Abstract
Phage display technology is an established technology particularly useful for the generation of monoclonal antibodies (mAbs). The isolation of phagemid-encoded mAb fragments depends on several features of a phage preparation. The aims of this study were to optimize phage display vectors, and to ascertain if different virion features can be optimized independently of each other.
Comparisons were made between phagemid virions assembled by g3p-deficient helper phage, Hyperphage, Ex-phage or Phaberge, or corresponding g3p-sufficient helper phage, M13K07. All g3p-deficient helper phage provided a similar level of antibody display, significantly higher than that of M13K07. Hyperphage packaged virions at least 100-fold more efficiently than did Ex-phage or Phaberge. Phaberge's packaging efficiency improved by using a SupE strain.
Different phagemids were also compared. Removal of a 56 base pair fragment from the promoter region resulted in increased display level and increased virion production. This critical fragment encodes a lacZ'-like peptide and is also present in other commonly used phagemids.
Increasing display level did not show statistical correlation with phage production, phage infectivity or bacterial growth rate. However, phage production was positively correlated to phage infectivity.
In summary, this study demonstrates simultaneously optimization of multiple and independent features of importance for phage selection.
Keywords: Filamentous phage, phage display, optimized assembly, monoclonal antibody, evaluation study
1. Introduction
Phage display is a powerful and versatile in vitro technology for isolating proteins with biological activity, such as antibody binding domains (Marks and Bradbury, 2004). Most commonly used phage display technologies employ phagemid vectors in which antibody fragments are expressed as fusion proteins with the minor coat protein III (capsid gene 3 product, g3p), transcriptionally controlled by the lac promoter. Through infection with helper phage, such phagemids can be packaged into filamentous phage particles displaying the g3p fusion protein. By affinity selection, or biopanning, it is possible to select phagemid clones encoding antibody binding domains of a desired specificity.
Although biopanning selection is a powerful approach for obtaining mAbs, it appears less efficient than in vivo selection of specific B-cell clones from an animal system. Using phage display-selection specific clones can typically be retrieved from non-immune phagemid libraries containing 109-1011 clones (Vaughan et al., 1996; Knappik et al., 2000; Edwards et al., 2003). In contrast a mouse with no more than 108 B-cells can mount an antibody response, and corresponding mAb-producing clones can be retrieved by hybridoma technology (Kohler and Milstein, 1975). In the case of phage display, different approaches have been used for more efficient retrieval of clones from phagemid libraries. First, repeat series of biopanning can be performed (Edwards et al., 2003). Second, antibody fragments can be displayed with increased efficiency on virions (McCafferty, 1996; Rondot et al., 2001). Third, increased numbers of biopanned clones can be tested by robotic high throughput screening (de Wildt et al., 2000; Konthur et al., 2005).
Although improved viral display of antibody fragments is helpful for isolating antibody binding domains, attempts to increase the expression of g3p-fusion protein by induction of transcription have sometimes led to bacterial stress (Hoogenboom et al., 1991; Krebber et al., 1996; Zahn et al., 1999). Increased host cell stress can make phagemid libraries unstable by providing a growth advantage to clones not encoding a full g3p-fusion protein (Beekwilder et al., 1999). Viral display of antibody fragments can be increased by using g3p-deficient helper phage (McCafferty, 1996; Rondot et al., 2001; Baek et al., 2002; Soltes et al., 2003) rather than employing the more commonly used g3p-sufficient helper phage. In the absence of helper phage-encoded gene 3, the only source of g3p is the phagemid-encoded g3p fusion protein, which will be efficiently incorporated into virions and displayed. However, in some cases g3p-deficient helper phage give decreased production of phagemid virions (McCafferty, 1996; Baek et al., 2002). A further approach to increasing the display level is lowering the temperature during virion production, however, this can also lead to lower yield of phagemid virions (Chappel et al., 1998). Finally, higher display level can be achieved by constructing phage display libraries in phage vectors rather than phagemid vectors (Griffiths et al., 1994; O'Connell et al., 2002). This vector format gives both high display level and efficient phage production. On the other hand, when cloning phage display libraries, the transformation efficiency is much lower for phage genomes than for phagemids (Harrison et al., 1996). This hampers the construction of large libraries. Furthermore, phage-based libraries are considered to be less stable than phagemid libraries (Scott and Barbas III, 2001), and high-level display can compromise the infectivity of virions (Parmley and Smith, 1988). To summarize, although increasing viral display of g3p fusion protein is desirable for improved selection of clones encoding antibody binding domains, this may have a negative impact on parameters of importance including library stability, virion production, and virion infectivity. It is uncertain if these restrictions are due to the host's biosynthetic capacity and therefore unavoidable, or if they can be addressed by improvement of phage display vectors. Although a number of efficient vector systems have been described (Bradbury and Marks, 2004; Hust and Dübel, 2004) and these are well suited for isolation of antibody binding domains, these systems have not been tested for all the critical parameters which may be important to the system. Analysis of the literature to compare vector systems is complicated by the very small numbers of systematic studies ((Kirsch et al., 2005) and references cited therein) and the lack of standardized test methodologies and the lack of common standards.
This study analyzes how several vector combinations affect different aspects of virion production. These vector combinations include a set of phagemids as well as three different g3p-deficient helper phage, Ex-Phage (Baek et al., 2002), Hyperphage (Rondot et al., 2001) and Phaberge (Soltes et al., 2003). The results of the study demonstrate that efficient virion production, high viral display level, high infectivity and minimal host cell stress can be obtained simultaneously by carefully selecting phagemid and helper phage types as well as the E. coli host strain.
2. Materials and Methods
2.1 Vectors
Phagemids pMAB77, pMAB136 and pMAB144 encoding the same human antibody Fab-fragment specific for tetanus toxoid (Esposito et al., 1994), linked to g3p are described below. Phagemid pMAB77 (GenBank accession no. AX744006) has been described (Soltes et al., 2003; Wiersma and Stewart, 2003). Phagemid pMAB136 (GenBank accession no. DQ119053) is a derivative of phagemid pFAB75.HUI obtained from Dr. Jan Engberg, The Danish University of Pharmaceutical Sciences, Copenhagen, Denmark. Phagemid pMAB144 was derived from pMAB77 by deleting a 56 bp fragment, as described in “Results”. The sequence of a version of pMAB144 not containing Fab genes is available, GenBank accession no. DQ62691. Phagemid pSEX81-PHOX(YOL) (Welschof et al., 1997) (herein called “pSEX81”) encodes an antibody scFv-insert specific for the hapten phenyl-oxazalone, which is linked to g3p. GenBank accession no. Y14584 is identical to pSEX81, except for the region encoding the scFv fragment. The helper phage M13K07 (GE Healthcare) has been described (Vieira and Messing, 1987). Hyperphage (Rondot et al., 2001) was derived from M13K07 by an in-frame deletion of gene 3, codons 8–406. Hyperphage is commercially available from RDI Inc, USA and Progen, Germany. Ex-phage (Baek et al., 2002) and Phaberge (Genbank accession no. AX744011) (Soltes et al., 2003) are derivatives of M13K07, having point mutations in gene 3. Ex-phage has two mutations, E2Amber and E14Amber, and Phaberge has the single mutation Q350Amber. Ex-phage is commercially available from IG Therapy Co, South Korea, and Phaberge is freely available from the corresponding author. Hyperphage was produced in E. coli strain DH5α/pIII, which supplies viral g3p from a chromosomal locus. DH5α/pIII can be obtained through licensing from BTMS GmbH, Germany. Ex-phage and Phaberge were produced in SupE E. coli hosts, TG1 (Stratagene) and XL-1 Blue MRF' (Stratagene) respectively, which allow read-through of the amber codons present in the helper phage gene 3.
2.2 Preparation of virions
Helper phage virions were prepared as described previously (Rondot et al., 2001; Baek et al., 2002; Soltes et al., 2003). M13K07, Ex-phage and Phaberge were prepared by growing infected bacteria in 2 xYT media (16 g bacto-tryptone, 10g yeast extract and 5 g NaCl per liter, pH 7.0-7.2) containing 10 μg tetracycline ml-1, and the resulting supernatant was used for infecting phagemid-containing E. coli. Hyperphage was prepared by growing infected bacteria in 2 xYT media containing 50 μg kanamycin ml-1, 0.5 mM IPTG and 100 mM glucose. Hyperphage was concentrated by to two rounds of PEG-precipitation, with a final resuspension in 10 mM TrisHCl , 20 mM NaCl, 2 mM EDTA, pH 7,5, and then used for infecting phagemid-containing E. coli.
Phagemid virions were prepared as previously described (Ausubel et al., 1997; Soltes et al., 2003), with the exception that the growth time after helper phage infection was shorter. Virions were prepared from TOP10F' host cells (Invitrogen, USA) unless otherwise indicated. Prior to helper phage infection, phagemid-containing TOP10F' cells were grown in 2 xYT, containing 1 % (w/v) glucose and 100 μg ampicillin ml-1. At midlog (A600 of 0.6-0.8) this culture was infected with helper phage, described above, at a multiplicity of infection of 15. To ascertain if E. coli were accidentally pre-infected with helper phage, each experimental set-up included a control mock-infection, where mid-log E. coli were incubated with liquid media instead of helper phage. After infection at 37°C for one hour, bacteria were pelleted by centrifugation and re-suspended in fresh liquid 2 xYT media containing 100 μg ampicillin ml-1 but no glucose. The volume of the infected culture was 50% larger than that of the pre-infected culture. Infected bacteria were grown for 4.5-5 hours at 30°C, and the supernatants recovered. Phagemid virions were purified by two consecutive precipitations with PEG-NaCl.
2.3 Analysis of phagemid virions
To quantify the number of infectious phagemid virions, CFU assay was performed as previously described (Ausubel et al., 1997; Soltes et al., 2003) except for a larger ratio of indicator bacteria. 10 μl of phagemid virions, in serial dilution, was mixed with 90 μl of mid-log XL-1 Blue MRF' indicator bacteria (Stratagene, USA), incubated for 30 minutes at 37°C and then plated on ampicillin-containing agar plates. CFU assay was performed with a serial dilution of each virion preparation. The titer was calculated scored from a dilution giving between 10 and 1000 colonies per plate, i.e. the multiplicity of infection was far less than one. CFU assay with kanamycin-containing agar plates indicated that less than 2% of the phagemid virions contained helper phage genomes.
Viral display of antibody fragments was determined by ELISA using either tetanus toxoid (TT) coated wells (Wiersma and Stewart, 2003) or wells coated with phenyl-oxazalone-BSA (phOX) as follows: BSA was coupled in 50 mM NaHCO3 buffer pH 8 with a 20-fold molar excess of phOX, and dialyzed. Microtiter wells were coated with 1 μg phOx-BSA per well, blocked with 2% (w/v) skim milk powder and incubated with serial dilutions of pSEX81 phage. The amount of virion particles was determined using ELISA plates coated with mouse-anti-fd/f1 monoclonal antibody (Research Diagnostics, USA) as previously described (Wiersma and Stewart, 2003). In all ELISAs control wells were plated with BSA to ascertain non-specific binding and in addition PEG-purified supernatants from mock-infected E. coli was also used as negative control. ELISA detection utilized either biotin-conjugated mouse anti-fd/f1 antibody followed by AP-conjugated streptavidin (Jackson ImmunoResearch, USA) or HRP-conjugated anti-M13 (Amersham Biosciences, Germany).
“Relative display level” is the average display of antibody fragment per infectious virion and was calculated from titer of anti-TT or anti-phOX ELISA and CFU assay as described previously (Soltes et al., 2003).
“Relative infectivity” measures the ability of an average phagemid virion to infect E. coli indicator cells, and was calculated using the infectivity of a virion preparation normalized for its content of virions:
| (Equation 1) |
The anti-phage titer is the reciprocal of the dilution of virions that gives 50% of maximal A405 in the anti-fd/f1 ELISA. The relative infectivity of a test sample was then expressed as a percentage of that of the reference sample, phage produced by the combination pMAB144, M13K07 and host TOP10F':
| (Equation 2) |
Atest is A from equation 1, calculated for the test sample, and Aref is A from equation 1, calculated for the reference sample. The reference sample is defined in Figure 2.
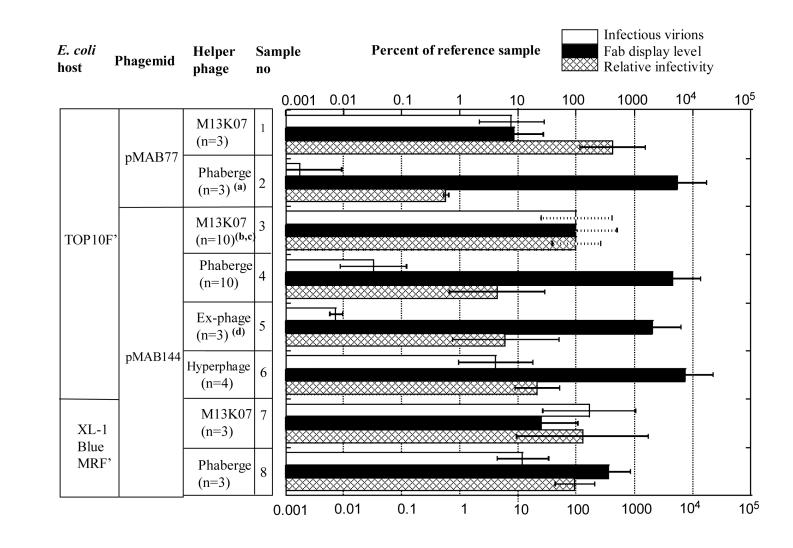
Figure 2.
Influence of E. coli host, phagemid and helper phage on production of phagemid virions.
- For sample 2, relative infectivity was determined in two out three experiments.
- The virion production for the standard sample correspond to 3×1010 CFU ml-1 of non-purified culture supernatant.
- Dotted error bars indicate that standard deviations were calculated differently than for other samples. If calculated as for other samples they would be zero since each value, by definition, would be 100%. A dotted error bar illustrates standard deviation for the standard sample, calculated vis a vis the geometric mean for independent repeat experiments.
- For sample 5, Fab display level was determined in two out of three experiments.
Western blot for viral g3p and Fab-g3p was performed as described (Soltes et al., 2003) using a mouse anti-g3p antibody, pSKAN3 (Mobitec GmbH, Göttingen, Germany). Chemiluminescence signals of serially diluted samples were captured and quantified by Luminescent Image Analyzer) according to manufacturer's instructions (Fuji Photo Film Canada Inc., Canada.
2.4 E. coli doubling time
A600 was measured for exponentially growing bacteria using a ND-100 spectrophotometer (Nanodrop, USA). Measurements were made at multiple time points in the interval 0.1≤ A600≤ 1.0. Log(A600) was plotted versus length of growth, and the doubling time was calculated from the k-value obtained by linear regression (KaleidaGraph 3.52, Synergy Software, USA). Only experiments with a high linear regression coefficient, R>0.98, were included.
2.5 Statistical analysis
Data for statistical analysis was from Figure 2 (virion production, viral display level and relative infectivity), Figures 3B (doubling time of non-infected E. coli) and 3C (doubling time of helper-phage infected E. coli), as well as additional data (not shown) for phagemids pMAB136 and pSEX81. A total of 12-16 measurements were obtained for each of the five parameters. Each measurement, “X”, was log-transformed, and the resulting “Y” value used for statistical analysis:
| (Equation 3) |
Possible correlations of these five parameters were analyzed by the SAS statistical software version 9.1.3 (SAS Institute Inc., Cary, NC, USA), using the program PROC CORR for correlation matrix analysis and PROC REG for multiple regression analysis.
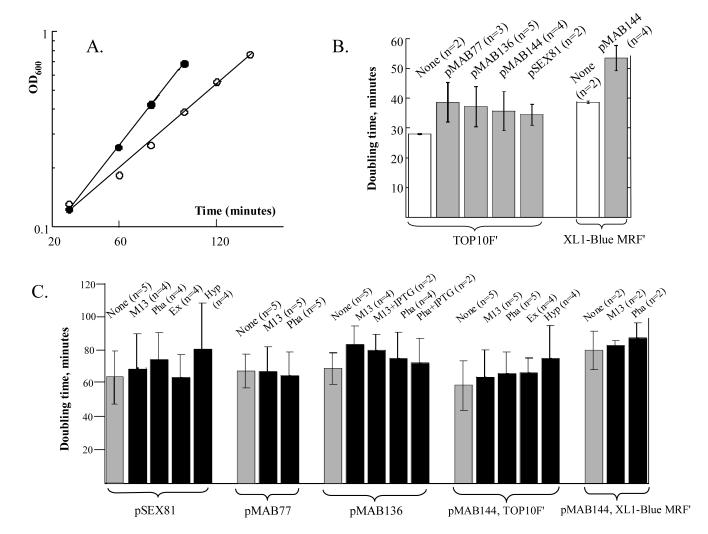
Figure 3.
- Example of growth rate measurement. TOP10F' (filled circles) and XL-1 Blue MRF' (open circles) not containing phagemid or helper phage, grown at 37°C in 2xYT containing 1% glucose, but not ampicillin.
- Influence of different phagemids on doubling time of TOP10F' and XL-1 Blue MRF'. Bacteria not containing phagemid (open bars) were grown at 37°C in 2xYT containing 1% glucose, but not ampicillin, whereas media for bacteria containing phagemid (grey bars) contained 100 μg ampicillin ml-1. This growth condition was used for E. coli propagation before helper phage infection.
- Influence of different helper phage on doubling times. All bacteria were grown under conditions used for production of phagemid virions, at 30°C, in 2xYT media containing 100 μg ampicillin ml-1 but not glucose. Black bars indicate the addition of a helper phage indicated by two or three letter abbreviation. Grey bars indicate that bacteria were grown under identical conditions, but were not infected by helper phage. In B and C each bar represents means of measurements from repeat experiments. Error bars indicate standard deviations.
3. Results
3.1 Comparison of different phagemids
To evaluate potential improvements of production of phage displaying antibody Fab fragments, a comparison of three different phagemid vectors was undertaken. These phagemids encode an identical tetanus toxoid-specific Fab fragment, but differ in other parts of the vectors (Figure 1). These vectors were housed in TOP10F' host cells, and virions were produced and analyzed under identical conditions, as detailed in Materials and Methods.
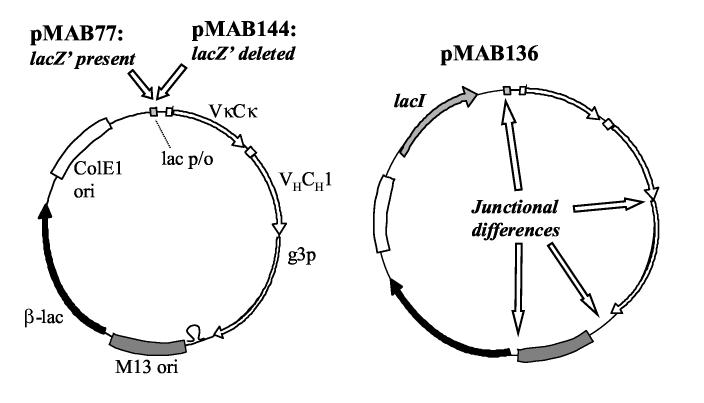
Figure 1.
Schematic illustration of phagemids pMAB77, pMAB136 and pMAB144.
The differences between pMAB77 and pMAB144 (left), and between pMAB77 and pMAB136 (right) are indicated by italic, bold font. pMAB77 contains a 56 base-pair fragment located between the lac promoter and the transcription start of VκCκ, absent in pMAB144. pMAB136 differs from pMAB77 in that it has a lacI gene, it lacks the 56 base-pair lacZ'-containing fragment preceding VκCκ, a 20 base-pair difference in the junction between VHCH1 and g3p, a 166 base-pair difference in the junction between g3p and M13 origin of replication and 255 base-pair difference in the junction between M13 origin and the β-lactamase gene. For full sequence information, please see references cited in Materials and Methods.
Phagemids pMAB136 and pMAB77 were compared. These phagemids differ as pMAB136 contains the lacI gene, and there are also differences in the junctions of some vector elements, as illustrated in Figure 1. When packaged with helper phage M13K07, the number of phagemid virions produced by pMAB136 was similar, 61%, of that produced by pMAB77, and the display level of Fab fragments was also similar, 54% of pMAB77 (data not shown). Addition of 1 mM IPTG after pMAB136 had undergone helper phage infection resulted in increased viral display of Fab fragments, 350% of that of pMAB77, and a somewhat reduced production of phagemid virions, 26% of pMAB77. Repeat experiments, performed without comparison to the pMAB77 + M13KO7 reference sample yielded a similar result (data not shown).
A vector was designed that contained potentially desirable elements from both pMAB77 and pMAB136, and was evaluated. A detailed sequence comparison of the phagemids showed that pMAB77 differs in the lac-promoter region from pMAB136 in that it contains a 56 bp DNA sequence, encoding a lacZ'-like peptide (MTMITPSLHANSISRRQS). To ascertain if this DNA segment is critical for Fab display, pMAB77 was modified to contain an identical lac-promoter as pMAB136 (vector pMAB144 (Figure 1)). pMAB144 demonstrated higher display levels than pMAB77 (Figure 2, samples 1 and 3), and compared well in display level with pMAB136. Surprisingly, the yield of pMAB144 phagemid virions was also increased beyond that seen with pMAB77, 3 ×1010 CFU per ml of non-purified culture supernatant.
3.2 Comparison of different helper phage and host strains
A comparison of different g3p-deficient helper phage was conducted to evaluate the potential of generating a further increase in display levels of antibody fragments. Hyperphage has a deletion of gene 3 (Rondot et al., 2001), and Ex-phage (Baek et al., 2002) and Phaberge (Soltes et al., 2003) both have suppressable stop mutations in gene 3. Each of these helper phage was previously found to provide higher viral display than M13K07, the corresponding g3p-sufficient helper phage, but have not been directly compared with each other. Bacteria carrying either phagemid pMAB77, or pMAB144 were infected with either Phaberge or M13K07. Phaberge consistently demonstrated higher viral display of Fab fragment than M13K07, but also showed lower production of phagemid virions (Figure 2, samples 1-4). A similar result was seen for pMAB136 (data not shown). Bacteria housing pMAB144 were infected with Hyperphage, Ex-phage, Phaberge, or M13K07. With Hyperphage, Ex-phage and Phaberge viral display levels were essentially comparable to each other, these levels being significantly higher than that of M13K07 (Figure 2, samples 3-6). Western blot analysis was used to estimate the display level of Fab-g3p for virions produced by the combination of TOP10F', pMAB144 and Hyperphage. From two separate phagemid virion preparations, we estimate that 2-12% of g3p is in the form of Fab-g3p (data not shown). Unexpectedly, there were large differences among helper phage in their ability to assemble phagemid virions. Whereas the virion production provided by Hyperphage was only moderately lower than that of M13K07, both Phaberge and Ex-phage gave approximately thousand fold less phagemid virions than M13K07. All four helper phage were also compared in combination with a different phagemid, pSEX81, encoding an antibody scFv-fragment specific for the hapten phenyl-oxazalone. The results with pSEX81 were comparable to those obtained with pMAB144 (data not shown). This indicates that the differences between the helper phage are not substantially influenced by the antibody format, scFv or Fab.
In a previous study (Soltes et al., 2003; Soltes et al., 2006) we found that Phaberge induced a similar production of phagemid virions to M13K07. We considered this to be due to the use of E. coli SupE XL-1 Blue MRF' as host strain in the previous study, rather than the non-SupE host TOP10F' used in the current study. To investigate, we compared the two E. coli strains for production of pMAB144 virions in a new set of side-by-side experiments using both M13KO7 and Phaberge helper phage. The results (Figure 2, samples 3, 4, 7 and 8) were consistent with those found in our previous study (Soltes et al., 2003; Soltes et al., 2006). While the combination TOP10F'/Phaberge gave a low production of phagemid virions, the combination XL-1 Blue MRF'/Phaberge gave a much increased production; compare samples 4 versus 8 in Figure 2. Unlike the previous study (Soltes et al., 2003; Soltes et al., 2006) the production by XL-1 Blue MRF'/Phaberge, sample 8, was somewhat less than that of XL-1 Blue MRF'/M13K07, sample 7. The reason for this is not yet clear, but might be due to using a different phagemid in this study than in the previous study, pMAB144 rather than pMAB77. In addition, the high display level seen by TOP10F'/Phaberge was attenuated when using the combination XL-1 Blue MRF'/Phaberge Figure 2 samples 4 versus 8). The attenuation seen with host XL-1 Blue MRF' is likely caused by read-through of the amber codon of Phaberge's gene 3. If the read-through was complete the amount of virions produced by XL-1 Blue MRF'/Phaberge, and their display level, should be comparable to virions produced by XL-1 Blue MRF'/M13K07. This was not the case: compare samples 7 and 8 in Figure 2. Instead, virions produced by XL-1 Blue MRF'/Phaberge had an intermediate phenotype of those produced by XL-1 Blue MRF'/M13KO7 (high amount, low display) and virion produced by TOP10 F'/ Phaberge (low amount, high display), implying that read-through was not complete. Unlike the virions produced by TOP10F'/Phaberge, the virions produced by XL-1 Blue MRF'/ Phaberge were in amounts suitable for biopanning.
3.3 Comparison of the effects on E. coli host growth
The effects of the different phagemids and helper phage on the growth rate of the E. coli host were evaluated. Potentially there could be effects on the library integrity and ability to select specific antigen binding domains. To quantify these effects bacterial doubling time was determined (Figure 3). At 37°C the E. coli host bacteria containing a phagemid grew slower than bacteria not containing a phagemid (Figure 3B). There was little difference in growth rate among bacteria housing different phagemids.
The impact of different helper phage was evaluated by comparing bacteria containing a phagemid alone with those containing both phagemid and a helper phage (Figure 3C). When bacteria housed both a helper phage and a phagemid the growth rate varied substantially in repeat experiments. This variability is likely due to compounded stress from both phagemid and helper phage. Nevertheless, all helper phage evaluated (M13K07, Phaberge, Ex-phage and Hyperphage) tended to cause some increase in doubling time and, in general, there were no great differences in the growth retardation caused by the different helper phage. Finally, it was noted that host strain XL-1 Blue MRF' grew slower than TOP10F' (Figures 3A-C).
3.4 Correlation between different virion features
The dataset including number of infectious virions produced, viral display levels, relative infectivity (Figure 2), and growth rate (Figure 3) was subjected to statistical analysis. To determine if any of these parameters were correlated to each other, a correlation analysis (Materials and Methods) was conducted and a correlation matrix obtained (Table 1). Two pair-wise significant correlations, p<0.05, were found. The strongest correlation, p=0.0001, was for virion production and relative infectivity. Although the correlation was strong, the impact was not: linear regression analysis indicates that for every ten-fold increase in virion production there was only a two-fold increase in infectivity. A more direct linear regression analysis, a log-log plot of CFU titer versus anti-phage ELISA, indicates the two parameters are highly correlated, R=0.95, and that for a ten-fold increase in ELISA-value there is a 14-fold increase in CFU titer. The second correlation, p=0.02 was for pre-infection doubling time versus post-infection doubling time, which were also positively correlated. All other matrix comparisons showed insignificant correlation.
Table 1.
Correlation between virion production parameters
|
Virion production |
Display level |
Relative infectivity |
Doubling time, pre- infection |
Doubling time, post- infection |
|
|---|---|---|---|---|---|
| Virion production |
1.000 | −0.501 (0.1) |
0.811 (0.0001) |
0.384 (0.1) |
0.302 (0.3) |
| Display level | 1.000 | −0.387 (0.2) |
−0.244 (0.4) |
−0.425 (0.2) |
|
| Relative infectivity |
1.000 | 0.487 (0.06) |
0.085 (0.8) |
||
| Doubling time, pre infection |
1.000 | 0.563 (0.02) |
|||
| Doubling time, post infection |
1.000 |
4. Discussion
This study provides insight into the optimization of production of phagemid virions, which is of importance for the overall success ofphage display technology.
Three different g3p-deficient helper phage were compared, Hyperphage (Rondot et al., 2001), Ex-phage (Baek et al., 2002) and Phaberge (Soltes et al., 2003). Each of these originate from the g3p-sufficient helper phage M13K07. Although each of the helper phage have been previously reported to confer higher viral display of antibody fragments than M13K07 no systematic cross-comparison has been conducted. In this study it was determined that Hyperphage, Ex-phage and Phaberge all exhibit comparable display levels, each at least ten-fold higher than M13K07. Unexpectedly, there were significant differences in the amounts of phagemid virions produced by the helper phage. Hyperphage packaged approximately ten-fold less phagemid virions than M13K07, and both Ex-phage and Phaberge packaged some thousand-fold less phagemid virions than M13K07. The amount of phagemid virions produced by Hyperphage is sufficient for biopanning large non-immune libraries, but the helper phage Phaberge and Ex-phage appear to have more limited utility. Hyperphage is structurally distinct from Ex-phage and Phaberge in that gene 3 in Hyperphage is largely deleted, necessitating it to be produced under somewhat different conditions. The observed differences in supporting phagemid production are considered due to the helper phage's structural differences and not the different conditions used for producing helper phage. Both Ex-phage and Phaberge contain full-length gene 3 that has been rendered non-functional by suppressable amber stop codons. It is considered that the poor support of virion production by Phaberge and Ex-phage in the non-SupE host is due to polar effects of the gene 3 amber codons on other essential helper phage genes. When Phaberge was used in a SupE host, which allows some degree of read-through of the amber codon, this defect was alleviated. A similar result was obtained when using Ex-Phage in a SupE host (Oh et al. 2006). Phaberge packages phagemid virions more efficiently in a SupE host, but display of antibody fragments was shown to be less efficient than in a non-SupE host. To obtain both high display level and relatively efficient production of phagemid virions Hyperphage appears to be the best choice.
Through comparison of phagemids it was determined that a motif located between the lac promoter and the genes encoding g3p fusion protein has a negative impact on phage production. Phagemid pMAB144, which lacks this motif, has several advantages over phagemid pMAB77, higher production of phagemid virions, increased display levels and better or similar propagation of E. coli host. This motif encodes a translatable lacZ'-peptide, and is an inadvertent remnant from the predecessor of pMAB77 plasmid pUC119 where it provides α-complementation (Vieira and Messing, 1987).
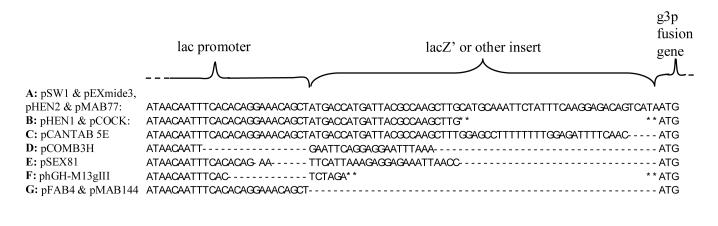
The negative effects of the lacZ'-encoding fragment could be due to interference with normal translation of the Fab-g3p fusion protein. Synthesis of Fab-g3p fusion protein would require that ribosomes re-initiate translation after translation of the lacZ'-like peptide has been completed. In addition, the negative effects could be due to the peptide having a toxic effect. A literature survey shows that some commonly used phagemids have a lacZ'-encoding fragment identical to that of pMAB77 (Figure 4), suggesting that their functionality could also be compromised. Other phagemids either have no insert or have different inserts, either with or without truncation of the lac promoter (Figure 4).
Figure 4.
Promoter-proximal sequences from published phagemid vectors.
Vectors on lines A-F include an insert between the lac promoter and the start of the g3p fusion protein. Vectors on line G has no insert. A dash indicates a missing nucleotide. A star indicates that this part of the sequence could not be found. The sequences are from the following references: pSW1 (Ward et al., 1989); pEXmide3 (Söderlind et al., 1993); pHEN2: http://www.mrc-cpe.cam.ac.uk/g1p.php?menu=1808; pMAB77: Genbank accession no. AX744006, (Soltes et al., 2003; Wiersma and Stewart, 2003); pHEN1 (Hoogenboom et al., 1991); pCOCK (Engelhardt et al., 1994); pCANTAB 5E: http://www4.amershambiosciences.com; pCOMB3H: Genbank accession no. AY254174, (Barbas III et al., 1991); pSEX81: Genbank accession no. ASY14584, (Welschof et al., 1997); phGH-M13gIII (Bass et al., 1990); pFAB4 (Orum et al., 1993); and pMAB144: present study.
Several phagemids, e.g. pHEN1 (Hoogenboom et al., 1991) and its derivatives, contain an amber stop codon in the fusion gene encoding antibody binding domains and gene 3. The advantage of such vectors is that scFv or Fab fragments can conveniently be expressed as either phage-bound fusion protein or soluble protein by utilizing either SupE or non-SupE host strains. In our phagemid system, expression of soluble scFv or Fab requires genetic alteration of the phagemid, which is somewhat more laborious. A potential disadvantage of amber-containing phagemids is that SupE-strains might only provide incomplete read-through of the stop codon, resulting in suboptimal display of scFv/Fab-g3p fusion protein.
There are examples described in the literature of where efficient viral display of g3p fusion protein leads to lower virion production, reduced viral infectivity or increased host cell stress. To analyse if these different virion-related parameters are linked, we performed statistical analysis. No significant correlations were found between high viral display level and lower virion production, reduced infectivity or decreased growth rate. This indicates that while increased display level may have negative consequences in some settings, this is not necessarily always the case. Two statistically significant correlations were found, the strongest was for efficient production of virions being correlated to virions having high infectivity. The other correlation, between pre-infection growth rate and post-infection growth rate was of lower statistical significance. The linkage between phage production and phage infectivity could be due to a balance among the components required for virion assembly. In the case of wildtype filamentous phage, virion assembly involves precise control mechanisms of the steady-state quantities of viral DNA and protein (Fulford et al., 1986). This control is almost certainly perturbed in systems involving helper phage and phagemids. Phage display systems can be even further compromised by extraneous vector elements, such as the phagemid-encoded lacZ'- fragments, which cause host cell stress without providing any advantageous function. The high-copy pUC origin of replication could also pose unnecessary strain. A study by Beekwilder et al. (Beekwilder et al., 1999) shows that if pUC origin is replaced by a low-copy pBR322 origin display level and virion production remain unchanged, but the host cell stress level is reduced.
The format of the displayed antibody, scFv or Fab, might also affects phage assembly parameters, but this has not been investigated in great detail. It has been reported that scFv-phage and Fab-phage have comparable display levels (Chapman et al., 1997), yet Fab-phage have been claimed to be superior for biopanning selection (Malone and Sullivan, 1996). Our study could not assess the impact of antibody format since the scFv-encoding phagemid, pSEX81, differs from the Fab-encoding phagemids by multiple sequence motifs.
In summary, we found that the preferred condition for producing phagemid virions were vector pMAB144 in combination with helper phage Hyperphage and host strain TOP10F'. This provided a high display level of antibody binding domains, as well as relatively high production and infectivity of phagemid virions. It is anticipated that the optimization described in this study will enable isolation of antibody binding domains from phagemid libraries with greater ease and efficiency.
Acknowledgements
We thank Dr. Jan Engberg, Denmark for providing vector pFAB75.HUI, Chris Druar, Cangene for performing phage ELISA studies, and AAI Development Services, Inc., MA, USA for performing statistical evaluation. This research was supported through funding from the Chemical, Biological, Radiological and Nuclear Research and Technology Initiative (CRTI) Project No 0087RD, by National Institutes of Health /National Institute of Allergy and Infectious Disease grant 1UO1AI056383-1, SMP “Antibody Factory” of the NGFN2 program of the German government, and by Cangene Corporation.
Abbreviations
- A600
absorbance at 600nm
- CFU
colony forming units
- ELISA
enzyme-linked immunosorbent assay
- Fab
antigen-binding fragment
- g3p
gene III protein
- lacZ
beta galactosidase gene
- mAb
monoclonal antibody
- SupE
mutant phenotype conferring read-through of amber stop codon
- phOX
phenyl-oxazalone
- scFv
single chain-Fv
- TT
tetanus toxoid
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ausubel F, Bent R, Kingston R, Moore D, Seidman J, Smith J, Struhl K. Current protocols in molecular biology. John Wiley & Sons, Inc; USA: 1997. [Google Scholar]
- Baek H, Suk KH, Kim YH, Cha S. An improved helper phage system for efficient isolation of specific antibody molecules in phage display. Nucleic Acids Res. 2002;30:e18. doi: 10.1093/nar/30.5.e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbas CF, III, Kang AS, Lerner RA, Benkovic SJ. Assembly of combinatorial antibody libraries on phage surfaces: the gene III site. Proc. Natl. Acad. Sci. USA. 1991;88:7978–82. doi: 10.1073/pnas.88.18.7978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bass S, Greene R, Wells JA. Hormone phage: an enrichment method for variant proteins with altered binding properties. Proteins. 1990;8:309–14. doi: 10.1002/prot.340080405. [DOI] [PubMed] [Google Scholar]
- Beekwilder J, Rakonjac J, Jongsma M, Bosch D. A phagemid vector using the E. coli phage shock promotor facilitates phage display of toxic proteins. Gene. 1999;228:23–31. doi: 10.1016/s0378-1119(99)00013-x. [DOI] [PubMed] [Google Scholar]
- Bradbury AR, Marks JD. Antibodies from phage antibody libraries. J. Immunol. Methods. 2004;290:29–49. doi: 10.1016/j.jim.2004.04.007. [DOI] [PubMed] [Google Scholar]
- Chapman CJ, Mockridge CI, Spellerberg MB, Zhu D, Stevenson FK. Phage surface expression for analysis of recognition sites of human autoantibodies: comparison of single chain Fv and Fab. Hum. Antibodies. 1997;8:124–8. [PubMed] [Google Scholar]
- Chappel JA, He M, Kang AS. Modulation of antibody display on M13 filamentous phage. J. Immunol. Methods. 1998;221:25–34. doi: 10.1016/s0022-1759(98)00094-5. [DOI] [PubMed] [Google Scholar]
- de Wildt RM, Mundy CR, Gorick BD, Tomlinson IM. Antibody arrays for high-throughput screening of antibody-antigen interactions. Nat. Biotechnol. 2000;18:989–994. doi: 10.1038/79494. [DOI] [PubMed] [Google Scholar]
- Edwards BM, Barash SC, Main SH, Choi GH, Minter R, Ullrich S, Williams E, DuFou L, Wilton J, Albert VR, Ruben SM, Vaughan TJ. The remarkable flexibility of the human antibody repertoire; isolation of over one thousand different antibodies to a single protein, BLyS. J. Mol. Biol. 2003;334:103–18. doi: 10.1016/j.jmb.2003.09.054. [DOI] [PubMed] [Google Scholar]
- Engelhardt O, Grabherr R, Himmler G, Ruker F. Two-step cloning of antibody variable domains in a phage display vector. BioTechniques. 1994;17:44–46. [PubMed] [Google Scholar]
- Esposito G, Scarselli E, Traboni C. Phage display of a human antibody against Clostridium tetani toxin. Gene. 1994;148:167–8. doi: 10.1016/0378-1119(94)90252-6. [DOI] [PubMed] [Google Scholar]
- Fulford W, Russel M, Model P. Aspects of the growth and regulation of the filamentous phages. Prog. Nucleic Acid Res. Mol. Biol. 1986;33:141–68. doi: 10.1016/s0079-6603(08)60022-7. [DOI] [PubMed] [Google Scholar]
- Griffiths AD, Williams SC, Hartley O, Tomlinson IM, Waterhouse P, Crosby WL, Kontermann RE, Jones PT, Low NM, Allison TJ, Prospero TD, Hoogenboom HR, Nissim A, Cox JPL, Harrison JL, Zaccolo M, Gherardi E, Winter G. Isolation of high affinity human antibodies directly from large synthetic libraries. EMBO J. 1994;13:3245–60. doi: 10.1002/j.1460-2075.1994.tb06626.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison J, Williams S, Winter G, Nissim A. Screening of phage antibody libraries. In: Abelson J, editor. Methods in Enzymology. Vol. 267. Academic Press; San Diego, New York, Boston, London, Sydney, Tokyo, Toronto: 1996. pp. 83–108. [DOI] [PubMed] [Google Scholar]
- Hoogenboom HR, Griffiths AD, Johnson KS, Chiswell DJ, Hudson P, Winter G. Multi-subunit proteins on the surface of filamentous phage: methodologies for displaying antibody (Fab) heavy and light chains. Nucleic Acids Res. 1991;19:4133–7. doi: 10.1093/nar/19.15.4133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hust M, Dübel S. Mating antibody phage display with proteomics. Trends Biotechnol. 2004;22:8–14. doi: 10.1016/j.tibtech.2003.10.011. [DOI] [PubMed] [Google Scholar]
- Kirsch M, Zaman M, Meier D, Dübel S, Hust M. Parameters affecting the display of antibodies on phage. J. Immunol. Methods. 2005;301:173–185. doi: 10.1016/j.jim.2005.04.017. [DOI] [PubMed] [Google Scholar]
- Knappik A, Ge L, Honegger A, Pack P, Fischer M, Wellnhofer G, Hoess A, Wolle J, Plückthun A, Virnekas B. Fully synthetic human combinatorial antibody libraries (HuCAL) based on modular consensus frameworks and CDRs randomized with trinucleotides. J. Mol. Biol. 2000;296:57–86. doi: 10.1006/jmbi.1999.3444. [DOI] [PubMed] [Google Scholar]
- Kohler G, Milstein C. Continuous cultures of fused cells secreting antibody of predefined specificity. Nature. 1975;256:495–497. doi: 10.1038/256495a0. [DOI] [PubMed] [Google Scholar]
- Konthur Z, Hust M, Dübel S. Perspectives for systematic in vitro antibody generation. Gene. 2005;364:19–29. doi: 10.1016/j.gene.2005.05.042. [DOI] [PubMed] [Google Scholar]
- Krebber A, Burmester J, Plückthun A. Inclusion of an upstream transcriptional terminator in phage display vectors abolishes background expression of toxic fusions with coat protein g3p. Gene. 1996;178:71–4. doi: 10.1016/0378-1119(96)00337-x. [DOI] [PubMed] [Google Scholar]
- Malone J, Sullivan MA. Analysis of antibody selection by phage display utilizing anti-phenobarbital antibodies. J. Mol. Recognit. 1996;9:738–45. doi: 10.1002/(sici)1099-1352(199634/12)9:5/6<738::aid-jmr333>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- Marks JD, Bradbury A. Selection of human antibodies from phage display libraries. Methods Mol. Biol. 2004;248:161–76. doi: 10.1385/1-59259-666-5:161. [DOI] [PubMed] [Google Scholar]
- McCafferty J. Phage display: Factors affecting panning efficiency. In: Kay BK, Winter J, McCafferty J, editors. Phage display of peptides and proteins. Academic Press Inc.; San Diego, CA, USA: 1996. pp. 261–76. [Google Scholar]
- O'Connell D, Becerril B, Roy-Burman A, Daws M, Marks JD. Phage versus phagemid libraries for generation of human monoclonal antibodies. J. Mol. Biol. 2002;321:49–56. doi: 10.1016/s0022-2836(02)00561-2. [DOI] [PubMed] [Google Scholar]
- Oh M, Joo H, Hur B, Jeong Y, Cha S. Inefficient read-through of amber codons at gIII of Ex12 helper phage in suppressive Escherichia coli host strains substantially favors the phage display of antibody fragments. 2006 Submitted. [Google Scholar]
- Orum H, Andersen PS, Oster A, Johansen LK, Riise E, Bjornvad M, Svendsen I, Engberg J. Efficient method for constructing comprehensive murine Fab antibody libraries displayed on phage. Nucleic Acids Res. 1993;21:4491–8. doi: 10.1093/nar/21.19.4491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parmley SF, Smith GP. Antibody-selectable filamentous fd phage vectors: affinity purification of target genes. Gene. 1988;73:305–18. doi: 10.1016/0378-1119(88)90495-7. [DOI] [PubMed] [Google Scholar]
- Rondot S, Koch J, Breitling F, Dübel S. A helper phage to improve single-chain antibody presentation in phage display. Nat. Biotechnol. 2001;19:75–8. doi: 10.1038/83567. [DOI] [PubMed] [Google Scholar]
- Scott JK, Barbas CF., III . Phage-display vectors. In: Barbas CF III, Burton DR, Scott JK, Silverman GJ, editors. Phage display. A laboratory manual. Cold Spring Harbor Laboratory Press; Cold Spring Harbor: 2001. pp. 2.1–2.19. [Google Scholar]
- Söderlind E, Simonsson-Lagerkvist AC, Duenas M, Malmborg A, Ayala M, Danielsson L, Borrebaeck CAK. Chaperonin assisted phage display of antibody fragments on filamentous bacteriophages. Biotechnology (NY) 1993;11:503–7. doi: 10.1038/nbt0493-503. [DOI] [PubMed] [Google Scholar]
- Soltes G, Barker H, Marmai K, Pun E, Yuen A, Wiersma EJ. A new helper phage and phagemid vector system improves viral display of antibody Fab fragments and avoids propagation of insert-less virions. J. Immunol. Methods. 2003;274:233–44. doi: 10.1016/s0022-1759(02)00294-6. [DOI] [PubMed] [Google Scholar]
- Soltes G, Barker H, Marmai K, Pun E, Yuen A, Wiersma EJ. Corrigendum to “A new helper phage and phagemid vector system improves viral display of antibody Fab fragments and avoids propagation of insert-less virions”. J. Immunol. Methods. 2006;311:226. doi: 10.1016/s0022-1759(02)00294-6. [DOI] [PubMed] [Google Scholar]
- Vaughan TJ, Williams AJ, Pritchard K, Osbourn JK, Pope AR, Earnshaw JC, McCafferty J, Hodits RA, Wilton J, Johnson KS. Human antibodies with sub-nanomolar affinities isolated from a large non-immunized phage display library. Nat. Biotechnol. 1996;14:309–14. doi: 10.1038/nbt0396-309. [DOI] [PubMed] [Google Scholar]
- Vieira J, Messing J. Production of single-stranded plasmid DNA. Methods Enzymol. 1987;153:3–11. doi: 10.1016/0076-6879(87)53044-0. [DOI] [PubMed] [Google Scholar]
- Ward ES, Gussow D, Griffiths AD, Jones PT, Winter G. Binding activities of a repertoire of single immunoglobulin variable domains secreted from Escherichia coli. Nature. 1989;341:544–6. doi: 10.1038/341544a0. [DOI] [PubMed] [Google Scholar]
- Welschof M, Terness P, Kipriyanov S, Stanescu D, Breitling F, Dorsam H, Dübel S, Little M, Opelz G. The antigen-binding domain of a human IgG-anti-F(ab')2 autoantibody. Proc. Natl. Acad. Sci. USA. 1997;94:1902–7. doi: 10.1073/pnas.94.5.1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiersma EJ, Stewart DIH. Phagemid display system. Cangene Corporation; 2003. pp. 1–86. [Google Scholar]
- Zahn G, Skerra A, Hohne W. Investigation of a tetracycline-regulated phage display system. Protein Eng. 1999;12:1031–4. doi: 10.1093/protein/12.12.1031. [DOI] [PubMed] [Google Scholar]