Abstract
The publication of a draft of the human genome and of large collections of transcribed sequences has made it possible to study the complex relationship between the transcriptome and the genome. In the work presented here, we have focused on mapping mRNA 3′ ends onto the genome by use of the raw data generated by the expressed sequence tag (EST) sequencing projects. We find that at least half of the human genes encode multiple transcripts whose polyadenylation is driven by multiple signals. The corresponding transcript 3′ ends are spread over distances in the kilobase range. This finding has profound implications for our understanding of gene expression regulation and of the diversity of human transcripts, for the design of cDNA microarray probes, and for the interpretation of gene expression profiling experiments.
[The following individuals kindly provided reagents, samples or unpublished information as indicated in the paper: G. Riggins, C. Ruegg, J.-B. Demoulin, P. Olsson, F. Funari, P. Schneider, L.F. Reis, and J.-C. Renauld]
Parallel to the sequencing of the human genome, a less heralded but nevertheless massive effort has been undertaken to document experimentally the portion of the genome that is transcribed into RNA, the transcriptome. It is only by comparing it with the transcriptome that the capacity of the genome to code for the RNAs and proteins that make up the cell machinery can be precisely defined (Burge 2001). Although the mapping of transcribed sequences to the genome has been used extensively to document the positions of genes (Caron et al. 2001), it has not yet been fully exploited to explore the complexity of the transcriptome. Of the three major mechanisms that contribute to this complexity, alternative initiation of transcription, splicing, and polyadenylation, the latter seemed most immediately amenable to analysis because of the wealth of data about transcript 3′ ends provided by the expressed sequence tag (EST) sequences generated by the NCI Cancer Genome Anatomy Project (Strausberg et al. 2000) (at Washington University, the NIH Intramural Sequencing Center, and Incyte Pharmaceuticals), the Merck Gene Index (Aaronson et al. 1996), and the NIH Mammalian Gene Collection (Strausberg et al. 1999). Although alternative polyadenylation of transcripts has been known to occur for a long time, the proportion of transcripts affected, the number of sites per transcript, and the distances over which alternative sites are spread have been explored exclusively using EST clustering techniques (Gautheret et al. 1998; Beaudoing and Gautheret 2001; Pauws et al. 2001) and have relied on the poly(A) being documented in the EST sequences. The most recent of these studies have concluded that >40% of human transcripts may undergo alternative polyadenylation, but that most of the observed variation is over a short range (<50 nt) and driven by a single polyadenylation signal (Beaudoing and Gautheret 2001; Pauws et al. 2001). Long-range variation (>1 kb) has so far been observed only experimentally. We show here that long-range variation is in fact extremely common, possibly affecting more than half of all genes.
RESULTS
To generate a transcript to genome map, we have exploited all publicly available human genome data (finished and draft) and transcriptome data (full-length mRNAs, partial mRNAs, ESTs, and electropherograms from EST projects). We also included reference human transcript sequences from the RefSeq database. The dataset that we have constructed comprises a set of alignments between transcript and genome sequences, documenting the position of the alignment on each sequence, and a set of poly(A)-proximal sequence tags aligned to the genome sequence. We visualized the complex relationships between the genome and full-length mRNAs, ESTs, and 3′ tags in the ACEDB environment (Durbin and Thierry-Mieg 1994). For chromosomes 21 and 22, which have been extensively annotated, we included the transcripts identified by the sequencing consortia (Dunham et al. 1999; Hattori et al. 2000); most of the examples described here were taken from chromosome 21, because they illustrate the additional information gained by using our methods relative to existing genome annotation procedures. We also developed a program called the Transcriptome Analyzer (tromer; C. Iseli, unpublished data) that uses the transcript to genome alignments to identify exon boundaries and analyzes the connectivity of these boundaries. The output of tromer can be used to reconstruct virtual transcripts from the underlying genomic sequence following a path from 3′ tags along experimentally verified exon boundaries.
One of the pillars of our strategy is the identification of trusted 3′ tags that provide unique identifiers for transcript 3′ ends. We chose to analyze the 50 nt immediately upstream of the poly(A) tail, as this should guarantee the uniqueness of the tag (there are 1030 possible tags of length 50, compared with 3 × 109 nt in the haploid genome) whereas keeping the effects of sequencing errors reasonably low (approximately a 50% chance of a single error in typical EST data). A set of candidate tags was selected by identifying runs of at least 10 A's or T's in the original electropherograms produced by the Merck, CGAP, and MGC sequencing projects, reverse complementing the sequence if necessary to end in poly(A) and extracting the 50 nt preceding the poly(A). This first set was reduced to unique tags and filtered to remove abundant human repeats and low complexity sequences. This “clean” set was then mapped onto the genome sequences and genome-linked tags were derived by combining the 50 nt 3′ tag with the 50 nt following the tag in the genome (the total genome-linked tag length is thus 100 nt). These combined tags were clustered to resolve short-range variations in the exact polyadenylation site between individual transcript sequences; it should be noted that this procedure eliminates from our analysis the short-range variation shown by Pauws et al. (2001). After clustering, 13% of the tags identified genome regions with nonidentical downstream sequences. The majority of these diverged in only one or two positions because of genetic polymorphisms or genome duplications (note that we used all available genome sequence data, not one of the nonredundant assemblies). Many others map to pseudogenes or retroposons. To distinguish bona fide mRNA 3′ends from poly(A)-containing sequences generated by internal priming or genomic contamination, we flagged tags that contained at least 10 A's or 11 A's and G's in the first 15 nt of downstream genome sequence; this is a conservative estimate of the minimal requirement for oligo(dT)-mediated priming and ensures that our tag collection is free of sequences derived from genomic or internal priming during cDNA library construction. Of 152,307 clustered, genome-matched candidates, 95,787 were judged to represent trusted 3′ tags by these criteria. The tag generation procedure is summarized in Table 1. The number of 3′ tags will most likely increase as more cell types, pathological conditions, and differentiation stages are sampled by cDNA cloning.
Table 1.
Statistics on the Generation of 3′ Tags
| Total no. of human trace files | 2,440,000 |
| Total no. of tags with poly(A) | 1,059,193 |
| Unique tags with poly(A) | 508,019 |
| Filtered tags (removes repeats, mitochondrial and low complexity) | 455,754 |
| Unique genome linked tags (including downstream sequence) | 248,017a |
| Unique tags matching the genome (excluding downstream sequence) | 196,056 |
| Unique tags with a trusted match to the genomeb | 141,548 |
| Genome-matched tag clusters | 152,308 |
| Trusted genome-matched tag clusters | 95,787 |
This number includes 158,794 tags with exact matches, 46,156 tags that differ by one or two nucleotides from this first set, and 43,067 that independently find inexact matches on the genome.
Trusted tags are those that do not contain a downstream poly(A) or poly(G).
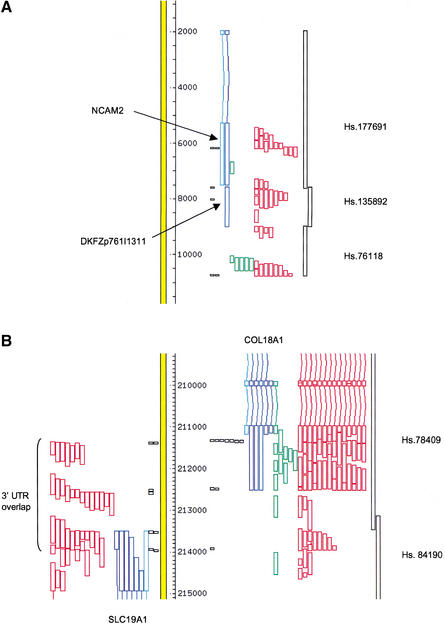
We examined the distribution on the genome of trusted 3′tags relative to cDNA sequences known or predicted to encode proteins. In about half of the genes, we observed intron-less EST matches downstream from the known polyadenylation site(s), many ending in trusted 3′ tags and forming clusters that connect them to the known extent of 3′ untranslated regions (UTRs). The regions containing the ESTs and 3′ tags can extend over several kilobases and contain multiple tags. The most likely explanation for the existence of these features is that transcripts originating from the gene localized upstream can be polyadenylated at many locations and thus contain long 3′ UTR. Figure 1a clearly illustrates this point in a well-annotated region of chromosome 21. The mRNA for the NCAM2 gene, which encodes a neural cell adhesion molecule, appears in the RefSeq database of full-length mRNA sequences (NM_004540). The known polyadenylation site is marked by a 3′ tag (2 ESTs), and an upstream site is also evident (3′ tag derived from 3 ESTs). The 3 kb downstream from these known sites are densely populated by intron-less ESTs (at least 26) and a cDNA clone of unknown function (DKFZp761I1311 from the German cDNA Consortium, EMBL/GenBank AL137344). This cDNA clone and a set of ESTs have been clustered together in the UniGene database, whereas other ESTs mapping downstream have been assigned to yet another cluster. It is clear from the transcript to genome map that all of the intron-less transcript sequences in this 3-kb region are in fact derived from the 3′ UTR of polyadenylation variants of the NCAM2 gene, and therefore that NCAM2 transcripts have been grouped in at least three distinct UniGene clusters (Hs.177691 for the NCAM2 cDNA, Hs.135892 for the DKFZ clone, and Hs.76118 for the 3′-most cluster of ESTs). There is a single gap in the EST sequence coverage of the region that is bridged by the assignment of two ESTs (N51204 and N47997) to the ends of the same IMAGE clone (281608).
Figure 1.
Examples of extended and overlapping 3′ untranslated region (UTR). Alignments of transcripts to the genome (yellow bar) were visualized using ACEDB. The direction of transcription is bottom to top on the left and top to bottom on the right. The direction of transcription of unspliced expressed sequence tags (ESTs) was based on their annotation, except for the unspliced ORESTES sequences, which were arbitrarily assigned to the right-hand strand. The orientation of 3′ tags was deduced from the polarity of the poly(A) tract. Light blue: RefSeq sequences; dark blue: full-length cDNA sequences; green: ORESTES sequences; and red: EST sequences. 3′ tags are represented by black boxes, with one box per cluster member. Regions covered by UniGene clusters are indicated on the right. (A) 3′ terminal exons of the NCAM2 gene. (B) Overlapping 3′ ends of the COL18A1 and SCL19A1 genes. Many ESTs derived from the COL18A1 gene were omitted for clarity.
Each gene with an extended 3′ UTR is a unique case, and therefore it is difficult to design a generally applicable automated procedure to detect such genes. To get a semiquantitative estimate of the proportion of genes that may be affected, we manually examined all genes in a relatively gene-rich region on chromosome 21q22.3 that spans 3.5 mb (NCBI contig NT_011515). A summary of the results is shown in Table 2, and an ACEDB database incorporating all transcripts and 3′ tags mapping to this contig can be downloaded from ftp://ftp.licr.org/pub/Genome_Research. Of a total of 52 genes currently annotated in this region, half (26) showed clear evidence of multiple polyadenylation sites spread over areas ranging from 300 nt to >15 kb. The existence of long-range alternative polyadenylation is independent of the size or the number of exons of the genes.
Table 2.
| Reference transcript | UniGene cluster | Description | No. of tags | Extent |
|---|---|---|---|---|
| D80001 | Hs.152629 | KIAA0179 protein | 6 | 11 kb |
| NM_003681 | Hs.38041 | Pyridoxal (pyridoxine, vitamin B6) kinase | 6 | 7 kb |
| NM_021941 | Hs.4746 | Hypothetical protein FLJ21324 | ||
| NM_000100 | Hs.695 | Cystatin B (stefin B) | 2 | 1.4 kb |
| AI885390 | Hs.49031 | ESTs | ||
| NM_003683 | Hs.110757 | NNP-1/Nop52, novel nuclear protein 1 | 2 | 1.2 kb |
| NM_020132 | Hs.324020 | 1-acylglycerol-3-phosphate O-acyltransferase 3 | 10 | 5.5 kb |
| AK026135 | Hs.173138 | FLJ22482 fis, clone HRC10859 | ||
| NM_031487 | Hs.284123 | hypothetical protein MGC4604 | ||
| NM_003274 | Hs.94479 | Transmembrane protein 1 | 5 | 2 kb |
| AF289028 | Hs.14155 | KIAA0653 protein, B7-like protein | 3 | 5 kb |
| NM_013369 | Hs.157237 | DNA (cytosine-5-)-methyltransferase 3-like | 3 | 0.9 kb |
| AI652007 | (cluster retired) | |||
| NM_000383 | Hs.129829 | Autoimmune regulator | 2 | 0.6 kb |
| NM_002626 | Hs.155455 | Phosphofructokinase, liverb | 4 | 14 kb |
| N22796 | Hs.12561 | ESTs | ||
| AA813142 | Hs.323133 | ESTs | ||
| NM_004928 | Hs.153452 | Chromosome 21 open reading frame 2 | 2 | 1.1 kb |
| AJ003549 | Hs.278715 | Chromosome 21 open reading frame 31 | 2 | 2.2 kb |
| AJ003544 | Hs.125774 | ESTs | ||
| T83849 | Hs.194595 | ESTs | ||
| NM_003343 | Hs.192853 | Ubiquitin-conjugating enzyme E2G 2 | 2 | 2.3 kb |
| NM_006936 | Hs.85119 | SMT3 (suppressor of mlf two 3, yeast) homolog | 5 | 1.2 kb |
| NM_004339 | Hs.111126 | Pituitary tumour-transforming 1 interacting protein | 5 | 1.5 kb |
| NM_001112 | Hs.85302 | Adenosine deaminase, RNA-specific, B1 | 2 | 4.2 kb |
| NM_015227 | Hs.22982 | KIAA0958 protein | 2 | 0.9 kb |
| AL390181 | Hs.170144 | CDNA DKFZp547J125 | 6 | 0.9 kb |
| NM_016214 | Hs.78409 | Collagen, type XVIII, alpha 1c | 3 | 2.6 kb |
| BF110399 | Hs.84190 | Solute carrier family 19 (folate transporter), member 1 | ||
| NM_003056 | Hs.84190 | Solute carrier family 19 (folate transporter), member 1 | 4 | 2.6 kb |
| NM_001848 | Hs.108885 | Collagen, type VI, alpha 1 | 2 | 1 kb |
| X15882 | Hs.159263 | Collagen, type VI, alpha 2 | 3 | 3 kb |
| NM_002340 | Hs.93199 | 2,3-oxidosqualene-lanosterol cyclase | 2 | 1 kb |
| NM_003906 | Hs.188481 | Minichromosome maintenance deficient 3-associated protein | 2 | 2.2 kb |
| AA527431 | Hs.178588 | ESTs | ||
| NM_006272 | Hs.83384 | S100 calcium-binding protein, beta (neural) | 2 | 0.3 kb |
| NM_001535 | Hs.235887 | HMT1 (hnRNP methyltransferase)-like 1d | >6 | ∼15 kb |
The distribution of transcript sequences and 3′ tags on the genome was visualized using ACEDB (see Fig. 1). The reference transcript was chosen from the NCBl curated RefSeq, EMBL human, or EST databases, in this order of preference. Transcripts matching multiple UniGene clusters are shown on multiple lines. The “Extent” column shows the distance separating the first from the last documented 3′ tag.
The 3′ UTR overlaps the entire extent of the C21orf2 gene on the other strand.
The 3′ UTR overlaps that of the folate transporter gene on the other strand. See the main text and Figure 1b for details.
This gene shows a very complex pattern of alternative splicing and polyadenylation that will require additional experimental data to sort out.
EST, expressed sequence tag; UTR, untranslated region.
If we extrapolate this small sample to the genome, long-range alternative polyadenylation could affect 15,000–20,000 genes. This is almost certainly an underestimate, because there are many transcripts for which 3′ tags are not available (see above) and in which EST coverage is insufficient to convincingly document the extent of the 3′ UTR. Although there have been numerous reports in the literature of transcripts undergoing alternative polyadenylation involving relatively distant sites (van Eyndhoven et al. 1998; Coy et al. 1999; Touriol et al. 1999), it was unexpected to observe it at this high frequency. Estimates gathered from EST clustering alone are significantly lower (Gautheret et al. 1998; Beaudoing and Gautheret 2001; Pauws et al. 2001). We experimentally verified our methodology by performing reverse transcriptase-polymerase chain reaction (RT-PCR) experiments on the predicted long 3′ UTRs of the WDR9 (WD repeat 9) and KCNJ5 (potassium inwardly rectifying channel 5) genes and found that they do indeed encode transcripts extending far downstream from the 3′ ends documented by “full-length” cDNA sequences, even though the intervening regions are not fully covered by ESTs (data not shown). Conversely, the TRAF3 gene on chromosome 14q32.3 has been shown experimentally to have two polyadenylation sites separated by 6 kb (van Eyndhoven et al. 1998); our data document this fact very clearly (not shown) and in addition mark a previously unrecognized site that explains the presence of 2.2 kb and 2.6 kb mRNA species observed in the original report. Northern blots very commonly show multiple bands when hybridized with a unique coding region probe. We informally collected Northern data from colleagues doing laboratory work. Probes for the FLG2, SCD, SLC20A1, PTEN, TNFRSF10C, IRF1, and IL9R genes all detect multiple bands; for all but SLC20A1 (which shows evidence of extensive alternative splicing), we were able to detect multiple polyadenylation sites documenting the origin of the major bands. A probe for ACAT2 detects only one band, and this gene has two polyadenylation sites only 150 nt apart, which would not be resolved into individual bands; β-actin (gene ACTB), commonly used as an internal control in Northerns and producing a single band, shows no evidence of long-range alternative polyadenylation despite an abundance of EST data. However, the fine structure of the ACTB polyadenylation site reveals six positions for the start of the poly(A) tail within 50 nt of each other.
One consequence of this hitherto unrecognized long-range variation in polyadenylation sites is that the extended 3′ UTR of unrelated genes transcribed in opposite directions occasionally overlap. A clear-cut case is provided by the SCL19A1 (folate transporter) and the COL18A1 (collagen type XVIII) genes, both located on the AL163302 segment of chromosome 21 (Fig. 1b). The known 3′ ends of the corresponding transcripts are separated by only 1 kb on the genome. The intervening region is densely populated by ESTs, which document unambiguously four polyadenylation sites for SCL19A1 and three for COL18A1; interestingly, the locations of the sites on the two strands are almost identical. The poly(A)-containing ESTs derived from one or the other gene can easily be distinguished from each other by their polarity on the genome; other ESTs can be oriented based on their annotation. The UniGene database assigns ESTs whose ends map before position ∼213,500 to the Hs.78409 cluster (COL18A1) and those mapping beyond that position to the Hs.84190 cluster (SLC19A1). Although these assignments are perfectly correct within the logic of EST clustering based on sequence overlaps, they fail to reflect the anatomy of the transcripts derived from this region.
In genes that are expressed at a sufficient level, the differential usage of polyadenylation sites can be estimated in silico by counting the ESTs documenting one or the other site or by extracting the corresponding serial analysis of gene expression (SAGE) tag counts from publicly available data (Lal et al. 1999). We counted poly(A)-containing ESTs and SAGE tags for the major polyadenylation sites of five of the genes in Table 2, and the results are shown in Table 3. Because many of the cDNA libraries from which ESTs were derived were normalized, the SAGE tag count is a better estimate of the relative abundance of the corresponding transcripts. It can be seen that in this limited sample the usage of individual polyadenylation sites is not related to their distance from the last splice acceptor. A larger scale study would be required to properly study the relationship between the abundance of transcripts and the length of their 3′ UTRs. Similarly, we did not attempt to determine from SAGE data whether some alternative polyadenylation sites were differentially used in libraries of different origin, because in most cases the tag numbers are not sufficient to make such comparisons.
Table 3.
Distribution of ESTs and SAGE Tags in Five Alternatively Polyadenylated Genesa
| Gene | Site no. | No. of ESTs | SAGE tag | No. of SAGE tags | Distance from site 1 |
|---|---|---|---|---|---|
| PDXK | 1 | 35 | AGTGTCCGGC | 186 | |
| Pyridoxal kinase | 2 | 9 | TAAACAGTTG | 16 | 2.8 kb |
| 3 | 23 | TGGATGTTTG | 12 | 6.2 kb | |
| CSTB | 1 | 130 | TTGGTGCTTT | 27 | |
| Cystatin B | 2 | 4 | CAAAACAGAA | 22b | 1.4 kb |
| AGPAT3 | 1 | 15 | GCGGAGCTGG | 12 | |
| 1-acylglycerol-3-phosphate | 2 | 14 | GGGTCTCCTG | 99 | 0.75 kb |
| O-acyltransferase 3 | 3 | 15 | AACACTCGCC | 52 | 1.9 kb |
| 4 | 13 | ACAGCCACTG | 62 | 2.5 kb | |
| TMEM1 | 1 | 5 | TAATTGTAGC | 12 | |
| Transmembrane protein 1 | 2 | 5 | GGATGTCCTA | 14 | 1.5 kb |
| 3 | 7 | GTTTACTATG | 12 | 1.8 kb | |
| SMT3H1 | 1 | 11 | CCGACCACAA | 121 | |
| SMT3 (suppressor of mlf two 3, | 2 | 11 | TAACCTCGGG | 57 | 0.85 kb |
| yeast) homolog | 3 | 85 | TTCTTCTCGT | 263 | 1.1 kb |
For five representative genes from Table 2, the number of EST contributing to the definition of the most prominent polyadenylation sites was counted, NIaIII SAGE tags immediately upstream of the polyadenylation sites were deduced, and the cumulative representation of the tags in SAGE libraries was determined for each of the major polyadenylation sites. The sites are listed in the 5′ to 3′ direction relative to the transcript, including the distance from the 5′ most site for those lying downstream.
This SAGE tag is not unique to the CSTB gene; it also occurs in a transcript from gene AF5Q31 (ALL1 fused gene from 5q31).
SAGE, serial analyses of gene expression.
DISCUSSION
The results presented here have important practical implications for the analysis of the human transcriptome, as well as those of other vertebrates. The cDNA or cRNA targets used in gene expression profiling experiments are almost always labeled after oligo(dT) primed cDNA synthesis by RT. Therefore, they are enriched in poly(A) proximal sequences, and as a general rule cDNA clones or oligonucleotides derived from the 3′ ends of transcripts have been chosen as probes for hybridization. The finding that in a significant proportion of genes polyadenylation sites are distributed over relatively long distances should have a significant impact on probe design, because ideally, sequences adjacent to each site should be included in the arrays. The parameters influencing polyadenylation site selection have not been studied for most genes, and the inclusion in cDNA arrays of multiple probes for each gene should allow one to readily address this question. The collection of trusted 3′ tags and the assignment of associated EST clusters to the 3′ UTR of validated genes will thus be a crucial resource for the rational design of hybridization arrays. Another important implication of the work described here is the need for more comprehensive tag to gene maps for SAGE experiments (Pauws et al. 2001). Current maps do not take into account the locations of trusted mRNA 3′ ends. In addition, tags derived from long 3′ UTR are often mapped to transcripts whose coding capacity has not been determined because they have not been linked to the correct upstream protein-coding region.
It has been argued that the relatively small number of genes present in the human genome encodes a much larger variety of transcripts, and that its true complexity cannot be deduced from a mere counting of genes. However, we are still far from comprehending the extent of this complexity, as evidenced by the results reported here. Alternative polyadenylation has been shown in numerous cases to give rise to transcripts with distinct biological properties by alteration of their protein-coding capacity (Chuvpilo et al. 1999), the regulation of their translation (Knirsch and Clerch 2000), their stability (Touriol et al. 1999), or their intracellular localization (Kislauskis et al. 1994). It is now evident that this contributes in a major way to the diversity of the human transcriptome.
METHODS
MegaBlast was used to identify pairwise similarities between all known transcript sequences and the draft genome sequence deposited in release 66 (March 2001) of the EMBL database. The transcript sequences analyzed include all human sequences deposited in the EST section, sequences identified as RNA in the HUM section, sequences available on May 15, 2001 from the NCBI curated RefSeq database (http://www.ncbi.nlm.nih.gov/LocusLink/refseq.html), and ∼700,000 ORESTES sequences from the LICR/FAPESP Human Cancer Genome project (Camargo et al. 2001). The transcript sequences were filtered of contaminants and repetitive elements were masked out using the PFP software package (Paracel). For the draft genome, we used human genomic sequences of a size >10 kb that are deposited in the HUM and HTG sections. Before analysis, we removed bacterial and other contaminants. Those HTG entries that had not been fully assembled were split into individual components. The human genome dataset we used is thus highly redundant but can easily be reduced to one of the available assemblies. For each pair of matching RNA and genomic sequence, local alignments were generated using sim4 (Florea et al. 1998) with the parameters W = 15, R = 0, A = 4, and P = 1. The output of sim4 was filtered to eliminate all alignments that did not contain at least one region (exon) matching with at least 95% identity >30 nt. ACEDB databases were generated directly from the filtered sim4 output files.
The tromer program attempts to automate the reconstruction of transcripts from transcript to genome mapping data. The output of sim4 is used to construct a set of oriented exons that are then merged if they share boundaries, thus reducing the redundancy of the EST data. The order of splice donors and acceptors is used to define the orientation of a transcript; for unspliced transcripts, the presence of a 3′ tag within 50 nt of the end (when available) is used instead. Full-length mRNA and RefSeq sequences are assumed to be derived from the coding strand. Virtual transcripts are reconstituted by following exon boundaries with known polarity; when several paths can be followed (i.e., when there is evidence for alternative splicing or polyadenylation), multiple transcripts can be generated. The output of tromer links each virtual transcript to a 3′ tag (if known) and to other virtual transcripts that share sequences derived from the same cDNA clones to flag potential gaps in EST sequence coverage.
Because poly(A) tracts documenting the position of mRNA 3′ ends have commonly been removed from the EST sequences deposited in the public databases, we analyzed the original trace files generated for each sequence. Sequences were extracted using the extract_seq (Staden et al. 2000) or phred (Ewing et al. 1998) programs; the longest poly(A) or poly(T) was identified, and if it was longer than 10 nt then the 50 nt immediately adjacent to it were recorded as a candidate tag (after obtaining the reverse complement for poly(T) tracts). Duplicate tags were eliminated, but information about the trace files containing them was retained. Tags matching LINE and Alu repetitive elements, ribosomal or mitochondrial sequences, and those containing simple repeats were eliminated. Exact matches for the remaining tags were mapped in the genome using ad hoc software, and the 50 nt found downstream from the match were also recorded. Those tags that did not find exact matches were mapped again, this time using a slower dynamic programming algorithm (Smith and Waterman 1981) allowing up to two mismatches. All 3′ tags that had found matches in the genome were clustered, again using ad hoc software, based on overlaps between the sequences of the tags (including the downstream genome sequence) and on their mapping positions in genomic clones; if two tags mapped within 50 nt they were considered to be part of the same cluster, and this procedure was iterated until no new members could be added. In this final collection, 3′ tags were tested for the occurrence of at least 10 A's or 11 A's and G's in the first 15 nt of downstream genomic sequence. A database of 3′ tag microclusters is available from ftp://ftp.licr.org/pub/databases/tags, including their sequence with the downstream sequence in the genome, the trace file(s) from which they were extracted, their position within genome segments, their offset relative to other members of the same microcluster, and information about the quality of the tag. Individual tags were incorporated into the ACEDB databases on the basis of their mapping coordinates on the genome segments.
WEB SITE REFERENCES
ftp://ftp.licr.org/pub/Genome_Research
http://cgap.nci.nih.gov/; Cancer Genome Anatomy Project.
http://mgc.nci.nih.gov/; Mammalian Gene Collection.
http://www.ncbi.nlm.nih.gov/LocusLink/refseq.html; NCBI curated RefSeq database.
http://www.ncbi.nlm.gov/UniGene/; UniGene database.
Acknowledgments
We thank Dr. Barbara Cohen for her thoughtful contributions during the preparation of this manuscript. We also thank Professor Ricardo Brentani, the director of the São Paulo Branch, and Dr. Lloyd Old, the scientific director of the LICR, for their constant encouragement and stimulation of this work. We gratefully acknowledge Dr. Greg Riggins for providing us with a curated collection of SAGE tags, and Drs. Curzio Ruegg, Jean-Baptiste Demoulin, Pär Olsson, Frank Funari, Pascal Schneider, Luiz Fernando Reis, and Jean-Christophe Renauld for sharing their Northern blot data.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.62002. Article published online before print in June 2002.
REFERENCES
- Aaronson JS, Eckman B, Blevins RA, Borkowski JA, Myerson J, Imran S, Elliston KO. Toward the development of a gene index to the human genome: An assessment of the nature of high-throughput EST sequence data. Genome Res. 1996;6:829–845. doi: 10.1101/gr.6.9.829. [DOI] [PubMed] [Google Scholar]
- Beaudoing E, Gautheret D. Identification of alternate polyadenylation sites and analysis of their tissue distribution using EST data. Genome Res. 2001;11:1520–1526. doi: 10.1101/gr.190501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burge CB. Chipping away at the transcriptome. Nat Genet. 2001;27:232–234. doi: 10.1038/85772. [DOI] [PubMed] [Google Scholar]
- Camargo AA, Samaia HPB, Dias-Neto E, Simão DF, Migotto IA, Briones MR, Costa FF, Nagai MA, Verjovski-Almeida S, Zago MA, et al. The contribution of 700,000 “ORF sequence tags” to the definition of the human transcriptome. Proc Natl Acad Sci. 2001;98:12103–12108. doi: 10.1073/pnas.201182798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caron H, van Schaik B, van der Mee M, Baas F, Riggins G, van Sluis P, Hermus MC, van Asperen R, Boon K, Voute PA, et al. The human transcriptome map: Clustering of highly expressed genes in chromosomal domains. Science. 2001;291:1289–1292. doi: 10.1126/science.1056794. [DOI] [PubMed] [Google Scholar]
- Chuvpilo S, Zimmer M, Kerstan A, Glockner J, Avots A, Escher C, Fischer C, Inashkina I, Jankevics E, Berberich-Siebelt F, et al. Alternative polyadenylation events contribute to the induction of NF- ATc in effector T cells. Immunity. 1999;10:261–269. doi: 10.1016/s1074-7613(00)80026-6. [DOI] [PubMed] [Google Scholar]
- Coy JF, Sedlacek Z, Bachner D, Delius H, Poustka A. A complex pattern of evolutionary conservation and alternative polyadenylation within the long 3′-untranslated region of the methyl- CpG-binding protein 2 gene (MeCP2) suggests a regulatory role in gene expression. Hum Mol Genet. 1999;8:1253–1262. doi: 10.1093/hmg/8.7.1253. [DOI] [PubMed] [Google Scholar]
- Dunham I, Shimizu N, Roe BA, Chissoe S, Hunt AR, Collins JE, Bruskiewich R, Beare DM, Clamp M, Smink LJ, et al. The DNA sequence of human chromosome 22. Nature. 1999;402:489–495. doi: 10.1038/990031. [DOI] [PubMed] [Google Scholar]
- Durbin R, Thierry-Mieg J. The ACEDB genome database. In: Suhai S, editor. Computational methods in genome research. NY: Plenum Press; 1994. pp. 45–55. [Google Scholar]
- Ewing B, Hillier L, Wendl MC, Green P. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 1998;8:175–185. doi: 10.1101/gr.8.3.175. [DOI] [PubMed] [Google Scholar]
- Florea L, Hartzell G, Zhang Z, Rubin GM, Miller W. A computer program for aligning a cDNA sequence with a genomic DNA sequence. Genome Res. 1998;8:967–974. doi: 10.1101/gr.8.9.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautheret D, Poirot O, Lopez F, Audic S, Claverie JM. Alternate polyadenylation in human mRNAs: A large-scale analysis by EST clustering. Genome Res. 1998;8:524–530. doi: 10.1101/gr.8.5.524. [DOI] [PubMed] [Google Scholar]
- Hattori M, Fujiyama A, Taylor TD, Watanabe H, Yada T, Park HS, Toyoda A, Ishii K, Totoki Y, Choi DK, et al. The DNA sequence of human chromosome 21. Nature. 2000;405:311–319. doi: 10.1038/35012518. [DOI] [PubMed] [Google Scholar]
- Kislauskis EH, Zhu X, Singer RH. Sequences responsible for intracellular localization of beta-actin messenger RNA also affect cell phenotype. J Cell Biol. 1994;127:441–451. doi: 10.1083/jcb.127.2.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knirsch L, Clerch LB. A region in the 3′ UTR of MnSOD RNA enhances translation of a heterologous RNA. Biochem Biophys Res Commun. 2000;272:164–168. doi: 10.1006/bbrc.2000.2754. [DOI] [PubMed] [Google Scholar]
- Lal A, Lash AE, Altschul SF, Velculescu V, Zhang L, McLendon RE, Marra MA, Prange C, Morin PJ, Polyak K, et al. A public database for gene expression in human cancers. Cancer Res. 1999;59:5403–5407. [PubMed] [Google Scholar]
- Pauws E, van Kampen AH, van de Graaf SA, de Vijlder JJ, Ris-Stalpers C. Heterogeneity in polyadenylation cleavage sites in mammalian mRNA sequences: Implications for SAGE analysis. Nucleic Acids Res. 2001;29:1690–1694. doi: 10.1093/nar/29.8.1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith TF, Waterman MS. Identification of common molecular subsequences. J Mol Biol. 1981;147:195–197. doi: 10.1016/0022-2836(81)90087-5. [DOI] [PubMed] [Google Scholar]
- Staden R, Beal KF, Bonfield JK. The Staden package, 1998. Methods Mol Biol. 2000;132:115–130. doi: 10.1385/1-59259-192-2:115. [DOI] [PubMed] [Google Scholar]
- Strausberg RL, Feingold EA, Klausner RD, Collins FS. The mammalian gene collection. Science. 1999;286:455–457. doi: 10.1126/science.286.5439.455. [DOI] [PubMed] [Google Scholar]
- Strausberg RL, Buetow KH, Emmert-Buck MR, Klausner RD. The cancer genome anatomy project: Building an annotated gene index. Trends Genet. 2000;16:103–106. doi: 10.1016/s0168-9525(99)01937-x. [DOI] [PubMed] [Google Scholar]
- Touriol C, Morillon A, Gensac MC, Prats H, Prats AC. Expression of human fibroblast growth factor 2 mRNA is post-transcriptionally controlled by a unique destabilizing element present in the 3′-untranslated region between alternative polyadenylation sites. J Biol Chem. 1999;274:21402–21408. doi: 10.1074/jbc.274.30.21402. [DOI] [PubMed] [Google Scholar]
- van Eyndhoven WG, Frank D, Kalachikov S, Cleary AM, Hong DI, Cho E, Nasr S, Perez AJ, Mackus WJ, Cayanis E, et al. A single gene for human TRAF-3 at chromosome 14q32.3 encodes a variety of mRNA species by alternative polyadenylation, mRNA splicing and transcription initiation. Mol Immunol. 1998;35:1189–1206. doi: 10.1016/s0161-5890(98)00099-6. [DOI] [PubMed] [Google Scholar]