Abstract
As more and more genomic DNAs are sequenced to characterize human genetic variations, the demand for a very fast and accurate method to genomically position these DNA sequences is high. We have developed a new mapping method that does not require sequence alignment. In this method, we first identified DNA fragments of 15 bp in length that are unique in the human genome and then used them to position single nucleotide polymorphism (SNP) sequences. By use of four desktop personal computers with AMD K7 (1 GHz) processors, our new method mapped more than 1.6 million SNP sequences in 20 hr and achieved a very good agreement with mapping results from alignment-based methods.
The SNP (Single Nucleotide Polymorphism) Consortium (http://snp.cshl.org) and laboratories around the world have generated millions of human SNP sequences (http://www.ncbi.nlm.nih.gov/SNP/). For these SNPs to become the next generation of genetic markers to transform biomedical research (Lander and Schork 1994; Lander 1996; Risch and Merikangas 1996; Kruglyak 1997; Collins et al. 1998; Schafer and Hawkins 1998), one must first know the location of each SNP in the genome. This is traditionally achieved by aligning numerous short DNA sequences, each typically of a few hundred nucleotides, one at a time with one very long DNA sequence, the genome. For example, to ensure that an SNP sequence is mapped to its cognate genomic position, the protocol published by the Whitehead Institute uses a double-BLAST (Altschuler et al. 1990) search strategy with stringent match criteria (http://snp.cshl.org/data/). By focusing on near-identity matches, faster DNA sequence aligning programs have also been developed (Chao et al. 1997; Zhang and Madden 1997; Florea et al. 1998; Delcher et al. 1999; Zhang et al. 2000; W. Gish at http://blast.wustl.edu). However, to the best of our knowledge and our experience in using BLAST, several minutes on a workstation are still required to align a single DNA fragment (expressed sequence tag [EST], SNP, etc.) with the human genome (see e.g., Florea et al. 1998; Bedell et al. 2000). At this speed, it would take months, if not years, to map a database, such as dbSNP (http://www.ncbi.nlm.nih.gov/SNP)—an impractical task for most laboratories.
We show here that the mapping of dbSNP can be performed in hours using only a few desktop personal computers. Our innovation lies in dispensing with the need to perform actual alignment for positional mapping; instead, fixed-length unique sequence markers, referred to as UniMarkers or UMs, were used to assign the genomic positions of SNP sites. By definition, every UM appears only once in the genome. Consequently, in the ideal situation of, for example, no sequence errors, a single UM match will suffice to locate an SNP sequence in the genome. The present UM method (schematically illustrated in Fig. 1) was developed on the basis of this simple premise; its performance in SNP mapping is compared below with that of the alignment-based assignment method reported at the National Center for Biotechnology Information (NCBI), hereafter referred to, for convenience, as the NCBI method.
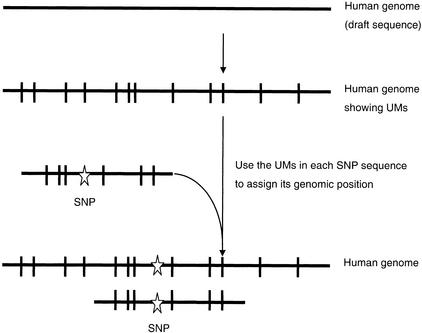
Figure 1.
A schematic overview of the UniMarker (UM) method for mapping single nucleotide polymorphism (SNP) sequences. Mapping was based solely on the identity and genomic position of the UMs found in each SNP sequence and not on sequence alignment.
RESULTS
UMs Using Different Length Markers
With a sufficiently large value of N, any N-mer DNA sequence will be unique in a genome. It is, however, computationally less efficient to work with longer N-mers. Thus, for example, in BLAST and BLAST-related methods for comparing DNA sequences (Altschul et al. 1990; Zhang and Madden 1997; Florea et al. 1998; Zhang et al. 2000), 10-, 11-, or 12-mers are usually used. For the present purpose of mapping SNPs using the UM method (see Methods), N needs to be larger such that the number of markers exceeds the number of SNPs. Indeed, as shown in Table 1, it is only when N is larger than 13 that considerable numbers of SNPs can be mapped. Table 1 furthermore indicates that, in the trade-off between computing costs and SNP-mapping capability, a value of N of 15 would be optimal. At this length, a UM exists, on average, every 36 bp, a density more than an order of magnitude higher than that estimated for the total number of SNPs (Taillon-Miller et al. 1998; Marth et al. 1999). In addition, at this length, 81.4% of the SNPs in dbSNP can be uniquely assigned in the genome by the UM method, compared with the NCBI's coverage of 75.7%. Because of these observations, we hereafter report only the results of 15-mer UMs. As one would expect, knowing that most genes are found in regions of nonrepetitive sequences (International Human Genome Sequencing Consortium 2001; Venter et al. 2001), the number of UMs for each chromosome was approximately proportional to the number of genes predicted (data not shown).
Table 1.
UniMarkers using Different Length Markers
| Mer | No. of UMs | Density (bp) | SNPs assigned (%) | Processing time (hr) |
|---|---|---|---|---|
| 13 | 2,440,788 | 2372.5 | 13.3 | 3 |
| 14 | 30,234,168 | 191.5 | 57.5 | 8 |
| 15 | 162,253,846 | 35.7 | 81.4 | 20 |
| 16 | 646,229,602 | 9.0 | 88.3 | 140 |
These are results using the draft sequence of the human genome (2,895,388,086 bases) and 1,655,188 nonredundant single nucleotide polymorphism (SNF) sequences. Mapping was performed on four AMD K7 (1GHz, 512 MB memory) personal computers running the FreeBSD operating system. The time shown includes both UniMarker (UM) iIdentification and SNP positional assignment, the former accounting for ∼50% of the total time in the case of the 15-mer assignment.
Comparison with NCBI Assignments
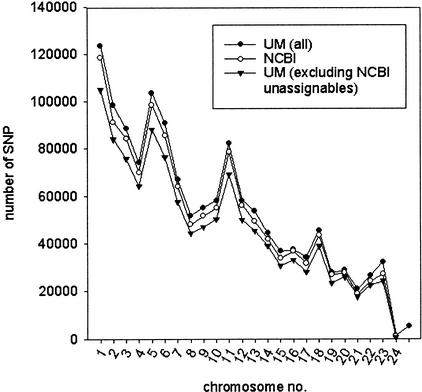
Tables 2 and 3, show, respectively, the results obtained using the UM method for those SNPs assigned or not assigned by NCBI to one of the 24 chromosomes. Except for the Y chromosome (chromosome 24), a gene desert (International Human Genome Sequencing Consortium 2001; Venter et al. 2001), with, consequently, considerably few UMs, statistical analysis indicated that the two independent SNP-mapping methods show little chromosomal bias. Namely, the percentage of SNPs that can be assigned to a particular chromosome was comparable for the first 23 chromosomes (Table 2), and no chromosome was particularly favored by one mapping method and disfavored by the other (Fig. 2). It is important that, for those SNPs successfully assigned by the UM method, the agreement with the NCBI method was very high (∼99.5%); although, in general, ∼7% of SNPs assigned by the NCBI method have no UM (at N = 15) and thus cannot be handled by the UM method (Table 2). However, ∼25% of SNPs assigned to multiple genomic positions by the NCBI method and >50% of those not assigned by the NCBI method could be uniquely assigned using the UM method (Table 3). These additional assignments allowed us to map substantially more SNPs than with the NCBI method (1,346,672 vs. 1,253,779 or 81.4% vs. 75.7% of the 1,655,188 nonredundant SNP entries [Build 22]). Notably, the additional assignments were not biased to particular chromosomes (Fig. 2).
Table 2.
Performance of the UM Method (Part I)
| chr | SNPs | Not assigned | % | Agreed | % | Disagreed | % | Multiple | % |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 118569 | 10054 | 8.5 | 104817 | 88.4 | 567 | 0.5 | 3131 | 2.6 |
| 2 | 91355 | 5364 | 5.9 | 83980 | 91.9 | 229 | 0.3 | 1782 | 2.0 |
| 3 | 84337 | 6129 | 7.3 | 75910 | 90.0 | 379 | 0.4 | 1919 | 2.3 |
| 4 | 70157 | 4336 | 6.2 | 64206 | 91.5 | 245 | 0.3 | 1370 | 2.0 |
| 5 | 98551 | 7170 | 7.3 | 88200 | 89.5 | 489 | 0.5 | 2692 | 2.7 |
| 6 | 85744 | 6722 | 7.8 | 76611 | 89.3 | 306 | 0.4 | 2105 | 2.5 |
| 7 | 64182 | 4886 | 7.6 | 57639 | 89.8 | 241 | 0.4 | 1416 | 2.2 |
| 8 | 48126 | 2590 | 5.4 | 44227 | 91.9 | 192 | 0.4 | 1117 | 2.3 |
| 9 | 51755 | 3230 | 6.2 | 47043 | 90.9 | 192 | 0.4 | 1290 | 2.5 |
| 10 | 54977 | 3156 | 5.7 | 50299 | 91.5 | 213 | 0.4 | 1309 | 2.4 |
| 11 | 78508 | 6616 | 8.4 | 69212 | 88.2 | 446 | 0.6 | 2234 | 2.8 |
| 12 | 56106 | 4285 | 7.6 | 49992 | 89.1 | 305 | 0.5 | 1524 | 2.7 |
| 13 | 49371 | 2646 | 5.4 | 45375 | 91.9 | 198 | 0.4 | 1152 | 2.3 |
| 14 | 41816 | 1981 | 4.7 | 38984 | 93.2 | 111 | 0.3 | 740 | 1.8 |
| 15 | 33836 | 2195 | 6.5 | 30628 | 90.5 | 150 | 0.4 | 863 | 2.6 |
| 16 | 36787 | 2598 | 7.1 | 32990 | 89.7 | 174 | 0.5 | 1025 | 2.8 |
| 17 | 31690 | 2442 | 7.7 | 28155 | 88.8 | 206 | 0.7 | 887 | 2.8 |
| 18 | 43458 | 3312 | 7.6 | 38842 | 89.4 | 179 | 0.4 | 1125 | 2.6 |
| 19 | 26757 | 2352 | 8.8 | 23536 | 88.0 | 137 | 0.5 | 732 | 2.7 |
| 20 | 27691 | 1099 | 4.0 | 25974 | 93.8 | 91 | 0.3 | 527 | 1.9 |
| 21 | 18826 | 743 | 3.9 | 17662 | 93.8 | 55 | 0.3 | 366 | 1.9 |
| 22 | 24192 | 1309 | 5.4 | 22367 | 92.5 | 103 | 0.4 | 413 | 1.7 |
| 23 | 27049 | 2186 | 8.1 | 24206 | 89.5 | 112 | 0.4 | 545 | 2.0 |
| 24 | 1031 | 364 | 35.3 | 600 | 58.2 | 18 | 1.7 | 49 | 4.8 |
| Total | 1264871 | 87765 | 6.9 | 1141455 | 90.2 | 5338 | 0.4 | 30313 | 2.4 |
The UM assignments were compared with the SNP positions reported in dbSNP for those assigned to chromosome 1–24 (see Methods for details).
Table 3.
Performance of the UM Method (Part II)
| SNPs | No. | Not assigned | % | Assigned | % | Multiple | % |
|---|---|---|---|---|---|---|---|
| NCBI-multi | 152677 | 105492 | 69.1 | 38304 | 25.1 | 8881 | 5.8 |
| NCBI-not | 272192 | 76863 | 28.2 | 161575 | 59.4 | 33754 | 12.4 |
The SNPs assigned by the UM method were National Center for Biotechnology Information (NCBI) unassignables: that is, those assigned to multiple genomic positions (NCBI-multi) and those not assigned (NCBI-not) at NCBI (see Methods for details).
Figure 2.
Chromosomal distribution of assigned SNP sequences. The last data point beyond chromosome 24 collects the contigs that have not yet been assembled into the genome.
On the other hand, because the strategies and criteria used by the UM method to assign SNPs are so different from the conventional use of BLAST scores (see Methods), the above statistics should not be taken to reflect an absolute advantage of the UM method. Indeed, many of the additional assignments achieved by the UM method were made on few UMs; they are consequently less reliable and could be ambiguous (see discussion in the following two sections). These assignments will mostly occur for SNPs that lie in regions of the genome that are abundant in repetitive or duplicated sequences. In such cases, the UM method presents an independent assignment with which the assignment of BLAST-based methods can be compared and evaluated.
Assessing the Quality of the UM Assignments
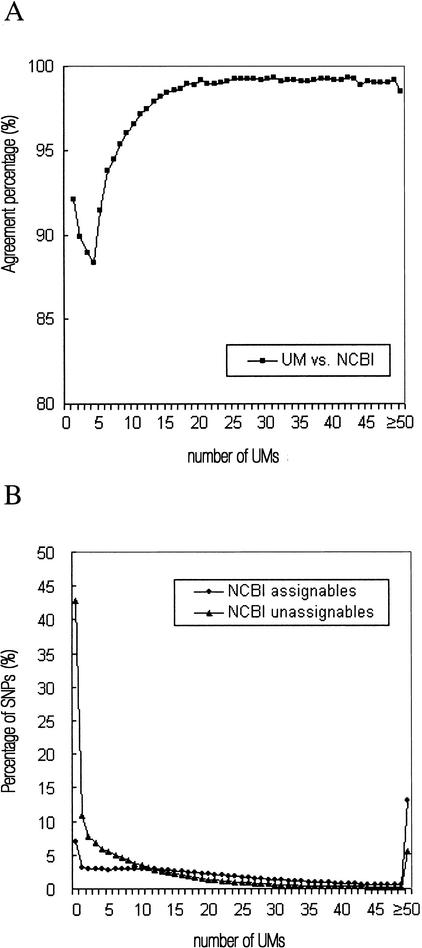
Although, in theory, a single UM would suffice to assign an SNP sequence, in practice, an SNP sequence may contain multiple UMs that do not all map to the same genomic region, resulting in ambiguities. In general, the more UMs that an SNP sequence contains, the easier and also the more reliable is the positioning of the SNP sequence. Thus, as shown in Figure 3A, the percentage agreement in the assignment of the SNP sequences between the two methods was lower when the SNP sequence contained a low number of UMs; for example, the percentage agreement does not reach 95% when the number of UMs is less than eight. It is, nevertheless, encouraging that the lowest percentage agreement (88.3%) was still high, and that at least 92% of NCBI-assignable SNP sequences containing a singleton UM received the same assignment as that produced by the NCBI method. The proportion of zero and singleton UM SNPs in NCBI-unassignable SNP sequences was comparatively much higher (Fig. 3B), reflecting the difficulties in mapping these groups of SNPs by any method. Interestingly, a residual percentage of disagreement in assignment (∼0.8%) persisted no matter how many UMs were contained in an SNP sequence, indicating the existence of cases that neither method can map with absolute confidence (see the example below).
Figure 3.
Distribution statistics on the number of UMs found in an SNP sequence: (A) the SNPs assigned by both UM and NCBI (National Center for Biotechnology Information); (B) the NCBI assignable/unassignable SNPs. “NCBI assignables” refers to the SNPs that were assigned to chromosomes 1–24 in dbSNP, and “NCBI unassignables” refer to those that were not assigned. The comparison between the UM and NCBI assignments was made using the NCBI assignables. Note that, according to the scheme of the UM method, SNPs with a single UM will be uniquely assigned and those with two, three, or four UMs will always be assigned as ‘multiple’ if they have more than one UM cluster (see Methods), explaining the dip in the percentage agreement plot.
Some Examples of Possible Ambiguities
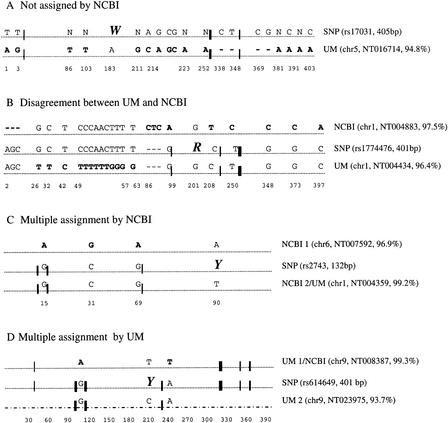
Figure 4 shows some examples illustrating how different assignments might arise from the UM method and an alignment-based method.
Figure 4.
Four examples of different assignments made using the UM and NCBI methods. The information in parentheses is the ID and length of the SNP sequences, chromosome number, contig number, and percent identity to the SNP sequence for the UM- or NCBI-matched genomic sequence. BLAST was used to align these sequences. Mismatches in the alignment are indicated above the dotted line; those differing from the SNP sequence are shown in bold type. The exception is the last sequence (UM 2 in case (D), which contains too many (29) mismatches for them to be clearly labeled; this sequence is therefore shown as a broken line. The SNP site is shown by a large italicized letter. UMs, represented by vertical lines of varying thickness to roughly indicate their amount, are shown on the sequence at the approximate locations. For example, in case (A), 10 UMs are shared by the SNP sequence and the UM-assigned genomic sequence, and of these, seven are found in a small region between 302 bp and 327 bp. In cases (B) and (C), 19 and 8 UMs are shared, respectively. In case (D), the UM assignments (UM 1 and UM 2) both shared 11 UMs with the SNP sequence but at different locations.
In case A, the SNP sequence contained many uncertain bases (labeled as N), making it difficult for the alignment method to find near identical genomic matches. In this SNP sequence, there were 15 UMs, of which 10 were clustered in three groups on a genomic segment identified by the UM method, which, overall, sees 94.8% sequence identity with the SNP sequence. The remaining five, which were found in three other chromosomes (chromosomes 4, 7, and 12) were, according to our scheme (see Methods), regarded as noise.
In case B, different contigs on the same chromosome were matched. Sequence alignment indicates that, excluding the first three bases, the first part of the sequence (up to position 63) was matched in the NCBI assignment, whereas the rest was matched in the UM assignment. The 19 UMs encompassing the SNP site allowed us to make a distinction between the two similarly homologous regions (97.5% and 96.4% homologous with the SNP sequence using the NCBI and UM assignment, respectively).
Case C is a multiple assignment using the NCBI method that was resolved using the UM method, whereas case D is the converse. In case C, the change of A to G at only two positions, 15 and 69, resulted in eight UMs, enabling a unique assignment by the UM method. In contrast, a substantial and comparable number of UMs in differing genomic regions may be found to match an SNP sequence, as illustrated in case D, in which the one with many more identity matches (99.3% vs. 93.7%) was selected by the NCBI method. It is, however, likely that the ambiguity arises from chimerization or sequencing errors.
DISCUSSION
Indexing a large dataset is an established means of facilitating the development of efficient algorithms for data analysis (Fleming and Halle 1989). Here, we showed that, even in the draft form, the human genome sequence can likewise be preprocessed to generate UMs to greatly enhance the efficiency of SNP mapping. As presented above, very good agreement with previous mapping results was achieved for most assignments, whereas in cases of disagreement there was often ambiguity about the precise genomic location, which will probably only be resolved by further sequencing. Indeed, particularly for duplicated regions, validating predicted SNPs poses considerable challenge, even for experimental investigations (Marth et al. 2001). At the expense of increasing computational cost, the present UM method can be improved by incorporating information from UMs of a longer marker length (N = 16, etc.) or markers found exactly two, three, or more times in the genome to resolve many of the mapping ambiguities. For example, using markers that occur twice, we were able to assign a further 18.3% of the UM unassignable SNPs to unique genomic positions (data not shown).
It is important that in addition to an increase in speed of orders of magnitude compared with conventional, alignment-based mapping, the UM method has other advantages: (1) the use of UMs effectively masks repetitive elements, a common practice preceding many genome-wide sequence comparisons (Bedell et al. 2000; A. Smit and P. Green at http://repeatmasker.genome.washington.edu), including SNP mapping (Wang et al. 1998; Altshuler et al. 2000; Mullikin et al. 2000; The International SNP Map Working Group 2001); (2) the use of base-calling programs (Ewing and Green 1998; Ewing et al. 1998; ) (another common practice) can also be eliminated, as poor quality sequences will be detected automatically by their incongruous UMs, which will probably be distributed randomly on different contigs or chromosomes. Although the present work has focused on SNP mapping, it is obvious that the same method can be applied to EST mapping, a topic of considerable interest that deals with a larger database riddled with experimental errors (Gemund et al. 2001). For this application, the UM method provides a much simpler way than analyzing sequencer traces (Ewing and Green 1998; Ewing et al. 1998; Mott 1998), which are not always available, to identify reliable regions for every EST (work in progress).
The sequencing of the human genome is a continuing project, and we can expect several releases of new versions in the months to come. Although, in a new version, there is a risk that every UM we have determined will lose its uniqueness and there will be new UMs; barring systematic errors in the genomic sequence, the odds of UMs collectively failing the UM method is slim. In addition, little cost is involved in updating the UMs and mapping results with each update of the genomic sequence. Furthermore, with refinement of the genomic sequence, the accuracy and sensitivity of UM-based analysis will be increased. Indeed, one may begin to envisage the use of UMs as a set of genome-wide, high-resolution genetic markers. UMs may also be used as filters and anchors to improve the efficiency of current sequence aligning schemes, thereby rendering the UM method applicable to most sequence analysis problems, provided that the sequence of the genome is available.
METHODS
Databases
The refSNP (reference SNP) sequences clustered by chromosome (ftp://ftp.ncbi.nlm.nih.gov/snp/human/rs_fasta) and the corresponding chromosomal reports (ftp://ftp.ncbi.nlm.nih.gov/snp/human/chr_rpts) of dbSNP (Build 22) were downloaded from NCBI. Excluding the 34,522 entries that have no flanking sequences reported and thus could not be mapped, this dataset contained a total of 1,655,188 nonredundant SNP sequences. The human genome draft sequence in the public domain (International Human Genome Sequencing Consortium 2001) as of July 18, 2001 (Build 22) (ftp://ftp.ncbi.nlm.nih.gov/repository/genomes/H_sapiens/) was used.
Finding UMs on the Human Genome
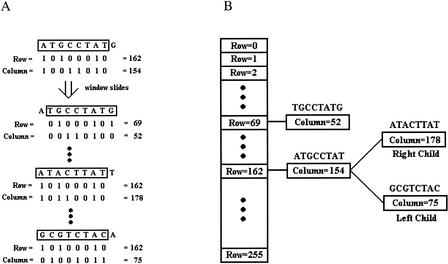
We moved an N-mer sliding window down the human genome draft sequence one base at a time to find all N-mers that occur only once in the genome. For this process, we digitized the genomic sequence with a binary code, using 11 for A, 10 for G, 01 for C, and 00 for T. In addition, the two binary bits were separated such that any N-mer DNA sequence was uniquely represented by two bit strings or two equivalent integers in the form of a row and a column, as illustrated in Figure 5A. Using this binary representation, we can process the DNA sequence using some of the bit operations used in computer science. For example, a left-shift operation adding 1 or 0, depending on the new nucleotide read in, will give the next N-mer DNA (Fig. 5A). Other operations to facilitate rapid searches of, say, complementary sequences should also be possible, although this was not explored in the present study.
Figure 5.
Binary encoding of DNA sequences. (A) Examples of how a DNA sequence, using 8-mers as an example, can be represented by two binary strings (row and column) or their corresponding integers. As an example, ATGCCTAT will be represented by the combination of two integers, 162 and 154, where 162 is the decimal-based value of the binary string 10100010 and 154 is that of 10011010; the two binary strings together encode the sequence ATGCCTAT because A is coded as (1,1), T as (0,0), G as (1,0), and C as (0,1) (see text). The first two sequences in the figure are neighboring 8-mers. (B) The data structure, sorted by row integers, used to store these fixed-length DNA sequences.
The N-mers were then placed in a binary tree (Fig. 5B) as tree nodes, along with their chromosome ID, contig ID, sequence position on the contig, and their occurrence count and links to left child and right child, respectively, for subsequently encountered N-mers with the same row value but a larger or smaller column value (Fig. 5B). By traversing every tree node after the genome scanning was completed, all UMs of length N along with their genomic location were identified; they are the nodes with the occurrence count equal to one. Note that in this process of identifying UMs, BLAST comparison is not required.
To take into account the fact that the orientation of some of the contigs is not yet certain, both strands of the genomic sequence were separately scanned, and the resulting UMs grouped accordingly: groups 1–24 for those occurring on the forward strand of chromosomes 1–24 (23 being the X chromosome and 24 the Y chromosome), group 25 for those on one strand of the contigs that have not yet been assembled into the genome, and group 26–50 for those on the reverse strand of each of the above groups, respectively. Thus, the UMs of group 1 and groups 26 are located on chromosome 1, the UMs of group 2 and group 27 are on chromosome 2, and so on. Our implementation was apparently correct, as the reverse sequence of every UM was also found to be a UM on the reverse strand.
SNP Mapping
We then parsed each SNP sequence to the UM database to record all the UMs contained in every SNP sequence and the position of each UM with reference to the SNP site. Frequently, the UMs found in an SNP sequence were from disparate genomic locations. To uniquely assign an SNP sequence, its UMs were first clustered to group those assigning an SNP to within 5 bp of the same genomic position. The SNP sequence was then assigned to the genomic position of the largest UM cluster if the number of UMs in the second largest cluster did not exceed 30% of that in the largest; otherwise, the assignment of the SNP sequence was termed ‘multiple.’ The choice of 30% was made to achieve the highest percentage of assignment agreement with NCBI under the condition that >90% of NCBI's assignable SNPs can also be assigned with the UM method.
In the comparison with the assignments reported in dbSNP, the 5-bp difference was also used to distinguish between ‘agreed’ and ‘disagreed’ SNPs. The choice of 5 bp was based on the statistical analysis showing that more than 96% of SNPs assigned to the same contig by the UM and NCBI methods were mapped to exactly the same nucleotide and that an additional ∼3% were mapped to within 5 bp (Table 4). Note that it is these base offsets, which are integer numbers and not sequence alignments, that were used to map SNPs using the UM method. Sample inspections indicated that some SNPs should have been assigned as ‘agreed’ but could not be as a result of, for example, a >5-base insertion in the SNP sequence. In addition, some of the SNPs assigned to chromosomes 1–24 were actually given multiple genomic positions in the chromosome report file of dbSNP; these SNPs were considered as ‘agreed’ as long as the UM assignment matched one of the dbSNP assignments by the criteria described above.
Table 4.
Positional Offsets between UM and NCBI Assignments
| Offset (bp) | SNPs | % |
|---|---|---|
| 0 | 11,044,80 | 96.71 |
| 1 | 23,942 | 2.10 |
| 2 | 6,735 | 0.59 |
| 3 | 2,862 | 0.25 |
| 4 | 2,470 | 0.22 |
| 5 | 966 | 0.08 |
| 6 | 264 | 0.02 |
| 7 | 88 | 0.01 |
| 8 | 118 | 0.01 |
| 9 | 39 | 0.00 |
| ≥10 | 47 | 0.00 |
The results shown include only those SNPs that were assigned to the same contig by both methods.
WEB SITE REFERENCES
http://snp.cshl.org; The SNP (Single Nucleotide Polymorphism) Consortium web site.
http://ftp.genome.washington.edu/RM/RepeatMasker.html; RepeatMasker web site.
http://blast.wustl.edu; Washington University Blast archives.
http://www.ncbi.nlm.nih.gov/SNP/); SNP database at National Center for Biotechnology Information.
Acknowledgments
We thank Richie Gan for assistance in setting up the computer system. We are grateful to the research freedom enabled by the funding from Taiwan's Academia Sinica, National Science Council, and National Health Research Institutes.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL mjhwang@ibms.sinica.edu.tw; FAX (886) 2-2788-7641.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.224502. Article published online before print in June 2002.
REFERENCES
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Altshuler D, Pollara VJ, Cowles CR, Van Etten WJ, Baldwin J, Linton L, Lander ES. An SNP map of the human genome generated by reduced representation shotgun sequencing. Nature. 2000;407:513–516. doi: 10.1038/35035083. [DOI] [PubMed] [Google Scholar]
- Bedell JA, Korf I, Gish W. MaskerAid: A performance enhancement to RepeatMasker. Bioinformatics. 2000;16:1040–1041. doi: 10.1093/bioinformatics/16.11.1040. [DOI] [PubMed] [Google Scholar]
- Chao KM, Zhang J, Ostell J, Miller W. A tool for aligning very similar DNA sequences. Comput Appl Biosci. 1997;13:75–80. doi: 10.1093/bioinformatics/13.1.75. [DOI] [PubMed] [Google Scholar]
- Collins FS, Brooks LD, Chakravarti A. A DNA polymorphism discovery resource for research on human genetic variation. Genome Res. 1998;8:1229–1231. doi: 10.1101/gr.8.12.1229. [DOI] [PubMed] [Google Scholar]
- Delcher AL, Kasif S, Fleischmann RD, Peterson J, White O, Salzberg SL. Alignment of whole genomes. Nucleic Acids Res. 1999;27:2369–2376. doi: 10.1093/nar/27.11.2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewing B, Green P. Base-calling of automated sequencer traces using Phred. II. Error probabilities. Genome Res. 1998;8:186–194. [PubMed] [Google Scholar]
- Ewing B, Hillier L, Wendl MC, Green P. Base-calling of automated sequencer traces using Phred. I. Accuracy assessment. Genome Res. 1998;8:175–185. doi: 10.1101/gr.8.3.175. [DOI] [PubMed] [Google Scholar]
- Fleming CC, Halle BV. Tune by Adding Index. In: In: Wollman K, editor. Handbook of relational database design. Reading, MA: Addison-Wesley; 1989. pp. 401–403. [Google Scholar]
- Florea L, Hartzell G, Zhang Z, Rubin GM, Miller W. A computer program for aligning a cDNA sequence with a genomic DNA sequence. Genome Res. 1998;8:967–974. doi: 10.1101/gr.8.9.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gemund C, Ramu C, Altenberg-Greulich B, Gibson TJ. Gene2EST: A BLAST2 server for searching expressed sequence tag EST databases with eukaryotic gene-sized queries. Nucleic Acids Res. 2001;29:1272–1277. doi: 10.1093/nar/29.6.1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Human Genome Sequencing Consortium. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Kruglyak L. The use of a genetic map of biallelic markers in linkage studies. Nat Genet. 1997;17:21–24. doi: 10.1038/ng0997-21. [DOI] [PubMed] [Google Scholar]
- Lander ES. The new genomics: Global views of biology. Science. 1996;274:536–539. doi: 10.1126/science.274.5287.536. [DOI] [PubMed] [Google Scholar]
- Lander ES, Schork NJ. Genetic dissection of complex traits. Science. 1994;265:2037–2048. doi: 10.1126/science.8091226. [DOI] [PubMed] [Google Scholar]
- Marth G, Yeh R, Minton M, Donaldson R, Li Q, Duan S, Davenport R, Miller RD, Kwok PY. Single-nucleotide polymorphisms in the public domain: How useful are they? Nat Genet. 2001;27:371–372. doi: 10.1038/86864. [DOI] [PubMed] [Google Scholar]
- Marth GT, Korf I, Yandell MD, Yeh RT, Gu Z, Zakeri H, Stitziel NO, Hillier L, Kwok PY, Gish WR. A general approach to single-nucleotide polymorphism discovery. Nat Genet. 1999;23:452–456. doi: 10.1038/70570. [DOI] [PubMed] [Google Scholar]
- Mott R. Trace alignment and some of its applications. Bioinformatics. 1998;14:92–97. doi: 10.1093/bioinformatics/14.1.92. [DOI] [PubMed] [Google Scholar]
- Mullikin JC, Hunt SE, Cole CG, Mortimore BJ, Rice CM, Burton J, Matthews LH, Pavitt R, Plumb RW, Sims SK, et al. An SNP map of human chromosome 22. Nature. 2000;407:516–520. doi: 10.1038/35035089. [DOI] [PubMed] [Google Scholar]
- Risch N, Merikangas K. The future of genetic studies of complex human diseases. Science. 1996;273:1516–1517. doi: 10.1126/science.273.5281.1516. [DOI] [PubMed] [Google Scholar]
- Schafer AJ, Hawkins JR. DNA variation and the future of human genetics. Nat Biotech. 1998;16:33–39. doi: 10.1038/nbt0198-33. [DOI] [PubMed] [Google Scholar]
- Taillon-Miller P, Gu ZJ, Li Q, Hillier L, Kwok PY. Overlapping genomic sequences: A treasure trove of single-nucleotide polymorphisms. Genome Res. 1998;8:748–754. doi: 10.1101/gr.8.7.748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The International SNP Map Working Group. A map of human genome sequence variation containing 1.42 million single nucleotide polymorphisms. Nature. 2001;409:928–933. doi: 10.1038/35057149. [DOI] [PubMed] [Google Scholar]
- Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO, Yandell M, Evans CA, Holt RA, et al. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- Wang DG, Fan JB, Siao CJ, Berno A, Young P, Sapolsky R, Ghandour G, Perkins N, Winchester E, Spencer J, et al. Large-scale identification, mapping, and genotyping of single-nucleotide polymorphisms in the human genome. Science. 1998;280:1077–1082. doi: 10.1126/science.280.5366.1077. [DOI] [PubMed] [Google Scholar]
- Zhang J, Madden TL. PowerBLAST: A new network BLAST application for interactive or automated sequence analysis and annotation. Genome Res. 1997;7:649–656. doi: 10.1101/gr.7.6.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Schwartz S, Wagner L, Miller W. A greedy algorithm for aligning DNA sequences. J Comput Biol. 2000;7:203–214. doi: 10.1089/10665270050081478. [DOI] [PubMed] [Google Scholar]