Abstract
Phospholipase C-β (PLC-β) isozymes (EC 3.1.4.11) hydrolyze the membrane phospholipid phosphatidylinositol-4,5-bisphosphate to generate intracellular second messenger signaling molecules inositol-1,4,5-trisphosphate (IP3) and diacylglycerol (DAG) in response to receptor activation and other cellular stimuli. PLCβ1 and PLCβ3 isozymes were previously demonstrated to bind the calcium-sensitive molecule calmodulin [1]. We have now shown through fluorescence anisotropy that calmodulin/PLCβ3 affinities increase with increasing calcium in a physiologically relevant concentration range. The bimolecular affinity constants for calmodulin interaction with PLCβ1 or PLCβ3 were estimated as 260 nM and 200 nM, respectively, from fluorescence anisotropy data. There was no effect of calmodulin on basal or Gαq-stimulated catalytic activity for either isozyme. However, the interaction between calmodulin and PLCβ3 leads to potentiation of activation by the G protein βγ dimer in an in vitro assay. 1321N1 cells treated with calmodulin inhibitors concurrent with and post–stimulation of muscarinic receptors significantly reduced [3H]PIP hydrolysis. Together these data are suggestive of cooperative role for calmodulin in the G protein βγ dimer-stimulated activity of PLCβ3.
Keywords: Phosphatidylinositol hydrolysis, Phospholipase C-beta, Gbetagamma, 1321N1 cells, Calmodulin, Fluorescence anisotropy
1. Introduction
Phosphatidylinositol phospholipid-specific phospholipase C (PLC)1 is a key intracellular signaling molecule that catalyzes the hydrolysis of PIP2 into IP3, a regulator of cytosolic calcium levels, and diacylglycerol, a well-characterized activator of protein kinase C [2]. The identified PLC isozymes have been classified by sequence homology into six families, β, γ, δ, ε, ζ, and η[3-5]. Each family has unique mechanisms of activation and regulation. The activity of the PLCβ family of isozymes is stimulated by membrane G-protein coupled receptors (GPCR) through heterotrimeric guanine nucleotide binding (G) proteins, which are composed of a GTP/GDP-binding α subunit and a βγ dimer.
There are four identified and characterized isoforms of PLCβ. numbered 1 to 4. PLCβ2 and PLCβ4 have limited tissue distributions, whereas PLCβ1 and PLCβ3 are nearly ubiquitous in human tissues; PLCβ1 being dominant in brain and PLCβ3 being dominant in heart and smooth muscle [2]. PLCβ1 and PLCβ3 are both activated by calcium and Gαq [6-8], but the PLCβ3 isoform is additionally sensitive to activation by Gβγ [9]. The mechanisms of regulation of PLCβ3 are incompletely understood despite the enzyme's importance in a variety of cellular processes [10-14]. Aberrancies in expression of PLCβ3 can lead to tumorigenesis [13, 15, 16], and PLCβ3 knockout mice show changes in μ-opioid response [12] or early embryonic lethality [17].
Calmodulin is an established, ubiquitous and abundant calcium-sensitive regulatory protein associated with a vast diversity of cellular functions including signal transduction [18, 19]. Calmodulin binds four molecules of calcium cooperatively, and undergoes a significant conformational change upon calcium binding that is important for its many calcium-sensitive regulatory functions. Calmodulin binding sites are nearly as diverse as the number of calmodulin binding proteins, but generally are amphipathic α-helices, typically 20-35 amino acids long, with basic and hydrophobic residues sorting to opposite sides on an α-helical projection. The list of various mechanisms by which calmodulin regulates proteins is growing nearly as fast as the list of calmodulin binding proteins [18, 20].
We previously reported that calmodulin directly interacts with PLCβ1 and PLCβ3, and that calmodulin inhibitors attenuate inositol phosphate (IP) accumulation in whole cells [1]. To further understand this interaction, we sought to determine the direct effect of calmodulin on PLC-β activity in vitro and to determine the binding affinity and calcium dependence of the PLCβ/calmodulin interaction.
2. Materials and Methods
2.1 Reagents
PLCβ-selective polyclonal rabbit antisera and alkaline phosphatase-conjugated goat anti-rabbit IgG secondary antibody were obtained from Santa Cruz Biotechnology Inc. (Santa Cruz, CA). SuperSignal™ chemiluminescent substrate was purchased from Pierce (Rockford, IL). Fatty-acid free bovine serum albumin (FAF-BSA), W-13 and carbamylcholine chloride (carbachol) were obtained from Calbiochem (San Diego, California). PIP Strip® phospholipid blots were obtained from Echelon Biosciences (Salt Lake City, UT). Synthetic and purified bovine brain PIP, PIP2, PS and PE were purchased from Avanti Polar Lipids (Alabaster, AL). Calmodulin was purified in the presence of calcium from bovine brain as previously described [21]. PLC-β1, PLC-β3, Gαq, Gβγ, and [3H]PIP2 were purified as previously described [22-24]. Alexa Fluor® 488 was acquired from Molecular Probes (Eugene, OR) and was conjugated to calmodulin according to the manufacturer's recommended procedure.
2.2 Fluorescence anisotropy
Fluorescence anisotropy and fluorescence emission spectra were recorded with a Perkin Elmer LS-50 Luminescence Spectrophotometer maintaining constant temperature (20°C). Measurements of fluorescence anisotropy for PLCβ1 and PLCβ3 with Alexa-calmodulin in buffer containing 0.1 M KCl and 30 mM MOPS, pH 7.2, were performed at an excitation wavelength of 490 nm (band-pass 5 mm) using a linear polarizer, and the fluorescence emission intensities at 525 nm (band-pass 20 mm) were monitored through a second linear polarizer. Anisotropy, r, was calculated according to the equation, , where IVV and IVH are the intensity of vertically or horizontally polarized emitted light, respectively, obtained with vertically polarized exciting light. Four readings were taken for each measurement and averages calculated and recorded. The value of G, 0.982, which corrects for unequal transmission of vertically and horizontally polarized emitted light, was obtained from tables prepared by the Anderson laboratory specific for their instrument (personal communication).
2.3 Reconstitution Assay
The catalytic activity of PLCβ1 and PLCβ3 was quantitated using [3H]PIP2 substrate as described previously [25]. Briefly, 45 ng of purified PLCβ3 or 15 ng of PLCβ1 in 20 μl of 50 mM HEPES pH 7.2, 3 mM EGTA, 80 mM KCl (Buffer 1) and 1mg/ml fatty acid free-BSA was added to 20 μl of Buffer 1 containing 15 μM PIP2, 135 μM phosphatidylethanolamine and 6-10,000 cpm [3H]PIP2. Ten μl of 50 mM HEPES pH 7.2, 1mM EDTA, 3 mM EGTA, 5 mM MgCl2, 100 mM NaCl, 1% cholate (Buffer 2) was added to each reaction containing either 30 ng Gαq or 50 ng Gβγ for G-protein stimulated activity of PLC. Basal PLC activity was quantitated with the same buffer with no added G protein. Buffer 1 with 9 mM CaCl2 (10 μl) was added to yield a final assay volume of 60 μl. The reaction proceeded at 30°C for 10 minutes and was terminated by the addition of 375 μL of ice cold mixture of chloroform, methanol and hydrochloric acid in a ratio of 80:40:1. Followed by addition of 125 μl chloroform and 125 μl 0.1N hydrochloric acid with vigorous mixing. The aqueous and organic phases were separated by centrifugation for 5 min. at 2000g. [3H]IP3 product release was quantitated by scintillation counting of 400 μl of the upper phase. Triplicate samples were run in three separate experiments.
2.4 Protein lipid overlay assays
Protein lipid overlay assays were conducted using either commercial phospholipid membrane arrays containing synthetic phospholipids (PIP-Strips®) or nitrocellulose membranes spotted in our laboratory with mammalian derived brain phospholipids. Nitrocellulose membrane phospholipid blots were prepared by spotting with chloroform containing 0, 50 or 100 pmol of phosphatidylserine, phosphatidylethanolamine or PIP2 in 1 μl and allowed to dry. Both purchased and nitrocellulose membranes were blocked with 3% (wt/vol) fatty acid-free BSA in TBST buffer (150 mM NaCl, 10 mM Tris·HCl, pH 8.0, and 0.1% (vol/vol) Tween-20) for 1 hour at 4°C. Blocked membranes were incubated overnight at 4°C with 0.5 μg/ml purified PLCβ3 or PLCβ1 in the presence or absence of 0.5 μg/ml calmodulin. The membranes were then washed three times for 10 minutes in TBST with 3% fatty acid-free BSA, followed by incubation with PLCβ-selective polyclonal antibodies (0.1 μg/ml in TBST with 1% fatty acid-free BSA) overnight at room temperature with gentle agitation. Following three washes over 30 minutes in TBST with 1% fatty acid-free BSA, the membranes were incubated for 1 hour with alkaline phosphatase-conjugated goat anti-rabbit antibodies (0.1 μg/ml) and visualized with SuperSignal™ alkaline phosphatase chemiluminescent substrate according to manufacturer's specifications.
2.5 Inositol phosphate assay
Whole cell inositol phosphate (IP) assays were performed with 1321N1 cells as previously described [26]. Prior to assay, 1321N1 cells were subcultured in 24 well plates until 80% confluent. Cells were then labeled overnight with [3H]myo-inositol, 1 μCi/0.5 ml/well, prepared in sterile inositol-free, bicarbonate-buffered DMEM without additives. Following radiolabeling, cells were pre-treated with 10 mM LiCl for 10 minutes in 20 mM Hepes-buffered DMEM, pH 7.4 (HDMEM) at 37°C in room air to halt degradation and allow for accumulation of inositol phosphates. Muscarinic receptors were then stimulated with 1 mM carbachol for 20 minutes. W-13 (a calmodulin inhibitor) was added concurrently with carbachol or 10 minutes post-carbachol stimulation. Following 20 minutes stimulation, cells were lysed and total inositol phosphates (IP, IP2, IP3) purified and quantitated as described [26]. Lipids were collected to quantitate residual [3H]inositol phospholipids. Percent (%) conversion was calculated as ([3H]inositol phosphates (dpm))/( [3H]inositol phospholipids (dpm) + [3H]inositol phosphates (dpm)) × 100. All assays were performed in triplicate and values reported reflect an average of at least three experiments ± SE. Students t-tests were performed to assess statistical significance where indicated.
2.6 Inositol trisphosphate accumulation assay
Prior to assay, 1321N1 cells were plated onto 6 well plates at a density of 0.5 × 106 cells/well and allowed to adhere overnight. Cells were then labeled with [3H]myo-inositol, 10 μCi/2 ml/well, prepared in inositol-free, bicarbonate-buffered DMEM without additives overnight. Following radiolabeling, cells were pre-treated with 10 mM LiCl for 10 minutes in 1 ml of 20 mM HDMEM, pH 7.4 at 37°C in room air in the presence or absence of 100 μM W-13. Wells were treated with vehicle (water) or 1 mM carbachol to stimulate muscarinic receptors for 30, 60 or 90 seconds. Following stimulation, the reaction was stopped and cells lysed by the addition of an equal volume of 15% trichloroacetic acid solution directly to the wells. The contents of the wells were transferred to glass tubes and spun at 1500 rpm in a table top centrifuge to pellet cell debris. The supernatant was then transferred to a clean glass tube and the TCA extracted 5 times with ether. After extraction the inositol phosphate species were separated as described previously [27]. Briefly, 2 ml dH20 was added to the extracted cell lysates and poured over Poly-prep columns containing 0.5 ml bed volume of AG1-X8 anion-exchange resin (Formate form, 200-400 mesh, Bio-Rad). Tubes were rinsed with 8 ml dH20 and applied to the columns. The columns were washed with 5 ml 25 mM sodium borate, 60 mM sodium formate, and eluted with 5 ml 0.1 M formic acid, 0.2 M ammonium formate (IP containing fraction) , 5 ml 0.1 M formic acid, 0.4 M ammonium formate (IP2 containing fraction) , and 5 ml 0.1 M formic acid, 1.0 M ammonium formate (IP3 containing fraction). The first 2.5 ml of each elution was collected and analyzed for incorporation of [3H]inositol into IP, IP2 and IP3 by liquid scintillation chromatography. The cell membrane pellets were solubilized with 1 ml of 1 M NaOH, which was used to wash the corresponding wells of the plate for residual labeled cell membrane. The membrane prep was neutralized with one ml of 1 N HCl. The membrane lipids were collected to quantitate total [3H]inositol phospholipids. Percent IP3 accumulation was calculated as ([3H]inositol trisphosphate (dpm))/( [3H]inositol phospholipids (dpm) + [3H]inositol phosphates (dpm)) × 100. All assays were performed in triplicate and values reported reflect an average of at least two experiments ± SE. Students t-tests were performed to assess statistical significance where indicated.
3. Results
3.1 Fluorescence anisotropy analysis of PLCβ3 and PLCβ1 binding to calmodulin
Having established direct binding between PLCβ1 or PLCβ3 and calmodulin in the context of the full-length proteins [1], we sought to determine the affinity of PLCβ for calmodulin. Fluorescence anisotropy depends on the rotational freedom of a fluorophore, which is influenced by its environment. By conjugating a fluorophore to calmodulin, we can estimate binding affinity by recording changes in the rotation of the fluorescent calmodulin in the presence of a potential binding partner, in this case PLCβ. Using calmodulin conjugated to Alexa Fluor® 488 we measured the fluorescence anisotropy of Alexa-calmodulin titrated with PLCβ1 and PLCβ3 in the presence of 1 mM free calcium. We can determine the degree of complex formation of CaM with the PLCβ by measuring the fluorescence anisotropy of the Alexa-calmodulin. PLCβ (isoenzyme 1 or 3) was titrated with Alexa-calmodulin in the presence and the absence of calcium, and Alexa-calmodulin fluorescence anisotropy was measured. These anisotropy titration curves are shown in Figure 1 (Panels A and B). In the presence of calcium, dissociation constants of 200 nM for Alexa-calmodulin/PLCβ3 and 280 nM for Alexa-calmodulin/PLCβ1 interaction were determined by non-linear curve-fitting of the experimental titration data shown in Figure 1 (Panel A and B). The fit curveline is corrected for a known and quantitated glycerol effect from the purified PLCβ storage buffer. In the presence of EDTA, titration of Alexa-calmodulin with PLCβ3 caused only small increases in anisotropy that failed to approach saturation over the concentration range covered, indicative of a non-specific interaction. To further validate the calcium dependence of the interaction between PLCβ3 and calmodulin, we performed a calcium titration curve for Alexa-calmodulin/PLCβ3 interaction using 10 nM Alexa-calmodulin and 30 nM PLCβ3. Calcium stimulated increased Alexa-calmodulin anisotropy in the presence of PLCβ3 (Figure 1, Panel C). The EC50 value for calcium dependence of the Alexa-calmodulin/PLCβ3 interaction can only be estimated as < 1 μM, a value that is within a physiologically relevant range. This apparent EC50 is necessarily greater than the true EC50 of the complex because saturating concentrations of PLCβ3 were not achievable with limited amounts of purified protein available to use. The profile of anisotropy versus Ca++ concentration would be concentration dependent until PLCβ3 concentration was greater than the Kd. However, these limited data suggest that increasing calcium concentrations following initial activation of the enzyme may support further interaction by increasing association of CaM with active PLCβ.
Figure 1.
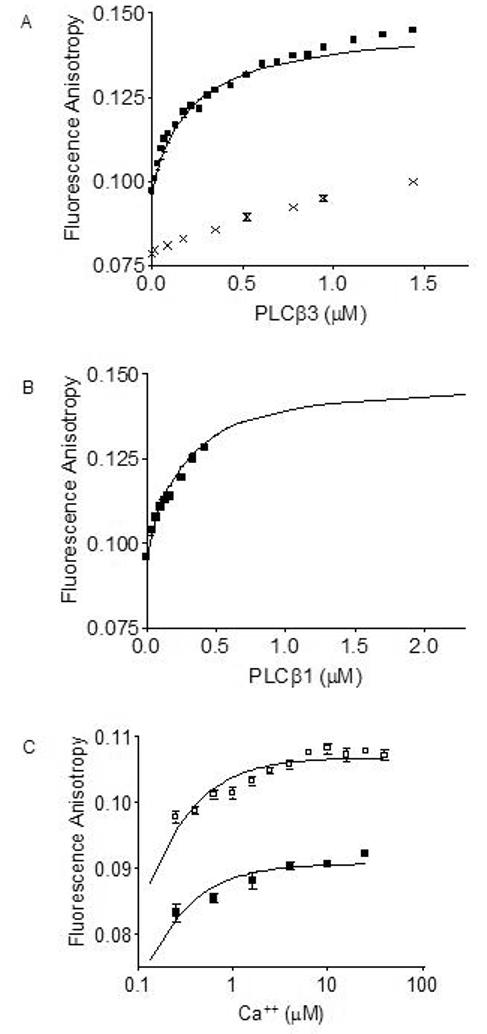
Binding of fluorescently-tagged calmodulin to PLCβ1 and PLCβ3 monitored by fluorescence anisotropy. Fluorescence anisotropy monitoring of 10 nM Alexa-calmodulin binding to increasing concentrations of (A) PLCβ3 (■) in the presence of 1 mM calcium, or presence of 1 mM EDTA (x). Lines were curvefit as described in Materials and Methods. Fluorescence anisotropy monitoring of 10 nM Alexa-calmodulin binding to increasing concentrations of (B) PLCβ1 (■) in the presence of 1 mM calcium. Lines were curvefit as described in Materials and Methods. KD values calculated are 260 nM for PLCβ1 interaction with calmodulin and 200 nM for PLCβ3 interaction with calmodulin. (C) Effect of increasing calcium concentrations on fluorescence anisotropy with 10 nM Alexa-calmodulin (■) or 10 nM Alexa-calmodulin plus 30 nM PLCβ3 (□). Fluorescence anisotropy (emission intensity) was measured at 525 nm with excitation at 490 nm at 20°C. Shown is representative data from two separate experiments. An apparent EC50 value of < 1 μM was calculated for calcium effects on PLCβ3/calmodulin interactions.
3.2 Calmodulin potentiates Gβγ activation of PLCβ3 in vitro
Pre-treatment with the calmodulin inhibitors W-13 and fluphenazine attenuated muscarinic receptor stimulated phosphoinositide (PI) hydrolysis in 1321N1 cells [1]. To determine whether the potentiation of PI hydrolysis by calmodulin could occur through direct stimulation of PLCβ1 or PLCβ3, we performed reconstitution assays with purified proteins and G protein subunits and purified substrate in vitro. PLCβ3 can be activated in vitro by either the Gαq or Gβγ dimer of the heterotrimeric G proteins, whereas PLCβ1 is sensitive to Gαq stimulation but relatively insensitive to Gβγ dimers [28]. Calmodulin did not affect basal or Gαq-stimulated PLCβ1 or PLCβ3 activities at any concentration tested (Figure 2, Panels A and B, respectively). However, calmodulin did potentiate Gβγ stimulation of PLCβ3 activity (Figure 2, Panel A, hatched bars). A titration of increasing Gβγ concentrations in the presence of 10 μM calmodulin showed an increase in PLCβ3 activity of almost 30% (Figure 2, Panel C).
Figure 2.
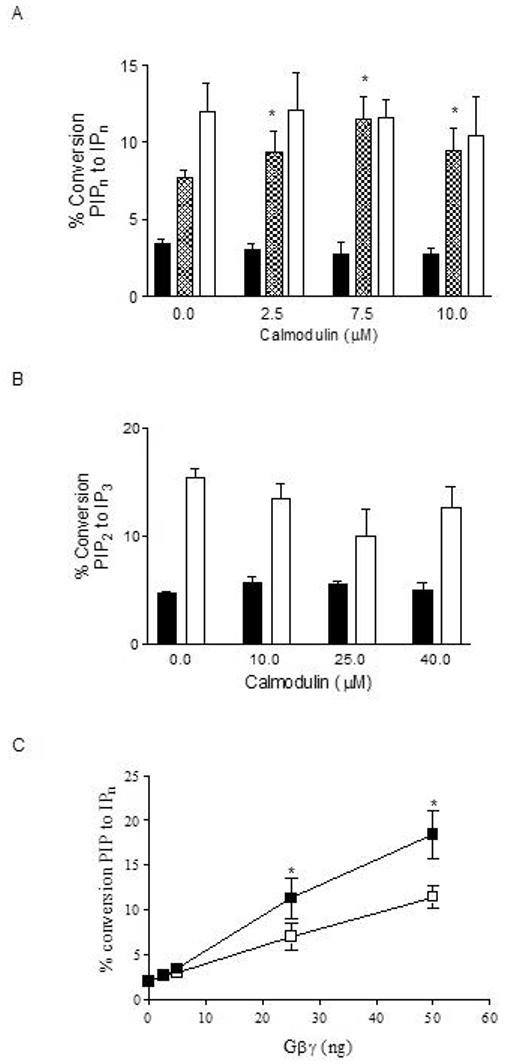
Effect of calmodulin on basal and G-protein-stimulated PLCβ activity in vitro. (A) Basal (solid bars), 50 ng (90 nM) Gβγ-stimulated- (hatched bars), and 30 ng (70 nM) GTPγSGαq-stimulated (white bars) PLCβ3 activity was quantitated in the presence of increasing concentrations of calmodulin. (B) Basal (solid bars) and 30 ng GTPγS-Gαq-stimulated (white bars) PLCβ1 activity was quantitated in the presence of increasing concentrations of calmodulin. Basal PLCβ activity was measured using PIP2 substrate in detergent/phospholipid vesicles. G-protein-stimulated PLCβ activity was measured in the presence of purified G protein subunits using PIP2 substrate in detergent/phospholipid vesicles. PLCβ activity was quantitated as percent conversion of [3H]PIP2 substrate to [3H]IPn (derivatives of inositol phosphate with the individual phosphate positions defined in accordance with the IUPAC convention) as described in Methods. (C) PLCβ3 activity was measured in the presence of increasing concentrations of Gβγ subunits with (■) or without (□) 10 μM calmodulin. Data shown are mean ± SEM of 3 to 5 separate experiments performed in triplicate. Asterisk (*) indicates treatment results significantly different from vehicle at p < 0.001.
3.3 Calmodulin does not affect lipid selectivity of PLCβ isozymes
Calmodulin regulates the activity of other signaling proteins via changes in membrane or lipid interactions [29, 30] and calmodulin binding sites share structural similarity to some phospholipid binding domains. We assessed the effect of calmodulin on the lipid selectivity of either PLCβ1 or PLCβ3 isozyme using lipid overlay assay with commercially available PIP Strips®. Calmodulin did not affect the lipid selectivity of PLCβ1 or PLCβ3 (Figure 3, Panels B and C). Although no changes in selectivity of lipid were seen in the presence or absence of calmodulin, we found that PLCβ1 and PLCβ3 bind to PIP3 in addition to PI, PIP, PIP2, PA, and PS (Figure 3, Panel B and C). To investigate the lipid binding specificity of PLCβs, we purchased mammalian derived phosphatidylinositol phospholipids for spotting onto nitrocellulose for additional lipid-protein overlay assays. Surprisingly, we found that PLCβ3 binding to synthetic phospholipids (single phospholipid species with 16:0 side-chains) did not mimic PLCβ3 binding to mammalian-derived phospholipids with a variety of chain lengths and saturations (Figure 3, Panel D). Side-chain specificity in PLCβ lipid binding has not, to our knowledge, been previously reported. The mammalian PIP2 contains the 16:0 side-chain species in only 7% of the total PIP2 species (manufacturer's analysis).
Figure 3.
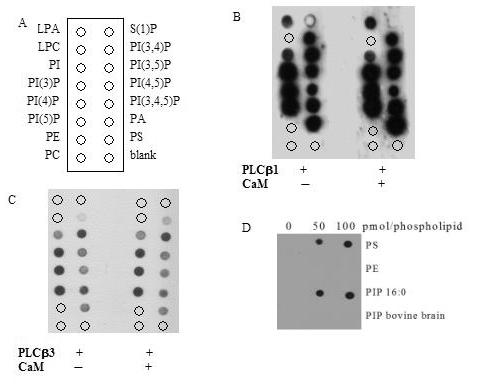
Protein lipid overlay. (A) PIP strips with the following membrane lipids; lysophosphatidic acid, LPA, lysophosphatidylcholine, LPC, phosphaptidylinositol, PI, phosphatidylinositol-3-phosphate, PI(3)P, phosphatidylinositol-4-phosphate, PI(4)P, phosphatidylinositol-5-phosphate, PI(5)P, phosphatidylethanolamine, PE, phosphatidylcholine, PC, sphingo-1-phosphate, S(1)P, phosphatidylinositol-3,4-phosphate, PI(3,4)P, phosphatidylinositol-3,5-phosphate, PI(3,5)P, phosphatidylinositol-4,5-diphosphate, PI(4,5)P, phosphatidylinositol-3,4,5-trisphosphate, PI(3,4,5)P, phosphatidic acid, PA, phosphatidylserine, PS, and no lipid blank. PIP strips incubated with 0.5ug/ml of (B) PLCβ1 or (C) PLCβ3 in the absence or presence of 0.5ug/ml calmodulin. (D) Nitrocellulose membranes spotted with indicated amounts of the listed phosphospholipids (pmol) and incubated with 0.5 μg/ml PLCβ3. PE and PS have been isolated from bovine brain. Nitrocellulose membranes and PIP strips were washed, incubated with anti-PLCβ isoenzymes, and detected by Western blotting procedures as described in Materials and Methods. Data shown is representative of three similar experiments.
3.4 Calmodulin inhibitors attenuate phosphatidylinositol phospholipids hydrolysis concurrent with muscarinic receptor activation
To further dissect the mechanism by which calmodulin potentiates phosphatidylinositol phospholipid hydrolysis in whole 1321N1 cells, we attempted to assess whether calmodulin inhibitors have differential abilities to attenuate phosphatidylinositol phospholipid hydrolysis under varying cytosolic calcium conditions. Previously, we pre-treated 1321N1 cells with CaM inhibitors prior to activation by carbachol and showed inhibition of IP3 accumulation [1]. Here we show the effect in 1321N1 cells of the calmodulin inhibitors when added concurrent with, and post stimulation of muscarinic receptors by carbachol to investigate the effect of W-13 attenuation of IP accumulation in actively signaling cells. W-13 attenuates carbachol-stimulated IP accumulation in all cellular activation states tested (Figure 4A) showing that calmodulin inhibitors effect phosphatidylinositol phospholipid turnover when the GPCR signaling cascade is active. The addition of W-13 concurrent with carbachol showed an attenuation of IP accumulation equal to that of pretreatment [1]. Furthermore, addition of W-13 10 min post-stimulation by carbachol inhibited any further IP accumulation resulting in values significantly lower than cells treated with vehicle alone. The effect of calmodulin inhibitor on the initial rate of PLCβ activation was investigated by pre-treating 1321N1 cells with LiCl and vehicle or W-13 for 10 minutes followed by carbachol stimulation for 30, 60 and 90 seconds. IP3 was separated from total IP. Accumulation of IP3 after carbachol stimulation was inhibited by W-13 at all time points (Figure 4B), supporting a role for calmodulin in PLCβ mediated phosphatidylinositol phospholipid hydrolysis.
Figure 4.
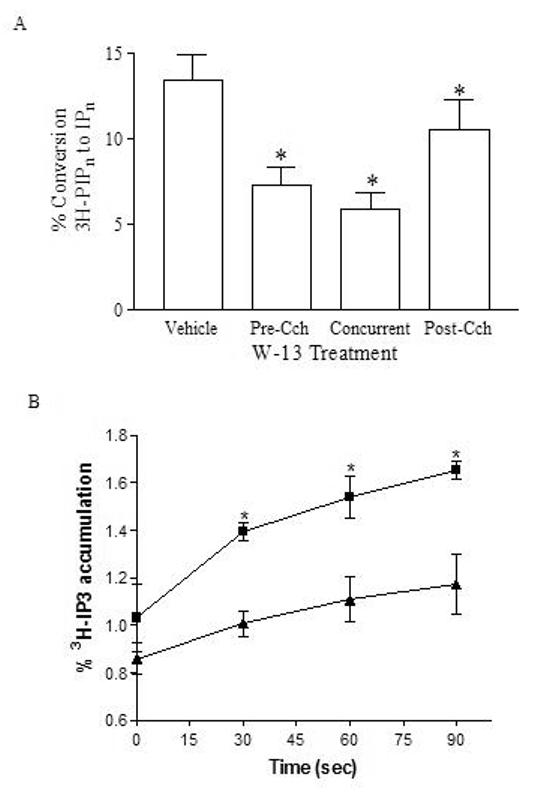
Calmodulin antagonist effects on carbachol-stimulated inositol phosphate accumulation under varying conditions of cell activation. (A) 1321N1 cells were stimulated for 20 minutes with 1 mM carbachol in the presence of 10 mM LiCl. Additionally, cells were either treated with dH20 (Vehicle) or treated with 100 μM W-13 for 20 minutes concurrent with carbachol addition (Concurrent), or treated for 10 minutes with 100 μM W-13 10 minutes after carbachol addition (Post-Cch). PLCβ activity was measured as percent of [3H]PIPn converted to IPn in whole cells, collected and quantitated as described in Materials and Methods. Shown is cumulative data from three experiments performed in triplicate. (B) 1321N1 cells were pre-treated with dH2O (Vehicle) or with 100 μM W-13 for 15 minutes. Cells were stimulated for 30, 60 and 90 seconds with 1 mM carbachol in the presence of 10 mM LiCl. PLCβ activity was measured as percent of [3H]PIPn converted to IP3 in whole cells, collected and quantitated as described in Materials and Methods. Shown is cumulative data from two experiments performed in triplicate. Asterisk (*) indicates W-13 pre-treatment results significantly different from vehicle pre-treatment at p < 0.001.
4. Discussion
Previously, we demonstrated direct interaction of calmodulin with PLCβ1 and PLCβ3 and the importance of calmodulin in regulating G protein-stimulated inositol phosphate accumulation by using calmodulin inhibitors in a whole cell assay. Additionally, calmodulin binding homology domains were identified in both PLCβ1 and PLCβ3 protein sequences. In our current study, we have characterized the affinity of the direct interaction between calmodulin and PLCβ3 or PLCβ1, quantitated the effect of calcium on the interaction, defined the effects of calmodulin on PLCβ3 activity in vitro, and further investigated the effects of calmodulin inhibitors in whole cells under varying conditions of activation. Cumulatively, these data support and expand the role for calmodulin in potentiating G protein-stimulated PLCβ activity, particularly Gβγ-stimulated PLCβ3 activity.
Fluorescence anisotropy data demonstrated 200 nM concentrations of both PLCβ1 and PLCβ3 bound calmodulin in the presence of 1 μM calcium. Because the number of known calmodulin binding proteins is vast and calmodulin binds proteins under both resting and calcium-activated conditions, the total amount of calmodulin in a cell is limited [31]. Therefore, it is predicted that protein must bind to calmodulin with Kd values in the sub-micromolar range to be expected to form physiologically relevant complexes [20], as found for calmodulins interaction with PLCβ isozymes. The Kd values for binding of PLCβ1 and PLCβ3 to calmodulin are also consistent with other calmodulin-binding signaling proteins such as the G-protein receptor kinases, GRK1, 2, and 5, which have affinities of 40 nM, and 2 μM, respectively, for calmodulin [32], and caldesmon which has an affinity for calmodulin of 1 μM [33]. The binding of calmodulin to PLCβ3 was sensitive to calcium concentrations over a physiological range with an apparent EC50 value of <1 μM (Figure 1), calculated using a non-saturating PLCβ3 concentration of 30 nM. This apparent EC50 is necessarily greater than the true EC50 of the complex because saturating concentrations of PLCβ3 were not achievable with limited amounts of purified protein available. The profile of Alexa-calmodulin anisotropy versus calcium concentration would be concentration dependent until PLCβ3 concentration was greater than the Kd. However, these data suggest that increasing cytosolic calcium concentrations following initial activation of PLCβ enzyme may support further activation by increasing association with calmodulin and potentiating Gβγ-stimulated PLCβ activity. Additionally the calcium dependence of the PLCβ3 isozyme is within the range of other known calcium/calmodulin dependent proteins with calcium dependent EC50 values of 0.3 μM, and 3 μM for GRK1 and GRK5, respectively [34]. Previous work in this lab with fragments from PLCβ3 protein suggested that the amino-terminal portion of the protein was a Ca++-independent calmodulin binding peptide. However, precipitation of full-length PLC-β from whole cell lysates with calmodulin-sepharose beads at physiologic calcium concentrations demonstrated that calmodulin and PLCβ can bind in the presence of calcium [1]. Clearly, the anisotropy data in this paper demonstrate that full length PLCβ1 and PLCβ3 bind calmodulin in a Ca++ sensitive manner. The EC50 value of 1 μM for calcium dependence of PLCβs interaction with calmodulin, although known to be a high estimate, is within physiological ranges. One limitation of our previous study employing protein fragments is that fragments lack the complete protein context which may confer additional functionalities or structural subtleties to a domain. While the binding data published previously using fragments of the PLCβ proteins identified a direct interaction between PLCβ and calmodulin, the anisotropy data presented herein should more accurately depict the calcium dependence of the full-length protein interaction. Calcium dependence is a hallmark of calmodulin binding protein interactions, and thus our current findings of calcium dependence are more expected rather than exceptional.
We have shown a direct effect of calmodulin in potentiating Gβγ activation of PLCβ3 (Figure 2, Panels A and C). One of the putative calmodulin binding sites of PLCβ3 overlaps with a site previously identified as important in Gβγ activation of PLCβ [35]. Additionally, the β subunit of Gβγ is a known calmodulin binding protein [36]. Thus calmodulin potentiation of PLCβ3 activation by Gβγ suggests the existence of a signaling complex that involves all three proteins, Gβγ, calmodulin and PLCβ3. The ability of calmodulin to potentiate activation of PLCβ3 is consistent with our data in whole cells whereby pre-treatment with calmodulin inhibitors attenuated muscarinic GPCR-stimulated inositol phosphate accumulation [1]. While muscarinic receptor-stimulated phosphatidylinositol hydrolysis in 1321N1 cells is characterized primarily as a Gαq /PLCβ1-mediated signaling event [37-39], the full response may involve Gβγ and PLCβ3. The potential for differential activation of PLCβ1 and PLCβ3 in varying spatial and temporal contexts is not at all well understood. Our data suggests that calmodulin is a co-factor in Gβγ-mediated PLCβ activation that may selectively increase PLCβ3 activity concurrently with temporal increases in intracellular calcium concentration. Possible mechanisms for potentiation of Gβγ-stimulated PLCβ activity by calmodulin include increased recruitment of Gβγ to PLCβ3 by calmodulin, or calmodulin potentiation of Gβγ-stimulated PLCβ3 hydrolytic activity by supporting the catalytic domain interaction with the membrane interface.
The role of calmodulin in regulating PLCβ1 remains elusive, despite our demonstration of direct binding. Calmodulin binding could potentiate PLCβ1 activation, independent of Gβγ, by serving as a temporal scaffold or recruiter of other signaling regulatory proteins. The functions of calmodulin are not limited to direct effects on activity. For example, calmodulin binding of Gβγ sterically interferes with binding to Gαi/o subunits [36]. In addition, the effect of calmodulin on GAP activities of the enzymes are unknown [40].
We determined that calmodulin does not alter the lipid selectivity profile of PLCβ3 or PLCβ1 for phospholipids bound to nitrocellulose (PIP strips®) in overlay assays. However, the PIP strip® overlay assays did reveal a previously unappreciated affinity of PLCβ3 and PLCβ1 for PIP3. PIP3 is a membrane phospholipid that has a well-deserved reputation as a regulatory lipid in a variety of cell signaling pathways [41-45], including a reciprocal association with calmodulin [29, 46] in other protein contexts. In addition to demonstrating an affinity of PLCβ3 and PLCβ1 for PIP3, the PIP strip® overlay assay also revealed that PLCβ isozymes have different affinities for synthetic and wild-type lipids. The single saturated chain species of synthetic phospholipids that are available on the commercial PIP Strip® membranes is very different from the wide array of lengths and saturations in side chains of lipids derived from mammalian sources that more accurately represent the composition of biological cell membranes [47]. The fatty acyl chain specificity of PLCβ isozymes has not yet been investigated, but has been reported in other proteins integral to GPCR signaling, such as RGS4 [29]. It is possible that calmodulin does contribute to lipid selectivity of PLCβs, but it is clear from the above data that chain length is an important factor in determining the role of calmodulin in lipid selectivity of PLCβ isozymes.
To further investigate the role of calmodulin in PLC activity in an actively signaling environment, we revisited the whole cell assays. Previously, we determined that pretreatment of 1321N1 cells with W-13 attenuates inositol phosphate accumulation after stimulation by carbachol. To determine if calmodulin inhibitors were disrupting a pre-activation function integral to PLCβ isozymes or eliciting a more direct effect in active signaling, we investigated the effects of W-13 on phosphatidylinositol phospholipid turnover in an actively signaling environment. We observed an effect of W-13 when added concurrently with carbachol equal to that of pre-treatment with W-13. Additionally, W-13 inhibited IP accumulation when added post-activation of muscarinic receptors. These results support a role for calmodulin in active signaling of PLCβs, suggesting that calmodulin does not simply act to pre-couple proteins necessary for activation. Because there are other PLCs in this signaling environment, in addition to other targets of calmodulin, we wanted to investigate early effects on IP3 accumulation in a time frame relevant to activation of PLCβs. W-13 attenuated IP3 accumulation at all time points, providing additional support for the hypothesis that the inhibition of calmodulin affects PLCβ signaling and not downstream pathways.
In conclusion, these studies further our understanding of the calmodulin/PLCβ interaction by showing that calmodulin can potentiate activation by the Gβγ dimer without affecting activation by Gαq. This work begins to detail the separate regulatory systems involving Gβγ and Gαq, and suggests cooperativity between Gβγ, calmodulin and PLCβ3; however, temporal and subcellular aspects of GPCR- and calcium-modulated cell signaling will also need to be better understood for a full understanding of calmodulin effects on PLCβ3-mediated signaling. Additionally, further research is required to demonstrate the importance of non-substrate phospholipids and membrane binding on the regulation of PLCβ3 activity.
Acknowledgements
This work was supported by the National Institutes of Health, National Institute of General Medical Sciences, PHS Grant GM61244 and an American Foundation for Pharmaceutical Education Predoctoral Fellowship (JSM).
Footnotes
Abbreviations: PLC, phosphatidylinositol phospholipid-specific phospholipase C; PE, phosphatidylethanolamine; PS, phosphatidylserine; PIP 16:0, synthetic phosphatidylinositol-4-phosphate with symmetric 16:0 saturated fatty acyl chains; PIP, phosphatidylinositol-4-phosphate; PIP2, phosphatidylinositol-4,5-bisphosphate; PIP3, phosphatidylinositol-3,4,5-trisphosphate; SDS-PAGE, sodium dodecyl phosphate polyacrylamide gel electrophoresis; W-13, N-(4-aminobutyl)-5-chloro-2-naphthalenesulfonamide; BSA, bovine serum albumin.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.McCullar JS, Larsen SA, Millimaki RA, Filtz TM. Calmodulin Is a Phospholipase C-{beta} Interacting Protein. J. Biol. Chem. 2003;278(36):33708–33713. doi: 10.1074/jbc.M301940200. [DOI] [PubMed] [Google Scholar]
- 2.Rebecchi MJ, Pentyala SN. Structure, Function, and Control of Phosphoinositide-Specific Phospholipase C. Physiological Reviews. 2000;80:1291–1335. [Google Scholar]
- 3.Hwang JI, Oh YS, Shin KJ, Kim H, Ryu SH, Suh PG. Molecular cloning and characterization of a novel phospholipase C, PLC-eta. Biochem J. 2005;389(Pt 1):181–6. doi: 10.1042/BJ20041677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Song C, Hu C-D, Masago M, Kariya K-i, Yamawaki-Kataoka Y, Shibatohge M, Wu D, Satoh T, Kataoka T. Regulation of a Novel Human Phospholipase C, PLCepsilon, through Membrane Targeting by Ras. J. Biol. Chem. 2001;276(4):2752–2757. doi: 10.1074/jbc.M008324200. [DOI] [PubMed] [Google Scholar]
- 5.Saunders CM, Larman MG, Parrington J, Cox LJ, Royse J, Blayney LM, Swann K, Lai FA. PLC{zeta}: a sperm-specific trigger of Ca2+ oscillations in eggs and embryo development. Development. 2002;129(15):3533–3544. doi: 10.1242/dev.129.15.3533. [DOI] [PubMed] [Google Scholar]
- 6.Lee CH, Park D, Wu D, Rhee SG, Simon MI. Members of the Gq alpha subunit gene family activate phospholipase C beta isozymes. J Biol Chem. 1992;267(23):16044–7. [PubMed] [Google Scholar]
- 7.Venkatakrishnan G, Exton JH. Identification of determinants in the alpha-subunit of Gq required for phospholipase C activation. J Biol Chem. 1996;271(9):5066–72. doi: 10.1074/jbc.271.9.5066. [DOI] [PubMed] [Google Scholar]
- 8.Wu DQ, Lee CH, Rhee SG, Simon MI. Activation of phospholipase C by the alpha subunits of the Gq and G11 proteins in transfected Cos-7 cells. J Biol Chem. 1992;267(3):1811–7. [PubMed] [Google Scholar]
- 9.Park D, Jhon D-Y, Lee C-W, Lee K-H, Rhee SG. Activation of phospholipase C isozymes by G protein βγ subunits. J.Biol.Chem. 1993;268:4573–4576. [PubMed] [Google Scholar]
- 10.Rhee SG, Bae YS. Regulation of phosphoinositide-specific phospholipase C isozymes. J.Biol.Chem. 1997;272(24):15045–15048. doi: 10.1074/jbc.272.24.15045. [DOI] [PubMed] [Google Scholar]
- 11.Lian L, Wang Y, Draznin J, Eslin D, Bennett JS, Poncz M, Wu D, Abrams CS. The relative role of PLC{beta} and PI3K{gamma} in platelet activation. Blood. 2005;106(1):110–117. doi: 10.1182/blood-2004-05-2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xie W, Samoriski GM, McLaughlin JP, Romoser VA, Smrcka A, Hinkle PM, Bidlack JM, Gross RA, Jiang H, Wu D. Genetic alteration of phospholipase C beta 3 expression modulates behavioral and cellular responses to {micro} opioids. PNAS. 1999;96(18):10385–10390. doi: 10.1073/pnas.96.18.10385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang H, Shen F, Herenyiova M, Weber G. Phospholipase C (EC 3.1.4.11): a malignancy linked signal transduction enzyme. Anticancer Res. 1998;18(3A):1399–404. [PubMed] [Google Scholar]
- 14.Yue C, Dodge KL, Weber G, Sanborn BM. Phosphorylation of serine 1105 by protein kinase A inhibits phospholipase Cbeta3 stimulation by Galphaq. J.Biol.Chem. 1998;273(29):18023–7. doi: 10.1074/jbc.273.29.18023. [DOI] [PubMed] [Google Scholar]
- 15.Stålberg P, Wang S, Larsson C, Weber G, Öberg K, Gobl A, Skogseid B. Suppression of the neoplastic phenotype by transfection of phospholipase C ? 3 to neuroendocrine tumor cells. FEBS Letters. 1999;450(3):210–216. doi: 10.1016/s0014-5793(99)00457-3. [DOI] [PubMed] [Google Scholar]
- 16.Weber G, Friedman E, Grimmond S, Hayward N, Phelan C, Skogseid B, Gobl A, Zedenius J, Sandelin K, Teh B. The phospholipase C beta 3 gene located in the MEN1 region shows loss of expression in endocrine tumours. Hum. Mol. Genet. 1994;3(10):1775–1781. doi: 10.1093/hmg/3.10.1775. [DOI] [PubMed] [Google Scholar]
- 17.Wang S, Gebre-Medhin S, Betsholtz C, Stalberg P, Zhou Y, Larsson C, Weber G, Feinstein R, Oberg K, Gobl A, Skogseid B. Targeted disruption of the mouse phospholipase C beta 3 gene results in early embryonic lethality. FEBS Lett. 1998;441:261–265. doi: 10.1016/s0014-5793(98)01518-x. [DOI] [PubMed] [Google Scholar]
- 18.Chin D, Means AR. Calmodulin: a prototypical calcium sensor. Trends Cell Biol. 2000;10(8):322–8. doi: 10.1016/s0962-8924(00)01800-6. [DOI] [PubMed] [Google Scholar]
- 19.Vetter SW, Leclerc E. Novel aspects of calmodulin target recognition and activation. Eur J Biochem. 2003;270(3):404–414. doi: 10.1046/j.1432-1033.2003.03414.x. [DOI] [PubMed] [Google Scholar]
- 20.Black DJ, Tran QK, Persechini A. Monitoring the total available calmodulin concentration in intact cells over the physiological range in free Ca2+ Cell Calcium. 2004;35(5):415–25. doi: 10.1016/j.ceca.2003.10.005. [DOI] [PubMed] [Google Scholar]
- 21.Malencik DA, Anderson SR. Dityrosine formation in calmodulin: cross-linking and polymerization catalyzed by Arthromyces peroxidase. Biochemistry. 1996;35(14):4375–86. doi: 10.1021/bi9526037. [DOI] [PubMed] [Google Scholar]
- 22.Filtz TM, Paterson A, Harden TK. Purification and G protein subunit regulation of phospholipase C-β from Xenopus laevis oocytes. J.Biol.Chem. 1996;271:31121–31126. doi: 10.1074/jbc.271.49.31121. [DOI] [PubMed] [Google Scholar]
- 23.Singer AU, Waldo GL, Harden TK, Sondek J. A unique fold of phospholipase C-mediates dimerization and interaction with Gq. Nature Structural Biology. 2002;9:32–36. [Google Scholar]
- 24.Paterson A, Harden TK. Expression, purification and reconstitution of recombinant phospholipase C-β isoenzymes. Methods in Neuroscience. 1996;29:246–263. [Google Scholar]
- 25.Smrcka AV, Hepler JR, Brown KO, Sternweis PC. Regulation of polyphosphoinositide-specific phospholipase C activity by purified Gq. Science. 1991;251:804–807. doi: 10.1126/science.1846707. [DOI] [PubMed] [Google Scholar]
- 26.Filtz TM, Li Q, Boyer JL, Nicholas RA, Harden TK. Expression of a cloned P2Y-purinergic receptor that couples to phospholipase C. Mol.Pharmacol. 1994;46:8–14. [PubMed] [Google Scholar]
- 27.Bishop CV, Stormshak F. Nongenomic Action of Progesterone Inhibits Oxytocin-Induced Phosphoinositide Hydrolysis and Prostaglandin F2{alpha} Secretion in the Ovine Endometrium. Endocrinology. 2006;147(2):937–942. doi: 10.1210/en.2005-0869. [DOI] [PubMed] [Google Scholar]
- 28.Runnels LW, Scarlata SF. Determination of the affinities between heterotrimeric G protein subunits and their phospholipase C-beta effectors. J.Biochem. 1999;38:1488–1496. doi: 10.1021/bi9821519. [DOI] [PubMed] [Google Scholar]
- 29.Popov SG, Krishna UM, Falck JR, Wilkie TM. Ca2+/Calmodulin Reverses Phosphatidylinositol 3,4,5-Trisphosphate-dependent Inhibition of Regulators of G Protein-signaling GTPase-activating Protein Activity. J. Biol. Chem. 2000;275(25):18962–24300. doi: 10.1074/jbc.M001128200. [DOI] [PubMed] [Google Scholar]
- 30.Wang J, Arbuzova A, Hangyas-Mihalyne G, McLaughlin S. The effector domain of myristoylated alanine-rich C kinase substrate binds strongly to phosphatidylinositol 4,5-bisphosphate. J Biol Chem. 2001;276(7):5012–9. doi: 10.1074/jbc.M008355200. [DOI] [PubMed] [Google Scholar]
- 31.Persechini A, Stemmer PM. Calmodulin is a limiting factor in the cell. Trends Cardiovasc Med. 2002;12(1):32–7. doi: 10.1016/s1050-1738(01)00144-x. [DOI] [PubMed] [Google Scholar]
- 32.Chuang TT, Paolucci L, De Blasi A. Inhibition of G protein-coupled receptor kinase subtypes by Ca2+/Calmodulin. J.Biol.Chem. 1996;271(45):28691–28696. doi: 10.1074/jbc.271.45.28691. [DOI] [PubMed] [Google Scholar]
- 33.Rhoads AR, Friedberg F. Sequence motifs for calmodulin recognition. FASEB J. 1997;11(5):331–340. doi: 10.1096/fasebj.11.5.9141499. [DOI] [PubMed] [Google Scholar]
- 34.Levay K, Satpaev D, Pronin A, Benovic J, VZ S. Localization of the sites for Ca2+-binding proteins on G protein-coupled receptor kinases. Biochemistry. 1998;37(39):13650–9. doi: 10.1021/bi980998z. [DOI] [PubMed] [Google Scholar]
- 35.Barr AJ, Ali H, Haribabu B, Snyderman R, Smrcka AV. Identification of a region at the N-terminus of phospholipase C-β3 that interacts with G protein βγ subunits. Biochem. 2000;39:1800–1806. doi: 10.1021/bi992021f. [DOI] [PubMed] [Google Scholar]
- 36.Liu M, Yu B, Nakanishi O, Wieland T, Simon M. The Ca2+-dependent Binding of Calmodulin to an N-terminal Motif of the Heterotrimeric G Protein β Subunit. J.Biol.Chem. 1997;272:18801–18807. doi: 10.1074/jbc.272.30.18801. [DOI] [PubMed] [Google Scholar]
- 37.Lechleiter J, Hellmiss R, Duerson K, Ennulat D, David N, Clapham D, Peralta E. Distinct sequence elements control the specificity of G protein activatio by musarnic acetylcholine receptor subtypes. EMBO. 1990;9:4381–4390. doi: 10.1002/j.1460-2075.1990.tb07888.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lechleiter J, Peralta E, Clapham D. Diverse functions of muscarinic acetylcholine receptor subtypes. Trends Pharmacol.Sci. 1989;(Supplement):34–38. [PubMed] [Google Scholar]
- 39.Peralta EG, Ashkenazi A, Winslow JW, Ramachandran J, Capon DJ. Differential regulation of PI hydrolysis and adenylyl cyclase by muscarinic receptor subtypes. Nature. 1988;334(6181):434–7. doi: 10.1038/334434a0. [DOI] [PubMed] [Google Scholar]
- 40.Chidiac P, Ross EM. Phospholipase C-beta 1 Directly Accelerates GTP Hydrolysis by Galpha q and Acceleration Is Inhibited by Gbeta gamma Subunits. J. Biol. Chem. 1999;274(28):19639–23699. doi: 10.1074/jbc.274.28.19639. [DOI] [PubMed] [Google Scholar]
- 41.Ching TT, Wang DS, Hsu AL, Lu PJ, Chen CS. Identification of multiple phosphoinositide-specific phospholipases D as new regulatory enzymes for phosphatidylinositol 3,4, 5-trisphosphate. J Biol Chem. 1999;274(13):8611–7. doi: 10.1074/jbc.274.13.8611. [DOI] [PubMed] [Google Scholar]
- 42.Pasquet J-M, Quek L, Stevens C, Bobe R, Huber M, Duronio V, Krystal G, Watson SP. Phosphatidylinositol 3,4,5-trisphosphate regulates Ca2+ entry via Btk in platelets and megakaryocytes without increasing phospholipase C activity. Embo Journal. 2000;19:2793–2802. doi: 10.1093/emboj/19.12.2793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Le Blanc C, Mironneau C, Barbot C, Henaff M, Bondeva T, Wetzker R, Macrez N. Regulation of Vascular L-type Ca2+ Channels by Phosphatidylinositol 3,4,5-Trisphosphate. Circ Res. 2004;95(3):300–307. doi: 10.1161/01.RES.0000138017.76125.8b. [DOI] [PubMed] [Google Scholar]
- 44.Menager C, Arimura N, Fukata Y, Kaibuchi K. PIP3 is involved in neuronal polarization and axon formation. Journal of Neurochemistry. 2004;89(1):109–118. doi: 10.1046/j.1471-4159.2004.02302.x. [DOI] [PubMed] [Google Scholar]
- 45.Czech MP. PIP2 and PIP3: complex roles at the cell surface. Cell. 2000;100(6):603–6. doi: 10.1016/s0092-8674(00)80696-0. [DOI] [PubMed] [Google Scholar]
- 46.Ishii M, Inanobe A, Kurachi Y. PIP3 inhibition of RGS protein and its reversal by Ca2+/calmodulin mediate voltage-dependent control of the G protein cycle in a cardiac K+ channel. PNAS. 2002;99(7):4325–4330. doi: 10.1073/pnas.072073399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Milne SB, Ivanova PT, DeCamp D, Hsueh RC, Brown HA. A targeted mass spectrometric analysis of phosphatidylinositol phosphate species. J. Lipid Res. 2005;46(8):1796–1802. doi: 10.1194/jlr.D500010-JLR200. [DOI] [PubMed] [Google Scholar]


