
For Western blot analysis, gel boxes, 10% NuPage bis-Tris gels, and buffers were purchased from Invitrogen and Immobilon-P membranes from Millipore. All blots were blocked in 5% skim milk in Tris-buffered saline-Tween (0.1%) at room temperature for 1 h. Phospho-Akt (Ser473, no. 9271), phospho-extracellular signal-regulated kinase 1/2 (ERK1/2) (Thr202/Tyr204, no. 9101), phospho-p38 mitogen-activated protein kinase (MAPK) (Thr180/Tyr182, no. 9211), phospho-IRF3 (Ser396, no. 4961), phospho-Jun N-terminal protein kinase (Thr183/Tyr185; no. 9251), phospho-IκBα (Ser32, no. 9241), phospho-p65 (Ser536, no. 3031), β-actin (no. 4967), and horseradish peroxidase-linked anti-rabbit immunoglobulin G (no. 7074) antibodies were from Cell Signaling. Total IRF3 antibody was from Santa Cruz Laboratories (no. SC-9082). All primary antibodies were used at a dilution of 1:1,000 in 5% bovine serum albumin in Tris-buffered saline, except β-actin antibody, which was used at a dilution of 1:2,000. Signals were visualized with an ECLplus kit (Amersham Biosciences).
Electrophoretic mobility shift assay (EMSA).
Cells were washed twice with ice-cold phosphate-buffered saline and disrupted in hypotonic buffer with NP-40 (10 mM HEPES [pH 7.9], 10 mM KCl, 0.1 mM EDTA, 0.1 mM EGTA, 1% NP-40) on a rocking platform for 10 min at 4°C. Nuclei were spun down, resuspended in 50 μl of hypertonic buffer (10 mM HEPES [pH 7.9], 400 mM NaCl, 0.1 mM EDTA, 0.1 mM EGTA), and incubated on ice for 30 min. Both buffers contained the same cocktail of protease and phosphatase inhibitors as described for Western blots. The final nuclear extracts were collected by centrifugation at 14,000 rpm for 20 min. The protein content in the extract was measured with the DC protein assay kit (Bio-Rad Laboratories). Gel shift assays were performed with the Promega gel shift assay system as instructed by the manufacturer, using 2 μg of nuclear proteins for each gel shift reaction. The sequence of the double-stranded consensus NF-κB-binding oligonucleotide was 5′-AGT TGA GGG GAC TTT CCC AGG C-3′. Probe labeling was carried out according to the manufacturer's instructions, using [α-32P]ATP (3,000 Ci/mmol, 10 mCi/ml; DuPont NEN Research Products). Reaction mixtures were separated on 6% DNA retardation gels (Invitrogen) at 150 V for 30 min at 4°C. Gels were dried and exposed on Kodak XAR film for 12 to 18 h.
RPA.
Total RNA was extracted with TRIzol reagent (Invitrogen) according to the manufacturer's protocol. For each sample, 5 μg of total RNA was used for RNase protection assay (RPA). The 32P-labeled RNA probe mix was prepared by in vitro transcription using mCK-3b (cytokines) or mCK5c (chemokines) multiprobe template sets (BD Pharmingen). The hybridized RNAs were treated with RNase, using an RPA kit (BD Pharmingen), and precipitated, and the protected fragments were resolved on vertical sequencing (10% acrylamide) gels. Following electrophoresis, the gels were dried and exposed to a PhosphorImager screen (Molecular Dynamics). The signals on the screen were analyzed by PhosphorImager software.
Enzyme-linked immunosorbent assay (ELISA).
Cells were seeded in 12-well tissue culture plates to give a final concentration of 2 × 106 cells/well in 1 ml of medium. At 36 h posttreatment the supernatants were harvested for determination of cytokines and IFN-α protein levels according to the manufacturer's instructions (R&D systems).
SYBR green I QRT-PCR.
For quantitative reverse transcription-PCR (QRT-PCR), total cellular RNA was isolated from 5 × 106 cells grown in 60-mm tissue culture plates using Tri-Reagent (Molecular Research Center) as instructed by the manufacturer. Amplifications were carried out in a total volume of 20 μl by using a one-step QuantiTect SYBR green kit (QIAGEN, Valenica, CA) in an Applied Biosystems Prism 7900H sequence detection system with SDS 2.1 software. Cycles consisted of an initial incubation at 95°C for 15 min followed by 35 cycles at 94°C for 15 s, 60°C for 30 s, and 72°C for 20 s, with a final incubation at 72°C for 7 min. All determinations were performed in triplicate. Nontemplate controls run with every assay consistently had no cycle threshold values before 35 cycles of PCR. The abundance of each mRNA was normalized to β-actin expression and compared to that in untreated cells to calculate the relative induction. Sequences of primers are available on request.
Statistical analysis.
Data were expressed as means ± standard errors of the means. Statistical analysis was performed with Student's t test. A P value of less than 0.05 was considered significant.
RESULTS
AdV activation of APC occurs in the absence of MyD88 or TRIF adaptor molecules.
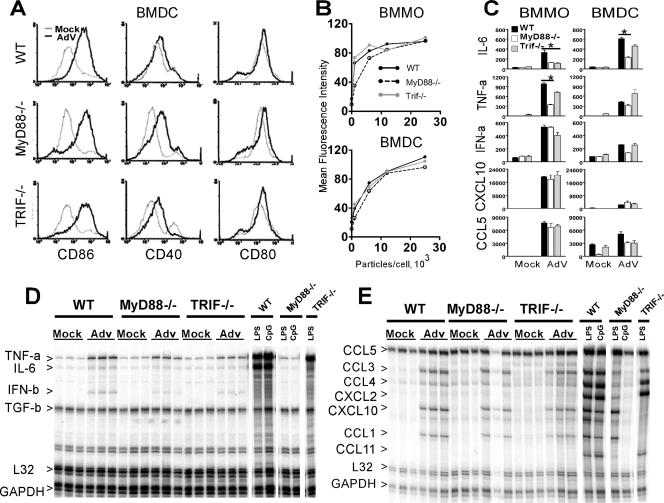
The immediate-early steps leading to AdV activation of APC are not well understood, leading us to investigate the molecular mechanisms of recognition and APC activation triggered by AdV. We first asked if the adaptor molecule MyD88 or TRIF contributes to the activation of BMDC and BMMO following exposure to AdV. WT, MyD88-deficient (MyD88−/−), and TRIF-deficient (TRIF−/−) cells were infected with 5,000 viral particles/cell, and upregulation of costimulatory molecules was examined at 36 h postinfection. AdV infection caused an increase in the expression of, and in the percentage of cells positive for, CD86, CD40, and CD80 that was independent of MyD88 and TRIF (BMDC are depicted in Fig. 1A). Furthermore, virus titration experiments conducted in both BMDC and BMMO revealed dose-dependent induction of CD86 with little distinction between the viral dose required for activating WT-, MyD88−/−-, or TRIF−/−-derived cells (Fig. 1B). Supernatants from AdV-infected BMMO and BMDC were used to quantify inflammatory cytokines and IFNs at 36 h postinfection by ELISA (Fig. 1C). AdV infection induced significant levels of IL-6, TNF-α, IFN-α, CXCL10, and CCL5 in cells derived from WT as well as MyD88−/− and TRIF−/− mice. For MyD88−/−-derived APC, there was a significant reduction in the levels of IL-6 in both BMMO and BMDC and of TNF-α from MyD88−/− BMMO compared to WT cells.
FIG. 1.
Role of MyD88 and TRIF in the innate immune response to AdV. (A) Expression of CD86, CD40, and CD80 on gated CD11c+ BMDC after 36 h of treatment with 5,000 AdV particles/cell (black) or mock treatment (gray). (B) CD86 expression in BMMO and BMDC from WT, MyD88−/−, and TRIF−/− mice incubated with various doses of AdV at 36 h postinfection. (C) Induction of cytokines in macrophages and DC derived from WT, MyD88−/−, and TRIF−/− mice. Cells were incubated with 25,000 AdV particles/cell for 36 h, and supernatant cytokine levels (pg/ml) were determined by ELISA. Error bars represent standard deviations for triplicate wells in a single experiment; data are representative of four independent experiments. Student's t tests were completed, and P values of <0.05 are indicated (*). (D and E) RPA analysis for cytokines (D) and chemokines (E) of total RNA isolated from BMDC of the indicated genotypes after 5 h of treatment with 25,000 AdV particles/cell, 10 ng/ml LPS, or 2 μM CpG DNA. The triplicate results shown for AdV are from independent infections and are representative of two separate experiments. GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
We next examined the influence of the MyD88−/− and TRIF−/− mutations on early AdV-mediated gene induction of a diverse array of inflammatory cytokines, chemokines, and IFNs in primary APC. BMDC were exposed to 25,000 AdV particles/cell, and total RNA was harvested at 5 h posttreatment. RPAs were performed to quantify mRNA levels for a panel of cytokines (Fig. 1D) and chemokines (Fig. 1E). For comparative purposes, APC were treated with 10 ng/ml LPS (TLR4 agonist, MyD88 and TRIF adaptor dependent) or 2 μM CpG DNA (TLR9 agonist, MyD88 adaptor dependent). The RNA protection profiles for TNF-α, IL-6, IFN-β, CCL5, CCL1, CCL3, CCL4, and IP-10 were comparable in WT, TRIF−/−, and MyD88−/− BMDC exposed to AdV. Transcripts corresponding to IL-6, TNF-α, and IFN-β were reduced in MyD88- and TRIF-deficient cells by 20% and 15%, respectively, compared to RNA from WT cells (phosphorimager quantification, data not shown). Similar RPA results were observed in BMMO (data not shown). The pattern of gene expression induced by AdV differed considerably from that of LPS or CpG DNA TLR agonists. AdV induced strong type I IFN but moderate chemokine and weak inflammatory cytokine responses, failing to induce CXCL2 and CCL11. In contrast, TLR agonists induced robust inflammatory cytokine and chemokine responses but undetectable levels of IFNs. These data indicate that a high level of MyD88- and TRIF-dependent activation occurs with known TLR agonists, whereas AdV results in a more modest APC response that is largely independent of MyD88 and TRIF adaptors.
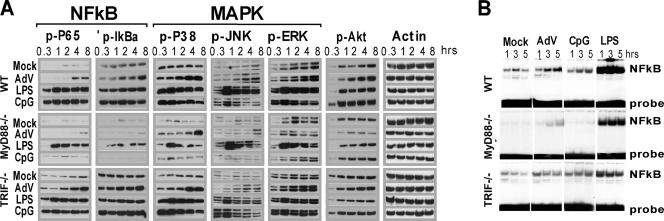
Pathogen recognition channeled through MyD88 or TRIF induces strong activation of NF-κB and MAPK signaling cascades and to a lesser degree involves the Akt signaling pathway (24). The distinct patterns of APC transcript induction elicited by AdV compared to TLR ligands led us to examine the activation profiles of these signaling cascades. BMMO were exposed to 25,000 AdV particles/cell, 10 ng/ml LPS, or 2 μM CpG DNA, and cell lysates were harvested at the indicated time points (Fig. 2A). Activation was measured by Western blotting using phospho-specific antibodies. Levels of β-actin were determined to confirm equal protein amounts in each sample, and mock-treated cells were harvested at each time point to establish a control for the experimental time course. Levels of phosphoprotein in all treated samples were compared to those found at the corresponding time points in the mock-treated samples to determine the levels of induction. Exposure of WT cells to AdV led to a modest induction of NF-κB (p65, IκBα), MAPK (p38, Jun N-terminal protein kinase, ERK1/2), and Akt that was detectable at 4 h postinfection and increased slightly at later time points. In contrast to AdV, LPS and CpG DNA treatments led to rapid (30 min posttreatment) and sustained accumulation of phosphorylated products (Fig. 2A).
FIG. 2.
Biochemical analysis of signaling responses to AdV, MyD88, and MyD88/TRIF agonists. (A) Western blot analysis of signal transduction using the indicated antibodies. At each indicated time point, whole-cell lysates from mock-treated macrophages were compared to lysates from macrophages stimulated with either 25,000 AdV particles/cell, 10 ng/ml LPS, or 2 μM CpG DNA. Data are representative of three independent experiments. (B) NF-κB gel shift assay. Nuclear extracts from macrophages stimulated for the indicated times were used in gel shift assay with an [α-32P]ATP-labeled probe containing a consensus NF-κB-binding sequence.
MyD88−/− BMMO were nonresponsive to CpG DNA but were responsive to LPS; however, the overall levels of phosphoprotein were significantly below those observed in WT-treated BMMO (Fig. 2A). Similarly, AdV induced phosphorylation of p65, IκBα, and p38 (compare 4-h time point to mock-treated cells), but the amount of phosphoprotein was lower than that achieved in WT cells. Importantly, background levels of phosphorylated signaling molecules, particularly NF-κB (p65, IκBα), were lower in MyD88−/−-derived extracts (compare mock treatments for WT and MyD88 in Fig. 2A). The differences in background were not due to differences in total NF-κB (Western blot of total p65, data not shown). Phospho-Akt and phospho-ERK were not detectable following AdV treatment of MyD88−/− BMMO. AdV-induced signaling in TRIF-deficient BMMO was largely consistent with WT induction patterns.
To clarify the involvement of the MyD88 and TRIF adaptor molecules in AdV activation of APC, an EMSA was used to examine the induction of NF-κB DNA binding activity induced by AdV, CpG DNA, and LPS. At 1, 3, and 5 h postinfection, nuclear extracts were prepared from WT, MyD88−/−, and TRIF−/− BMMO and used in an NF-κB-specific gel shift assay (EMSA) (Fig. 2B). In WT BMMO, AdV induced a detectable but modest NF-κB binding activity that increased throughout the 5-hour experimental time course. A much stronger and more rapid NF-κB gel shift was associated with LPS treatments, and CpG DNA induced modest levels of NF-κB activation (Fig. 2B). As observed with the Western blot analysis of phosphoproteins, background levels of active NF-κB were greatly reduced in MyD88−/−-derived BMMO (compare mock lanes in Fig. 2B). Both AdV and LPS induced an identifiable NF-κB gel shift product, and the time course of induction was consistent with those found in WT BMMO. CpG DNA did not induce an NF-κB gel shift product in MyD88−/− BMMO.
The data indicate that AdV stimulation of NF-κB, MAPK, and Akt occurs in BMMO at very modest levels, with kinetics that are delayed compared to those for LPS or CpG DNA agonists. They demonstrate a lack of TRIF involvement in AdV signaling and diminished but significant levels of activation in MyD88−/− BMMO. This compromise in activation may reflect a minor contribution of MyD88 to a primary activation event, it may be due to MyD88 involvement in a secondary (autocrine) pathway of APC stimulation, or, alternatively, the attenuated levels of response may be attributed to inherent differences in the MyD88 mutant strain (54, 56) that are unrelated to the direct function of this protein as an adaptor molecule.
AdV induce a robust primary type I IFN response in APC.
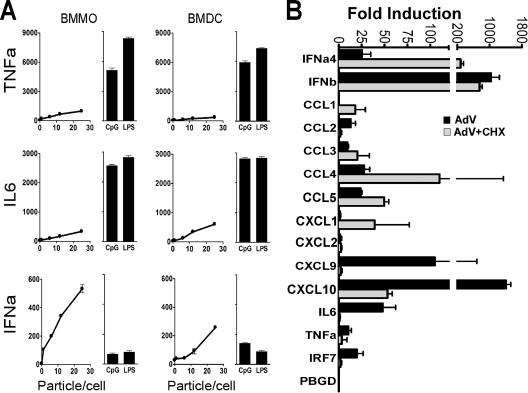
The data presented led us to examine in greater detail the cytokine response triggered by AdV infection of macrophages and DC. BMMO and BMDC were stimulated with increasing concentrations of AdV, 10 ng/ml LPS, or 2 μM CpG DNA, and supernatants were collected after 36 h. ELISAs were used to determine the levels of proinflammatory cytokines (IL-6 and TNF-α) and type I IFN (IFN-α). AdV-induced expression of IL-6, TNF-α, and IFN-α was dose dependent in both APC types. AdV induced smaller amounts of IL-6 and TNF-α than the NF-κB agonists (LPS and CpG DNA) (Fig. 3A). However, Ad infection caused a major induction of IFN-α compared to LPS and CpG DNA. Type I IFNs have been shown to augment cytokine and chemokine synthesis in an autocrine and paracrine manner (14). To determine whether induction was a direct consequence of AdV stimulation and not the result of autocrine IFN stimulation, BMMO were exposed to CHX at 1 h prior to treatment with 25,000 particles/cell of AdV and were harvested for total RNA at 5 h postinfection. Using real-time RT-PCR, we examined the influence of protein synthesis inhibition on an array of cellular transcripts, including cytokine, chemokine, and IFN genes (Fig. 3B). Samples were standardized with β-actin and the fold increase calculated against mock-infected controls. Induction of IRF7 requires the type I IFN-dependent transcription factor ISGF3 (42), and suppression of IRF7 induction by CHX treatment is consistent with suppression of autocrine-induced pathways that require de novo protein synthesis (Fig. 3B). Similarly, CHX treatment resulted in diminished levels of AdV-induced CXCL9, CXCL10, IL-6, and TNF-α, suggesting that an autocrine cascade may also contribute to induction of these genes. However, type I IFNs (IFN-α4 and IFN-β) and the chemokine CCL5 were largely insensitive to downregulation by CHX treatment, indicating that induction of these transcripts was a primary response to Ad infection. Type I IFN and CCL5 are regulated, in part, by the transcriptional activator IRF3 (5, 12). Therefore, further experiments were conducted to investigate the possible involvement of IRF3 in the activation of the innate immune response to AdV.
FIG. 3.
AdV induce a primary type I IFN and chemokine response in APC. (A) Cytokines produced by BMMO and BMDC after stimulation for 36 h with various doses of AdV, 10 ng/ml LPS, or 2 μM CpG DNA. (B) QRT-PCR analysis of type I IFNs and proinflammatory cytokines induced in BMMO after 5 h of treatment with buffer or 25,000 AdV particles/cell. White and black bars represent the fold induction in the absence and in the presence of 50 μg/ml CHX, respectively. Cell treatments were performed in triplicate. β-Actin levels were quantified in all total RNA samples for normalization of the data. The fold change in gene expression was determined by comparison to the mock-treated samples. IRF7 was monitored to assess CHX effectiveness, and the porphobilinogen deaminase (PBGD) housekeeping gene was used as negative control. Two independent real-time PCR experiments gave comparable results.
AdV potently activate IRF3.
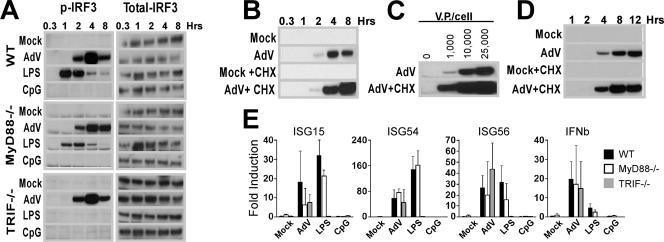
IRF3 is a phosphoprotein that is constitutively expressed in its inactive form in the cytoplasm of most cell types (33). Activation of IRF3 has been associated with the phosphorylation of a cluster of Ser/Thr residues (Ser385, Ser386, Ser396, Ser398, Ser402, Ser405, and Thr402 in human or their equivalents Ser378, Ser379, Ser388, Ser390, Ser394, Ser397, and Thr396 in mouse) at the C-terminal end of the protein (14, 39). To determine whether IRF3 activation occurs as a result of AdV infection, BMMO were infected with AdV and harvested over a time course of 8 hours (Fig. 4A). Whole-cell extracts were immunoblotted with phospho-Ser396-specific IRF3 antibody. The TLR ligands LPS and CpG DNA served as control inducers for comparison. AdV led to strong MyD88- and TRIF-independent C-terminal phosphorylation of IRF3, with maximal activation achieved at 4 h postinfection. LPS induced strong MyD88-independent, TRIF-dependent C-terminal phosphorylation, while CpG DNA did not activate IRF3 (Fig. 4A).
FIG. 4.
AdV activate an IRF3-mediated innate immune response. (A to D) Ser396P-IRF3 levels in whole-cell lysates from BMMO. AdV stimulations were performed at 25,000 viral particles per cell unless otherwise indicated. (A) WT, MyD88-KO, and TRIF-KO BMMO were stimulated with buffer, AdV, 10 ng/ml LPS, or 2 μM CpG DNA for the indicated times. After Ser396P-IRF3 determination, membranes were stripped and reprobed using a pan-IRF3 specific antibody. (B) WT macrophages were stimulated with buffer or AdV with and without CHX; whole-cell lysates were prepared at the indicated times. (C) Macrophages were stimulated with various doses of AdV in the presence or absence of CHX for 4 h. (D) IRF3 phosphorylation from primary lung fibroblasts treated with or without 25,000 particles AdV/cell with or without CHX over a 12-h time course. (E) Induction of IRF3-dependent gene expression in BMMO. Total RNA from BMMO infected at 25,000 AdV particles/cell for 5 h in the presence of CHX (to avoid ISGF3 gene induction) was analyzed by QRT-PCR. Cell treatments were performed in triplicate. β-Actin levels were quantified for normalization of the data. The fold change in gene expression was determined by comparison to the mock-treated samples. Data are representative of at least three independent experiments.
To determine whether IRF3 phosphorylation was a primary antiviral response, BMMO were infected after CHX pretreatment. IRF3 activation levels were enhanced by treatment with the protein synthesis inhibitor (Fig. 4B). AdV induction of BMMO is dose dependent (Fig. 1B and 3A); accordingly, AdV mediated a dose-dependent increase in Ser396 IRF3 phosphorylation (Fig. 4C). To determine whether AdV can stimulate IRF3 phosphorylation in other primary cell types, we characterized activation of IRF3 in early-passage primary lung fibroblasts. Lung fibroblast treatment with 25,000 particles/cell resulted in IRF3 phosphorylation (Fig. 4D); however, the induction was delayed compared to that after a similar treatment of BMMO.
Ser396 phosphorylation is necessary but not sufficient to confer transcriptional activity to IRF3 (39, 49). If AdV stimulates transcriptionally functional IRF3, then IRF3-dependent transcripts should undergo specific induction following AdV treatment. BMMO from WT, MyD88−/−, and TRIF−/− animals were infected in the presence of CHX to eliminate secondary stimulation by type I IFNs. Real-time RT-PCR analysis of four IRF3-driven genes, those for ISG15, ISG54, ISG56, and IFN-β (42, 43), revealed AdV-mediated induction in WT as well as MyD88−/− and TRIF−/− BMMO (Fig. 4E). LPS-induced expression of IRF3-dependent transcripts occurs in a MyD88-independent, TRIF-dependent manner, whereas the TLR9-MyD88-dependent agonist CpG DNA had no effect on the IRF3-dependent transcripts (Fig. 4E). Therefore, AdV-mediated C-terminal phosphorylation of IRF3 directly correlates with transcriptional activity of IRF3-dependent genes.
The IRF3 pathway is triggered by an element in the internalized virion.
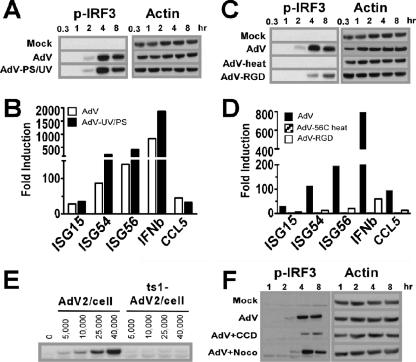
Activation of IRF3-driven genes is associated with host cell recognition of viral dsRNA that is generated by transcription and replication following infection by RNA viruses (53). High-multiplicity AdV infection has been associated with dsRNA formation (36) and has been implicated as a contributor to the activation of type I IFNs by Ad. To determine if viral transcription and RNA production contribute to activation of the IRF3 cascade in BMMO, we used psoralen/UV cross-linking to specifically inhibit transcription of the viral genome. Psoralen/UV inactivation had no effect on the ability of BMMO to phosphorylate IRF3 in response to AdV infection (Fig. 5A). Real-time RT-PCR of RNA isolated from AdV and psoralen/UV AdV-treated virus revealed that high levels of IRF3-dependent transcripts were induced under both conditions (Fig. 5B). That AdV induction of IRF3 phosphorylation occurs in the absence of viral gene expression implies that recognition of AdV takes place during an early stage of virus infection independent of dsRNA formation.
FIG. 5.
Viral entrance but not viral transcription or nuclear localization is required for AdV activation of IRF3. (A) Ser396P-IRF3 and actin Western blots of lysates from macrophages stimulated with buffer, AdV, or UV/psoralen-inactivated AdV (multiplicity of infection, 25,000 particles/cell) (B) Real-time PCR analysis comparing the induction of IRF3-dependent genes by AdV (white bars) and by UV/psoralen-inactivated AdV (black bars) at a multiplicity of infection of 25,000 particles/cell. Total RNA from BMMO was harvested after 5 h. β-Actin levels were quantified for normalization of the data. The fold change in gene expression was determined by comparison to the mock-treated samples. Data are representative of at least three independent experiments. (C) Ser396P-IRF3 and actin Western blots of lysates from macrophages stimulated with buffer, AdV, heat-inactivated (1 h at 56°C) AdV, or penton base-RGD mutant AdV (AdV-RGD) at a multiplicity of infection of 25,000 particles/cell. (D) Real-time PCR analysis comparing the induction of IRF3-dependent genes by AdV (black bars), 56°C AdV (gray bars), and AdV-RGD (white bars). (E) Ser396P-IRF3 and actin Western blots of lysates from macrophages infected with 25,000 particles/cell UV/psoralen-inactivated WT Ad2 or ts1 mt Ad2 and harvested at the indicated times. (F) Ser396P-IRF3 and actin Western blots of lysates from macrophages stimulated with buffer or AdV in the presence of cytochalasin D (CCD) or nocodazole (Noco) for the indicated times. Western blot data are representative of three independent experiments.
To determine whether an intact viral particle is required for BMMO activation, AdV was treated for 1 h at 56°C, a condition known to inactivate virus entrance by causing a dissociation of penton base-fiber complex from the capsid body but leaving the capsid proteins intact (48). When heat-inactivated virus was used to stimulate BMMO, we did not detect IRF3 phosphorylation (Fig. 5C). Real time RT-PCR analysis of RNA isolated from BMMO revealed a complete absence of ISGs, IFN-α, and CCL5 transcripts following treatment with heat inactivated virus (Fig. 5D). We next determined if a virus mutated in the RGD motif of penton base was compromised in phosphorylation of IRF3 and gene expression. BMMO treated with AdVΔRGD showed delayed and diminished IRF3 phosphorylation (Fig. 5C), while mRNA levels for ISGs, IFN-α, and CCL5 were reduced by more than 80% (Fig. 5D).
To investigate whether triggering of the IRF3 pathway occurs in the endosome or requires viral escape into the cytoplasm, macrophages were infected with an Ad carrying a thermosensitive mutation in the L3/p23 protease. Mutant Ad2 viruses lacking a functional p23 protease (ts1-Ad2) are taken up through endosomes but fail to escape into the cytosol (8). Since the ts1 virus is an E1-containing WT Ad, it was made transcriptionally inactive by psoralen/UV cross-linking. A WT Ad2 virus similarly inactivated was used as a positive control. Consistent with a need for AdV to escape from the endocytic particle, the ts1 virus failed to activate IRF3 (Fig. 5E). Endocytic internalization of Ad depends on an intact actin cytoskeleton (32). When primary macrophages were treated with the actin polymerization blocker cytochalasin D, AdV-mediated IRF3 activation was significantly reduced (Fig. 5F). Following entry into the cell and release into the cytoplasm, AdV translocates to the nucleus through a microtubule-dynein-dependent transport mechanism (31, 57). When macrophages were treated with nocodazole, a microtubule depolymerization agent that inhibits Ad particle translocation to the nuclear pore, activation of the IRF3 pathway was detected. Taken together, these data indicate that AdV recognition by macrophages requires viral entrance into the cytosol but not nuclear translocation.
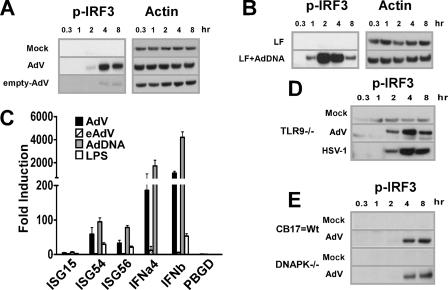
Viral DNA is the IRF3-activating ligand of Ad.
Infectious Ad consists of two main elements, the viral capsid and the dsDNA Ad genome. To determine whether either the capsid or the viral nucleic acid stimulates the IRF3 pathway, we isolated empty viral capsids (intact virions devoid of genomic material and core proteins) and protein-free viral DNA. Infection of BMMO with empty adenoviral particles did not induce Ser396 phosphorylation (Fig. 6A). In contrast, AdV DNA transfected with Lipofectamine, but not Lipofectamine alone, resulted in induction of IRF3 phosphorylation (Fig. 6B). AdV dsDNA but not empty virions triggered the transcription of ISGs and type I IFNs in BMMO (Fig. 6C). These data indicate that AdV dsDNA serves as the key mediator of APC recognition through IRF3 activation. This observation led us to examine whether another dsDNA virus, HSV-1, would also stimulate the IRF3 cascade. We used TLR9-deficient macrophages in this experiment to rule out any potential contribution of TLR9 as a foreign DNA sensor. TLR9-deficient BMMO underwent IRF3 induction by both AdV and HSV-1 with identical activation kinetics (Fig. 6D). The catalytic subunit of DNA-dependent protein kinase (DNA-PKcs) can stimulate IRF3 phosphorylation (20). To determine whether the DNA-PKcs mutation influenced AdV activation of IRF3, BMMO from DNA-PKcs-deficient animals (SCID) were exposed to AdV. We found a strong IRF3 phosphorylation response (Fig. 6E), indicating that the DNA-PKcs pathway does not influence the state of IRF3 following AdV activation. These results demonstrate that AdV DNA is a central activating ligand of IRF3 phosphorylation and IRF3-mediated gene transcription, and they provide evidence in support of the existence of a TLR-independent pathway that senses cytosolic DNA (17). Importantly, the data presented reveal the importance of this recognition pathway in response to infection by DNA viruses.
FIG. 6.
Viral DNA and not capsid protein is the IRF3-activating ligand of Ad. (A, B, D, and E) Western blots showing levels of Ser396P-IRF3 and corresponding β-actin in lysates from BMMO that were stimulated for the times indicated. (A) Macrophages were infected with buffer, AdV, or eAdV. (B) BMMO were transfected with buffer or AdV genomic DNA. (C) Total RNA was isolated from BMMO 5 h after stimulation with AdV or eAdV or transfection with AdV DNA. β-Actin levels were quantified for normalization of the data, and the fold change in gene expression (compared to a mock-treated sample) was quantified by QRT-PCR. (D) BMMO from TLR9-defective mice were infected with buffer, AdV, or HSV-1 (multiplicity of infection, 5). (E) BMMO from SCID mice (DNA-PKcs defective) and from CB-17 controls were treated with buffer or AdV.
Essential role of IRF3 in AdV recognition by macrophages.
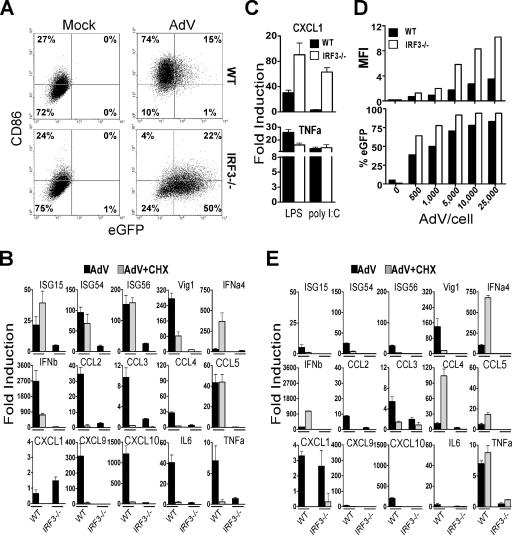
To examine the contribution of the IRF3 pathway to BMMO activation by AdV, primary macrophages from IRF3-deficient mice (50) were infected with 25,000 AdV particles/cell, and upregulation of CD86 was measured 36 h later. The total percentage of CD86-positive cells (27%) following infection of IRF3−/− BMMO was equivalent to the percentage found in mock-infected WT or IRF3−/− cells (26%), whereas AdV infection of WT cells results in 89% CD86-positive cells (Fig. 7A, sum of upper right and left quadrants), indicating that CD86 upregulation is strictly dependent on IRF3 activation. Previous studies have shown that expression of the virally encoded enhanced green fluorescent protein (eGFP) transgene inversely correlates with the ability of the host cell to mount an antiviral state (10, 21). Thus, AdV-infected BMMO were also analyzed for expression of the virally encoded eGFP transgene. Seventy-two percent of the IRF3−/− macrophage population was positive for eGFP, compared to 16% of WT cells (Fig. 7A). Therefore, the data are consistent with IRF3 fulfilling a central role in the antiviral response to AdV. An assessment of the IRF3 contribution to eGFP expression was also carried out with primary lung fibroblasts. Primary lung fibroblasts were more permissive to AdV infection than BMMO; whereas 25,000 AdV particles/BMMO results in 16% of cells being GFP positive, 500 AdV particles/lung fibroblast resulted in 35% of exposed cells being positive for GFP at 36 h postinfection (compare Fig. 7A and D). Even in the more permissive fibroblast cell, the per-cell fluorescence and the percentage of the population positive for eGFP was much higher in IRF3−/− than in WT primary lung fibroblasts (Fig. 7D), confirming that loss of IRF3 results in a more permissive viral transduction environment.
FIG. 7.
IRF3 Is essential for innate immune responses against AdV. (A) BMMO from WT and IRF3−/− mice were stimulated for 36 h with buffer or AdV at 25,000 particles/cell. CD86 upregulation and eGFP expression were analyzed by flow cytometry. The number in each quadrant represents the percentage of positive cells. (B) QRT-PCR analysis of total RNA isolated from macrophages derived from WT and IRF3−/− mice 5 h after stimulation with buffer or AdV at 25,000 particles/cell in the presence or absence of CHX. (C) mRNA levels of CXCL1 and TNF-α in macrophages stimulated with 10 ng/ml LPS or 10 μg poly (I·C) with Lipofectamine 2000. (D) Mean fluorescence intensity (MFI) and percentage of eGFP-positive lung fibroblasts infected for 36 h with the indicated doses of AdV. (E) QRT-PCR analysis of mRNA levels induced in lung fibroblasts derived from WT and IRF3-KO mice. The experiments were performed similarly to those for panel B except that the total RNA was harvested at 8 h after stimulation. β-Actin levels were quantified for normalization of the data. The fold change in gene expression was determined by comparison to the mock-treated samples. Data are representative of at least three independent experiments.
Next, using real-time RT-PCR, we found that IRF3 plays a critical role not only in the induction of type I IFNs and IFN-inducible genes following AdV infection but also in the production of proinflammatory mediators in cell types as diverse as macrophages (Fig. 7B) and lung fibroblasts (Fig. 7E). Thus, induction of IFN-α4, IFN-β, ISG15, ISG54, ISG56, Vig1, IL-6, TNF-α, CXCL10, CCL5, CCl2, CCL3, CCL4, and CXCL9 in response to AdV was reduced by more than 90% in IRF3−/− cells compared to WT controls. In addition, direct induction of these genes was totally abrogated in IRF3−/− cells infected with AdV in the presence of CHX (Fig. 7B and E). In contrast, when IRF3-defective macrophages were treated with the MyD88/TRIF-dependent ligand LPS or poly(I·C), transcripts strictly dependent on MyD88, such as CXCL1 and TNF-α, were induced in both WT and IRF3−/− cells (Fig. 7C). Not all responses to AdV were absent in IRF3−/− cells. AdV induced IRF3−/− macrophages and fibroblasts to produce the CXCL1 chemokine in small quantities comparable to those in WT cells, where AdV induction of CXCL1 was largely dependent on de novo protein synthesis (Fig. 7B and E). Residual mRNA upregulation in IRF3−/− macrophages that was inhibited by CHX treatment most likely represents a contribution of IRF7 to the AdV innate immune surveillance system, as previously mentioned.
Finally, we found that the genes primarily induced by AdV infection in macrophages (ISG15, ISG54, ISG56, Vig1, IFN-α4, IFN-β, and CCL5) only partially overlapped with those primarily induced in fibroblasts (IFN-α4, IFN-β, CCL4, CCL5, and TNF-α), suggesting clear differences in the antiviral program induced in nonimmune cells compared to APC. Further work will be required to establish the functional relevance of each cell type in orchestrating the host immune response against Ad vectors.
DISCUSSION
Recognition of a viral infection triggers the antiviral innate immune response, which in turn contributes to activation of antiviral adaptive immunity. Both the innate and adaptive immune responses to AdV are well documented, yet the molecular mechanisms associated with recognition of the nonenveloped DNA viral vector are poorly understood. We have established that APC recognition of AdV occurs through a TLR-independent viral DNA sensing mechanism that targets phosphorylation of IRF3 as a master antiviral transcriptional switch. The results presented in this study are consistent with the following model of viral recognition: (i) virus engagement of the APC is mediated primarily through penton-base RGD motif binding to membrane integrins; (ii) virus engagement stimulates endocytosis, particle internalization, and endosomal escape; and (iii) coincident with or following endosomal escape, viral DNA is exposed to a cellular sensor, leading to phosphorylation of IRF3. Phosphorylated IRF3 translocates to the nucleus and contributes to APC activation and induction of the antiviral transcription profile.
In contrast to the view that AdV capsid plays a major role in triggering the innate immune response in APC (41, 44), our current model has the viral capsid functioning mainly as a necessary delivery vehicle presenting the viral genome to the intracellular sensor. AdV capsid proteins influence the binding and uptake of virus to a variety of cell types, including APC. We have demonstrated that APC are nonresponsive to heat-inactivated virus; therefore, an intact capsid is required for APC recognition. Mutation of the penton base RGD motif greatly reduced the activation of BMMO by AdV; however, residual IRF3 phosphorylation was detected, albeit with a greater delay in activation than for WT AdV. Comparison between whole virus, empty capsid, and viral DNA revealed that AdV DNA recapitulates the whole particle's ability to activate IRF3 and to induce inflammatory and IFN-associated responses, whereas APC were refractory to activation by AdV empty capsid, which includes the intact penton (penton base-fiber) necessary for binding and internalization of the virion. These data argue against the structural proteins of AdV being the principal target sensory ligand; rather, they indicate that the capsid plays an important role in viral genome delivery.
Neither the MyD88−/− nor the TRIF−/− genotype abolished the APC activation responses, nor did they affect the kinetics of NF-κB and MAPK activation. In contrast, deletion of IRF3 results in a drastic and generalized defect in multiple cell types, indicating that IRF3 is essential for AdV induction of innate immune responses. The IRF3 pathway is activated in response to AdV in nonpermissive macrophages (Fig. 4) as well as in permissive lung fibroblasts (Fig. 4), and based on eGFP expression, IRF3 activation leads to diminished transgene expression from the viral genome. LPS and transfected DNA induce maximal phospho-IRF3 within 1 to 2 h, whereas AdV maximal induction of phospho-IRF3 occurs at 4 h in macrophages and at 8 h in lung fibroblasts. Similarly, it has been recently reported that macrophage recognition of DNA from intracellular bacteria approaches maximal levels after 4 h (11). Differences in the kinetics of IRF3 phosphorylation may reflect cell-specific trafficking differences that influence exposure of pathogenic DNA to the DNA sensor. In permissive nonimmune cells such as lung fibroblasts, more efficient delivery of viral DNA to the nucleus may result in delayed or diminished exposure to the DNA sensor, whereas transient DNA transfections deliver significant concentrations of DNA directly to the target sensor.
The best-characterized sensor of foreign DNA is TLR9. It functions as an intracellular detector of unmethylated CpG DNA (9) and has been implicated in recognition of several members of the herpesvirus family (13, 27, 35). In this report, we provide several lines of evidence showing that TLR9 is not the DNA sensor responsible for turning on the IRF3 pathway in response to AdV. First, TLR9-mediated stimulation by CpG DNA was confirmed to be a MyD88-dependent event that leads to strong NF-κB but undetectable IRF3 activation in BMMO. Second, we demonstrate that two dsDNA viruses, AdV and HSV-1, induce IRF3 phosphorylation in BMMO from TLR9-deficient mice. Recent studies indicate the existence of a TLR-independent cytosolic surveillance system for transfected DNA that elicits type I IFN gene induction via TBK-1/Ikkɛ→IRF3 (17, 55). The current study demonstrates that dsDNA virus infection of APC and fibroblasts stimulates antiviral recognition by way of a cellular dsDNA sensor.
The demonstration that IRF3 activation by the AdV genome provides a molecular mechanism supports previous studies that have characterized an antiviral type I IFN response to Ad and provides a framework to understand the viral strategy for suppressing induction of the antiviral state (reviewed in reference 63). The E1A immediate-early gene of Ad is involved in regulating viral and host gene expression. E1A is also involved in suppressing induction of type I IFNs and IFN-stimulated genes (19, 47). Studies from the laboratory of P. Pitha (18) extended these observations to demonstrate that Ad E1A inhibits induction of ISGs that are dependent on IRF3. They proposed that inhibition occurred through competition between IRF3 and E1A for the coactivator CBP/p300. That Ad has evolved immediate-early gene products that target the type I IFNs and specifically the ISGs induced by IRF3 offers a biological argument supporting the importance of the IRF3 cascade to host protection from dsDNA viruses. Similarly, the herpesvirus family of enveloped dsDNA viruses which express a large array of immune modulators (reviewed in reference 40) produce several viral proteins that target inhibition of IRF3 activation, most notably the immediate-early gene product ICPO (34, 37).
The immune response generated against AdV following systemic administration is a composite response generated by a variety of cell types that recognize and respond to AdV (reviewed in reference 41). We have utilized primary bone marrow-derived murine APC as our model system for characterizing immediate-early innate recognition of AdV. In this relatively simple system, it is clear that following primary recognition of AdV, autocrine and paracrine responses make major contributions to the final outcome of macrophage activation. We have also shown that macrophages and primary lung fibroblasts both respond to AdV through IRF3 activation, but the end response is qualitatively cell specific. It is also consistent with the redundant nature of the immune system to anticipate that additional recognition pathways not featured in our studies of primary macrophages may operate in other cell types, including TLR-dependent pathways. Further investigation will be required to fully understand how the antiviral IRF3 cascade contributes to the innate and adaptive immune responses to AdV in vivo.
Acknowledgments
We thank Franklin Roth for editorial assistance, Carolina Lopez at Mt. Sinai and Patricia Fontan at UMDNJ for helpful discussions, and Mary Murphy and Marko Stankic for their technical assistance.
This work was supported by NIH grants AI-63142 to E.F.-P. and KO1 HL70438-01 to M.N.
GenVec has licensing agreements with Cornell Research Foundation for technology developed in collaboration with E. Falck-Pedersen.
Footnotes
Published ahead of print on 24 January 2007.
REFERENCES
- 1.Bergelson, J. M., J. A. Cunningham, G. Droguett, E. A. Kurt-Jones, A. Krithivas, J. S. Hong, M. S. Horwitz, R. L. Crowell, and R. W. Finberg. 1997. Isolation of a common receptor for Coxsackie B viruses and adenoviruses 2 and 5. Science 275:1320-1323. [DOI] [PubMed] [Google Scholar]
- 2.Bieback, K., E. Lien, I. M. Klagge, E. Avota, J. Schneider-Schaulies, W. P. Duprex, H. Wagner, C. J. Kirschning, V. Ter Meulen, and S. Schneider-Schaulies. 2002. Hemagglutinin protein of wild-type measles virus activates toll-like receptor 2 signaling. J. Virol. 76:8729-8736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Compton, T., E. A. Kurt-Jones, K. W. Boehme, J. Belko, E. Latz, D. T. Golenbock, and R. W. Finberg. 2003. Human cytomegalovirus activates inflammatory cytokine responses via CD14 and Toll-like receptor 2. J. Virol. 77:4588-4596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Covert, M. W., T. H. Leung, J. E. Gaston, and D. Baltimore. 2005. Achieving stability of lipopolysaccharide-induced NF-kappaB activation. Science 309:1854-1857. [DOI] [PubMed] [Google Scholar]
- 5.Genin, P., M. Algarte, P. Roof, R. Lin, and J. Hiscott. 2000. Regulation of RANTES chemokine gene expression requires cooperativity between NF-kappa B and IFN-regulatory factor transcription factors. J. Immunol. 164:5352-5361. [DOI] [PubMed] [Google Scholar]
- 6.Gitlin, L., W. Barchet, S. Gilfillan, M. Cella, B. Beutler, R. A. Flavell, M. S. Diamond, and M. Colonna. 2006. Essential role of mda-5 in type I IFN responses to polyriboinosinic:polyribocytidylic acid and encephalomyocarditis picornavirus. Proc. Natl. Acad. Sci. USA 103:8459-8464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Greber, U. F., M. Suomalainen, R. P. Stidwill, K. Boucke, M. W. Ebersold, and A. Helenius. 1997. The role of the nuclear pore complex in adenovirus DNA entry. EMBO J. 16:5998-6007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Greber, U. F., P. Webster, J. Weber, and A. Helenius. 1996. The role of the adenovirus protease on virus entry into cells. EMBO J. 15:1766-1777. [PMC free article] [PubMed] [Google Scholar]
- 9.Hemmi, H., O. Takeuchi, T. Kawai, T. Kaisho, S. Sato, H. Sanjo, M. Matsumoto, K. Hoshino, H. Wagner, K. Takeda, and S. Akira. 2000. A Toll-like receptor recognizes bacterial DNA. Nature 408:740-745. [DOI] [PubMed] [Google Scholar]
- 10.Hensley, S. E., W. Giles-Davis, K. C. McCoy, W. Weninger, and H. C. Ertl. 2005. Dendritic cell maturation, but not CD8+ T cell induction, is dependent on type I IFN signaling during vaccination with adenovirus vectors. J. Immunol. 175:6032-6041. [DOI] [PubMed] [Google Scholar]
- 11.Hirotani, T., M. Yamamoto, Y. Kumagai, S. Uematsu, I. Kawase, O. Takeuchi, and S. Akira. 2005. Regulation of lipopolysaccharide-inducible genes by MyD88 and Toll/IL-1 domain containing adaptor inducing IFN-beta. Biochem. Biophys. Res. Commun. 328:383-392. [DOI] [PubMed] [Google Scholar]
- 12.Hiscott, J., P. Pitha, P. Genin, H. Nguyen, C. Heylbroeck, Y. Mamane, M. Algarte, and R. Lin. 1999. Triggering the interferon response: the role of IRF-3 transcription factor. J. Interferon Cytokine Res. 19:1-13. [DOI] [PubMed] [Google Scholar]
- 13.Hochrein, H., B. Schlatter, M. O'Keeffe, C. Wagner, F. Schmitz, M. Schiemann, S. Bauer, M. Suter, and H. Wagner. 2004. Herpes simplex virus type-1 induces IFN-alpha production via Toll-like receptor 9-dependent and -independent pathways. Proc. Natl. Acad. Sci. USA 101:11416-11421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Honda, K., H. Yanai, A. Takaoka, and T. Taniguchi. 2005. Regulation of the type I IFN induction: a current view. Int. Immunol. 17:1367-1378. [DOI] [PubMed] [Google Scholar]
- 15.Huang, S., T. Kamata, Y. Takada, Z. M. Ruggeri, and G. R. Nemerow. 1996. Adenovirus interaction with distinct integrins mediates separate events in cell entry and gene delivery to hematopoietic cells. J. Virol. 70:4502-4508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Inaba, K., M. Inaba, N. Romani, H. Aya, M. Deguchi, S. Ikehara, S. Muramatsu, and R. M. Steinman. 1992. Generation of large numbers of dendritic cells from mouse bone marrow cultures supplemented with granulocyte/macrophage colony-stimulating factor. J Exp. Med. 176:1693-1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ishii, K. J., C. Coban, H. Kato, K. Takahashi, Y. Torii, F. Takeshita, H. Ludwig, G. Sutter, K. Suzuki, H. Hemmi, S. Sato, M. Yamamoto, S. Uematsu, T. Kawai, O. Takeuchi, and S. Akira. 2006. A Toll-like receptor-independent antiviral response induced by double-stranded B-form DNA. Nat. Immunol. 7:40-48. [DOI] [PubMed] [Google Scholar]
- 18.Juang, Y. T., W. Lowther, M. Kellum, W. C. Au, R. Lin, J. Hiscott, and P. M. Pitha. 1998. Primary activation of interferon A and interferon B gene transcription by interferon regulatory factor 3. Proc. Natl. Acad. Sci. USA 95:9837-9842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kalvakolanu, D. V., S. K. Bandyopadhyay, M. L. Harter, and G. C. Sen. 1991. Inhibition of interferon-inducible gene expression by adenovirus E1A proteins: block in transcriptional complex formation. Proc. Natl. Acad. Sci. USA 88:7459-7463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Karpova, A. Y., M. Trost, J. M. Murray, L. C. Cantley, and P. M. Howley. 2002. Interferon regulatory factor-3 is an in vivo target of DNA-PK. Proc. Natl. Acad. Sci. USA 99:2818-2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kato, H., S. Sato, M. Yoneyama, M. Yamamoto, S. Uematsu, K. Matsui, T. Tsujimura, K. Takeda, T. Fujita, O. Takeuchi, and S. Akira. 2005. Cell type-specific involvement of RIG-I in antiviral response. Immunity 23:19-28. [DOI] [PubMed] [Google Scholar]
- 22.Kato, H., O. Takeuchi, S. Sato, M. Yoneyama, M. Yamamoto, K. Matsui, S. Uematsu, A. Jung, T. Kawai, K. J. Ishii, O. Yamaguchi, K. Otsu, T. Tsujimura, C. S. Koh, C. Reis e Sousa, Y. Matsuura, T. Fujita, and S. Akira. 2006. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature 441:101-105. [DOI] [PubMed] [Google Scholar]
- 23.Kawai, T., O. Adachi, T. Ogawa, K. Takeda, and S. Akira. 1999. Unresponsiveness of MyD88-deficient mice to endotoxin. Immunity 11:115-122. [DOI] [PubMed] [Google Scholar]
- 24.Kawai, T., and S. Akira. 2006. Innate immune recognition of viral infection. Nat. Immunol. 7:131-137. [DOI] [PubMed] [Google Scholar]
- 25.Kawai, T., and S. Akira. 2006. TLR signaling. Cell Death Differ. 13:816-825. [DOI] [PubMed] [Google Scholar]
- 26.Kawai, T., O. Takeuchi, T. Fujita, J. Inoue, P. F. Muhlradt, S. Sato, K. Hoshino, and S. Akira. 2001. Lipopolysaccharide stimulates the MyD88-independent pathway and results in activation of IFN-regulatory factor 3 and the expression of a subset of lipopolysaccharide-inducible genes. J. Immunol. 167:5887-5894. [DOI] [PubMed] [Google Scholar]
- 27.Krug, A., G. D. Luker, W. Barchet, D. A. Leib, S. Akira, and M. Colonna. 2004. Herpes simplex virus type 1 activates murine natural interferon-producing cells through toll-like receptor 9. Blood 103:1433-1437. [DOI] [PubMed] [Google Scholar]
- 28.Kumar, H., T. Kawai, H. Kato, S. Sato, K. Takahashi, C. Coban, M. Yamamoto, S. Uematsu, K. J. Ishii, O. Takeuchi, and S. Akira. 2006. Essential role of IPS-1 in innate immune responses against RNA viruses. J. Exp. Med. 203:1795-1803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kurt-Jones, E. A., M. Chan, S. Zhou, J. Wang, G. Reed, R. Bronson, M. M. Arnold, D. M. Knipe, and R. W. Finberg. 2004. Herpes simplex virus 1 interaction with Toll-like receptor 2 contributes to lethal encephalitis. Proc. Natl. Acad. Sci. USA 101:1315-1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kurt-Jones, E. A., L. Popova, L. Kwinn, L. M. Haynes, L. P. Jones, R. A. Tripp, E. E. Walsh, M. W. Freeman, D. T. Golenbock, L. J. Anderson, and R. W. Finberg. 2000. Pattern recognition receptors TLR4 and CD14 mediate response to respiratory syncytial virus. Nat. Immunol. 1:398-401. [DOI] [PubMed] [Google Scholar]
- 31.Leopold, P. L., G. Kreitzer, N. Miyazawa, S. Rempel, K. K. Pfister, E. Rodriguez-Boulan, and R. G. Crystal. 2000. Dynein- and microtubule-mediated translocation of adenovirus serotype 5 occurs after endosomal lysis. Hum. Gene Ther. 11:151-165. [DOI] [PubMed] [Google Scholar]
- 32.Li, E., D. Stupack, G. M. Bokoch, and G. R. Nemerow. 1998. Adenovirus endocytosis requires actin cytoskeleton reorganization mediated by Rho family GTPases. J. Virol. 72:8806-8812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lin, R., C. Heylbroeck, P. M. Pitha, and J. Hiscott. 1998. Virus-dependent phosphorylation of the IRF-3 transcription factor regulates nuclear translocation, transactivation potential, and proteasome-mediated degradation. Mol. Cell. Biol. 18:2986-2996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lin, R., R. S. Noyce, S. E. Collins, R. D. Everett, and K. L. Mossman. 2004. The herpes simplex virus ICP0 RING finger domain inhibits IRF3- and IRF7-mediated activation of interferon-stimulated genes. J. Virol. 78:1675-1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lund, J., A. Sato, S. Akira, R. Medzhitov, and A. Iwasaki. 2003. Toll-like receptor 9-mediated recognition of herpes simplex virus-2 by plasmacytoid dendritic cells. J. Exp. Med. 198:513-520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maran, A., and M. B. Mathews. 1988. Characterization of the double-stranded RNA implicated in the inhibition of protein synthesis in cells infected with a mutant adenovirus defective for VA RNA. Virology 164:106-113. [DOI] [PubMed] [Google Scholar]
- 37.Melroe, G. T., N. A. DeLuca, and D. M. Knipe. 2004. Herpes simplex virus 1 has multiple mechanisms for blocking virus-induced interferon production. J. Virol. 78:8411-8420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Morelli, A. E., A. T. Larregina, R. W. Ganster, A. F. Zahorchak, J. M. Plowey, T. Takayama, A. J. Logar, P. D. Robbins, L. D. Falo, and A. W. Thomson. 2000. Recombinant adenovirus induces maturation of dendritic cells via an NF-κB-dependent pathway. J. Virol. 74:9617-9628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mori, M., M. Yoneyama, T. Ito, K. Takahashi, F. Inagaki, and T. Fujita. 2004. Identification of Ser-386 of interferon regulatory factor 3 as critical target for inducible phosphorylation that determines activation. J. Biol. Chem. 279:9698-9702. [DOI] [PubMed] [Google Scholar]
- 40.Mossman, K. L., and A. A. Ashkar. 2005. Herpesviruses and the innate immune response. Viral Immunol. 18:267-281. [DOI] [PubMed] [Google Scholar]
- 41.Muruve, D. A. 2004. The innate immune response to adenovirus vectors. Hum. Gene Ther. 15:1157-1166. [DOI] [PubMed] [Google Scholar]
- 42.Nakaya, T., M. Sato, N. Hata, M. Asagiri, H. Suemori, S. Noguchi, N. Tanaka, and T. Taniguchi. 2001. Gene induction pathways mediated by distinct IRFs during viral infection. Biochem. Biophys. Res. Commun. 283:1150-1156. [DOI] [PubMed] [Google Scholar]
- 43.Peters, K. L., H. L. Smith, G. R. Stark, and G. C. Sen. 2002. IRF-3-dependent, NFkappa B- and JNK-independent activation of the 561 and IFN-beta genes in response to double-stranded RNA. Proc. Natl. Acad. Sci. USA 99:6322-6327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Philpott, N. J., M. Nociari, K. B. Elkon, and E. Falck-Pedersen. 2004. Adenovirus-induced maturation of dendritic cells through a PI3 kinase-mediated TNF-alpha induction pathway. Proc. Natl. Acad. Sci. USA 101:6200-6205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Reference deleted.
- 46.Rassa, J. C., J. L. Meyers, Y. Zhang, R. Kudaravalli, and S. R. Ross. 2002. Murine retroviruses activate B cells via interaction with toll-like receptor 4. Proc. Natl. Acad. Sci. USA 99:2281-2286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Reich, N., R. Pine, D. Levy, and J. E. Darnell, Jr. 1988. Transcription of interferon-stimulated genes is induced by adenovirus particles but is suppressed by E1A gene products. J. Virol. 62:114-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Russell, W. C., R. C. Valentine, and H. G. Pereira. 1967. The effect of heat on the anatomy of the adenovirus. J. Gen. Virol. 1:509-522. [DOI] [PubMed] [Google Scholar]
- 49.Sarkar, S. N., K. L. Peters, C. P. Elco, S. Sakamoto, S. Pal, and G. C. Sen. 2004. Novel roles of TLR3 tyrosine phosphorylation and PI3 kinase in double-stranded RNA signaling. Nat. Struct. Mol. Biol. 11:1060-1067. [DOI] [PubMed] [Google Scholar]
- 50.Sato, M., H. Suemori, N. Hata, M. Asagiri, K. Ogasawara, K. Nakao, T. Nakaya, M. Katsuki, S. Noguchi, N. Tanaka, and T. Taniguchi. 2000. Distinct and essential roles of transcription factors IRF-3 and IRF-7 in response to viruses for IFN-alpha/beta gene induction. Immunity 13:539-548. [DOI] [PubMed] [Google Scholar]
- 51.Schaack, J., W. Y. Ho, P. Freimuth, and T. Shenk. 1990. Adenovirus terminal protein mediates both nuclear matrix association and efficient transcription of adenovirus DNA. Genes Dev. 4:1197-1208. [DOI] [PubMed] [Google Scholar]
- 52.Schoggins, J. W., and E. Falck-Pedersen. 2006. Fiber and penton base capsid modifications yield diminished adenovirus type 5 transduction and proinflammatory gene expression with retention of antigen-specific humoral immunity. J. Virol. 80:10634-10644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Servant, M. J., N. Grandvaux, B. R. tenOever, D. Duguay, R. Lin, and J. Hiscott. 2003. Identification of the minimal phosphoacceptor site required for in vivo activation of interferon regulatory factor 3 in response to virus and double-stranded RNA. J. Biol. Chem. 278:9441-9447. [DOI] [PubMed] [Google Scholar]
- 54.Shi, S., C. Nathan, D. Schnappinger, J. Drenkow, M. Fuortes, E. Block, A. Ding, T. R. Gingeras, G. Schoolnik, S. Akira, K. Takeda, and S. Ehrt. 2003. MyD88 primes macrophages for full-scale activation by interferon-gamma yet mediates few responses to Mycobacterium tuberculosis. J. Exp. Med. 198:987-997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stetson, D. B., and R. Medzhitov. 2006. Recognition of cytosolic DNA activates an IRF3-dependent innate immune response. Immunity 24:93-103. [DOI] [PubMed] [Google Scholar]
- 56.Sun, D., and A. Ding. 2006. MyD88-mediated stabilization of interferon-gamma-induced cytokine and chemokine mRNA. Nat. Immunol. 7:375-381. [DOI] [PubMed] [Google Scholar]
- 57.Suomalainen, M., M. Y. Nakano, S. Keller, K. Boucke, R. P. Stidwill, and U. F. Greber. 1999. Microtubule-dependent plus- and minus end-directed motilities are competing processes for nuclear targeting of adenovirus. J. Cell Biol. 144:657-672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tenoever, B. R., and T. Maniatis. 2006. Parallel pathways of virus recognition. Immunity 24:510-512. [DOI] [PubMed] [Google Scholar]
- 59.Tillman, B. W., T. D. de Gruijl, S. A. Luykx-de Bakker, R. J. Scheper, H. M. Pinedo, T. J. Curiel, W. R. Gerritsen, and D. T. Curiel. 1999. Maturation of dendritic cells accompanies high-efficiency gene transfer by a CD40-targeted adenoviral vector. J. Immunol. 162:6378-6383. [PubMed] [Google Scholar]
- 60.Trevejo, J. M., M. W. Marino, N. Philpott, R. Josien, E. C. Richards, K. B. Elkon, and E. Falck-Pedersen. 2001. TNF-alpha-dependent maturation of local dendritic cells is critical for activating the adaptive immune response to virus infection. Proc. Natl. Acad. Sci. USA 98:12162-12167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wickham, T. J., E. J. Filardo, D. A. Cheresh, and G. R. Nemerow. 1994. Integrin alpha v beta 5 selectively promotes adenovirus mediated cell membrane permeabilization. J. Cell Biol. 127:257-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wiethoff, C. M., H. Wodrich, L. Gerace, and G. R. Nemerow. 2005. Adenovirus protein VI mediates membrane disruption following capsid disassembly. J. Virol. 79:1992-2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wold, W. S., T. W. Hermiston, and A. E. Tollefson. 1994. Adenovirus proteins that subvert host defenses. Trends Microbiol. 2:437-443. [DOI] [PubMed] [Google Scholar]
- 64.Yamamoto, M., S. Sato, H. Hemmi, K. Hoshino, T. Kaisho, H. Sanjo, O. Takeuchi, M. Sugiyama, M. Okabe, K. Takeda, and S. Akira. 2003. Role of adaptor TRIF in the MyD88-independent toll-like receptor signaling pathway. Science 301:640-643. [DOI] [PubMed] [Google Scholar]
- 65.Yang, Y., F. A. Nunes, K. Berencsi, E. E. Furth, E. Gonczol, and J. M. Wilson. 1994. Cellular immunity to viral antigens limits E1-deleted adenoviruses for gene therapy. Proc. Natl. Acad. Sci. USA 91:4407-4411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang, Y., N. Chirmule, G. P. Gao, R. Qian, M. Croyle, B. Joshi, J. Tazelaar, and J. M. Wilson. 2001. Acute cytokine response to systemic adenoviral vectors in mice is mediated by dendritic cells and macrophages. Mol. Ther. 3:697-707. [DOI] [PubMed] [Google Scholar]