Abstract
Here we describe a high-throughput screen to isolate transcripts with spatially restricted patterns of expression in early embryos. Our approach utilizes robotic automation for rapid analysis of sequence-selected cDNAs in a whole-mount in situ hybridization assay. We determined the spatial distribution of a random collection of 778 different genes from an embryonic cDNA library and show that a significant fraction of these exhibit patterned profiles of expression. In addition, gene ontology studies revealed groups of gene products exhibiting shared expression patterns, providing new insights into the largely overlooked effector molecules that function in development. As described in this paper, automated hybridization to whole-mount embryos in situ proved to be straightforward and provided us with a very powerful method for the global survey of gene expression in early embryos. From the perspective of biological significance, our finding that many spatially restricted transcripts correspond to loci encoding novel transcripts that have not been previously identified in nearly saturating genetic screens for maternal effect and zygotic lethals is particularly notable.
[Supplementary material available online at http://www.genome.org. The following individuals kindly provided reagents, samples, or unpublished information as indicated in the paper: N. Brown]
Pattern formation represents an initial event in the development of multicellular organisms. Subsequent organismal complexity is generated from translation of the early embryonic pattern into differentiated cell types and tissues. Our current understanding of the molecular mechanisms of patterning in Drosophila embryogenesis derives in large part from an analysis of genes corresponding to embryonic lethal mutants that display defects in larval cuticular morphology (Jurgens et al. 1984; Nusslein-Volhard et al. 1984; Wieschaus et al. 1984; Schupbach and Wieschaus 1989). Molecular characterization of genes essential for embryonic patterning has revealed that these genes code almost exclusively for components of signal transduction cascades and their associated transcription factors. Taken together, molecular and genetic studies have furnished us with a comprehensive understanding of how patterned transcription can arise in Drosophila embryogenesis. From a developmental perspective, the importance of patterned transcription is clear: Placement of gene products in a subset of embryonic cells causes these cells to assume fates different from their nonexpressing neighbors.
In contrast to our comprehensive understanding of how embryonic patterns are established in Drosophila, our understanding of the molecular mechanisms that are employed in translating pattern into differentiated tissues and cell types is more limited. It is notable that many of the transcriptionally regulated targets of the pattern-establishing signaling cascades have escaped detection using standard methods of genetic surveillance. It has been suggested that patterned expression of single genes belonging to subgroups that are expressed in overlapping domains may promote establishment of the final differentiated state without functioning as an absolute determinant of differentiation (Wieschaus 1996). Effects of mutation in genes such as these are expected to be subtle or transient.
As a complement to classical genetic screens, reverse genetic approaches have been exploited to identify a broader assortment of developmentally important loci in Drosophila. By definition, the output of a reverse genetic screen is not limited by phenotype(s). More importantly, the demonstration that gene expression patterns often presage an essential function for the corresponding gene product within the spatially restricted domain of gene expression validates the use of reverse genetics for the identification of developmentally regulated gene products. Genes essential for both pattern establishment and differentiation might be spatially and/or temporally regulated and therefore will be recovered in expression screens.
Enhancer detection screens have traditionally represented the reverse genetic method of choice in Drosophila (O'Kane and Gehring 1987; Bellen et al. 1989; Bier et al. 1989; Torok et al. 1993). The large collections of P-element enhancer detection insertions that are now widely available have had a tremendous impact on all aspects of Drosophila biology. In particular, P-element enhancer detection lines have facilitated identification and characterization of numerous developmentally regulated genes and cell-specific markers in Drosophila. There are, however, some caveats associated with enhancer detection as a reverse genetic screening method. These studies require a substantial investment of personnel and resources in the generation of founder fly lines, the identification of P-element enhancer detection carriers, and the maintenance of fly lines. In addition, gene identification in P-element enhancer detection lines can be difficult and oftentimes impossible because many P-element enhancer detection fly lines harbor multiple P-element insertions. Finally, anecdotal evidence suggests that expression of the reporter gene accurately reflects the expression of a nearby gene in as few as 50% of the enhancer detection lines.
An alternative approach for identifying genes with spatially or temporally regulated expression during embryonic development is the analysis of mRNA expression patterns in situ using randomly isolated cDNAs as hybridization probes (Gawantka et al. 1998; Kopczynski et al. 1998; Liang and Biggin 1998; Kudoh et al. 2001). This approach has three important advantages over traditional enhancer detection screens: (1) Reagent stocks are cDNAs, thus eliminating the labor and cost associated with maintaining thousands of fly lines; (2) probes correspond to individual cDNAs, thus leading to immediate gene identification; and (3) expression patterns correspond to endogenous mRNAs, thus establishing physiological relevance. Until now, fundamental difficulties associated with the availability and processing of large numbers of sequence-verified cDNAs have limited the utility of expression studies such as these. We have employed our own sequencing studies and the Drosophila genome project (Adams et al. 2000) as an immediate solution to the first of these problems and a robotic screening method to provide a solution to the second.
In this paper, we describe a high-throughput screen for the identification of spatially restricted transcripts. Exploiting robotic automation, we analyzed 778 sequence-selected genes from a 0- to 4-h embryonic cDNA library by hybridization in situ. As expected, we identified several previously characterized genes with documented roles in patterning of the early Drosophila embryo. More exciting was our discovery that many novel and uncharacterized genes exhibit spatially restricted patterns of expression in developing embryos, suggestive of their special roles as determinants and/or markers of tissue-specific fates. Taken together, our studies represent a first step in an important phase in translational genomic studies—positioning new gene products in already characterized embryonic patterning pathways.
RESULTS AND DISCUSSION
Sequence Analyses
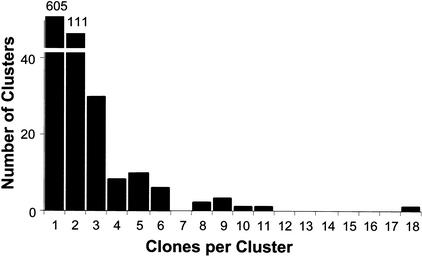
We sequenced the 5′ termini of 1117 randomly selected cDNAs from a 0- to 4-h embryonic library. Based on similarity searches against themselves and publicly available databases, the cDNAs condensed to a nonoverlapping set of 778 different cDNAs (Fig. 1); most of these are full length with respect to the presence of the defined ATG initiating codon. The majority of genes in our original collection (605 genes; 78%) were represented by a single clone. In contrast, the gene coding for heat shock cognate protein 70 (Hsc4), a chaperone protein, was represented by 18 cDNAs. Other abundant genes coded for a mitochondrial cytochrome-c oxidase subunit (n = 11), a chymotrypsin-like serine protease (n = 10), and the arrest (aret) gene product (n = 9). Several genes encoding ribosomal proteins L22 (RpL22) and L7A (RpL7a), stubarista (sta), and Qm were also represented by multiple cDNAs (n = 8 or 9). Our 778-member unigene set represents 6% of all predicted Drosophila genes (Adams et al. 2000) and a significant fraction (13%) of the Berkeley Drosophila Genome Project (BDGP) 5849-member unigene collection.
Figure 1.
Nonoverlapping ESTs. The ESTs were assembled into clusters of singletons (605) and contigs (174). The number of clusters (Y-axis) is plotted versus the number of clones in each cluster (X-axis). EST clusters containing at least eight clones included those coding for heat shock protein cognate 70 (18), mitochondrial cytochrome-c oxidase subunit III (11), chymotrypsin-like serine protease (10), Arrest (9), RpL22 (9), RpL7A (9), Qm (8), and Stubarista (8).
We compared the 5′ sequences of the 778 cDNA set to Drosophila nucleotide databases made available by BDGP using the BLASTN search algorithm (v. 2.0; Altschul et al. 1990). The searched databases included curated genes, repetitive elements, and transposable elements. We found 682 cDNAs (88%) showing close matches to defined Drosophila ESTs. Our cutoff value (E values < 10−5) was determined empirically to include sequences with short overlaps but perfect matches. Five transcripts (<1%) represented repetitive or transposable elements.
In an attempt to assign identities to the remaining 91 sequences, we used the BLASTX algorithm to search the nonredundant database at the National Center for Biotechnology Information (NCBI). A 5-member subset of the unassigned sequences matched ESTs not yet described in Drosophila but found in other organisms: worm (3 clones; E < 10−10), plant (1 clone; E = 10−13), and rat (1 clone; E = 10−16). However, most of our unassigned cDNA sequences (n = 86, corresponding to 11% of our unigene set) are not included in any of the NCBI EST databases nor do they match predicted fly genes. The majority of these (n = 73) correspond to transcriptions we can detect in hybridizations to whole mount embryos in situ. While the remaining 13 clones may represent artifacts particular to EST analyses, including splicing intermediates, genomic contamination, or illegitimate transcription from intergenic regions, at least one other report suggests that the estimate of 13,601 Drosophila genes (Adams et al. 2000) is too conservative (Andrews et al. 2000). In an analysis of 1560 cDNAs derived from Drosophila testes, 47% failed to align with existing ESTs and a subset of these, consisting of 16%, failed to align with predicted genes in the current genome release. Our results support the conclusion that some genes have been overlooked in other EST collections (Rubin et al. 2000), perhaps due to their temporally or spatially restricted patterns of expression.
Gene Ontology Determinations
After assigning gene identities, we used the reported gene ontologies (BDGP, Celera, and Gene Ontology Consortium [GO; http://genome-www.stanford.edu/GO] ) to assign predicted functions to 423 of the 778 cDNAs in our unigene set (Table 1). Consistent with results from whole-genome sequencing studies (Adams et al. 2000), a significant fraction of the genes in our unigene set (355 genes; 46%) reveal no significant functional homologies. In general, the distribution of gene functions expressed 0–4 h after egg lay (AEL) is similar to that observed in the entire fly genome. There are, however, a few notable exceptions. Using Fisher's exact test (two-sided), we identified nine functional classes that are represented at significantly different levels (P < 0.02) in the cDNA and genomic samples.
Table 1.
Relative Representation of Gene Functions in the Genome and Early Embryos
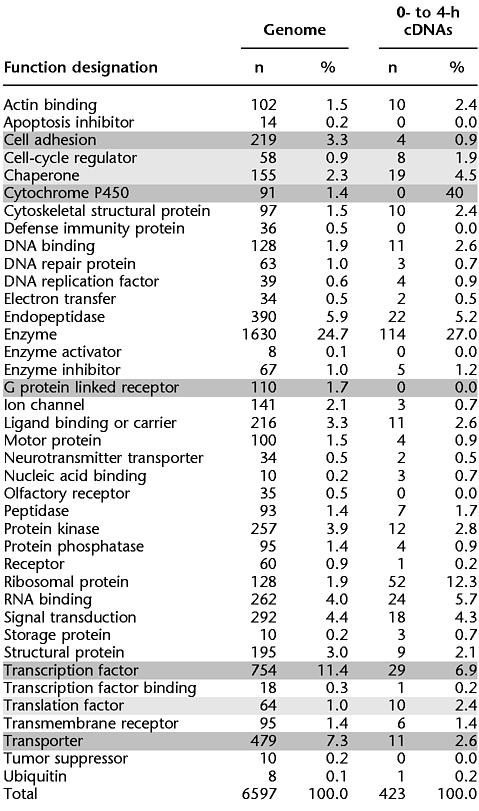 |
cDNA gene ontologies underrepresented in comparison to the genome are darkly shaded; overrepresented cDNA gene ontologies are lightly shaded.
Four functions are overrepresented in the 0- to 4-h library relative to the whole genome. These include ribosomal proteins (sixfold enriched; P < 10−9), protein chaperones (twofold enriched; P = 0.01), translation factors (twofold enriched; P = 0.01), and cell-cycle regulators (twofold enriched; P = 0.02). Enrichment of these proteins in early embryos is consistent with previous reports (Santon and Pellegrini 1980) and likely reflects the fact that the early embryo is poised for protein production and synchronous mitotic cell cycles.
At the other end of the spectrum, five functional classes are underrepresented in the 0- to 4-h library relative to the whole genome. These include G-protein-linked receptors (at least ninefold reduced; P = 0.002), cytochrome P450s (at least sevenfold reduced; P = 0.006), cell-adhesion molecules (threefold reduced; P = 0.004), transporters (threefold reduced; P < 0.0001), and transcription factors (twofold reduced; P = 0.007). The fact that the early Drosophila blastoderm is a syncitium, the nuclei of which are not completely cellularized until 3 h AEL, may account for the observed underrepresentation of these protein classes in the earliest hours of embryogenesis. This delay in transcription and/or maternal loading of transcripts encoding proteins not required until cellularization may reflect an efficient utilization of limited metabolic resources.
Gene Expression Studies
The 778-member unigene set was re-arrayed into nine 96-well plates, with a single clone representing each gene, and we determined the spatial expression pattern of each gene in the unigene set by RNA hybridization to whole-mount embryos in situ (Fig. 2). Because most genes are expressed in dynamic patterns throughout early embryonic development, we often recorded several staining patterns for individual genes. Only patterns observed in multiple similarly staged embryos were recorded.
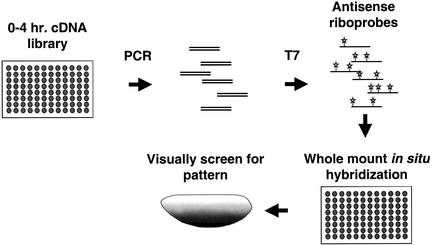
Figure 2.
Profiling Patterned Transcripts in Drosophila. Bacterial cultures of individual clones from a 0 to 4 h cDNA library were used to seed PCR reactions driven by oligonucleotides that are complementary to sequences that flank the cDNA insertion site. The cDNA products were then purified on glass fiber filters in a 96-well format. This DNA was used to program transcription from a T7 promoter that resides just 3′ to the cDNA insert. Digoxigenin-UTP was included to label the antisense RNA probes produced. These probes were used subsequently in hybridizations to whole mount embryos in situ. The embryos were then visually screened using standard light microscopy to identify patterns of expression. All steps of the process were performed using a robotics workstation. The antisense RNA probes can be stored for extended periods of time and provide a reusable resource since only a small fraction of the probe is needed to subsequent steps. The slowest step in the process was the hybridization in situ, which took nearly 16 hours to complete. However, due to extended incubations, four plates could be staggered through the process simultaneously, yielding an effective throughput of 384 clones for each overnight run.
A significant fraction (118 genes; 15%) of the randomly selected genes included in this study exhibit spatially restricted patterns of expression in early embryogenesis, suggestive of their special roles in development (Table 2). Furthermore, many of these genes manifest related patterns of expression, suggestive of their shared gene regulation and/or their functional symbiosis. Several patterned transcripts (22 genes; 3%) were observed in blastoderm stages. Many more, however, were confined during gastrulation to the developing germ layers, either along the anterioposterior (segment polarity class: 10 genes; 1%) or dorsoventral (germ layer restricted: 64 genes; 8%) axes, and subsequently to their derivatives, which include myoblasts, neuroblasts, and tracheal pits (germ layer derivative: 48 genes; 6%).
Table 2.
Gene Expression Patterns
| Pattern | Stage | n | % |
|---|---|---|---|
| Dorsal-ventral | 0–8 | 7 | 5% |
| Anterior-posterior | 0–8 | 5 | 3% |
| Pole cell | 0–8 | 8 | 5% |
| Gap | 0–8 | 1 | <1% |
| Pair-rule | 0–8 | 1 | <1% |
| Segment polarity | 9–11 | 10 | 7% |
| Germ layer restricted | 9–11 | 64 | 44% |
| Germ layer derivative | 9–11 | 48 | 33% |
| Total patterns scored | all stages | 144† |
(118 different cDNAs).
To get a broader picture of the role of patterned transcription in embryogenesis, we considered our blastoderm and dorsoventral (DV) patterning data within the context of a previously published screen, similar in scale to the one described here (Kopczynski et al. 1998). In this earlier screen of 2518 secreted and transmembrane embryonic cDNAs, just 14 (0.6%) exhibited blastoderm patterns, and only one of these (0.04%) was restricted along the DV axis. The three- to fivefold differences in recovery of cDNAs restricted in blastoderm stages and along the DV axis in Kopczynski's and our screen was foreseen because the secreted and transmembrane molecules selected for study in the former screen are not expected to function at precellular stages of embryogenesis. Assuming that the Drosophila genome codes for ∼5000 different blastoderm-stage transcripts, our results suggest that ∼40 mRNAs are spatially restricted along the dorsoventral axis at this stage of embryogenesis; fewer than a dozen of these have been identified in screens for embryonic lethal mutations affecting larval cuticle pattern.
In another study of patterned gene expression in Drosophila, Liang and Biggin (1998) documented 199 cDNA expression patterns. These investigators found that 100% (n = 16) of cDNAs from a 0 to 4 hour library were distributed uniformly in the earliest stages of development, consistent with their definition as maternal transcripts. In contrast, a significant fraction of these (31%) were patterned in later developmental stages (stages 5 to 10). Transcripts from a 4–8 h library (n = 84) and from an 8 to 12 hour library (n = 99) exhibited a similar patterning profile: most were relatively uniform in precellular stages (80% and 71% respectively), but patterned in later stage embryos (74% and 88%, respectively). Although we too observed a shift to patterned transcription as development proceeds, overall we found relatively fewer patterned transcripts in our screen of 0 to 4 cDNAs (15% compared to 31%). In fact, this two-fold difference is not significant because Liang and Biggin examined only 16 cDNAs from the 0 to 4 hour library. Moreover, this two-fold difference in estimates for patterned transcription in early embryos likely reflects the different approaches we brought to our studies: Liang and Biggin focused on segmental patterning in later stage embryos than those which we considered here. Put another way, our study represents a snapshot of early development—in particular, the molecular prepatterns that precede segmentation.
Parallel Analyses of Gene Sequence and Gene Expression
In contrast to previous studies of fruit fly gene expression in which gene identity was unknown, the in situ hybridization studies described here exploited a set of sequenced gene probes. By integrating our sequence and expression data, we found that transcripts exhibiting patterned expression correspond to well-characterized developmental regulators, as well as to genes not yet described in the literature.
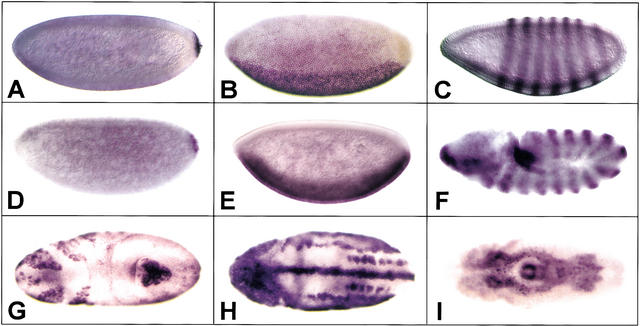
The parallel analysis of gene sequence and gene expression revealed the quality of our automated expression studies to be quite high. In this regard, our unigene set includes cDNAs with spatial patterns of expression that have been documented previously, and our independent identification of these cDNAs validated the effectiveness of automated hybridization in situ as a screening method. We correctly identified blastoderm-stage patterned cDNAs included in our unigene set: Our expression screen yielded oskar and cyclin B (Lehmann and Nusslein-Volhard 1986; Whitfield et al. 1989) as posterior-pole-restricted transcripts, neuralized and delta as dorsoventrally restricted transcripts (Haenlin et al. 1990; Boulianne et al. 1993), and odd-skipped as a pair-rule transcript (Fig. 3A–C; Coulter et al. 1990). In no case did a gene that is known to be expressed in blastoderm-stage embryos fail to give the expected pattern.
Figure 3.
Patterned transcription. Hybridization of cDNA probes to whole-mount embryos in situ reveals patterned expression in staged embryos. (A) cyclin B; (B) neuralized; (C) odd-skipped; (D–I) novel transcripts. Lateral views of embryos are shown in A–F, with dorsal up and anterior to the left. In G–I; ventral views are shown, with anterior to the left.
More exciting from a biological perspective was our identification of novel and/or genetically uncharacterized transcripts displaying analogous patterns of transcription in early-stage embryos (e.g., Fig. 3D–F). Overall, our survey of gene expression revealed a wide range of predicted functions encoded by transcripts asymmetrically distributed along the DV and AP (anterioposterior) axes in stage 0–8 embryos. These transcripts coded for three enzymes, two RNA-binding proteins, and three unknown proteins, as well as for single isolates of a ligand-binding or carrier protein, a transcription factor, a DNA-binding protein, a DNA replication factor, and a ribosomal protein. Pole-cell-restricted transcripts coded for proteins with a similarly broad range of predicted functions, including two chaperones, a cell-cycle regulator, a DNA-binding protein, and a serine threonine kinase. Two transcripts coding for proteins with unknown functions were also localized to pole cells. Collectively, the genes isolated in this expression screen afford a striking contrast to those identified in earlier genetic screens. Whereas genes coding for signal transducers represented a significant fraction of loci identified in screens for patterning mutants, our screen for patterned expression revealed a very different repertoire of gene products. In blastoderm-stage embryos, patterned transcripts correspond in large part to specific components of protein complexes dedicated to transcription and protein biosynthesis.
Transcripts exhibiting segmental restriction in germ-band extended-stage embryos (as in Fig. 3F) showed a somewhat tighter clustering of functions than those identified in earlier embryos. Patterned transcripts in this subgroup fell predominantly into two major protein classes: proteins involved in gene regulation and proteins involved in cell–cell communication. We identified multiple transcription factors (3), cell-adhesion proteins (2), and transporters (2), in addition to single isolates belonging to the chaperone, cyclin-dependent protein kinase inhibitor and RNA-binding protein classes.
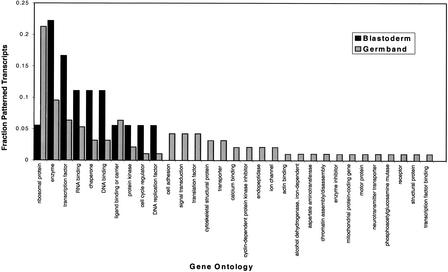
Patterned transcripts emerging from our studies of stage 9–11 embryos exhibited the broadest range of tissue specificities and functions. As examples of patterns, we identified novel markers of mitotic domains (Fig. 3G), the nervous system (Fig. 3H), and gut (Fig. 3J). Where gene identities could be established (Fig. 4), we found that in addition to the 10 functional classes correlated with patterned transcription in blastoderm-stage embryos, 21 additional functions corresponded to patterned transcripts in the later and more complex stages of embryogenesis included in this study. Our observation that patterned transcript complexity increases as development progresses from maternal cues to visible regional and cellular differences is consistent with previous studies (Liang and Biggin 1998) and with long-held views of evolution and development. The tenet of these models—that organismal complexity mirrors molecular complexity—has not withstood the tests of genome sequencing studies; for example, the human genome is only about twice as complex as that of the fruit fly. An amended view, contending that a higher proportion of the genome's coding potential is expressed in more complex organisms, is supported, however, by the data we present here. With particular respect to embryonic development and genetic hierarchies functioning in the fruit fly, we suggest that the patterned transcripts documented here represent markers of regional and cellular differentiation. Accordingly, these gene products correspond to the direct and indirect transcriptionally regulated targets of maternally defined signaling cascades that function to establish pattern at the time of egg lay.
Figure 4.
Gene ontology of patterned transcripts. Patterned transcripts were grouped by developmental stage, either blastoderm (stages 0–8) or germ band (stages 9–11). Predicted transcript functions were determined based on reported gene ontologies (GO; http:/genome-www.stanford.edu/GO). The fraction of patterned transcripts with a predicted function is shown for each developmental stage.
Intriguingly, during the latest stages examined, 21% (n = 10) of patterned transcripts were encoded by ribosomal genes. A handful of ribosomal proteins have been implicated in extraribosomal functions that affect development (Wool 1996 and references therein). However, given the more general role of ribosomes in protein production, it is likely that the patterned transcripts we observed represent de facto markers of proliferating cells. Translating a patterning signal into a cell biological “decision” to divide or arrest has significant implications for morphogenesis (Duronio 1999). An alternative and perhaps more parsimonious explanation of the patterned expression of ribosomal protein transcripts is that an expressing cell may simply have a higher capacity for protein production than its nonexpressing neighbor. Consequently, the two cells possess different capacities to respond to subsequent intrinsic or extrinsic signals.
Overall, robotic methods for transcript detection are robust. We were unable to detect transcripts corresponding to only 36 cDNAs (5% of our unigene set) in hybridizations to whole-mount embryos in situ. Because genes with previously defined embryonic expression patterns are not represented in this subset, we propose that the non-cross-hybridizing cDNAs correspond to early embryonic transcripts that are expressed at levels below the sensitivity threshold of our assay. Consistent with this postulate is our demonstration that none of these cDNAs correspond to highly expressed genes (e.g., those encoding housekeeping function). In fact, many of these cDNAs (n = 13, corresponding to 38%) code for proteins with unassigned identities, prompting us to speculate that little is known about the rarest transcripts functioning in Drosophila development.
Finally, and as expected for classification of cDNAs derived from 0- to 4-h embryos, a large fraction (n = 681; 88%) correspond to maternal transcripts that are distributed uniformly in precellular blastoderm stage embryos. Many of these transcripts (n = 103) show refined patterns of expression later in development; others (n = 516) continue to be expressed ubiquitously. The maternally restricted transcripts that we identified (n = 62) correspond to ∼8% of cDNAs in our unigene set. Because we did not assay expression after stage 11 of embryogenesis, this estimate likely represents an upper limit for the cDNA fraction that corresponds to strictly maternal transcripts. Indeed, our observation that many of the cDNAs we identified as maternally restricted have documented postembryonic developmental roles (e.g., larval cuticle protein IV precursor [Lcp4] and topoisomerase 2 [Top2]) is consistent with this idea. Only four of the maternally restricted transcripts correspond to novel cDNAs. The majority of the 58 previously characterized transcripts code for proteins with well-documented roles in DNA replication and protein biosynthesis and homeostasis.
Summary
Automated hybridization to whole-mount embryos in situ provides a powerful method for the global survey of gene expression in early embryos. Sequence data sets, described here and elsewhere (Adams et al. 2000), coupled with long-established genetic databases (FlyBase 1999) proved invaluable in extending expression screening to a mechanistic level. In many cases, we were able to assign molecular and/or genetic functions to patterned transcripts. More notable was our finding that many spatially restricted transcripts correspond to loci-encoding novel transcripts that have not previously been identified in nearly saturating screens for maternal effect and zygotic lethals. This discovery has two important implications. First, developmental processes in Drosophila may be dependent upon redundant gene functions. Second, developmental processes in Drosophila may be dependent upon signaling pathways that branch at their terminal stages so that mutations in target genes (1) do not mutate to lethality or (2) do not produce defects strong enough to recognize using well-characterized anatomical markers. Clearly, genetic methods have been useful for identifying many developmentally important molecules. However, as we move to refining our underestimate of downstream targets of genetically defined pathways, there is a need for assays that will identify those target genes with redundant or overlapping gene functions. We show here that automated RNA expression screening provides a powerful and rapid method for the identification of patterned transcripts. In light of published estimates that for flies and other animals, less than one-third of genes lead to obvious phenotypes when mutated (Caenorhabditis elegans Sequencing Consortium 1998; Thatcher et al. 1998; Ashburner et al. 1999), parallel studies of gene sequence and gene expression such as the one described here complement earlier genetic studies in yielding new sets of genes with symbiotic roles in development.
METHODS
cDNA Library
Because we required a high-complexity cDNA library to examine low-abundance transcripts, Dr. Nick Brown, Wellcome Trust and Cancer Research Institute generously supplied us with aliquots of DNA from the initial plating of a high-quality 0- to 4-h embryonic plasmid library (Brown and Kafatos 1988). After bacterial transformation, ampicillin-resistant colonies were picked randomly and used to generate single-clone cultures in 96-well microtiter plates. After growth overnight at 37°C, glycerol was added to a final concentration of 15% and cultures were stored at −80°C.
Sequencing
Plasmid templates for sequencing were generated by the alkaline lysis method in 96-well microtiter plates. Clones lacking inserts were identified by double digestion with NotI and HindIII restriction enzymes and analysis by agarose gel electrophoresis; these clones were excluded from further analysis. cDNAs were sequenced from their 5′ ends using standard dye primer chemistry (ABI, Perkin Elmer) and an ABI Prism 377 sequencer (ABI, Perkin Elmer).
Sequence Data Analysis
Sequences were compiled and clustered using the Gap4 package for sequence analysis (Bonfield et al. 1995). High-quality reads were trimmed manually. Drosophila nucleotide sequence databases gadfly.dros, predictedCDS.dros, te.dros, and re.dros were downloaded from the Berkeley Drosophila Genome Project web site (http://www.fruitfly.org/) and searched locally using the BLASTN algorithm (v. 2.0; Altschul et al. 1990). Additional databases used for our analyses included the NCBI, the gene ontologies database (Ashburner et al. 2000), and the euGenes database (http://iubio.bio.indiana.edu:8089/).
Digoxigenin-Labeled Antisense RNA Probes
Templates for transcription in vitro were generated robotically: cDNA inserts were amplified from bacterial cultures diluted 1:50 in ddH2O by the polymerase chain reaction (PCR). Primers, NB1 (5′CGTTAGAACGCGGCTACAAT3′) and NB2 (5′TGCAGCTGGCTTATCGAAAT 3′), specific for the pNB40 vector (Brown and Kafatos 1988) were designed to amplify cDNA inserts and include the vector's T7 RNA polymerase promoter, which is adjacent to the insert's 3′ terminus. Reaction products were purified using glass fiber multiscreen plates (Millipore MAFB NOB). Briefly, 100 μL 5.3 M guanidium-HCl at pH 4.8 was added to the PCR reaction, and the reaction was transferred to the glass fiber plate, prewetted with the same guanidium-HCl solution. Fluid was removed by vacuum manifold, and the glass fiber plates were washed three times in 80% ethanol. After the final wash, residual fluid was removed by centrifugation at 750 g for 10 min. PCR reaction products were eluted in 30 μL Tris-HCl at pH 7.5 by centrifugation at 750 g for 15 min.
Antisense probes labeled with digoxigenin-UTP were generated using 5 μL of the amplified reaction product as template in a 10-μL T7 in vitro transcription reaction (Boehringer Mannheim). The addition of 2.5 μL 4 M LiCl terminated transcription. RNA was precipitated by the addition of 75 μL of 100% ethanol and centrifugation at 750 g for 30 min. A 40× probe stock was generated by resuspending the probe pellet in 100 μL hybridization buffer. Excess probe was stored at −80°C.
Automated Whole-Mount RNA in situ Hybridization
We adapted a well-established protocol for RNA in situ hybridization to whole-mount Drosophila embryos (Tautz and Pfeifle 1989) for automation and processing in 96-well microtiter plates. For automation, we employed an integrated system consisting of two robotic workstations, a Beckman Biomek 2000 and a Hewlett-Packard ORCA. In the integrated robotic system, an articulated arm with plate grippers (ORCA) moves plates about the workstation along a 2-m rail, whereas a pipetting workstation (Biomek 2000) dispenses and aspirates liquids. Additional components of the workstation essential for these assays included an incubator, a plate shaker, a multivalve peristaltic pump, and a house vacuum. Detailed descriptions of the equipment and its uses can be found at http://metherall.genetics.utah.edu.
Embryos (n ∼ 150), probes, and hybridization buffer were aliquotted manually into 96-well microtiter plates and incubated for 6 h at 56°C. All subsequent steps were performed robotically. Solution exchange was achieved by removing 50% of the fluid volume from each well by vacuum aspiration and dispensing fresh solution via a peristaltic pump attached to a multichannel pipettor. After each solution exchange, the plate was moved to an orbital shaker and gently agitated for 30 sec. Following probe removal, embryos were washed in hybridization buffer at 56°C for 3 h, with wash buffer exchanges every 20 min. Subsequent rinses in phosphate buffered saline 1% TritonX-100 (PBT) were repeated 13 times. Embryos were then incubated for 1 h in alkaline phosphatase conjugated anti-DIG antibody (Boehringer Mannheim) diluted 1:2000, followed by 13 rinse cycles in PBT. For detection, processed embryos were incubated with chromagenic substrates 5-bromo-4-chloro-3-indolyl phosphate (BCIP) and nitroblue tetrazolium (NBT) for 1 h at 22°C. The color reaction was terminated with a series of 13 PBT washes. Embryos were cleared for 24 h in 40% glycerol, 1 mM EDTA. The protocol for a single 96-well plate was completed in an unattended 16-h overnight run.
Scoring and Imaging
Stained embryos were mounted on glass slides, cured overnight, and then scored using stereomicroscopy. An antisense probe corresponding to zerknüllt (zen) was included as a positive control on each plate. Embryos were classified by developmental stage (Campos-Ortega and Hartenstein 1997): pre-blastoderm and blastoderm (stages 0–5), early gastrula (stages 6–8), and germ band extended (stages 9–11). Expression patterns were scored by at least two independent investigators and without prior knowledge of cDNA sequence identities. Images were captured on an Axiophot (Zeiss) microscope using standard 35mm film or a Sensys digital camera (Photometrics) and a PowerMac G3 computer (Apple).
WEB SITE REFERENCES
http:/genome-www.stanford.edu/GO; Reported gene ontologies.
http://iubio.bio.indiana.edu:8089/; Web stie for the euGenes database.
http://metherall.genetics.utah.edu; Detailed descriptions of robotic workstations.
http://www.fruitfly.org/; Drosophila Genome Project web site, Berkeley, CA.
Acknowledgments
We thank members of our laboratories for valuable discussions; Susan Mango and Tom Vogt for critical reading of the manuscript; Nick Brown (Wellcome Trust and Cancer Research Institute, Cambridge, England) for the generous gift of the 0- to 4-h cDNA library; and Ryan Stokes, Suzanna Lewis, and Don Gilbert for their assistance with database analyses, and Diana Lim for help with figures. This work was supported by research grants to A. Letsou from the Huntsman Cancer Institute, American Cancer Society (RPG-99-078-01 DDC), and NIH (ROI GM61972). K. Simin and A. Scuderi were the recipients of predoctoral fellowship support from NIH training grants (5T32 HD07491 and 5T32 HG00043, respectively).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL aletsou@genetics.utah.edu; FAX (801) 581-7796.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.84402.
REFERENCES
- Adams MD, Celniker SE, Holt RA, Evans CA, Gocayne JD, Amanatides PG, Scherer SE, Li PW, Hoskins RA, Galle RF, et al. The genome sequence of Drosophila melanogaster. Science. 2000;287:2185–2195. doi: 10.1126/science.287.5461.2185. [DOI] [PubMed] [Google Scholar]
- Altschul S, Gish W, Miller W, Myers E, Lipman D. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Andrews J, Bouffard G, Cheadle C, Lu J, Becker K, Oliver B. Gene discovery using computational and microarray analysis of transcription in the Drosophila melanogaster testis. Genome Res. 2000;10:2030–2043. doi: 10.1101/gr.10.12.2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M, Misra S, Roote J, Lewis SE, Blazej R, Davis T, Doyle C, Galle R, George R, Harris N, et al. An exploration of the sequence of a 2.9-Mb region of the genome of Drosophila melanogaster: The Adh region. Genetics. 1999;153:179–219. doi: 10.1093/genetics/153.1.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellen H, O'Kane C, Wilson C, Grossniklaus U, Pearson R, Gehring W. P-element-mediated enhancer detection: A versatile method to study development in Drosophila. Genes & Dev. 1989;3:1288–1300. doi: 10.1101/gad.3.9.1288. [DOI] [PubMed] [Google Scholar]
- Bier E, Vaessin H, Shepherd S, Lee K, McCall K, Barbel S, Ackerman L, Carretto R, Uemura T, Grell E. Searching for pattern and mutation in the Drosophila genome with a P-lacZ vector. Genes Dev. 1989;3:1273–1287. doi: 10.1101/gad.3.9.1273. [DOI] [PubMed] [Google Scholar]
- Bonfield JK, Kf S, Staden R. A new DNA sequence assembly program. Nucleic Acids Res. 1995;23:4992–4999. doi: 10.1093/nar/23.24.4992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulianne GL, de la Concha A, Campos-Ortega JA, Jan LY, Jan YN. The Drosophila neurogenic gene neuralized encodes a novel protein and is expressed in precursors of larval and adult neurons. EMBO J. 1993;12:2586. doi: 10.1002/j.1460-2075.1993.tb05914.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown N, Kafatos F. Functional cDNA libraries from Drosophila embryos. J Mol Biol. 1988;203:425–437. doi: 10.1016/0022-2836(88)90010-1. [DOI] [PubMed] [Google Scholar]
- Caenorhabditis elegans Sequencing Consortium. Genome sequence of the nematode C. elegans: A platform for investigating biology. Science. 1998;282:2012–2018. doi: 10.1126/science.282.5396.2012. [DOI] [PubMed] [Google Scholar]
- Campos-Ortega JA, Hartenstein V. The embryonic development of Drosophila melanogaster. New York: Springer Verlag; 1997. [Google Scholar]
- Coulter D, Swaykus E, Beran-Koehn M, Goldberg D, Wieschaus E, Schedl P. Molecular analysis of odd-skipped, a zinc finger encoding segmentation gene with a novel pair-rule expression pattern. EMBO J. 1990;9:3795–3804. doi: 10.1002/j.1460-2075.1990.tb07593.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duronio RJ. Establishing links between developmental signaling pathways and cell-cycle regulation in Drosophila. Curr Opin Genet Dev. 1999;9:81–88. doi: 10.1016/s0959-437x(99)80012-4. [DOI] [PubMed] [Google Scholar]
- FlyBase. The FlyBase Database of the Drosophila Genome Projects and community literature. Nucleic Acids Res. 1999;27:85–88. doi: 10.1093/nar/27.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gawantka V, Pollet N, Delius H, Vingron M, Pfister R, Nitsch R, Blumenstock C, Niehrs C. Gene expression screening in Xenopus identifies molecular pathways, predicts gene function and provides a global view of embryonic patterning. Mech Dev. 1998;77:95–141. doi: 10.1016/s0925-4773(98)00115-4. [DOI] [PubMed] [Google Scholar]
- Haenlin M, Kramatschek B, Campos-Ortega JA. The pattern of transcription of the neurogenic gene Delta of Drosophila melanogaster. Development. 1990;110:905–914. doi: 10.1242/dev.110.3.905. [DOI] [PubMed] [Google Scholar]
- Jurgens G, Wieschaus E, Nusslein-Volhard C, Kluding H. Mutations affecting the pattern of the larval cuticle in Drosophila melanogaster. II. Zygotic loci on the third chromosome. Roux's Arch Dev Biol. 1984;193:283–295. doi: 10.1007/BF00848157. [DOI] [PubMed] [Google Scholar]
- Kopczynski CC, Noordermeer JN, Serano TL, Chen WY, Pendleton JD, Lewis S, Goodman CS, Rubin GM. A high-throughput screen to identify secreted and transmembrane proteins involved in Drosophila embryogenesis. Proc Natl Acad Sci. 1998;95:9973–9978. doi: 10.1073/pnas.95.17.9973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudoh T, Tsang M, Hukriede NA, Chen X, Dedekian M, Clarke CJ, Kiang A, Schultz S, Epstein J A, Toyama R, et al. A gene expression screen in zebrafish embryogenesis. Genome Res. 2001;11:1979–1987. doi: 10.1101/gr.209601. [DOI] [PubMed] [Google Scholar]
- Lehmann R, Nusslein-Volhard C. Abdominal segmentation, pole cell formation, and embryonic polarity require the localized activity of oskar, a maternal gene in Drosophila. Cell. 1986;47:141–152. doi: 10.1016/0092-8674(86)90375-2. [DOI] [PubMed] [Google Scholar]
- Liang Z, Biggin MD. Eve and ftz regulate a wide array of genes in blastoderm embryos: The selector homeoproteins directly or indirectly regulate most genes in Drosophila. Development. 1998;125:4471–4482. doi: 10.1242/dev.125.22.4471. [DOI] [PubMed] [Google Scholar]
- Nusslein-Volhard C, Wieschaus E, Kluding H. Mutations affecting the pattern of the larval cuticle in Drosophila melanogaster. I. Zygotic loci on the second chromosome. Roux's Arch Dev Biol. 1984;183:267–282. doi: 10.1007/BF00848156. [DOI] [PubMed] [Google Scholar]
- O'Kane C, Gehring W. Detection in situ of genomic regulatory elements in Drosophila. Proc Natl Acad Sci. 1987;84:9123–9127. doi: 10.1073/pnas.84.24.9123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin GM, Hong L, Brokstein P, Evans-Holm M, Frise E, Stapleton M, Harvey DA. A Drosophila complementary DNA resource. Science. 2000;287:2222–2224. doi: 10.1126/science.287.5461.2222. [DOI] [PubMed] [Google Scholar]
- Santon JB, Pellegrini M. Expression of ribosomal proteins during Drosophila early development. Proc Natl Acad Sci. 1980;77:5649–5653. doi: 10.1073/pnas.77.10.5649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schupbach T, Wieschaus E. Female sterile mutations on the second chromosome of Drosophila melanogaster. I. Maternal effect mutations. Genetics. 1989;121:101–117. doi: 10.1093/genetics/121.1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tautz D, Pfeifle C. A nonradioactive in situ hybridization method for the localization of specific RNAs in Drosophila embryos reveals translational control of the segmentation gene hunchback. Chromosoma. 1989;98:81–85. doi: 10.1007/BF00291041. [DOI] [PubMed] [Google Scholar]
- Thatcher JW, Shaw JM, Dickinson WJ. Marginal fitness contributions of nonessential genes in yeast. Proc Natl Acad Sci. 1998;95:253–257. doi: 10.1073/pnas.95.1.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torok T, Tick G, Alvarado M, Kiss I. P-lacW insertional mutagenesis on the second chromosome of Drosophila melanogaster: Isolation of lethals with different overgrowth phenotypes. Genetics. 1993;135:71–80. doi: 10.1093/genetics/135.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitfield WG, Gonzalez C, Sanchez-Herrero E, Glover DM. Transcripts of one of two Drosophila cyclin genes become localized in pole cells during embryogenesis. Nature. 1989;338:337–340. doi: 10.1038/338337a0. [DOI] [PubMed] [Google Scholar]
- Wieschaus E. Embryonic transcription and the control of developmental pathways. Genetics. 1996;142:5–10. doi: 10.1093/genetics/142.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieschaus E, Nusslein-Volhard C, Jurgens G. Mutations affecting the pattern of the larval cuticle in Drosophila melanogaster. III. Zygotic loci on the X-chromosome and fourth chromosome. Roux's Arch Dev Biol. 1984;193:296–307. doi: 10.1007/BF00848158. [DOI] [PubMed] [Google Scholar]
- Wool IG. Extraribosomal functions of ribosomal proteins. Trends Biochem Sci. 1996;21:164–165. [PubMed] [Google Scholar]