Abstract
We have developed a new tool to visualize expression data on metabolic pathways and to evaluate which metabolic pathways are most affected by transcriptional changes in whole-genome expression experiments. Using the Fisher Exact Test, the method scores biochemical pathways according to the probability that as many or more genes in a pathway would be significantly altered in a given experiment by chance alone. This method has been validated on diauxic shift experiments and reproduces well known effects of carbon source on yeast metabolism. The analysis is implemented with Pathway Analyzer, one of the tools of Pathway Processor, a new statistical package for the analysis of whole-genome expression data. Results from multiple experiments can be compared, reducing the analysis from the full set of individual genes to a limited number of pathways of interest. The pathways are visualized with OpenDX, an open-source visualization software package, and the relationship between genes in the pathways can be examined in detail using Expression Mapper, the second program of the package. This program features a graphical output displaying differences in expression on metabolic charts of the biochemical pathways to which the open reading frames are assigned.
[Supplementary materials are available at http://www.cgr.harvard.edu/cavalieri/pp.html and http://www.genome.org.]
New technologies in biology such as DNA microarrays, oligonucleotide arrays, and serial analysis of gene expression (SAGE) are generating massive data sets, describing biological function in terms of whole-genome expression profiles. The challenge now is how to extract a comprehensive overview from this huge amount of information. To do this it is necessary to develop new bioinformatic tools to automatically connect expression data with the increasing biological information on the function of single open reading frames (ORFs) and their interaction in metabolic networks.
Yeast is currently the ideal model for developing new tools for genome analysis and for understanding networks of gene interactions, because of the detailed information about its genetics and molecular and cellular biology available in databases such as the Saccharomyces genome database (SGD) [http://genome-www.stanford.edu/Saccharomyces/)], the yeast proteome database (YPD) [http://www.proteome.com/databases/YPD/YPDsearch-quick.html], and the Kyoto Encyclopedia of Genes and Genomes (KEGG) [http://www.genome.ad.jp/kegg/].
Efforts have also been made to integrate functional genomic information into the Saccharomyces databases (Ermolaeva 1998; Kaneisha and Goto 2000; Nakao et al. 1999; Ball et al. 2000, 2001; Costanzo et al. 2000), and databases of expression profiles are available for large-scale yeast deletion and mutational analyses (Winzeler and Davis 1997; Winzeler et al. 1999; Hughes et al. 2000; Sherlock et al. 2001).
A number of software packages for the analysis of microarray data are available. Most of the currently available programs use cluster algorithms (Eisen et al. 1998), self-organizing maps (SOM), or principal-component analysis (PCA; Tamayo et al. 1999). These approaches cluster together genes irrespective of their function and without reference to the valuable amount of biological information available in public databases. An extensive list of such software, reviewed by Gardiner-Garden and Littlejohn (2001), can be found at: http://www.ncgr.org/genex/other_tools.html.
Many investigators have manually mapped transcriptional changes to metabolic charts (De Risi et al. 1997; Cavalieri et al. 2000), and others have tried to develop automatic methods to assign genes showing expression variation to functional categories, focusing on single pathways (Zien et al. 2000), or to link array target sequences with NCBI's Entrez retrieval system, and KEGG pathway views (Ermolaeva et al. 1998; Nakao et al. 1999). An innovative approach describing interactions in a cellular pathway has also been discussed by Ideker et al. (2001), integrating DNA microarrays, quantitative proteomics, and databases of known physical interactions. Nevertheless, none of the methods currently available include a statistical test to determine in an automatic way the probability that the genes of any of a large number of pathways are significantly altered in a given experiment, nor do they provide a user-friendly interface to automatically associate expression changes with genes organized into metabolic maps. Here we report an automatic statistical method to determine which pathways are most affected by transcriptional changes and to map expression data from multiple experiments on metabolic pathways.
RESULTS AND DISCUSSION
Pathway Processor is a new statistical package for the analysis of whole-genome expression data which allows the visualization of expression data on metabolic pathways and the evaluation of which metabolic pathways are most affected by transcriptional changes in whole-genome expression experiments. Pathway Processor consists of two programs, Pathway Analyzer and Expression Mapper.
Pathway Analyzer implements a method that uses the Fisher Exact Test to score biochemical pathways according to the probability that as many or more genes in a pathway would be significantly altered in a given experiment by chance alone. Expression Mapper, the second program of the package, features a graphical output displaying differences in expression on metabolic charts of the biochemical pathways to which the ORFs are assigned, enabling a detailed analysis of the relationship between genes in the pathways.
We used the first version of Pathway Processor to interpret results from whole-genome expression analysis in the budding yeast S. cerevisiae, using the fold-change values obtained from hybridization experiments. The results can be obtained from competitive hybridizations on DNA microarrays or from comparison of results from individual hybridization experiments carried out with the Affymetrix Genechip® system. Studies of S. cerevisiae have provided the foundation for much of our current understanding of the fundamental mechanisms of cell biology. This organism has also provided the test bed for the development of DNA microarrays and for their applications to the understanding of intracellular signaling networks.
We tested the utility of Pathway Processor with the data from the diauxic shift experiments (De Risi et al. 1997), which have become the “gold standard” for the application of expression arrays to the study of metabolism. The experiment investigates the temporal program of gene expression accompanying the metabolic shift from fermentation to respiration that occurs when fermenting yeast cells, inoculated into a rich medium containing glucose (20 g/L), turn to aerobic utilization of the ethanol produced during the fermentation after the fermentable sugar is exhausted. De Risi et al. (1997) made whole-genome hybridization experiments comparing gene expression at seven timepoints (T1–T7) to characterize the changes in gene expression that take place during the diauxic shift.
We used Pathway Analyzer to rank the statistical significance of the changes observed in the genes organized according to the logic of the 92 KEGG metabolic pathways during the diauxic shift. The results of the comparison of the seven timepoints are visualized as tables using Microsoft Excel. Pathway Analyzer employs the Fisher Exact Test to measure the probability that a pathway is significantly altered, for any specified threshold. The signed Fisher Exact Test value can be used to compare results from different experiments. The comparison of results of the Signed Fisher Exact Test for the seven timepoints of the diauxic shift experiments (Table 1) shows little alteration of the cellular metabolic pathways from timepoint 1 to 4, which is in agreement with Figure 4 of the De Risi paper (De Risi et al. 1997) and with the observation that during exponential growth in glucose-rich medium, the global pattern of gene expression is remarkably stable (De Risi et al. 1997). Interestingly, the P- value for the most significantly affected pathways increases from timepoints 5 to 7, indicating that an increasing number of genes are altered significantly in expression.
Table 1.
Comparison of the Results of Selected Signed Fisher Exact Test Analyses from Experiment 1
| Pathway | T1 | T2 | T3 | T4 | T5 | T6 | T7 |
|---|---|---|---|---|---|---|---|
| Oxidative phosphorylation map190 | 1.00 | 1.00 | 0.10 | 0.06 | 0.00 | 0.00 | 0.00 |
| Citrate cycle (TCA cycle) map20 | 1.00 | 1.00 | 1.00 | 0.05 | 0.03 | 0.00 | 0.00 |
| Electron Transport System, Complex II map3150 | 1.00 | 1.00 | 1.00 | 0.02 | 0.00 | 0.00 | 0.00 |
| Electron Transport System, Complex IV map3130 | 1.00 | 1.00 | 1.00 | 1.00 | 0.00 | 0.00 | 0.00 |
| Pyruvate metabolism map620 | 1.00 | 1.00 | 0.00 | 0.01 | 0.04 | 0.03 | 0.00 |
| Pentose phosphate cycle map30 | 1.00 | 1.00 | 0.07 | 1.00 | 0.03 | 0.00 | 0.00 |
| Reductive carboxylate cycle (CO2 fixation) map720 | 1.00 | 1.00 | 1.00 | 0.06 | 0.12 | 0.00 | 0.00 |
| Carbon fixation map710 | 1.00 | 1.00 | 0.04 | 0.13 | 0.07 | 0.12 | 0.00 |
| Glyoxylate and dicarboxylate metabolism map630 | 1.00 | 1.00 | 1.00 | 0.11 | 0.05 | 0.07 | 0.00 |
| ATP Synthase map3110 | 1.00 | 1.00 | 0.01 | 1.00 | 0.38 | 0.05 | 0.00 |
| Electron Transport System, Complex III map3140 | 1.00 | 1.00 | 1.00 | 0.03 | 1.00 | 0.00 | 0.01 |
| Glycolysis/Gluconeogenesis map10 | 1.00 | 1.00 | 0.01 | 0.16 | 0.06 | 0.49 | 0.01 |
| Alanine and aspartate metabolism map252 | 1.00 | 1.00 | 0.00 | 0.26 | 0.19 | −0.22 | 0.02 |
| beta-Alanine metabolism map410 | 1.00 | 1.00 | 1.00 | 0.00 | 0.01 | 0.15 | 0.07 |
| Glutamate metabolism map251 | 1.00 | 1.00 | 0.08 | 0.01 | −0.05 | −0.16 | 0.07 |
| Butanoate metabolism map650 | 1.00 | 1.00 | 0.10 | 0.02 | 0.07 | 0.07 | 0.07 |
| Taurine and hypotaurine metabolism map430 | 1.00 | 1.00 | 1.00 | 1.00 | 0.00 | 0.06 | 0.07 |
| Prostaglandin and leukotriene metabolism map590 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 0.06 | 0.07 |
| Fructose and mannose metabolism map51 | 1.00 | 1.00 | 1.00 | 0.03 | 0.59 | 0.53 | 0.09 |
| Fatty acid metabolism map71 | 1.00 | 1.00 | 1.00 | 0.13 | 0.07 | −0.26 | −0.09 |
| Glutathione metabolism map480 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 0.10 | 0.11 |
| Cysteine metabolism map272 | 1.00 | 1.00 | 1.00 | 1.00 | −0.01 | −0.46 | −0.11 |
| Glycine, serine and threonine metabolism map260 | 1.00 | 1.00 | 0.17 | 0.43 | 0.07 | −0.10 | −0.08 |
| Synthesis and degradation of ketone bodies map72 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | −0.07 |
| Pantothenate and CoA biosynthesis map770 | 1.00 | 1.00 | 1.00 | −0.04 | −0.08 | −0.15 | −0.07 |
| Valine, leucine and isoleucine biosynthesis map290 | 1.00 | 1.00 | 0.04 | −0.11 | −0.21 | −0.07 | −0.04 |
| Selenoamino acid metabolism map450 | 1.00 | 1.00 | 1.00 | 1.00 | 0.00 | −0.02 | −0.01 |
| Methionine metabolism map271 | 1.00 | 1.00 | 1.00 | 1.00 | −0.01 | −0.03 | 0.00 |
| Aminoacryl-tRNA biosynthesis map970 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 0.00 | 0.00 |
| RNA polymerase map3020 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | −0.12 | 0.00 |
| Pyrimidine metabolism map240 | 1.00 | 1.00 | 1.00 | 1.00 | −0.53 | −0.17 | 0.00 |
| Purine metabolism map230 | 1.00 | 1.00 | 0.01 | −0.82 | 0.09 | −0.03 | 0.00 |
| Ribosome map3010 | 1.00 | 1.00 | 1.00 | 1.00 | −0.99 | 0.00 | 0.00 |
(T0 vs T1) to 7 (T0 T7) (T1 = 9 hours, T2 = 11 hours, T3 = 13 hours, T4 = 15 hours, T5 = 17 hours, T6 = 19 hours, T7 = 21 hours) The results have been sorted according to their ranking in the T7 column. The table shows only pathways with an absolute P-value of the Fisher exact test greater than or equal to 0.1; the complete results are reported in Supplementary material, Table 2.xls.
The comparison between the Fisher Exact Test values of the seven experiments has been visualized with OpenDX [http://www.opendx.org], an open-source visualization software package.
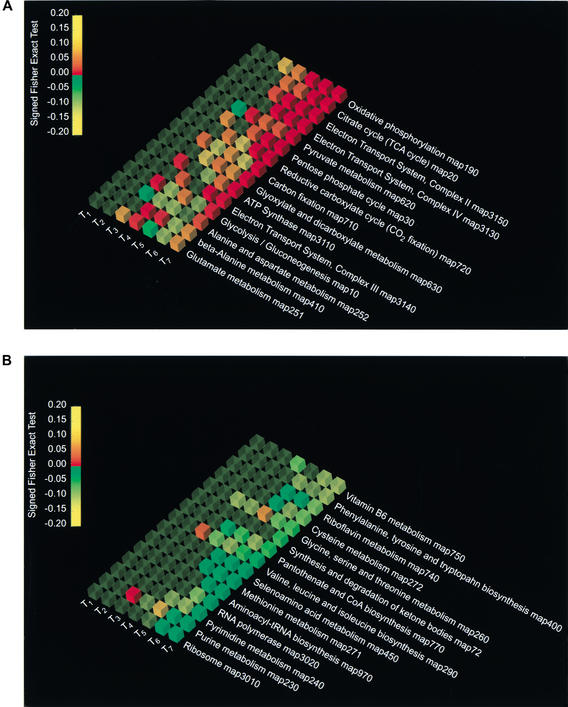
The graphic representation of the results from Pathway Analyzer (Fig. 1A) indicates that the main positively affected pathways during the diauxic shift, from timepoint 5 to timepoint 7 are oxidative phosphorylation, the citrate cycle, the electron transport system complexes II and IV, and pyruvate metabolism. The negative values of the genes for ribosomal proteins and RNA polymerase (Fig. 1B) are also in agreement with the progressive reduction in cellular metabolism, DNA and RNA synthesis, and entry into stationary phase, which are expected with the exhaustion of the sugars and alternative carbon sources.
Figure 1.
Pathway Analyzer results showing the 15 most activated pathways (A) and the 15 most repressed pathways (B) for the seven timepoints of the diauxic shift experiments. The columns from T1 to T7 report the P-values of the Signed Fisher Exact Test, obtained from the comparison of relative expression at T0 to that at the different timepoints of the diauxic shift experiments, with T1 = 9 h, T2 = 11 h, T3 = 13 h, T4 = 15 h, T5 = 17 h, T6 = 19 h, and T7 = 21 h. The data have been sorted according to their ranking in the last experiment, T7 versus T0. The pathways are visualized using OpenDX. The color of the cube indicates the sign of the variation, with red being up-regulated, green down-regulated, and yellow no change. The opacity represents the statistical significance of the variation: the greater the opacity, the smaller the P-value. The color of the cube depends on the P-value in the following way: from 1 to 0.15 the color remains yellow, from 0.15 to 0 with overexpression (+) it goes from yellow to red, and from 0.15 to 0 with underexpression (−) it goes from yellow to green.
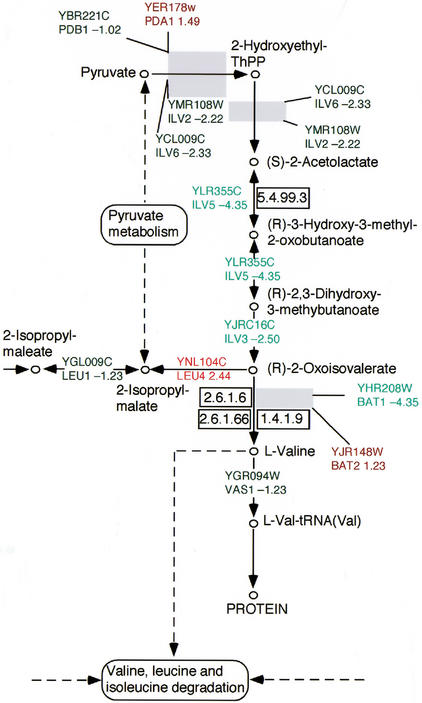
The Expression Mapper analysis confirms the agreement of our results with previous interpretations, and also yields additional insights beyond those that are apparent from the expression levels of individual ORFs. The results shown for the TCA cycle (Map20, supplementary material available online at http://www.cgr.harvard.edu/cavalieri/pp.html and http://www.genome.org. ) report, in the context of a wider network of interactions, the differences in expression between T0 and T7, which in previous analyses were mapped manually on the metabolic charts to which the ORFs are assigned (De Risi et al. 1997). Furthermore, aminoacid metabolic pathways such as the valine, leucine, isoleucine, and methionine biosynthetic pathways are repressed. Interestingly, one gene in the leucine pathway (Fig. 2), LEU4 (Yor104c), is upregulated in T7 (+2.2) with respect to T0, although all the other genes of the pathways are generally repressed. This apparent contradiction is in fact in agreement with the observation that LEU4 is highly expressed under leucine deprivation. The caloric restriction is also consistent with the repression of the biosynthesis of methionine, an amino acid whose synthesis is very costly from a metabolic point of view (Map271, supplementary material available online), and repression of the biosynthesis of valine, leucine, and isoleucine, the most abundant amino acids in the cell. This and the repression of the genes for the aminoacyl-tRNA biosynthetic enzymes (Map 970 supplementary material available online) suggest that residual pyruvate and acetyl-Coa are channeled into the citrate cycle (up-regulated) rather than in the amino acid-producing pathways. The results in Map 190 (supplementary material available online) exploit the graphics available showing both the metabolic network and the cellular localization of the differentially expressed genes. The results show the coregulation of all the genes in the electron transport and oxidative phosphorylation complex 2,3,4, which is consistent with the switch to aerobic metabolism in conditions of caloric restriction.
Figure 2.
Part of the Expression Mapper output for valine, leucine, and isoleucine metabolism, adapted from KEGG map 290. The text is colored red if the relative change in gene expression is ≥1, and green if it is <1. The intensity of the color is proportional to the magnitude of the differential expression. The presence of a gray box indicates that the corresponding step in the biochemical pathway requires multiple gene products.
We also implemented a version of the program to analyze whole-genome B.subtilis expression data, and applied the program to the genome-wide analysis of the general stress response in B. subtilis described by Price et al. (2001) (supplementary information available online at http://www.cgr.harvard.edu/cavalieri/pp.html and http://www.genome.org.). The program can be adapted to analyze expression data from any set or subset of interacting genes of any other organism for which the relationships between the names of ORFs and the enzymes in metabolic pathways are provided, the limit being proper and unique ORF annotation.
We have demonstrated the utility, efficiency and versatility of this approach on the diauxic shift experiments (De Risi et al. 1997) and have further shown its potential to help interpret the results from one or more experiments, by examining differential expression.
Pathway Processor provides a powerful and user-friendly tool for the integration of expression profiling with the functional roles of gene products that are increasingly becoming available in public databases. The program efficiently organizes the data according to the logic of metabolic networks and enables the user to examine the expression patterns of all genes for metabolic enzymes simultaneously, thus facilitating a genomic approach to the understanding of fundamental biological processes. Patterns of differential expression can also be detected in discrete classes of genes, such as those involved in intermediary metabolism, the cytoskeleton, cell-division control, apoptosis, membrane transport, sexual reproduction, and so forth.
The use of KEGG as reference pathway is motivated not only by its exhaustive organization, but also for the possibility of simple graphical representation.
The 92 KEGG pathways are interconnected, sharing common intermediates. A consequence of this interconnection is that, although the nominal P values from the Fisher Exact Test cannot be taken literally (because there are multiple simultaneous statistical tests), there is no known way to correct the nominal P values because the multiple tests are not statistically independent.
METHODS
Pathway Processor
Pathway Processor is an ordering and visualization device that organizes profiles of gene expression according to the metabolic pathways that are affected, and it features a unique graphical output. The package consists of two programs, Pathway Analyzer and Expression Mapper.
Pathway Analyzer
Pathway Analyzer implements the statistical method in Java, automatically identifying which metabolic pathways are most affected by differences in gene expression observed in an experiment. The method associates an ORF with a given biochemical step according to the information contained in 92 pathway files from KEGG [http://www.genome.ad.jp/kegg/]. Pathway Analyzer scores KEGG biochemical pathways, measuring the probability that the genes of a pathway are significantly altered in a given experiment. The factors taken into account are (1) the number of ORFs whose expression is altered in each pathway, (2) the total number of ORFs contained in the pathway, and (3) the proportion of the ORFs in the genome contained in a given pathway.
In the first step of the analysis, the user specifies the magnitude of the difference in ORF expression that is to be regarded as above background. The relative change in gene expression is the multiplier by which the level of expression of a particular ORF is increased or decreased in an experiment. For each ORF considered separately and without regard to other information, a cutoff of 2 for the relative change in gene expression is appropriate given current technology, but probably a little conservative, in particular when assessing differential expression of genes that function in the same metabolic pathway, and when the experiment has been repeated. Thus Pathway Analyzer affords the researcher the opportunity to examine differences that are somewhat smaller than twofold (for example 1.8), but consistent in that they affect a statistically significant number of ORFs in a particular metabolic pathway. Consistent differential expression of a number of ORFs in the same pathway can have important biological implications—for example, it may signify the existence of a set of coordinately regulated ORFs. The program then uses the Fisher Exact Test to calculate the probability that differences in ORF expression in each of the 92 pathways could be due to chance alone. A statistically significant probability means that a particular pathway contains more affected ORFs than would be expected by chance. The program allows the user to choose different cutoffs for the Fisher Exact Test.
The analysis performed using the Fisher Exact Test provides a quick and user-friendly way of determining which pathways are the most strongly affected. The one-sided Fisher Exact Test calculates a P-value, based on the number of genes that exceeds the cutoff in a given pathway. This P-value is the probability that the pathway would contain as many or more affected genes as actually observed, on the null hypothesis being that the relative changes in gene expressions of the genes in the pathway are a random subset of those observed in the experiment as a whole. The resulting set of P-values for all pathways is then used to rank the pathways according to the magnitude and direction of the effects, in order to select those pathways to examine more closely with Expression Mapper.
Two tab-delimited text files are generated from the comparison files. One of them contains all the genes that pass the cutoff, organized by pathway. The other file contains the summary of the statistics for each pathway, which can be imported into Microsoft Excel to enable the user to sort the results according to various columns.
The “Signed Fisher Exact Test” column allows sorting of up-regulated or down-regulated pathways. The value in this column is composed of two distinct parts. The first part consists of the sign + or −, indicating whether the particular pathway contains genes that tend to be up-regulated or down-regulated. The second part of each entry is a positive real number in [0,1] that corresponds to the P-value of the Fisher Exact Test for the pathway. The sign is determined by subtracting the mean relative expression of all genes that pass the cutoff and are in the pathway from the mean relative expression of the genes that pass the cutoff and are not within the pathway (up-regulation/down-regulation column). If there are no genes above the cutoff in a pathway, the sign is arbitrarily set to +. This is for convenience only, as the P-values for such pathways will always be nonsignificant. Sorting for the Signed Fisher Exact Test is done so that the most significant values are at the top for the up-regulated pathways and at the bottom for the down-regulated pathways. In the middle are the least significant pathways. The values of the Fisher Exact Test vector can be used to compare different experiments using Microsoft Excel (Table1), and the comparison among the different experiments can be represented graphically.
Graphic Representation Using OpenDX
Data from the Excel worksheet can be visualized with the open-source visualization software OpenDX [http://www.opendx.org]. This visualization program allows a detailed examination of the expression levels observed in the experiment according to pathways.
The input of the program consists of three files: one with the pathway names, another with the Signed Fisher Exact Test, and a third with the header row. The program represents each value graphically as a cube. The color of the cube indicates the extent of the variation, based on the magnitude of the P-values and the sign, with red being up-regulated, green down-regulated, and yellow no change. The color of the cube depends on the P-value in the following way: from 1 to 0.15 the color remains yellow; from 0.15 to 0 with overexpression (+) it goes from yellow to red; from 0.15 to 0 with underexpression (−) it goes from yellow to green. To allow the eye to focus on the most significant results, we also changed the opacity so that the greater the significance of the variation, the greater the opacity (Fig. 1A,B).
A detailed description of the program is reported in the Manual. The pathways identified as of greatest interest with Pathway Analyzer can be visualized using Expression Mapper.
Expression Mapper
Expression Mapper is a Java program that creates a visual representation of the data, displaying the differences in expression on metabolic charts of the biochemical pathways to which the ORFs are assigned (Fig. 2). The program has been implemented using the KEGG nomenclature. When the map number of the pathway of interest is typed in the Expression Mapper dialog box, the program parses an HTML file corresponding to the KEGG map number and plots differential gene expression onto the map. The text is colored red if the relative change in gene expression is ≥1, or green if it is <1. The intensity of the color is proportional to the magnitude of the differential expression. The presence of a gray box indicates that the corresponding step in the biochemical pathway requires multiple gene products, the individual components of which can be accessed by click-and-drag from the gray box. The pathway diagrams can be saved as JPEG files.
The metabolic maps can easily be adapted to the user's preferences, integrating expression-profiling results with visualization of the interactions among different but functionally related genes.
Downloading Files
Academic implementations of Pathway Processor with a detailed Instruction Manual are freely available for downloading from the Duccio Cavalieri CGR website via URL http://www.cgr.harvard.edu/cavalieri/pp.html or by contacting Duccio Cavalieri. (dcavalieri@cgr.harvard.edu) or Paul Grosu (paul_grosu@harvard.edu). For the analysis of the diauxic shift experiment, we downloaded the publicly available results from the Web via URL [http://cmgm.stanford.edu/pbrown/explore/array.txt].
AKNOWLEDGMENTS
This work could not have been possible without the support of the staff of the Harvard Bauer Center for Genomics Research. We thank Laura Garwin, Andrew Murray, Reddi Gali, Hans Hofmann, Deborah Marks, and Chris Sander for critical analysis and useful comments on the manuscript.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
WEB SITE REFERENCES
http://www.cgr.harvard.edu/cavalieri/pp.html, Duccio Cavalieri CGR web site.
http://www.genome.ad.jp/kegg/, The Kyoto Encyclopedia of Genes and Genomes (KEGG) home page.
http://www.proteome.com/databases/YPD/YPDsearch-quick.html, The yeast proteome database (YPD) home page.
http://www.opendx.org, The open-source visualization software, OpenDX
http://genome-www.stanford.edu/Saccharomyces/, The Saccharomyces Genome database (SGD) home page.
http://www.ncgr.org/genex/other_tools.html. Gene X, Gene expression home page at the National Center for Genome Resources.
http://cmgm.stanford.edu/pbrown/explore/array.txt, The Pat Brown Laboratory web site
Footnotes
E-MAIL dcavalieri@cgr.harvard.edu; FAX 1 (617) 495-2196.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.226602.
REFERENCES
- Ball CA, Dolinski K, Dwight SS, Harris MA, Issel-Tarver L, Kasarskis A, Scafe CR, Sherlock G, Binkley G, Jin H, et al. Integrating functional genomic information into the Saccharomyces genome database. Nucleic Acids Res. 2000;28:77–80. doi: 10.1093/nar/28.1.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ball CA, Jin H, Sherlock G, Weng S, Matese JC, Andrada R, Binkley G, Dolinski K, Dwight SS, Harris MA, et al. Saccharomyces Genome Database provides tools to survey gene expression and functional analysis data. Nucleic Acids Res. 2001;29:80–81. doi: 10.1093/nar/29.1.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalieri D, Townsend JP, Hartl DL. Manifold anomalies in gene expression in a vineyard isolate of Saccharomyces cerevisiae revealed by DNA microarray analysis. Proc Natl Acad Sci. 2000;97:12369–12374. doi: 10.1073/pnas.210395297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costanzo MC, Hogan JD, Cusick ME, Davis BP, Fancher AM, Hodges PE, Kondu P, Lengieza C, Lew-Smith JE, Lingner C, et al. The yeast proteome database (YPD) and Caenorhabditis elegans proteome database (WormPD): Comprehensive resources for the organization and comparison of model organism protein information. Nucleic Acids Res. 2000;28:73–76. doi: 10.1093/nar/28.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeRisi JL, Iyer VR, Brown PO. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science. 1997;278:680–686. doi: 10.1126/science.278.5338.680. [DOI] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci. 1998;95:863–868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ermolaeva O, Rastogi M, Pruitt KD, Schuler GD, Bittner ML, Chen Y, Simon R, Meltzer P, Trent JM, Boguski MS. Data management and analysis for gene expression arrays. Nat Genet. 1998;20:19–23. doi: 10.1038/1670. [DOI] [PubMed] [Google Scholar]
- Gardiner-Garden M, Littlejohn TG. A comparison of microarray databases. Brief Bioinform. 2001;2:143–158. doi: 10.1093/bib/2.2.143. [DOI] [PubMed] [Google Scholar]
- Hughes TR, Marton MJ, Jones AR, Roberts CJ, Stoughton R, Armour CD, Bennett HA, Coffey E, Dai H, He YD, et al. Functional discovery via a compendium of expression profiles. Cell. 2000;102:109–126. doi: 10.1016/s0092-8674(00)00015-5. [DOI] [PubMed] [Google Scholar]
- Kanehisa M, Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ideker T, Thorsson V, Ranish JA, Christmas R, Buhler J, Eng JK, Bumgarner R, Goodlett DR, Aebersold R, Hood L. Integrated genomic and proteomic analyses of a systematically perturbed metabolic network. Science. 2001;292:929–934. doi: 10.1126/science.292.5518.929. [DOI] [PubMed] [Google Scholar]
- Nakao M, Bono H, Kawashima S, Kamiya T, Sato K, Goto S, Kanehisa M. Genome-scale gene expression analysis and pathway reconstruction in KEGG. Genome Inform Ser Workshop Genome Inform. 1999;10:94–103. [PubMed] [Google Scholar]
- Price CW, Fawcett P, Ceremonie H, Su N, Murphy CK, Youngman P. Genome-wide analysis of the general stress response in Bacillus subtilis. Mol Microbiol. 2001;41:757–774. doi: 10.1046/j.1365-2958.2001.02534.x. [DOI] [PubMed] [Google Scholar]
- Sherlock G, Hernandez-Boussard T, Kasarskis A, Binkley G, Matese JC, Dwight SS, Kaloper M, Weng S, Jin H, Ball CA, et al. The Stanford Microarray Database. Nucleic Acids Res. 2001;29:152–155. doi: 10.1093/nar/29.1.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamayo P, Slonim D, Mesirov J, Zhu Q, Kitareewan S, Dmitrovsky E, Lander ES, Golub TR. Interpreting patterns of gene expression with self-organizing maps: Methods and application to hematopoietic differentiation. Proc Natl Acad Sci. 1999;96:2907–2912. doi: 10.1073/pnas.96.6.2907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winzeler EA, Davis RW. Functional analysis of the yeast genome. Curr Opin Genet Dev. 1997;7:771–776. doi: 10.1016/s0959-437x(97)80039-1. [DOI] [PubMed] [Google Scholar]
- Winzeler EA, Shoemaker DD, Astromoff A, Liang H, Anderson K, Andre B, Bangham R, Benito R, Boeke JD, Bussey H, et al. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science. 1999;285:901–906. doi: 10.1126/science.285.5429.901. [DOI] [PubMed] [Google Scholar]
- Zien A, Kuffner R, Zimmer R, Lengauer T. Analysis of gene expression data with pathway scores. Proc Int Conf Intell Syst Mol Biol. 2000;8:407–417. [PubMed] [Google Scholar]