Abstract
Helicases use the energy derived from nucleoside triphosphate hydrolysis to unwind double helices in essentially every metabolic pathway involving nucleic acids. Earlier crystal structures have suggested that DNA helicases translocate along a single-stranded DNA in an inchworm fashion. We report here a series of crystal structures of the UvrD helicase complexed with DNA and ATP hydrolysis intermediates. These structures reveal that ATP binding alone leads to unwinding of 1 base pair by directional rotation and translation of the DNA duplex and ADP and Pi release leads to translocation of the developing single strand. Thus DNA unwinding is achieved by a two-part power stroke in a combined wrench-and-inchworm mechanism. The rotational angle and translational distance of DNA define the unwinding step to be 1 base pair per ATP hydrolyzed. Finally, a gateway for ssDNA translocation and an alternative strand displacement mode may explain the varying step sizes previously reported.
Introduction
The discovery of DNA double helix immediately presented the challenge of separating the intertwined strands for replication. DNA helicases were first isolated and characterized in the 1970s as DNA-dependent ATPases (Abdel-Monem et al., 1977; Richet and Kohiyama, 1976; Wickner et al., 1974). Since then a large variety of DNA and RNA helicases have been discovered and characterized. They are implicated in processes ranging from replication to translation (Gorbalenya and Koonin, 1993; Singleton and Wigley, 2002; von Hippel and Delagoutte, 2003) and more recently in ATP-dependent chromatin remodeling (Becker and Horz, 2002). In a broad sense, helicases can be viewed as motor proteins that translocate along double or single stranded nucleic acids. Based on sequence analysis, helicases have been grouped into six families: three superfamilies (SF 1 to 3), two small families (Rho and DnaB-like), and a branch in the AAA+ family (Gorbalenya and Koonin, 1993; Iyer et al., 2004). UvrD, originally known as DNA helicase II in E. coli (Hickson et al., 1983), is the founding member of SF1 and unwinds DNA in the 3′[###] 5′ direction (Matson and George, 1987). UvrD plays a critical role in replication, recombination, and repair of ultraviolet (UV) damage and mismatched base pairs (Arthur and Lloyd, 1980; Bruand and Ehrlich, 2000; Dao and Modrich, 1998; Ogawa et al., 1968; van de Putte et al., 1965; Veaute et al., 2005).
SF1 and SF2 members are distinct from other helicases by sharing seven conserved sequence motifs (Gorbalenya and Koonin, 1993; Hodgman, 1988). These motifs are involved in nucleoside triphosphate (NTP) binding and are located at the interface between two RecA-like domains in the structures of PcrA, Rep and RecBCD of SF1, and RecG, RecQ, UvrB, eIF4A, Swi2/SNF2 and viral NS3 helicases of SF2 (Bernstein et al., 2003; Durr et al., 2005; Singleton et al., 2004; Singleton and Wigley, 2002). The crystal structures of a UvrD homolog, PcrA, complexed with DNA-SO4 2- or DNA-AMPPNP (an ATP analog) (Velankar et al., 1999) showed that (1) single-stranded DNA (ssDNA) binds across the surface of the two RecA-like domains (1A and 2A), (2) 1A and 2A rotate towards each other when bound to ATP and open up after ATP hydrolysis, and (3) with the domain movement PcrA translocates along the ssDNA like an inchworm with alternating tight and loose interactions at two contact points. Structures of the Rep and NS3 helicases reveal a similar arrangement of the ATP-binding domains and single-stranded nucleic acid (Kim et al., 1998; Korolev et al., 1997). Kinetic studies of PcrA (Dillingham et al., 2000) support the structural finding that the step size of ssDNA translocation is one nucleotide advanced per ATP hydrolyzed.
Despite extensive biochemical and structural characterization of SF1 and other helicases, the mechanism for unwinding a DNA or RNA duplex remains highly controversial. To unwind a double helix, contacts between the helicase and the duplex region of DNA and a rotational movement are likely required in addition to ssDNA translocation, but no DNA rotation has yet been detected. PcrA and Rep helicases contain domains 1B and 2B in addition to 1A and 2A. Domain 2B of PcrA, which undergoes a ∼150° rotation upon binding to a double-and-single stranded (ds-ss) DNA junction and transforms the helicase from an “open” to “closed” state (Movie S1), has been shown to be essential for duplex binding and unwinding in solution (Soultanas et al., 2000). In the PcrA-DNA-AMPPNP co-crystal structure, the dsDNA is contacted by domains 1B and 2B, but the base pairs adjacent to the ds-ss junction are disrupted, and in the PcrA-DNA-SO4 2- structure this portion of the dsDNA is disordered (Velankar et al., 1999). The protein-dsDNA contact was therefore proposed to melt duplex and facilitate ssDNA translocation (Soultanas et al., 2000). However, when domain 2B is deleted in the homologous Rep helicase, the mutant Rep retains helicase activity both in vitro and in vivo (Cheng et al., 2002). In fact, the 2B domain can assume “open” or “closed” conformations when Rep is bound to a ssDNA (Korolev et al., 1997). These differences not only highlight the puzzling role of the 2B domain but the absence of a complete model of how DNA duplex is unwound. Furthermore, although PcrA is functional as a monomer, single-turnover kinetic studies of UvrD suggest that a dimeric form is required for DNA unwinding (Maluf et al., 2003). Finally, with different types of kinetic assay and data analysis, different values of step size (defined as the number of base pairs unwound and translocated per ATP hydrolyzed) have emerged, varying from 1 to 6 bps for a given helicase (Ali and Lohman, 1997; Lohman et al., 2003; Lucius and Lohman, 2004; Roman and Kowalczykowski, 1989). Even with the simplified ssDNA translocation analyses of monomeric UvrD, step sizes of 2 and 4 were reported by a single group (Fischer et al., 2004; Lohman et al., 2003).
We have determined 10 crystal structures of UvrD-DNA complexes at atomic resolution, which represent three distinct ATP hydrolysis states: (1) binary complexes with or without a bound SO4 2-, (2) ternary complexes with a non-hydrolyzable ATP analog (AMPPNP), and (3) ternary complexes with an ATP hydrolysis intermediate (ADP·MgF3). Each ds-ss DNA junction is bound by one UvrD monomer, and each structural state presents different interactions between UvrD and the single- and double-stranded regions of DNA. Together they reveal a previously unknown uni-directional rotation and translation of dsDNA and a gateway for ssDNA translocation. The structures and accompanying functional studies lead to the proposal of a combined wrench-and-inchworm mechanism for DNA unwinding by UvrD at the step size of one base pair.
Results and Discussion
Structures of UvrD-DNA complexes
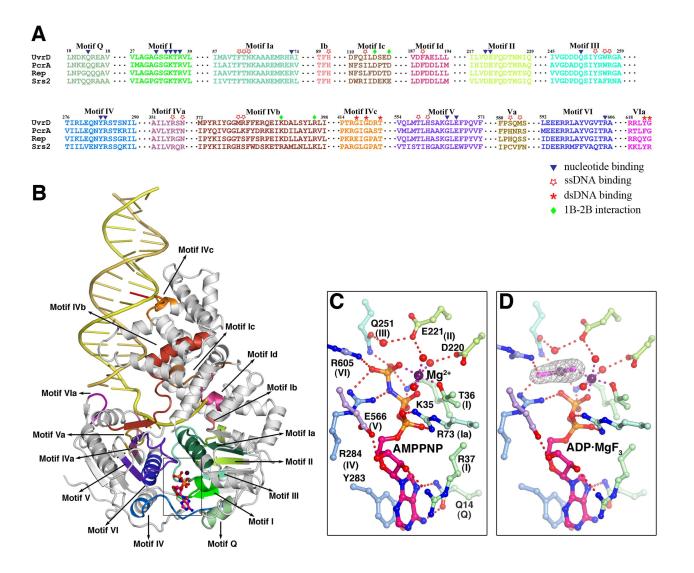
E. coli UvrD was crystallized with multiple self-complementary oligonucleotides that form 18 to 28 base pair (bp) duplexes flanked by 3′ 7-nucleotide (nt) overhangs and, in some instances, 5′ 1-nt overhangs (Figure S1). In each case, the crystallographic asymmetric units were composed of one such DNA and two bound UvrD monomers, one at each ds-ss junction. Crystals of UvrD-DNA binary complexes (28 bps + 7 nts), UvrD-DNA-AMPPNP (1nt + 18 bps + 7 nts) and the UvrD-DNA-ADP·MgF3 ternary complexes (18 bps + 7 nts) diffracted X-rays to 3.0, 2.6 and 2.2 Å, respectively, the highest resolution structures determined thus far for helicase-DNA complexes (Figure 1, Table 1). Residues 1-646 of UvrD, the entire duplex region of DNA, and the first 5 or 6 nts from the ds-ss junction are modeled in each structure. Residues 647-662 of UvrD form random coils but are often traceable in the ternary complex structures. In all UvrD-DNA co-crystal structures, the two UvrD molecules in an asymmetric unit are essentially identical, with rmsd’s of all Cαatoms varying between 0.3-0.5 Å. In addition, each reaction state was observed in multiple crystal lattices to verify that a particular conformation is independent of crystal packing (Figure S1). The differences between the binary and two ternary complexes are substantial (Figure 1).
Figure 1.
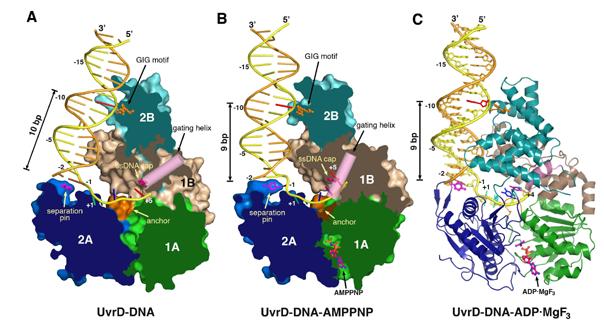
Crystal structures of UvrD-DNA complexes. (A) binary complex, (B) AMPPNP and (c) ADP·MgF3 ternary complexes. Domain 1A, 1B, 2A and 2B are colored green, beige, blue and cyan and shown in molecular surface with the front of molecule removed to exposed the bound DNA in (A) and (B), and in ribbon diagram in (C). Functionally important regions are highlighted in orange (the GIG motif and anchor) and hot pink (the separation pin, ssDNA cap and gating helix). DNAs are shown as tubular models in (A) and (B) and stick models in (C). The translocated strand is colored yellow and the partner strand orange. The DNA base contacting the GIG motif is highlighted in red.
Table 1.
Data collection and refinement statistics
| Sulfate-bound form | Nt-free | AMPPNP | ADP·MgF3 | |
|---|---|---|---|---|
| Data collection | ||||
| Space group | P21212 | P212121 | P21 | P21 |
| Resolution (Å)1 | 50 - 2.9 (3.00 - 2.90) | 50 - 3.0 (3.11 - 3.00) | 50 - 2.6 (2.69 - 2.6) | 50 - 2.2 (2.28 - 2.2) |
| Completeness (%)1 | 99.2 (92.5) | 87.0 (57.9) | 89.7 (51.5) | 93.5 (63.0) |
| Rmerge 1,2 | 8.8 (58.4) | 7.7 (46.9) | 6.1 (20.6) | 6.8 (29.3) |
| I / σ(I) 1 | 21.7 (2.17) | 22.8 (2.56) | 23.3 (4.1) | 30.1 (2.67) |
| Refinement | ||||
| Unique reflections | 48,037 | 37,494 | 61,012 | 102,202 |
| Protein + DNA atoms | 10,815 | 11,484 | 10,969 | 11,214 |
| Metal + Solvent atoms | 87 | 23 | 182 | 577 |
| R-factor (Rfree) (%)1,3 | 23.7 (29.6) | 23.0 (28.5) | 21.5 (25.7) | 21.0 (24.0) |
| Average B-factor (Wilson) (Å2)4 | 55.31 (32.36) | 76.27 (61.35) | 77.04 (46.42) | 56.05 (42.29) |
| R.m.s. deviations | ||||
| Bonds (Å) | 0.0080 | 0.0080 | 0.0067 | 0.0060 |
| Angles (°) | 1.38 | 1.41 | 1.23 | 1.13 |
Values for the highest resolution shell are indicated in parentheses.
, where I(h) is the intensity of reflection h, Σh is the sum over all reflections, and Σi is the sum over i measurements of reflection h.
, where Fo and Fc are the observed and calculated structure factor amplitudes. Rfree is calculated for a randomly chosen 10% of reflections which were not used for structure refinement and R-factor is calculated for the remaining reflections.
UvrD shares 42% sequence identity with PcrA and 37% with Rep. Like these two SF1 helicases, UvrD contains four structural domains 1A (1-89, 215-280 aa), 1B (90-214 aa), 2A (281-377, 551-647 aa), and 2B (378-550 aa), and adopts the “closed” conformation observed for the PcrA-DNA complexes (Velankar et al., 1999). Domains 1A and 2A form the core of the helicase responsible for ATP binding and hydrolysis (Figure 1). The ds-ss DNA junction associated with each UvrD molecule assumes the “L” shape, and the duplex and single stranded portions are roughly orthogonal to each other. The 3′-ssDNA tail is bound across domains 1A and 2A at their interface with 1B and 2B. Domains 1B and 2B interact with the DNA duplex along one side, covering 14 or 16 bps in the ternary and binary complexes, respectively (Figure 1). When the duplex is only 18 bps in length as in the AMPPNP ternary complexes, the central 10 bps are sandwiched between two non-contacting UvrD molecules (Figure S1A). Surprisingly, all DNA duplexes bound to UvrD are fully base paired and in regular B-form with no sign of melting or distortion.
The seven sequence motifs (I, Ia, II-VI) (Gorbalenya and Koonin, 1993) and the recently identified Q motif (Tanner et al., 2003) conserved among SF1 and SF2 are involved in ATP binding as reported (Theis et al., 1999; Velankar et al., 1999) (Figure 2). Motifs Ia, III and V are also involved in ssDNA binding. In addition to these eight motifs and motif IVa reported to be unique in SF1 (Korolev et al., 1998), we have identified seven new sequence motifs conserved among UvrD homologs including yeast Srs2. They are Ib, Ic, Id, IVb, IVc, Va and VIa (Figure 2A). These conserved residues participate in DNA binding or domain 1B and 2B interactions (Figure 2B). The unprecedented high-resolution of the UvrD structures allows examination of the atomic details of ATP binding and hydrolysis induced protein and DNA movement.
Figure 2.
The conserved sequence motifs among UvrD, PcrA, Rep and Srs2. (A) Sequence alignment of the 8 ATPase motifs (in different shades of green (domain 1A) and blue and purple (2A), and 8 newly found DNA binding and domain interaction motifs (in brown to red colors). (B) The 16 motifs are mapped onto the UvrD-DNA-AMPPNP structure using the same color scheme as in (A). (C) and (D) Coordination of the AMPPNP and ADP·MgF3 by the 8 helicase motifs. Carbon atoms of the UvrD side chains are colored light green (domain 1A) and light blue and purple (2A). Nitrogen atoms are shown in blue, oxygen in red, and Mg2+ in dark purple. Water molecules are shown as red spheres. The Fo-Fc electron density of MgF3 is superimposed onto the model.
ATP induced domain rotation
The ATP analog, AMPPNP, is bound in the cleft between domains 1A and 2A (Figure 2). The adenine base is sandwiched between Y283 (motif IV) and R37 (motif I) and is specifically selected by Q14 (Q motif) through bifurcated hydrogen bonds. GTP does not support helicase activity (Figure S2) (Matson and Kaiser-Rogers, 1990). The 3′ OH of the ribose is hydrogen bonded to E566 (motif V), but the 2′ OH is only weakly hydrogen bonded to R37, which explains why UvrD can use both ATP and dATP (Figure S2) (Matson and Kaiser-Rogers, 1990). Four basic residues, K35 (motif I or Walker A), R73 (Ia), R284 (IV) and R605 (VI), coordinate the triphosphate moiety, in particular the γ-phosphate (Figure 2C). A single Mg2+ ion essential for ATP hydrolysis is coordinated directly by the βand γphosphates and T36 (I), and by D220 and E221 (motif II or Walker B) through water. E221 is also well positioned to serve as a general base to deprotonate the water molecule that is oriented by Q251 (III) for the in-line nucleophilic attack (Figure 2C). Substitutions of the equivalent of Q251 in PcrA resulted in reduced ATPase activity (Dillingham et al., 1999).
In the attempt to crystallize UvrD-DNA-ADP complexes, we fortuitously made ADP·MgF3 complexes after NaF was added to improve the crystal growth. NaF reacted with MgCl2 and ADP in the crystallization solution and formed ADP·MgF3, which produced the best diffracting crystals of UvrD-DNA complexes (Table 1, Figure S1). MgF3 mimics the planar structure of the pentacovalent phosphate in the transition state, and the distances between the Mg (P mimic) and the apical oxygen atoms (attacking and leaving groups) are 1.9-2.0 Å (Figure 2D). MgF3 is believed to form a more authentic transition state analog than AlF(x) or BeF(x), of which x is either 3 or 4 (Graham et al., 2002). The arrangement of UvrD and DNA in the AMPPNP versus ADP·MgF3 bound state is almost identical except for the linkers between domains 2A and 2B and the 3′ end of ssDNA (see details later).
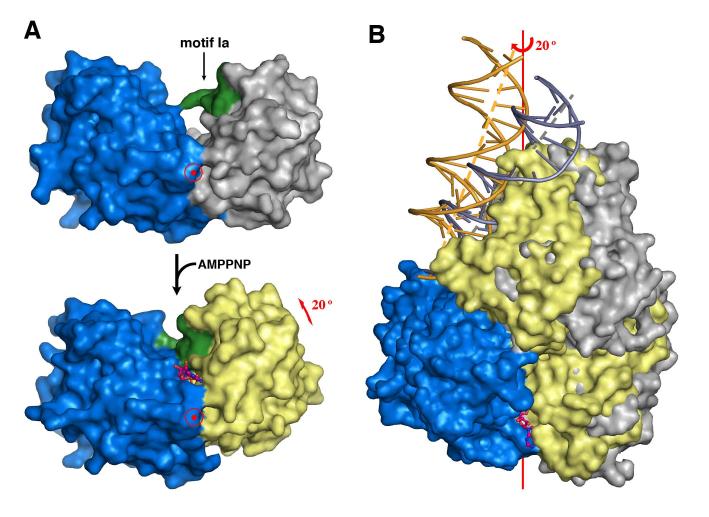
AMPPNP binding induces a ∼ 20° rotation between domain 2A and the remaining three domains (1A, 1B and 2B), and the distance between the respective centers of mass is reduced by 3.3 Å (Figure 3). Domains 1A and 1B are linked by a shared hydrophobic core, and domains 1B and 2B by salt bridges (K389 to D115 and R396 to D118). The three domains of UvrD move as one unit (Figure S3). In contrast, the movement of domain 2B in PcrA is uncoupled from that of domains 1A and 1B (Velankar et al., 1999). The fully ordered dsDNA in the UvrD structures likely stabilizes the 2B domain and 2B-1B interactions (Figure 1). Local structural changes between the binary and ternary complexes also occur in the ATP-binding loops and the loops that contact DNA. After ATP hydrolysis, the domain rotation is presumably reversed during release of ADP and Pi. The domain rotation axis passes through the Cαof W256 (motif III, located at the 1A and 2A interface) and is 15Å from the helical axis of dsDNA at a ∼ 25° angle (Figure 3).
Figure 3.
ATP-dependent domain rotation. (A) Domains 2A (blue) and 1A (grey or yellow) shown in molecular surface rotate towards each other by 20° upon binding of AMPPNP. For convenience, domain 2A is held stationary. Motif 1a is highlighted in green as a reference point. AMPPNP is shown as pink and orange ball-and-sticks. The view is down the rotation axis marked by the target sign. (B) An orthogonal view from (A) with the superimposed full UvrD-DNA binary (blue and silver) and ternary (blue, yellow and gold) complex structures. The DNA helical axis is shown as a grey (binary complex) or yellow (ternary complex) dashed line.
Rotation and translation of the dsDNA
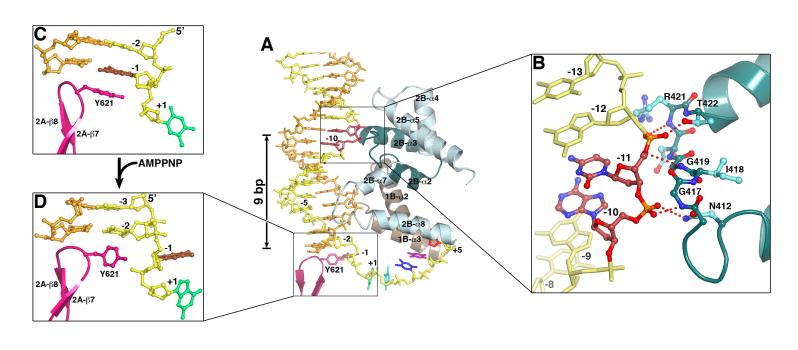
The domain movement of UvrD is coupled with the directional movement of the DNA duplex. Three helix-loop-helix (HLH) structures emanating from domain 2B (α2-α3, α4-α5, and α7-α8) alternately contact each DNA strand in the duplex, and the N-terminus of the second helix of each HLH is in close contact with the DNA backbone (Figure 4A). The first HLH (2B-α2-α3), which contains the PxxGIGxxT sequence (motif IVc or GIG), is conserved among UvrD homologs (Figure 2A). The backbone amide groups of the two Gly’s form tight hydrogen bonds with the DNA backbone (Figure 4B). Interestingly, the first Gly of GIG is replaced by Glu in Rep (Figure 2A), and Rep binds duplex DNA poorly (Myong et al., 2005). Near the ds-ss junction, a fourth HLH from domain 1B (α2-α3) is in van der Waals contact with both DNA strands in the minor groove (Figure 4A). Upon AMPPNP binding, the duplex moves with domains 1A/1B/2B towards 2A. The movement includes a 3.3 Å translation and a ∼20° left-handed rotation that untwists the double helix (Figures 1, 3).
Figure 4.
Interactions between UvrD and dsDNA. (A) The four HLH structures from domains 2B (light and dark cyan) and 1B (brown) interact with 14 to 16 bps. These interactions are similar between the binary and ternary complexes, while the ternary complex structure is shown. The separation pin (Y621) buttresses the end of DNA duplex. (B) A close-up view of the GIG motif and dsDNA interactions. (C) Stacking of Y621 with the-1 bp in the binary complex. (D) Unwinding of the-1 bp in the ternary complex, and the accompanying side chain conformational change of Y621.
At the ds-ss junction, a β-hairpin (614-626 aa, motif VIa) in the 2A domain, which we call the separation pin, buttresses the end of duplex. Y621 on its tip forms a π-ring stack with the first base pair (-1) of the DNA duplex in the binary complex (Figure 4C). When pressed against the separation pin in the AMPPNP or ADP·MgF3 ternary complexes, the-1 base pair becomes unpaired (Movie S2), and the side chain of Y621 rotates to a vertical position as if to facilitate a newly unpaired base to flip out (Figure 4D). The nucleotide opposite it on the partner strand is disordered. As the-1 bp is unpaired, the number of base pairs between the GIG motif and separation pin is reduced by one, from 10 to 9, in the ternary complexes (Figures 1, 4).
Movement of the ssDNA
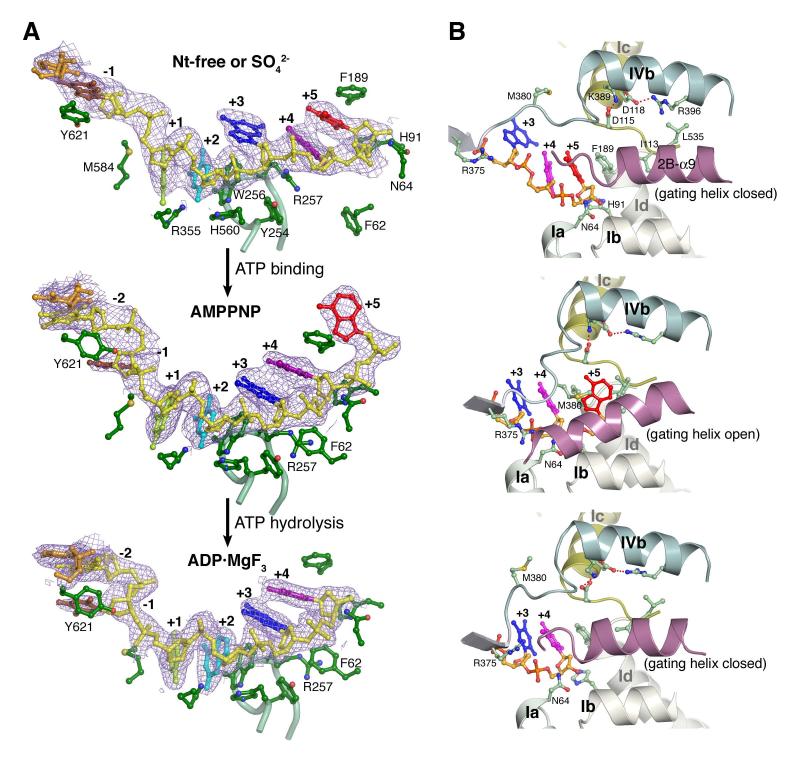
Upon binding of AMPPNP, the first three nucleotides of the existing ssDNA (numbered +1 onwards from the ds-ss junction) retain the same positions as in the binary complexes, so the newly unpaired base (-1) on the same strand bulges out (Figures 1, 5). The +1 to +3 nts interact extensively with domain 2A (M584, R355, H560) and 1A (Y254 and W256 of motif III) (Figure 5A). Since W256 intersects the domain rotation axis, it remains immobile throughout the ATPase cycle. W256 stacks with the bases of the +1 and +2 nts and buttresses the +3 base (Figure 5A). Motif III, which is essential for the helicase activity (Dillingham et al., 1999), functions as an anchor for ssDNA during the transition from the binary to the ternary complex.
Figure 5.
Interactions between UvrD and ssDNA. (A) The interactions in the three distinct states of UvrD-DNA complexes. The backbone of -1 to +5 nts of the translocated strand is shown in yellow and the bases in distinct colors. A 2Fo-Fc electron density map corresponding to the ssDNA is shown in light purple. Carbon atoms of the protein side chains are shown in green, oxygen in red, nitrogen in blue and sulfur in yellow. The Cα trace of Motif III is shown as a light green tube. (B) Diagram of the gating helix conformation with the salt bridges at the domain 1B and 2B interface shown. Each corresponds to the nucleotide-binding state shown on the left.
Movement of the ssDNA upon AMPPNP binding occurs at the +4 and +5 nts. In the binary complexes, the +4 and +5 bases are stacked with the side chains of Y254 and R257 forming a π-cation-πladder with ∼3.4 Å spacing, and the ladder is capped after the +5 nt by F189 (motif Id) and H91 (motif Ib) on the base and deoxyribose, respectively (Figure 5A). The domain rotation of 1A/1B/2B induced by AMPPNP brings the ssDNA cap (F189 and H91) towards the anchor (W256) and breaks the π-cation-πstack between Y254 and the F189. The guanidinium group of R257 moves out of the way and stacks with F62 (motif Ia) (Figure 5A). Concomitantly, the +4 base shifts up and stacks between the +3 base and F189; the +5 nt moves beyond F189 and exits from the helicase (Movie S3). Among 8 crystal structures of UvrD-DNA complexed with AMPPNP or ADP·MgF3 (Figure S1C), exiting of the +5 nt is either complete, so it becomes untraceable like the +6 and +7 nts in the binary complexes, or is caught in the process when the gateway (see below) is stuck open (Figure 5B).
The “gating helix” regulates ssDNA translocation
The ssDNA gateway is formed by the linker between domain 2A and 2B and the last helix in domain 2B (2B-α9), which we term the “gating helix”. In the binary complex, the gating helix is held in the “closed” conformation against domain 1B (residue 110-118 or motif Ic) by van der Waals contacts and a water-mediated hydrogen bond (Figure 5B). When closed, it leaves a small aperture that permits the phosphosugar backbone of ssDNA to thread through but blocks the passage of a whole nucleotide. In two AMPPNP and two ADP·MgF3 UvrD ternary complexes (Figure S1C), the gating helix is reoriented and “open”, and the transiting +5 nt is sandwiched between the gating helix and motif Ic. The conserved N64 (motif Ia) helps to orient the gating helix in both “closed” and “open” conformations by forming a hydrogen bond with alternate carbonyl oxygens at its C-terminus (Figure 5B). The gating function of this helix was not observed previously because it remained closed in the PcrA-DNA-AMPPNP ternary complexes (Velankar et al., 1999). Similarly, it is closed in four other UvrD-DNA ternary complexes (Figures 1C, 5B, S1C). However, the gateway for the ssDNA passage must be transiently open in the AMPPNP or ADP·MgF3 bound state to let the +5 nt pass through.
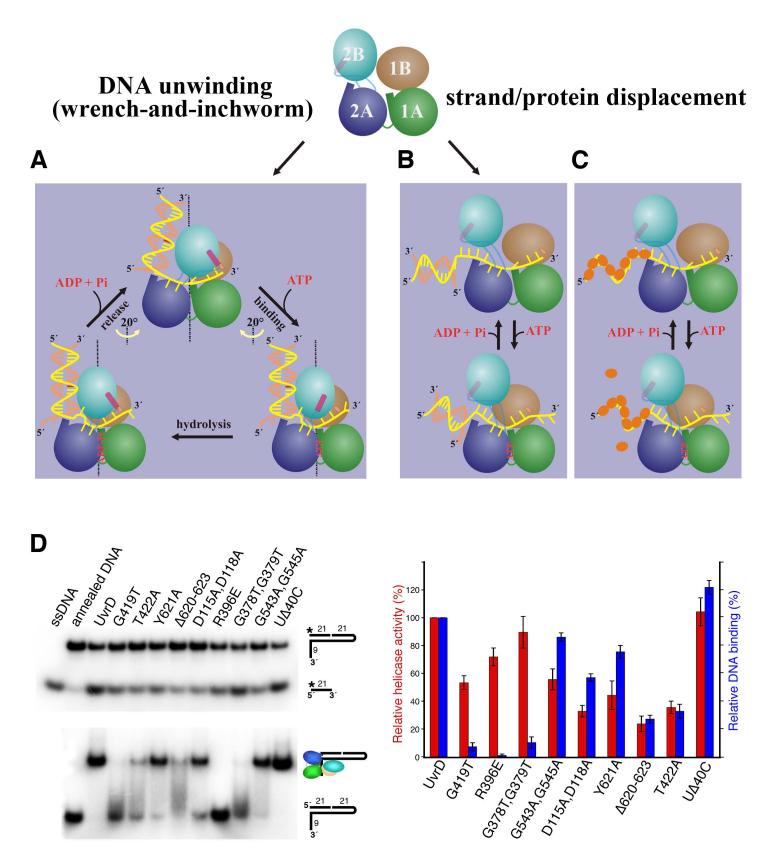
DNA unwinding by a combined “wrench-and-inchworm” mechanism
ATP binding-induced domain closing between 2A and 1A/1B/2B, which leads to separation of the first base pair (-1) at the ds-ss junction (Figures 1, 4), is only the first part of the ATPase-driven power stroke. During this step, the rearrangement of the +4 nt and exit of the +5 nt prepare the ssDNA for transloction. After ATP hydrolysis, release of ADP and Pi leads to domain opening and ssDNA translocation, which constitute the second part of the power stroke and complete the unwinding and translocation of 1 bp. During this step, the ssDNA cap (F189 and H91) must be fixed on the +4 nt (or the new +5), so the nucleotides preceding it, which are conveniently stacked in a spiral (Figure 5A), can be “pulled” by the domain rotation and each translocates by 1 nt relative to the anchor (motif III) (Figure 5, Movie S4). This straightens the bulged out-1 nt and leads it to the +1 position. Meanwhile, domains 2B and 1B slide up along the duplex curvature by 1 bp and rotate 20° in the right-handed direction, so UvrD is ready to unwind the next base pair. The two 20° rotations associated with the ATP binding and ADP release occur in opposite directions and total ∼40°, which approximates the twisting angle of one base pair in the B form. The combined rotation angle and the translocation distance define the UvrD step size to be 1 bp. The screw-like movement of dsDNA observed in the UvrD structures complements the ssDNA translocation depicted by the “inchworm” mechanism (Velankar et al., 1999). We name this two-part power stroke of DNA unwinding (Movies S5, S6) the “wrench-and-inchworm” mechanism.
Four protein-DNA contact points are critical for the “wrench-and-inchworm” mechanism. During the ATP binding step, the contacts made by the GIG motif with the duplex and by the anchor point with the ssDNA are fixed, while the contacts at the separation pin and ssDNA gateway (gating helix and ssDNA cap) are flexible thereby permitting 1 bp (-1) to unwind and 1 nt (+5) to exit. During the ADP and Pi release step, however, these four contact points reverse their roles in “holding” and “letting go” of DNA. For instance, the “gating helix” must be closed and the “separation pin” is stiff to prevent the ssDNA from sliding backward and the-1 bp from re-annealing. Meanwhile, the contact between the GIG motif and the DNA duplex is presumably loosened. Although the GIG-DNA duplex interactions appear similar in the binary and ternary complexes (Figure 1), studies of PcrA-DNA interactions revealed that the duplex is better protected from nuclease digestion in the presence of AMPPNP than in its absence (Soultanas et al., 2000). In a third of UvrD-DNA ternary complexes, the base pair number between the separation pin and GIG motif deviates from 9 bps (Figure S1C). This may be attributed to the crystal lattice contacts that supersede the weak GIG-dsDNA interactions. The alternating tight and loose associations at these four contact points thereby define the step size and 3′ to 5′ direction of DNA unwinding.
Mutagenic probing of the 2B domain and UvrD-dsDNA interactions
The “wrench-and-inchworm” mechanism predicts that the 2B domain is essential for binding and unwinding the duplex region of DNA (Figures 4, 6A) (Velankar et al., 1999). In addition, the “gating helix” located in domain 2B regulates ssDNA translocation only when the 2B domain assumes the closed conformation (Movie S1, Figure 6A). Otherwise the “gating helix” is more than 20Å away from the 3′ ssDNA. Yet domain 2B was shown to be unnecessary and even inhibitory for Rep to translocate along ssDNA and separate duplex DNA (Cheng et al., 2002). To probe the role of the 2B domain in dsDNA binding and helicase activity, several mutations were introduced into UvrD. G419T and T422A in the GIG motif and Y621A and Δ620-623 (deletion of residues 620 to 623) in the separation pin were constructed to directly perturb the UvrD-duplex interactions (Figure 4). Since only in the “closed” conformation can domain 2B interact with the dsDNA (Figure 1, 6A) (Rasnik et al., 2004; Velankar et al., 1999), G378T/G379T and G543A/G545A in the linkers between domains 2A and 2B and R396E and D115A/D118A at the domain 1B-2B interface (Figure 5B) were constructed to destabilize the “closed” conformation.
Figure 6.
Structural and functional duality of UvrD. (A) Cartoon presentations of the “wrench-and-inchworm mechanism” for DNA unwinding. Each domain is color-coded and the gating helix is highlighted in pink. (B) and (C) A mechanism for strand and protein displacement. The small orange ovals represent RecA-like proteins. (D) Assays of WT and mutant UvrDs in DNA unwinding (top left) and dsDNA binding (bottom left) using the nicked hairpin DNA substrate as diagramed. The relative helicase and DNA binding activities of each mutant UvrD are averaged from 3 measurements and plotted on the right in red (helicase) and blue (DNA binding).
These UvrD variants retained normal ATPase activity (data not shown). Furthermore, similar to wildtype UvrD, each mutant protein was able to bind ssDNA as indicated by greatly increased ATPase activity in the presence of ssDNA (data not shown). Such behavior was expected since the amino acid substitutions were distant from the ATP- and ssDNA-binding sites. dsDNA binding and helicase activities were measured using a specially designed nicked hairpin substrate, which consists of two contiguous 21-bp duplexes and a 9-nt 3′ tail (Figure 6D). With this DNA substrate, wildtype UvrD is able to maintain the 2B-dsDNA interactions and the “closed” conformation when unwinding the first 21 bps.
Five mutant UvrDs showed reductions in both dsDNA binding and helicase activity (Figure 6D). The Y621A, G537A/G539A and D115A/D118A UvrD variants showed only slight loss of both activities. This is not surprising since G537 and G539 are not conserved among UvrD homologs and Y621A and D115A/D118A substitutions remove one or two functional groups without perturbing the protein structure. In contrast, UvrD containing the T422A or Δ620-623 mutation showed moderately reduced dsDNA binding and severely diminished helicase activity. The correlation between duplex binding and helicase activity supports the “wrench-and-inchworm” mechanism that we are proposing based on the crystal structures.
The remaining three mutant proteins (G419T, R396E and G378T/G379T) exhibited severely reduced dsDNA binding yet robust helicase activity (Figure 6D). UvrD with the G378T/G379T (motif IVb) and R396E substitutions were expected to retain the “open” conformation due to the restricted φand Ψ angles of Thr’s that replacing the two Gly’s in the 2A-2B linker and the repulsion between 1B and 2B domain by the charge reversal of R396E. The diminished dsDNA binding supported this prediction. The replacement of the second Gly in the GIG motif by Thr (G419T) (Figure 4B) likely distorted the protein backbone conformation, and dsDNA binding was barely detectible. The robust helicase activity of these three UvrD variants implies that dsDNA binding is not necessary for UvrD to separate DNA duplexes efficiently. These observations resemble that of the Rep helicase without the 2B domain (Cheng et al., 2002) and appear inconsistent with the “wrench-and-inch” mechanism.
“Strand displacement” as an alternative mode of duplex separation
The presence of helicase activity that is independent of the 2B domain and dsDNA binding led us to consider a “strand displacement” mode as an alternative to the “wrench-and-inchworm” mechanism. When a helicase translocates along one strand of a duplex, it may displace the partner strand like a wire-stripper. In this mode of duplex separation, only the ability to translocate along ssDNA is essential; dsDNA binding is unnecessary. The 2B domain may even be inhibitory for ssDNA translocation (Brendza et al., 2005) because when not associated with the duplex it assumes either the “open” or “closed” conformation (Korolev et al., 1998). When the 2B domain is “closed’, the gating helix may also be closed and inhibit ssDNA translocation.
The robust helicase activity of the R396E and G378T/G379T UvrD variants as well as a previously identified mutant UvrD with the D403A/D404A substitutions at the domain 1B-2B interface (Zhang et al., 1998) supports the idea that the ”open“ conformation is favorable for strand displacement. The lack of dsDNA binding by the G419T UvrD variant probably allows this mutant protein to intermittently assume the ”open“ conformation and separate DNA duplexes in the ”strand displacement“ mode (Figure 6B, 6D). In contrast, the nearby T422A substitution, which results in poor helicase activity (Figure 6D), may cause defects in both modes of duplex separation. Unwinding of dsDNA by the T422A UvrD is likely impeded due to its loosened grip on dsDNA (Figure 4B), yet the remaining interactions with dsDNA may keep the 2B domain in the ”closed“ conformation and slow down ssDNA translocation. Similarly, inefficient dsDNA unwinding and the ”closed“ conformation of the Δ620-623 UvrD variant, which lacks the separation pin, may lead to defects in both modes of duplex separation and the low helicase activity (Figure 6D).
One may consider that ssDNA translocation without dsDNA binding in the ”strand displacement“ mode is equivalent to half of the “wrench-and-inchworm” movement. However, the conformational requirement for the 2B domain and the role of the “gating helix” are distinctly different in the two modes. The “wrench-and-inchworm” mechanism requires a “closed” conformation of 2B, so the GIG motif controls the screw-like motion of dsDNA and the “gating helix” regulates the step size of ssDNA translocation. In the “strand displacement” mode, the “open” conformation and a disengaged “gating helix” appear to work most efficiently. Given a DNA substrate with the duplex portion sufficient to bind the 2B domain, wildtype UvrD is likely to unwind it by the “wrench-and-inchworm” mechanism. The “strand displacement” mode may be adopted in the absence of dsDNA or by mutant UvrDs that are unable to bind dsDNA.
Reconciliation of multimerization state and different step sizes of UvrD
The established helicase activity assays usually do not distinguish whether a duplex is separated by the “wrench-and-inchworm” or “strand displacement” mechanism. Given a DNA with a 20 bp-plus duplex and a 3′ tail, UvrD is likely to unwind it by the “wrench-and-inchworm” mechanism at the beginning, but when the duplex portion is too short (<14 bps) to interact with the 2B domain, the helicase has to switch to the “strand displacement” mode. The single turnover kinetic analysis has led to the proposal that a dimeric UvrD is required to unwind dsDNA (Maluf et al., 2003). To date, only monomeric forms of SF1 and SF2 helicases have been observed by crystallography with or without DNA substrate (Bernstein et al., 2003; Durr et al., 2005; Singleton and Wigley, 2002). The heterotrimeric RecBCD is exceptional, but it contains only a single copy of each subunit (Singleton et al., 2004). The co-crystal structures of PcrA and UvrD with DNA show that no more than one helicase molecule can be accommodated at a ds-ss DNA junction. These observations concur with the biochemical analyses in solution that monomeric UvrD and PcrA are active helicases (Dillingham et al., 2000; Mechanic et al., 1999; Soultanas et al., 2000). In the single turnover experiment (Maluf et al., 2003), the first UvrD molecule may have difficulty in switching from the “wrench-and-inchworm” to the “strand displacement” mode and dissociate from DNA, and a second UvrD molecule may be needed to facilitate the conformational change or complete the duplex separation (Byrd and Raney, 2005).
The two alternative modes of duplex separation and mixed “open” and “closed” conformations of the 2B domain without dsDNA binding may also complicate kinetic analyses of the step size of UvrD-like helicases. Each kinetic measurement may represent an ensemble of multiple structural and functional states, and data interpretation may not account for all the possibilities. Moreover, in the “strand displacement” mode the GIG motif and the gating helix are likely disengaged, which may lead to deregulation of the direction and step size of ssDNA translocation (Figure 6B). When bound to Rep and UvrD, the bases on the ssDNA are stacked neatly in a spiral (Figure S4) and can slide in either direction more than 1 nt at a time without the gating helix. PcrA is exceptional as it has a binding pocket for the +4 nt, which precludes it from forming the ssDNA spiral (Figure S4). This feature may limit PcrA to translocating along ssDNA 1 nt at a time (Dillingham et al., 2000). In contrast, the step size of UvrD translocating along ssDNA is reported to be 2 to 4 nts (Fischer et al., 2004; Lohman et al., 2003).
Implications for DNA repair and recombination
Why do the UvrD-like helicases have this structural and mechanistic duality (Figure 6A, B)? UvrD is required to unwind hundreds of base pairs during DNA mismatch repair. Unwinding most probably occurs by the “wrench-and-inchworm” mechanism. For example, the D403A/D404A UvrD with the destabilized 1B-2B domain interface causes deficiencies in DNA repair in vivo despite the robust helicase activity in vitro (Zhang et al., 1998). UvrD is also required to dismantle RecA filaments on ssDNA and prevent unwanted recombination (Arthur and Lloyd, 1980; Veaute et al., 2005). Removal of RecA from ssDNA is mechanically similar to strand displacement. Structural and mechanistic duality may allow UvrD to serve both cellular functions efficiently (Figure 6A-C). Interestingly, the dual conformational states of the 2B domain can interfere and suppress both functions, which may be important to keep the helicase inactive until needed. UvrD without the 2B domain is cytotoxic (Cheng et al., 2002), and E. coli cells harboring the G378T/G379T UvrD variants grew very poorly (data not shown). To overcome the auto-inhibition, UvrD has to be activated by MutS and MutL during mismatch repair (Dao and Modrich, 1998). Unwinding of 100 bps was undetectable when UvrD and DNA were mixed in an equimolar ratio, and addition of MutL significantly improved the helicase activity (Guarné et al., 2004). MutL may stabilize the “closed” conformation of UvrD to facilitate dsDNA unwinding. Conversely, to displace RecA efficiently, stabilization of the “open” conformation may be required. SUMOylated PCNA may serve such a role in recruitment and stimulation of the yeast UvrD homolog, Srs2, to disrupt the RAD51 filament (RecA homolog) (Macris and Sung, 2005; Pfander et al., 2005).
Concluding remarks
Our analyses of DNA unwinding by UvrD for the first time reveal two separate parts of the power stroke delivered by a motor protein. Large conformational changes upon ATP binding have been observed in many ATPases. Apparently the free energy of ATP binding alone is sufficient to unwind one base pair, but to translocate the newly unpaired base in the developing single strand requires ATP hydrolysis and ADP and Pi release. The 1 bp step size of unwinding appears to be inefficient given that hydrolysis of one ATP releases ∼8 kcal/mole. The excess of energy may be spent partly on the conformational changes of the helicase and partly on coupling such conformational changes with the uni-directional unwinding of DNA. Similar ATPase domains found in SF1 and SF2 helicases must be the heart of the engine that couples protein domain rotation with the movement of ds, ss or ds-ss forms of DNA or RNA and alters the nucleic acid conformation for appropriate biological processes.
Experimental Procedures
Preparation of protein and DNA
The expression vector for the C-terminal 40 residue truncated UvrD (U∆40C) (pWY2068) was derived from pET11d-UvrD (George et al., 1994) using QuikChange (Stratagene). Mutant UvrD, G419T (pWY2072), Y621A (pWY2073), Δ620-623 (pWY2074), G378T/G379T (pWY2075), G543A/G545A (pWY2076), T422A (pWY2077), R396E (pWY2078), and D115A/D118A (pWY2079), were also derived from pET11d-UvrD and are full-length. U∆40C and mutant UvrD were expressed and purified as described (Guarné et al., 2004). Oligonucleotides U01 composed of a 10 mer (5′-CGAGCACTGC-3′) and a 17 mer (5′-GCAGTGCTCGTTTTTTT-3′), self-complementary oligonucleotides, U18 (5′-CGAGCACTGCACTCGAGTGCAGTGCTCGTTGTTAT-3′ and U13 (5′-CGAGCACTGCAGTGCTCGTTGTTAT-3′ were synthesized by the Keck Oligonucleotide Synthesis Facility (Yale University) and purified using HPLC. U38 (5′-TCGAGCACTGCAGTGCTCGTTGTTTA-3′) were synthesized in-house and gel purified. After submitting this manuscript, we became aware that the pET11d-UvrD contains the A399V mutation and so are all UvrD clones subsequently made. The mutation was correct in pET11d-UvrD, and fortunately the ATPase, DNA binding and helicase activity of the A399V and wildtype UvrD are indistinguishable.
Crystallization
U∆40C complexed with the U01 oligos was purified by gel-filtration chromatography in buffer A (20 mM Tris (pH 8.0), 150 mM KCl, 0.1 mM EDTA, 5 mM MgCl2, 1 mM DTT and 5% glycerol) and concentrated in a Centricon to a final protein concentration of 6 mg/ml. Crystals were obtained using the sitting-drop vapor-diffusion method at 4 °C with PEG3350 as a precipitant and improved by soaking in 0.6-1.0 mM ethylmercury phosphate. U∆40C and self-complementary U13, U18 or U38 were mixed at a 2:1 molar ratio in buffer A and concentrated to ∼ 6 mg/ml of UvrD. UvrD-U18 complexes were crystallized in 12-16 % PEG 3350 and 0.2 M potassium formate; UvrD-U13-ADP·MgF3 in 18% PEG 3350, 0.15 M NaF and 50 mM Tris (pH 8.8); and UvrD-U38-AMPPNP in 17% PEG 3350 and 0.2 M NaF. Crystals were cryo-protected by 20% (v/v) glycerol and flash frozen in liquid nitrogen.
Data collection and structure determination
Diffraction data were collected at the SER-CAT beam-line 22-ID in the Advanced Photon Source, Argonne National Laboratories. The data were processed using HKL2000 (Otwinowski and Minor, 1997). The structure of UvrD-U01 complex was solved by molecular replacement (Navaza, 2001) using the PcrA structure (PDB: 2PJR) as a search model. Two UvrD-DNA complexes were placed in each asymmetric unit and packed in the head-to-head fashion (Figure S1A). Subsequently, this model was used to solve the structures of nucleotide-free (Nt-free), AMPPNP and ADP·MgF3 complexes. The structures were refined using COOT and CNS (Brünger et al., 1998; Emsley and Cowtan, 2004) (Table 1).
Helicase and DNA-binding activity assays
Helicase activity of UvrD was assayed in buffer B (20 mM Tris pH7.5, 80 mM NaCl, 0.1 mg/ml BSA, 2.6 mM MgCl2, 1 mM DTT) with 2 mM ATP and 4 nM of the nicked hairpin DNA substrate, which is made of a 21 mer (32P 5′-end labeled) and 75 mer forming a 42bp dsDNA with a nick in the center and 3-nt hairpin loop followed by 9 nts at the 3′ end (Figure 6D). Reactions were initiated by addition of 40 nM UvrD to the final 10 μl reaction mixture and stopped by the addition of 5 μl of 9 mM EDTA, 0.3% SDS and 150μg/ml proteinase K (New England Biolabs) after incubation at room temperature for 20 min. Unwinding products were analyzed on a 20% TBE gel by electrophoresis and quantified using phosphorimaging plates and TYPHOON 8600.
DNA binding by UvrD was examined by gel mobility shift assays. Reactions were in 10 μl buffer B with 5% glycerol, 40 nM UvrD, and 4 nM 32P-end labeled DNA at the room temperature for 10 minutes. Samples were then electrophoresed at 80V for 1 hr in 6% TBE gel and quantified using phosphorimaging plates and TYPHOON 8600.
Coordinates
Atomic coordinates and structure factors of the UvrD-DNA complexes have been deposited in the Protein Data Bank. The accession codes are 2IS2 for the Nt-free and 2IS1 for the SO4 2--bound binary complex, and 2IS4 for the AMPPNP and 2IS6 for the ADP·MgF3 ternary complex.
Supplementary Material
Acknowledgement
We thank D. Leahy, R. Craigie and P. Kwong for reviewing the manuscript, P. Friedhoff for finding the A399V mutation, and M. Dillingham for insightful discussion. This research was supported fully by the Intramural Research Program of the NIH, NIDDK.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errorsmaybe discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abdel-Monem M, Chanal MC, Hoffmann-Berling H. DNA unwinding enzyme II of Escherichia coli. 1. Purification and characterization of the ATPase activity. Eur J Biochem. 1977;79:33–38. doi: 10.1111/j.1432-1033.1977.tb11780.x. [DOI] [PubMed] [Google Scholar]
- Ali JA, Lohman TM. Kinetic measurement of the step size of DNA unwinding by Escherichia coli UvrD helicase. Science. 1997;275:377–380. doi: 10.1126/science.275.5298.377. [DOI] [PubMed] [Google Scholar]
- Arthur HM, Lloyd RG. Hyper-recombination in uvrD mutants of Escherichia coli K-12. Mol Gen Genet. 1980;180:185–191. doi: 10.1007/BF00267368. [DOI] [PubMed] [Google Scholar]
- Becker PB, Horz W. ATP-dependent nucleosome remodeling. Annu Rev Biochem. 2002;71:247–273. doi: 10.1146/annurev.biochem.71.110601.135400. [DOI] [PubMed] [Google Scholar]
- Bernstein DA, Zittel MC, Keck JL. High-resolution structure of the E.coli RecQ helicase catalytic core. Embo J. 2003;22:4910–4921. doi: 10.1093/emboj/cdg500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brendza KM, Cheng W, Fischer CJ, Chesnik MA, Niedziela-Majka A, Lohman TM. Autoinhibition of Escherichia coli Rep monomer helicase activity by its 2B subdomain. Proc Natl Acad Sci U S A. 2005;102:10076–10081. doi: 10.1073/pnas.0502886102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruand C, Ehrlich SD. UvrD-dependent replication of rolling-circle plasmids in Escherichia coli. Mol Microbiol. 2000;35:204–210. doi: 10.1046/j.1365-2958.2000.01700.x. [DOI] [PubMed] [Google Scholar]
- Brünger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54(Pt 5):905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- Byrd AK, Raney KD. Increasing the length of the single-stranded overhang enhances unwinding of duplex DNA by bacteriophage T4 Dda helicase. Biochemistry. 2005;44:12990–12997. doi: 10.1021/bi050703z. [DOI] [PubMed] [Google Scholar]
- Cheng W, Brendza KM, Gauss GH, Korolev S, Waksman G, Lohman TM. The 2B domain of the Escherichia coli Rep protein is not required for DNA helicase activity. Proc Natl Acad Sci U S A. 2002;99:16006–16011. doi: 10.1073/pnas.242479399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dao V, Modrich P. Mismatch-, MutS-, MutL-, and helicase II-dependent unwinding from the single-strand break of an incised heteroduplex. J Biol Chem. 1998;273:9202–9207. doi: 10.1074/jbc.273.15.9202. [DOI] [PubMed] [Google Scholar]
- Dillingham MS, Soultanas P, Wigley DB. Site-directed mutagenesis of motif III in PcrA helicase reveals a role in coupling ATP hydrolysis to strand separation. Nucleic Acids Res. 1999;27:3310–3317. doi: 10.1093/nar/27.16.3310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillingham MS, Wigley DB, Webb MR. Demonstration of unidirectional single-stranded DNA translocation by PcrA helicase: measurement of step size and translocation speed. Biochemistry. 2000;39:205–212. doi: 10.1021/bi992105o. [DOI] [PubMed] [Google Scholar]
- Durr H, Korner C, Muller M, Hickmann V, Hopfner KP. X-ray structures of the Sulfolobus solfataricus SWI2/SNF2 ATPase core and its complex with DNA. Cell. 2005;121:363–373. doi: 10.1016/j.cell.2005.03.026. [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Fischer CJ, Maluf NK, Lohman TM. Mechanism of ATP-dependent translocation of E.coli UvrD monomers along single-stranded DNA. J Mol Biol. 2004;344:1287–1309. doi: 10.1016/j.jmb.2004.10.005. [DOI] [PubMed] [Google Scholar]
- George JW, Brosh RM, Jr., Matson SW. A dominant negative allele of the Escherichia coli uvrD gene encoding DNA helicase II. A biochemical and genetic characterization. J Mol Biol. 1994;235:424–435. doi: 10.1006/jmbi.1994.1003. [DOI] [PubMed] [Google Scholar]
- Gorbalenya AE, Koonin EV. Helicases: amino acid sequence comparisons and structure—function relationships. Curr Opin Struct Biol. 1993;3:419–429. [Google Scholar]
- Graham DL, Lowe PN, Grime GW, Marsh M, Rittinger K, Smerdon SJ, Gamblin SJ, Eccleston JF. MgF (-) (3) as a transition state analog of phosphoryl transfer. Chem Biol. 2002;9:375–381. doi: 10.1016/s1074-5521(02)00112-6. [DOI] [PubMed] [Google Scholar]
- Guarné A, Ramon-Maiques S, Wolff EM, Ghirlando R, Hu X, Miller JH, Yang W. Structure of the MutL C-terminal domain: a model of intact MutL and its roles in mismatch repair. Embo J. 2004;23:4134–4145. doi: 10.1038/sj.emboj.7600412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickson ID, Arthur HM, Bramhill D, Emmerson PT. The E. coli uvrD gene product is DNA helicase II. Mol Gen Genet. 1983;190:265–270. doi: 10.1007/BF00330649. [DOI] [PubMed] [Google Scholar]
- Hodgman TC. A new superfamily of replicative proteins. Nature. 1988;333:22–23. doi: 10.1038/333022b0. [DOI] [PubMed] [Google Scholar]
- Iyer LM, Leipe DD, Koonin EV, Aravind L. Evolutionary history and higher order classification of AAA+ ATPases. J Struct Biol. 2004;146:11–31. doi: 10.1016/j.jsb.2003.10.010. [DOI] [PubMed] [Google Scholar]
- Kim JL, Morgenstern KA, Griffith JP, Dwyer MD, Thomson JA, Murcko MA, Lin C, Caron PR. Hepatitis C virus NS3 RNA helicase domain with a bound oligonucleotide: the crystal structure provides insights into the mode of unwinding. Structure. 1998;6:89–100. doi: 10.1016/s0969-2126(98)00010-0. [DOI] [PubMed] [Google Scholar]
- Korolev S, Hsieh J, Gauss GH, Lohman TM, Waksman G. Major domain swiveling revealed by the crystal structures of complexes of E. coli Rep helicase bound to single-stranded DNA and ADP. Cell. 1997;90:635–647. doi: 10.1016/s0092-8674(00)80525-5. [DOI] [PubMed] [Google Scholar]
- Korolev S, Yao N, Lohman TM, Weber PC, Waksman G. Comparisons between the structures of HCV and Rep helicases reveal structural similarities between SF1 and SF2 super-families of helicases. Protein Sci. 1998;7:605–610. doi: 10.1002/pro.5560070309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohman TM, Hsieh J, Maluf NK, Cheng W, Lucius AL, Fischer CJ, Brendza KM, Korolev S, Waksman G. DNA helicases, motors that move along nucleic acids: lessons from the SF1 helicase superfamily. In: Hackney DD, Tamanoi F, editors. Energy coupling and molecular motors (The Enzymes) Elsevier Academic Press; San Diego: 2003. pp. 304–364. [Google Scholar]
- Lucius AL, Lohman TM. Effects of temperature and ATP on the kinetic mechanism and kinetic step-size for E.coli RecBCD helicase-catalyzed DNA unwinding. J Mol Biol. 2004;339:751–771. doi: 10.1016/j.jmb.2004.04.010. [DOI] [PubMed] [Google Scholar]
- Macris MA, Sung P. Multifaceted role of the Saccharomyces cerevisiae Srs2 helicase in homologous recombination regulation. Biochem Soc Trans. 2005;33:1447–1450. doi: 10.1042/BST0331447. [DOI] [PubMed] [Google Scholar]
- Maluf NK, Fischer CJ, Lohman TM. A Dimer of Escherichia coli UvrD is the active form of the helicase in vitro. J Mol Biol. 2003;325:913–935. doi: 10.1016/s0022-2836(02)01277-9. [DOI] [PubMed] [Google Scholar]
- Matson SW, George JW. DNA helicase II of Escherichia coli. Characterization of the single-stranded DNA-dependent NTPase and helicase activities. J Biol Chem. 1987;262:2066–2076. [PubMed] [Google Scholar]
- Matson SW, Kaiser-Rogers KA. DNA helicases. Annu Rev Biochem. 1990;59:289–329. doi: 10.1146/annurev.bi.59.070190.001445. [DOI] [PubMed] [Google Scholar]
- Mechanic LE, Hall MC, Matson SW. Escherichia coli DNA helicase II is active as a monomer. J Biol Chem. 1999;274:12488–12498. doi: 10.1074/jbc.274.18.12488. [DOI] [PubMed] [Google Scholar]
- Myong S, Rasnik I, Joo C, Lohman TM, Ha T. Repetitive shuttling of a motor protein on DNA. Nature. 2005;437:1321–1325. doi: 10.1038/nature04049. [DOI] [PubMed] [Google Scholar]
- Navaza J. Implementation of molecular replacement in AMoRe. Acta Crystallogr D Biol Crystallogr. 2001;57:1367–1372. doi: 10.1107/s0907444901012422. [DOI] [PubMed] [Google Scholar]
- Ogawa H, Shimada K, Tomizawa J. Studies on radiation-sensitive mutants of E. coli. I. Mutants defective in the repair synthesis. Mol Gen Genet. 1968;101:227–244. doi: 10.1007/BF00271625. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Pfander B, Moldovan GL, Sacher M, Hoege C, Jentsch S. SUMO-modified PCNA recruits Srs2 to prevent recombination during S phase. Nature. 2005;436:428–433. doi: 10.1038/nature03665. [DOI] [PubMed] [Google Scholar]
- Rasnik I, Myong S, Cheng W, Lohman TM, Ha T. DNA-binding orientation and domain conformation of the E. coli rep helicase monomer bound to a partial duplex junction: single-molecule studies of fluorescently labeled enzymes. J Mol Biol. 2004;336:395–408. doi: 10.1016/j.jmb.2003.12.031. [DOI] [PubMed] [Google Scholar]
- Richet E, Kohiyama M. Purification and characterization of a DNA-dependent ATPase from Escherichia coli. J Biol Chem. 1976;251:808–812. [PubMed] [Google Scholar]
- Roman LJ, Kowalczykowski SC. Characterization of the adenosinetriphosphatase activity of the Escherichia coli RecBCD enzyme: relationship of ATP hydrolysis to the unwinding of duplex DNA. Biochemistry. 1989;28:2873–2881. doi: 10.1021/bi00433a019. [DOI] [PubMed] [Google Scholar]
- Singleton MR, Dillingham MS, Gaudier M, Kowalczykowski SC, Wigley DB. Crystal structure of RecBCD enzyme reveals a machine for processing DNA breaks. Nature. 2004;432:187–193. doi: 10.1038/nature02988. [DOI] [PubMed] [Google Scholar]
- Singleton MR, Wigley DB. Modularity and specialization in superfamily 1 and 2 helicases. J Bacteriol. 2002;184:1819–1826. doi: 10.1128/JB.184.7.1819-1826.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soultanas P, Dillingham MS, Wiley P, Webb MR, Wigley DB. Uncoupling DNA translocation and helicase activity in PcrA: direct evidence for an active mechanism. EMBO J. 2000;19:3799–3810. doi: 10.1093/emboj/19.14.3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanner NK, Cordin O, Banroques J, Doere M, Linder P. The Q motif: a newly identified motif in DEAD box helicases may regulate ATP binding and hydrolysis. Mol Cell. 2003;11:127–138. doi: 10.1016/s1097-2765(03)00006-6. [DOI] [PubMed] [Google Scholar]
- Theis K, Chen PJ, Skorvaga M, Van Houten B, Kisker C. Crystal structure of UvrB, a DNA helicase adapted for nucleotide excision repair. Embo J. 1999;18:6899–6907. doi: 10.1093/emboj/18.24.6899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Putte P, van Sluis CA, van Dillewijn J, Rorsch A. The location of genes controlling radiation sensitivity in Escherichia coli. Mutat Res. 1965;2:97–110. doi: 10.1016/0027-5107(65)90041-2. [DOI] [PubMed] [Google Scholar]
- Veaute X, Delmas S, Selva M, Jeusset J, Le Cam E, Matic I, Fabre F, Petit MA. UvrD helicase, unlike Rep helicase, dismantles RecA nucleoprotein filaments in Escherichia coli. Embo J. 2005;24:180–189. doi: 10.1038/sj.emboj.7600485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velankar SS, Soultanas P, Dillingham MS, Subramanya HS, Wigley DB. Crystal structures of complexes of PcrA DNA helicase with a DNA substrate indicate an inchworm mechanism. Cell. 1999;97:75–84. doi: 10.1016/s0092-8674(00)80716-3. [DOI] [PubMed] [Google Scholar]
- von Hippel PH, Delagoutte E. Macromolecular complexes that unwind nucleic acids. Bioessays. 2003;25:1168–1177. doi: 10.1002/bies.10369. [DOI] [PubMed] [Google Scholar]
- Wickner S, Wright M, Hurwitz J. Association of DNA-dependent and - independent ribonucleoside triphosphatase activities with dnaB gene product of Escherichia coli. Proc Natl Acad Sci U S A. 1974;71:783–787. doi: 10.1073/pnas.71.3.783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G, Deng E, Baugh L, Kushner SR. Identification and characterization of Escherichia coli DNA helicase II mutants that exhibit increased unwinding efficiency. J Bacteriol. 1998;180:377–387. doi: 10.1128/jb.180.2.377-387.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.