Abstract
Exosomes are complexes containing 3′ → 5′ exoribonucleases that have important roles in processing, decay and quality control of various RNA molecules. Archaeal exosomes consist of a hexameric core of three active RNase PH subunits (ribosomal RNA processing factor (Rrp)41) and three inactive RNase PH subunits (Rrp42). A trimeric ring of subunits with putative RNA-binding domains (Rrp4/cep1 synthetic lethality (Csl)4) is positioned on top of the hexamer on the opposite side to the RNA degrading sites. Here, we present the 1.6 Å resolution crystal structure of the nine-subunit exosome of Sulfolobus solfataricus and the 2.3 Å structure of this complex bound to an RNA substrate designed to be partly trimmed rather than completely degraded. The RNA binds both at the active site on one side of the molecule and on the opposite side in the narrowest constriction of the central channel. Multiple substrate-binding sites and the entrapment of the substrate in the central channel provide a rationale for the processive degradation of extended RNAs and the stalling of structured RNAs.
Keywords: exosome, RNA degradation, RNase PH, PNPase, exoribonuclease
Introduction
The exosome is a protein complex that participates in a large number of RNA metabolic pathways (reviewed in Houseley et al, 2006). The complex can either completely degrade or partly trim a given RNA substrate by ribonuclease cleavage from the 3′ end. In eukaryotes, the exosome in the nucleus mediates the maturation of stable RNA precursors and the fast decay of defective messenger RNA precursors (Mitchell et al, 1997; Briggs et al, 1998; Allmang et al, 1999; Bousquet-Antonelli et al, 2000; van Hoof et al, 2000). In the cytoplasm, it is involved in the general turnover of messenger RNAs, and in quality control pathways that remove aberrant messenger RNAs without a stop codon or with a premature stop codon (Anderson & Parker, 1998; van Hoof & Parker, 2002). Although several proteins and protein complexes have been found to associate with the exosome in a compartment-specific manner (reviewed by Houseley et al, 2006), the core exosome consists of the same ten subunits both in the nucleus and cytoplasm. Nine subunits are homologous to the components of bacterial polynucleotide phosphorylase (PNPase; a bacterial phosphorolytic exoribonuclease) and the tenth subunit is homologous to bacterial RNase II (a bacterial hydrolytic exoribonuclease).
The exosome is not only present in eukaryotes but also in several archaeal organisms (Koonin et al, 2001; Evguenieva-Hackenberg et al, 2003). The archaeal exosome is a PNPase-like molecule, but lacks the hydrolytic exoribonuclease activity characteristic of eukaryotic exosomes. It consists of four different core subunits: ribosomal RNA processing factor (Rrp)41, Rrp42, Rrp4 and cep1 synthetic lethality (Csl)4. Structural studies of the exosomes of Sulfolobus solfataricus and Archaeoglobus fulgidus have shown that three copies of the Rrp41–Rrp42 heterodimer are arranged in a ring-like structure around a central channel (Buttner et al, 2005; Lorentzen et al, 2005). Although both Rrp41 and Rrp42 adopt the RNase PH-fold characteristic of phosphorolytic 3′ → 5′ exoribonucleases, only Rrp41 has catalytic activity (Lorentzen et al, 2005). The active sites of the three Rrp41 subunits are located on the same side of the hexameric ring. The opposite side of the Rrp41–Rrp42 ring associates with a trimer of either Rrp4 or Csl4 subunits—or possibly a mixed trimer of Rrp4 and Csl4—both of which contain an S1 domain typical of RNA-binding proteins (Buttner et al, 2005). The S1 domain proteins form a cap around the entrance of the central channel and their presence markedly increases the affinity for RNA (Oddone et al, 2007).
The crystal structure of the PNPase-like core of a eukaryotic exosome—the human Rrp41–Rrp45–Rrp46–Rrp43–Mtr3 (mRNA transport regulator)–Rrp42–Csl4–Rrp4–Rrp40 complex—shows an overall architecture similar to its simpler archaeal counterpart, albeit with significant differences (Liu et al, 2006). First, in the eukaryotic exosome, the RNase PH-like ring does not exist as a separate subcomplex, but requires the S1-domain-containing proteins to form a stable assembly. Second, only one of the RNase PH-like subunits of the human exosome (Rrp41) has phosphorolytic activity (Liu et al, 2006). This is in contrast to the yeast exosome, which has diverged further to lose all phosphorolytic activity and has retained only hydrolytic activity of the tenth subunit, Rrp44 (also known as Dis3; Liu et al, 2006; Dziembowski et al, 2007). Given the homology, the molecular mechanism of single-stranded RNA recognition and phosphorolytic cleavage observed in the archaeal complex (Lorentzen & Conti, 2005) can be extrapolated both to bacterial PNPase and to the human exosome.
The exosome degrades single-stranded RNA in a processive manner but stalls when stable secondary structures are encountered. What is the mechanism of substrate selection that results in either complete degradation or partial trimming of the RNA? Circumstantial evidence, including the positions of the RNA-binding domains around the central channel of the RNase PH-like ring as well as RNase protection assays, suggests that the constricted central channel acts as the access route for RNA substrates to reach the active sites (Buttner et al, 2005; Lorentzen & Conti, 2005). Here, the high-resolution structure of the 270 kDa S. solfataricus exosome—Rrp41—Rrp42–Rrp4 complex—is reported. By using a catalytically dead mutant, we determined the structure of the complex in the presence of an RNA substrate with a stable secondary structure designed to stall at the entrance of the channel. These results provide direct evidence that RNA binds at the entrance of the channel, supporting the idea that threading the substrate through a constricted hole is the basis for the discrimination between structured and unstructured RNAs.
Results And Discussion
Quality of the structures
The 1.6 Å resolution crystal structure of a catalytically dead mutant of the unliganded S. solfataricus exosome—Rrp41 (Asp182Ala (D182A))–Rrp42–Rrp4—was solved by molecular replacement using the previously determined structure of the Rrp41–Rrp42 complex (Lorentzen et al, 2005). The complex crystallized with one copy of Rrp41–Rrp42–Rrp4 in the asymmetric unit, with the full complex being generated by crystallographic threefold symmetry. The structure was refined to an Rfree of 24.9% and an Rwork of 21.6%, with good geometry (see Table 1 for further data collection and refinement statistics).
Table 1.
Data collection and refinement statistics
| Ss exosome (D182A) Space group P213 | Apo | Mn2+ | WO42− | RNA stem–loop 15A tail | RNA stem–loop 15 Iodo-U tail |
|---|---|---|---|---|---|
| Data collection | |||||
| Wavelength (Å) | 0.9790 | 1.550 | 1.023 | 0.9801 | 1.550 |
| Unit cell (Å), a=b=c | 135.5 | 133.6 | 134.9 | 135.9 | 134.2 |
| Resolution (Å) | 50–1.6 (1.7–1.6) | 50–2.4 (2.55–2.40) | 50–2.5 (2.7–2.5) | 50–2.3 (2.4–2.3) | 50–2.5 (2.7–2.5) |
| Rsym | 0.096 (0.681) | 0.065 (0.443) | 0.125 (0.644) | 0.081 (0.677) | 0.074 (0.461) |
| I/σ(I) | 11.7 (2.2) | 17.9 (2.7) | 18.2 (4.4) | 14.8 (2.8) | 20.9 (3.0) |
| Completeness | 1.00 (0.999) | 0.995 (0.976) | 1.00 (1.00) | 0.959 (0.665) | 0.100 (0.999) |
| Redundancy | 7.9 (4.8) | 5.5(2.9) | 12.8 (12.5) | 6.2 (6.1) | 7.4 (3.9) |
| Refinement | |||||
| Resolution (Å) | 50–1.6 | 50–2.4 | 50–2.3 | ||
| No. of reflections | 106,599 | 29,796 | 34,014 | ||
| Rwork | 0.216 (0.404) | 0.198 (0.230) | 0.204 (0.264) | ||
| Rfree | 0.249 (0.427) | 0.258 (0.311) | 0.243 (0.330) | ||
| R.m.s deviations | |||||
| Bond lengths (Å) | 0.014 | 0.009 | 0.012 | ||
| Bond angles (deg) | 1.5 | 1.3 | 1.4 |
The highest resolution shell is shown in parentheses.
Iodo-U tail, 5-iodo-uridine; r.m.s.d., root mean square deviation.
In addition, a structure of the complex with a structured RNA containing a 3′ poly(A) extension was determined at 2.3 Å resolution (Rfree 24.3%). To confirm the RNA-binding mode observed in this structure, either tungstate or a structured RNA containing a 3′ tail of 5-iodo-uridine nucleotides was diffused into the crystals and anomalous diffraction data sets were collected to a resolution of 2.5 Å. Finally, manganese ions were added and the structure was determined to a resolution of 2.4 Å (Rfree of 25.8%).
Overall exosome structure of S. solfataricus
The overall structure of the S. solfataricus (Ss) exosome is similar to that of the A. fulgidus (Af; Buttner et al, 2005) and Homo sapiens (Hs) exosomes (Liu et al, 2006), with a hexameric core of three heterodimers capped by a trimer of S1-domain-containing proteins (Fig 1). The highest similarity between the exosome structures is found for the RNase PH rings, which superpose over more than 70% of all atoms with a root mean square deviation (r.m.s.d.) of 1.6 Å (Ss and Af rings) and 2.7 Å (Ss and Hs rings). By contrast, the trimeric caps of the S1-containing subunits are less similar: r.m.s.d. of 5.4 Å over 77% of all atoms for the archaeal exosomes and 9.2 Å over 57% of all atoms between the human and the S. solfataricus exosome.
Figure 1.
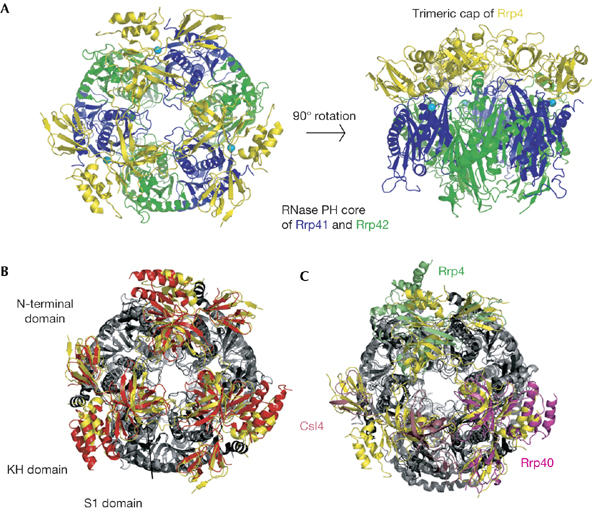
Structure of the complete 270 kDa Sulfolobus solfataricus exosome. (A) Two views of the complex, with Rrp41 in blue, Rrp42 in green and Rrp4 in yellow. Manganese ions are shown in cyan. The views are rotated by 90° around the horizontal axis. This figure and all others representing structures were generated with the program PYMOL (http://pymol.sourceforge.net, Warren L. DeLano). (B) View of the S. solfataricus exosome (Rrp4 in yellow, and Rrp41 and Rrp42 in light grey) superposed on the A. fulgidus exosome using the RNase PH cores (Rrp4 in red, Rrp41 and Rrp42 in dark grey). The structures are viewed as in (A), left. The three domains of Rrp4 (N-terminal, S1 and KH) are indicated. (C) View of the S. solfataricus exosome superposed on the human exosome using the RNase PH cores (light grey for S. solfataricus and dark grey for human RNase PH). Ss-Rrp4 is shown in yellow, Hs-Rrp40 in magenta, Hs-Rrp4 in green and Hs-Csl4 in pink. Csl4, cep1 synthetic lethality; Hs, Homo sapiens; Rrp, ribosomal RNA processing factor; Ss, Sulfolobus solfatarious.
As seen for Af-Rrp4, Hs-Rrp4 and Hs-Rrp40, Ss-Rrp4 consists of three domains: an amino-terminal domain that mediates most of the interactions with the hexameric core, a central S1 domain and a carboxy-terminal KH domain. The S1 and KH domains interact closely and seem to form a single structural unit. The N-terminal domain of Rrp4 is well ordered, with lower temperature factors than the S1/KH domains (BN−term=50 Å2 and BS1/KH=67 Å2, compared with 31 Å2 for the RNase PH core). The flexibility of the S1/KH domains has also been observed in the case of PNPase (Symmons et al, 2000), and might reflect the need to bind to and accommodate various different RNA substrates.
Binding of a stalled RNA substrate
Each Rrp41–Rrp42 dimer is able to bind to four nucleotides of single-stranded RNA at the active site, with the most 3′ nucleotide positioned near the catalytic phosphate-binding site and the most 5′ nucleotide positioned near the central channel (Lorentzen & Conti, 2005). Attempts to visualize RNA molecules within the channel by soaking Rrp41 (D182A)–Rrp42–Rrp4 crystals with longer single-stranded RNA molecules—up to 30 nucleotides in length—failed to yield interpretable electron density for additional binding pockets. To trap RNA in the channel, an RNA molecule containing a stable stem–loop structure at the 5′ end and a 3′ poly(A) tail of 15 nucleotides was diffused into the crystals. The prediction was that the single-stranded 3′ extension would enter the central channel and bind at the active sites, whereas the bulky 5′ end would stall at the entrance of the channel.
The resulting electron density map shows well-ordered electron density for one nucleotide in the central channel, in addition to the four nucleotides at the active sites (Fig 2A,B). No well-ordered electron density is observed between the two binding sites, which are separated by a distance of 30 Å, which would be spanned by five ribonucleotides of RNA in an extended conformation. In addition, no well-ordered density is observed for the 5′ stem–loop structure that should be located in the S1 entry pore of the Rrp4 subunits, but seems not to interact with it in a well-ordered manner.
Figure 2.
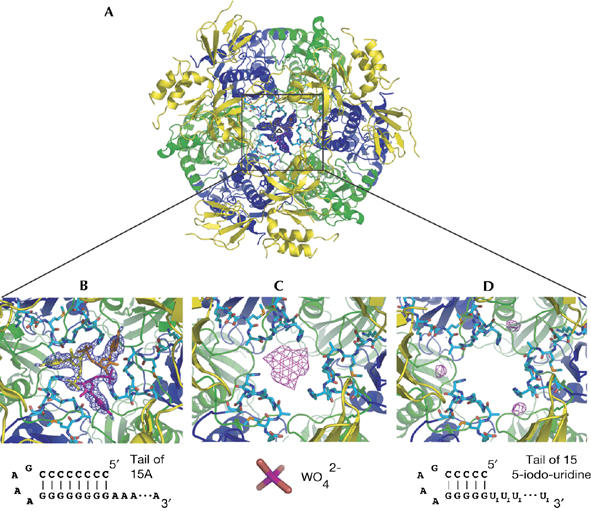
RNA binding in the central channel of the archaeal exosome. (A) An overview showing the electron density of the ribonucleotide bound at the entrance of the channel (2.3 Å resolution Fo−Fc map, contoured at 2σ) with the final model superimposed. The complex crystallized with one unique heterotrimer in the asymmetric unit, with the full complex generated by crystallographic threefold symmetry (indicated by the triangular symbol). The map is unbiased as it is calculated using model phases before any ligands were introduced. (B) Close-up of the density and model at the entrance of the channel. The loop of Rrp41 (residues 62–70) is shown in cyan forming the constriction where the ribonucleotide binds. Schematics of the ligands used for the soaking experiments are shown below the pictures. (C) Anomalous electron density map showing the binding of tungstate at the narrow constriction of the central channel. The map is calculated to 4 Å resolution (no useful anomalous signal is present beyond 4 Å), contoured at 4σ and is calculated using model phases after restrained refinement against all data extending to 2.5 Å resolution. (D) Similar view for the anomalous electron density obtained by soaking a structured RNA substrate with an iodinated 3′ poly(U) tail. The anomalous map is calculated as in (C). Rrp, ribosomal RNA processing factor.
RNA recognition in the central channel
The ribonucleotide for which we observed well-ordered density in the central channel is bound at the narrowest constriction, formed by a loop comprising residues 62–70 of the three Ss-Rrp41 subunits (Fig 2B). The constriction is 10 Å in diameter and is large enough to accommodate only one RNA molecule at a time. As the exosome structure presented here has perfect threefold symmetry, it shows three identical binding sites for the RNA in the central channel. As a result, each of the three binding sites has an occupancy of one-third (Fig 2B). This is in agreement with the stoichiometry measured by isothermal titration calorimetry (Oddone et al, 2007).
The nucleotide in the central channel is positioned so that the 3′ end points towards the active sites. The RNA is bound by the Rrp41 loop by several hydrogen bonds between the backbone of residues 65–68 and the phosphate, the 2′ hydroxyl and the adenine base of the nucleotide. In addition, His68 has a dual role in RNA binding, as its side chain forms a hydrogen bond with the phosphate part and engages in stacking interactions with the adenine base (Fig 2B). To confirm the binding mode of this ribonucleotide, we diffused either tungstate or a stem–loop RNA with a 3′ tail of 5-iodo-uridines into the crystals and collected anomalous diffraction data. The tungstate, which mimics the RNA phosphate groups, clearly bound more towards the centre of the constriction, whereas the iodine-labelled uracil base was located more peripherally (Fig 2C,D). This is consistent with the density observed for the RNA nucleotide and suggests a similar binding mode for adenine and uridine nucleotides at this site.
Conserved features of the Rrp41 RNA-binding loop
The Rrp41 loop that forms the constriction is well conserved among archaeal Rrp41 sequences and adopts a similar structure in the A. fulgidus exosome, where the loop also binds tungstate (Buttner et al, 2005). Mutation of Arg 65 in the loop of Af-Rrp41 reduces the in vitro RNA degradation activity of the complex by more than 90%. The corresponding residue in Ss-Rrp41 (Arg 67) contacts RNA by means of its main chain, whereas the side chain has no ordered electron density.
In the structure of the human exosome, only the corresponding loop of Hs-Rrp41 (residues 56–64) is well ordered and is in a conformation similar to that in Ss-Rrp41 (supplementary Fig S1 online; Liu et al, 2006). Furthermore, this loop contains several arginine residues, suggesting that it could be involved in binding RNA in the central channel of the human exosome in a manner similar to that observed in the archaeal complex. The binding of single-stranded RNA to this loop, and also at the active site of Hs-Rrp41, explains the substrate requirements detected in RNA protection assays with the Hs-Rrp41–Rrp45 heterodimer (Liu et al, 2006).
In bacterial PNPase, this loop is conserved neither in sequence nor in structure. Instead, a different loop with an FFRR sequence motif characteristic of PNPase (S. antibioticus residues 78–92) constricts the central channel and probably interacts with RNA substrates passing through the channel (Symmons et al, 2000). Together with data from RNA protection assays (Spickler & Mackie, 2000), this indicates that the archaeal exosome and PNPase both recruit substrates through the central channel, although the details of the substrate recognition are likely to differ.
A structural metal-binding site
The finding of a manganese-binding site in the yeast exosome subunit Rrp40 (Oddone et al, 2007) prompted us to investigate whether the archaeal S. solfataricus exosome also binds to divalent cations. Crystals of the Rrp41(D182A)–Rrp42–Rrp4 complex were soaked in a solution containing manganese and X-ray diffraction data were collected at a wavelength of 1.55 Å, where manganese has a considerable anomalous signal. The resulting anomalous electron density map clearly shows a manganese ion (20σ peak) bound at the interface between Rrp41 and Rrp4 (Fig 3A). The manganese-binding site is far from the catalytic site and the central channel, and is also in a different position from the manganese-binding site detected in yeast Rrp40. The metal ion seems to have a structural role rather than a functional role, a result also supported by RNase activity assays (supplementary Fig S2 online).
Figure 3.
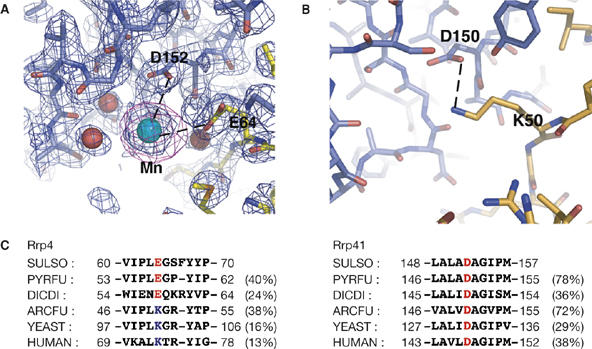
Metal ions mediate subunit interactions. (A) A close-up view showing the structural manganese (Mn) ion-binding site in the Sulfolobus solfataricus (Ss) exosome, located at the interface between Ss-Rrp41 (blue) and Ss-Rrp4 (yellow). A 2.4 Å resolution 2Fo−Fc electron density map contoured at 1σ is shown in blue and an anomalous map at 4 Å resolution contoured at 4σ is shown in magenta. Residues that coordinate the metal ion are labelled. (B) The equivalent region of the Archaeglobus fulgidus exosome structure showing that the metal ion-binding site found in the S. solfataricus exosome is replaced by a direct salt-bridge (dotted line). (C) Sequence alignment showing that the Rrp41–Rrp4 contacts are probably mediated either by divalent metal ions or by a direct salt bridge in exosomes from different organisms. Sequences included are: S. solfataricus (SULSO), Pyrococcus furiosus (PYRFU), Dictyostelium discoideum (DICDI), A. fulgidus (ARCFU), Saccharomyces cerevisiae (YEAST) and Homo sapiens (HUMAN). Numbers in parentheses denote overall percentage identity of the full-length proteins to the S. solfataricus sequence. D, aspartic acid; E, glutamic acid; K, lysine; Rrp, ribosomal RNA processing factor.
The manganese ion is coordinated by Asp (D) 152 of Ss-Rrp41, Glu (E) 64 of Ss-Rrp4 and by several well-ordered water molecules (Fig 3A). In both the A. fulgidus and the human complexes the two corresponding residues are an aspartic acid and a lysine (K), which form a direct salt bridge rather than a metal-binding site. Sequence comparisons show that Asp 152 of Ss-Rrp41 is well conserved among archaeal and eukaryotic Rrp41 subunits, whereas the position Glu 64 of Ss-Rrp4 accommodates both negatively and positively charged residues in Rrp41 protein sequences from different organisms (Fig 3C). Thus, in exosomes from different organisms the Rrp41–Rrp4 interaction is probably mediated by either a direct salt bridge, as in the A. fulgidus and the human complexes, or divalent metal ions, as in the S. solfataricus complex.
Conclusions
Two different binding sites for RNA are observed in the substrate-bound structure presented here: one at the active site of the Rrp41 subunit, and another at the entrance of the central channel of the exosome. As the narrow constriction of the central channel allows only one RNA molecule to enter at a time, it seems most likely that the RNA observed at the two sites belongs to the same molecule. Given the two binding sites, a likely path for RNA substrates is depicted in Fig 4. In the emerging model for phosphorolytic degradation of RNA by the exosome, the substrate is initially recruited by the S1-domain-containing subunits—Rrp4, Rrp40 and Csl4—and then channelled into the central cavity to reach the active sites. The narrow constriction of the central channel provides a molecular basis for why the phosphorolytic activity of the exosome can readily degrade single-stranded RNA but stalls when stable secondary structure is encountered.
Figure 4.
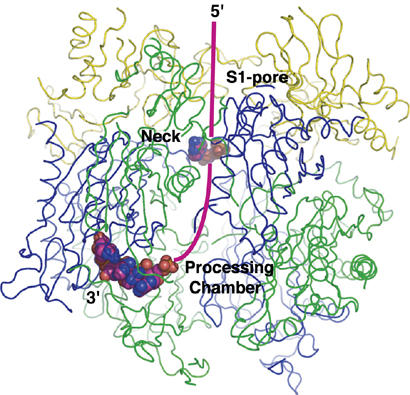
Ribbon representation of the Sulfolobus solfataricus exosome including a Corey–Pauling–Koltun model of the RNA bound in the structure. A possible path for RNA substrates is indicated by a magenta-coloured line. In this model, the 3′ end of the RNA substrate is recruited by the S1 pore of the Rrp4/Csl4 cap and then threaded through the neck of the central channel to the processing chamber, where it binds at one of the active sites and is degraded in a processive manner. Csl4, cep1 synthetic lethality; Rrp, ribosomal RNA processing factor.
The lack of interpretable electron density for the RNA between the two binding sites indicates that most of the central channel of the hexameric core does not bind to RNA in a well-ordered manner. Thus, in addition to its role in binding to RNA, the central channel might be important for trapping RNA substrates and ensuring that RNA degradation occurs in a processive manner. Furthermore, the flexibility of the RNA in the central channel could allow RNA degradation by any of the three active sites or even allow the same RNA molecule to rebind at a different active site if prematurely released. The process of channelling substrates to the sites of degradation is also used by the proteasome, which degrades polypeptides in a processive manner. This has the advantage of allowing selective substrate recruitment and avoiding indiscriminative degradation (reviewed by Buttner et al, 2006; Lorentzen & Conti, 2006). How different classes of substrates are recruited and which additional activators are needed for RNA degradation in vivo by the archaeal exosome are questions for future research.
Methods
Protein expression and crystallization. A tricistronic Rrp41–Rrp42–Rrp4 construct was cloned into a pETMCN vector and the Rrp41 D182A mutation introduced by standard methods. The protein complex was purified by heat precipitation (30 min at 75°C) followed by Ni2+-nitrilotriacetic acid (Ni-NTA) affinity chromatography and size-exclusion chromatography on a Superdex 200 column. After concentrating the complex to 10 mg/ml in a buffer containing 10 mM Tris–HCl pH 7.6 and 150 mM NaCl, crystallization was performed by sitting drop vapour diffusion against a well solution containing 40% PEG 400 buffered with 50 mM Tris–HCl at pH 8.0. Crystals grew within 2 days at 18°C.
Data collection and structure determination. All X-ray diffraction data were collected at cryogenic temperatures by using the PX beam line X06SA at the Swiss Light Source (SLS; Villigen, Switzerland) equipped with a MarCCD 225 mm detector and processed with the program XDS (Kabsch, 1993). The structures were solved by molecular replacement with the program Phaser (Storoni et al, 2004), by using the previously determined structure of the Rrp41–Rrp42 heterodimer as a search model. The Rrp4 subunit was manually built in COOT (Emsley & Cowtan, 2004) and the structure refined in REFMAC (Murshudov et al, 1997), with each domain defined as a translation-liberation-screw (TLS) group—treating the S1 and KH domains of Rrp4 as one group—in the modelling of anisotropy. The crystals contained one Rrp41–Rrp42–Rrp4 heterotrimer in the asymmetric unit and the full exosome complex was generated by crystallographic threefold symmetry. All electron density maps shown in the figures were calculated with model phases following restrained refinement of the complete protein model before any ligands were introduced. For the RNA-bound structure, the entire ligand was given an occupancy of either one-third or one in separate refinements. Only with an occupancy of one-third did the B-factors of the RNA match those of the surrounding protein atoms. Consistently, a one-third occupancy was also found for the RNA in isothermal calorimetry titration experiments (Oddone et al, 2007).
The RNA-bound structures were obtained by soaking crystals of Rrp41(D182A)–Rrp42–Rrp4 in mother liquor containing 0.3 mM RNA for 16 h. The structure with bound tungstate was determined from a crystal soaked for 5 min in 100 mM Na2WO4 and the manganese-bound structure from a crystal soaked for 5 min in 30 mM MnCl2.
PDB codes: The coordinates and structure factors have been deposited to the Protein Data Bank with accession codes 2je6 (apo), 2jea (RNA bound) and 2jeb (Mn bound).
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
Supplemental Material
Acknowledgments
We thank the staff at SLS (Villigen) for assistance with data collection and C. Romier (Institut de Génétique et de Biologie Moléculaire et Cellulaire (IGBMC)-Strasbourg) for the polycistronic pETMCN vector. We also thank A. Oddone, P. Brick and members of the laboratory for critical reading of the manuscript, P. Walter and E. Hackenberg for many valuable discussions, and M. Kjeldgaard and H. von Moeller for the contribution of important ideas regarding the choice of RNA substrates. This project was supported by the European 6th Framework Programme, and E.L. by a fellowship from Carlsbergfondet.
References
- Allmang C, Kufel J, Chanfreau G, Mitchell P, Petfalski E, Tollervey D (1999) Functions of the exosome in rRNA, snoRNA and snRNA synthesis. EMBO J 18: 5399–5410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson JS, Parker RP (1998) The 3′ to 5′ degradation of yeast mRNAs is a general mechanism for mRNA turnover that requires the SKI2 DEVH box protein and 3′ to 5′ exonucleases of the exosome complex. EMBO J 17: 1497–1506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bousquet-Antonelli C, Presutti C, Tollervey D (2000) Identification of a regulated pathway for nuclear pre-mRNA turnover. Cell 102: 765–775 [DOI] [PubMed] [Google Scholar]
- Briggs MW, Burkard KT, Butler JS (1998) Rrp6p, the yeast homologue of the human PM-Scl 100-kDa autoantigen, is essential for efficient 5.8S rRNA 3′ end formation. J Biol Chem 273: 13255–13263 [DOI] [PubMed] [Google Scholar]
- Buttner K, Wenig K, Hopfner KP (2005) Structural framework for the mechanism of archaeal exosomes in RNA processing. Mol Cell 20: 461–471 [DOI] [PubMed] [Google Scholar]
- Buttner K, Wenig K, Hopfner KP (2006) The exosome: a macromolecular cage for controlled RNA degradation. Mol Microbiol 61: 1372–1379 [DOI] [PubMed] [Google Scholar]
- Dziembowski A, Lorentzen E, Conti E, Seraphin B (2007) A single subunit, Dis3, is essentially responsible for yeast exosome core activity. Nat Struct Mol Biol 14: 15–22 [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr D 60: 2126–2132 [DOI] [PubMed] [Google Scholar]
- Evguenieva-Hackenberg E, Walter P, Hochleitner E, Lottspeich F, Klug G (2003) An exosome-like complex in Sulfolobus solfataricus. EMBO Rep 4: 889–893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houseley J, LaCava J, Tollervey D (2006) RNA-quality control by the exosome. Nat Rev Mol Cell Biol 7: 529–539 [DOI] [PubMed] [Google Scholar]
- Kabsch W (1993) Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J Appl Crystallogr 26: 795–800 [Google Scholar]
- Koonin EV, Wolf YI, Aravind L (2001) Prediction of the archaeal exosome and its connections with the proteasome and the translation and transcription machineries by a comparative-genomic approach. Genome Res 11: 240–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Greimann JC, Lima CD (2006) Reconstitution, activities, and structure of the eukaryotic RNA exosome. Cell 127: 1223–1237 [DOI] [PubMed] [Google Scholar]
- Lorentzen E, Conti E (2005) Structural basis of 3′ end RNA recognition and exoribonucleolytic cleavage by an exosome RNase PH core. Mol Cell 20: 473–481 [DOI] [PubMed] [Google Scholar]
- Lorentzen E, Conti E (2006) The exosome and the proteasome: nano-compartments for degradation. Cell 125: 651–654 [DOI] [PubMed] [Google Scholar]
- Lorentzen E, Walter P, Fribourg S, Evguenieva-Hackenberg E, Klug G, Conti E (2005) The archaeal exosome core is a hexameric ring structure with three catalytic subunits. Nat Struct Mol Biol 12: 575–581 [DOI] [PubMed] [Google Scholar]
- Mitchell P, Petfalski E, Shevchenko A, Mann M, Tollervey D (1997) The exosome: a conserved eukaryotic RNA processing complex containing multiple 3′ → 5′ exoribonucleases. Cell 91: 457–466 [DOI] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D 53: 240–255 [DOI] [PubMed] [Google Scholar]
- Oddone A, Lorentzen E, Basquin J, Gasch A, Rybin V, Conti E, Sattler M (2007) Structural and biochemical characterization of the yeast exosome component Rrp40. EMBO Rep 8: 63–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spickler C, Mackie GA (2000) Action of RNase II and polynucleotide phosphorylase against RNAs containing stem–loops of defined structure. J Bacteriol 182: 2422–2427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storoni LC, McCoy AJ, Read RJ (2004) Likelihood-enhanced fast rotation functions. Acta Crystallogr D 60: 432–438 [DOI] [PubMed] [Google Scholar]
- Symmons MF, Jones GH, Luisi BF (2000) A duplicated fold is the structural basis for polynucleotide phosphorylase catalytic activity, processivity, and regulation. Struct Fold Des 8: 1215–1226 [DOI] [PubMed] [Google Scholar]
- van Hoof A, Parker R (2002) Messenger RNA degradation: beginning at the end. Curr Biol 12: R285–R287 [DOI] [PubMed] [Google Scholar]
- van Hoof A, Lennertz P, Parker R (2000) Yeast exosome mutants accumulate 3′-extended polyadenylated forms of U4 small nuclear RNA and small nucleolar RNAs. Mol Cell Biol 20: 441–452 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Material


