Abstract
It has been claimed that complete genome sequences would clarify phylogenetic relationships between organisms, but up to now, no satisfying approach has been proposed to use efficiently these data. For instance, if the coding of presence or absence of genes in complete genomes gives interesting results, it does not take into account the phylogenetic information contained in sequences and ignores hidden paralogies by using a BLAST reciprocal best hit definition of orthology. In addition, concatenation of sequences of different genes as well as building of consensus trees only consider the few genes that are shared among all organisms. Here we present an attempt to use a supertree method to build the phylogenetic tree of 45 organisms, with special focus on bacterial phylogeny. This led us to perform a phylogenetic study of congruence of tree topologies, which allows the identification of a core of genes supporting similar species phylogeny. We then used this core of genes to infer a tree. This phylogeny presents several differences with the rRNA phylogeny, notably for the position of hyperthermophilic bacteria.
Though it seems sensible to consider that genes remain associated in genomes for long periods in Eukaryotes, recent data suggest that this is not the case in Prokaryotes, where a large number of horizontal transfers is believed to have occurred. Methods using comparisons of base or codon composition have revealed that up to 17% of the genes of bacterial genomes maybe of alien origin, with only a few of them identifiable as mobile elements (Ochman et al. 2000). However, it was recently shown that alternative mechanisms may explain biases in nucleotide composition (Guindon and Perriere 2001; Koski et al. 2001; Wang 2001) and that unexpected sequence patterns may not be proofs of alien origin. Moreover, the numerous intrinsic methods tend to give very different estimations of the pool of laterally transferred genes (Ragan 2001).
An objective proof of alien origin should be given by phylogenetic analysis. However, this raises other problems such as reconstruction artifacts and hidden paralogies, and though phylogeneticists steadily warn against these problems (Philippe and Laurent 1998; Glansdorff 2000), the difficulty of obtaining congruent gene phylogenies is often seen as a result of lateral exchanges. Thus, another problem regarding phylogenetic detection of lateral transfers is the existence of a reliable reference phylogeny. Ribosomal RNA is often considered the best tool to infer prokaryotic phylogeny because it is thought to be one of the most constrained and ubiquitous molecules available, and thus the most informative (Woese 1987). However, several examples of likely lateral transfers concern molecules that are constrained and ubiquitous (Brochier et al. 2000; Brown et al. 2001). It is therefore desirable to base a reference prokaryotic phylogeny on evidence derived from a large number of genes.
The prokaryotic world is now often seen as a “genome space” (Bellgard et al. 1999) in which horizontal transfers between organisms appear to be the rule. However, transfers probably do not concern every type of gene in the same way. For example, Jain et al. (1999) reported evidence that informational genes—which are thought to have more macromolecular interactions than operational genes—are less likely to be transferred. It is thus possible that a core of genes remains more closely associated over a long period through evolution than the rest of the genome. If so, a tree of bacterial species remains possible, and phylogeny could be used as a systematic tool to identify lateral transfers with respect to this reference.
Thus, there is a need for an efficient way to transcribe all available genome data into pertinent phylogenetic information (Eisen 2000a). Several methods have been proposed to build genome trees, or to test whether this concept makes sense for bacterial species. Among them, a recent work by Brown et al. (2001) proposes a phylogeny based on the concatenation of 23 genes from 45 species. However, after removing genes that have very likely undergone at least one lateral transfer between bacteria and another domain, only 14 genes remained available for this analysis, and the support of the topology decreased in the same proportion. This result raises the problem of including phylogenetic information contained in nonubiquitous genes.
Here we present our study of the congruence of gene phylogenies for 45 organisms, with particular emphasis on Bacteria, for which an abundance of data is available. We found evidence in Bacteria of a core of genes that have undergone less lateral transfers. We then used the results of this study to infer a topology for the tree of life, based on the matrix representation using the parsimony (MRP) method proposed by Baum (1992) and Ragan (1992) (Fig. 1). This method was used to infer a phylogeny of Eutheria (Liu et al. 2001) but it has never been applied to the study of completely sequenced organisms. The results of our analysis are partially in agreement with the rRNA reference; however, some important differences raise questions about bacterial phylogeny.
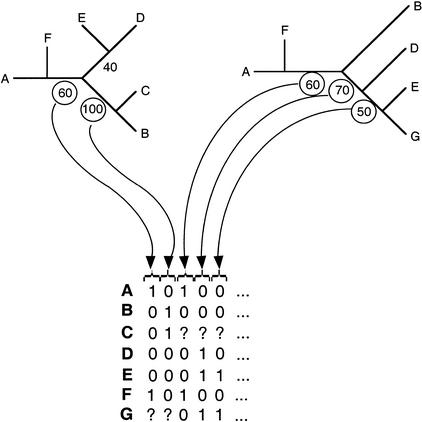
Figure 1.
Construction of supertrees by MRP with bootstrap weighting. Each tree obtained for a set of species from a single orthologous gene family was coded into a binary matrix of informative sites. Only branches having a bootstrap value (or RELL-BP value for ML trees) over 50% were coded. The matrices obtained were concatenated into a supermatrix in which species absent from a gene family are encoded as unknown state (“?”). The supertree was calculated on the supermatrix using DNAPARS with all default options, and 500 replicates of bootstrap were made using SEQBOOT.
RESULTS
The Supertree Based on 730 Genes
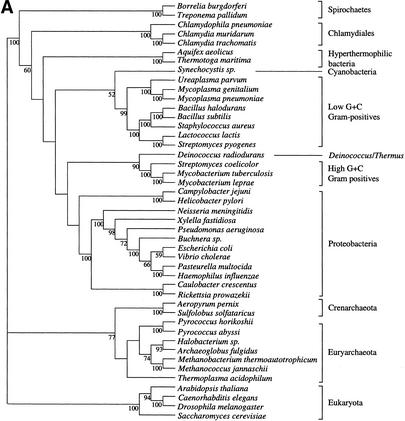
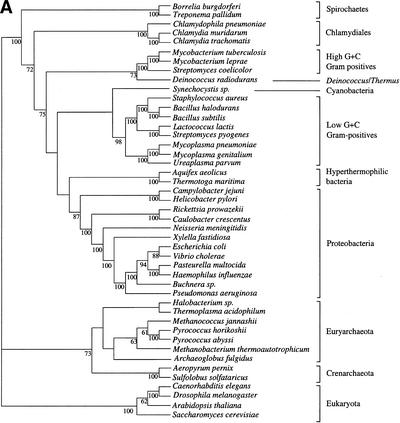
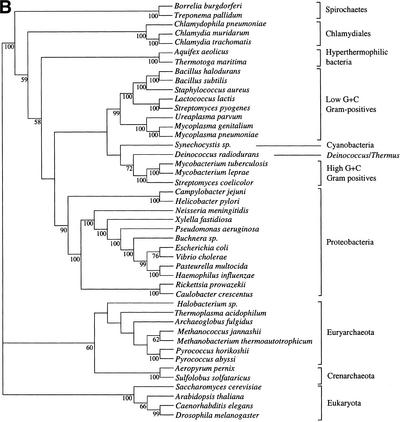
We first built the supertree using 730 trees selected as described in the Methods section. We used the MRP method, coding only nodes with a bootstrap value higher than 50% (Fig. 1). Figure 2 shows the supertrees obtained from elementary trees built with BIONJ and gamma-corrected distance, and those obtained using maximum likelihood (ML). These supertrees strongly support the monophyly of the three domains of life, that is, Archaea, Eukarya, and Bacteria. The Archaeal part is well resolved in the supertree based on ML trees, and shows monophyly of Crenarchaeota and of Euryarchaeota. Relations between archaea appear to be less clear in the supertree based on gamma distances. In addition, the eukaryotic part of both supertrees presents a basal position for fungi. Finally, the bacterial part of the trees is very poorly resolved for deep branches, but gives strong support for the monophyly of Chlamydiales, Spirochaetes, low G+C Gram-positives, high G+C Gram-positives, and (α,β,δ)-Proteobacteria. More surprising is the strong support given to the grouping of Deinococcus and high G+C Gram-positives. The ɛ-Proteobacteria (i.e., Helicobacter and Campilobacter) are grouped with other Proteobacteria in the gamma distance-based supertree, although with relatively low support. The remainder of the tree is only weakly supported, and presents an atypical topology, notably concerning the species present at the base of the bacteria. However, the ML-based supertree tends to have a more aberrant topology since ɛ-Proteobacteria have a very basal position. This difficulty of resolving deep branches may be related to the increasing probability of lateral transfers, hidden paralogies, and long branch artifacts with separation time. Thus, it is necessary to determine whether genes give completely incompatible phylogenetic information or whether a common signal can be extracted from bacterial phylogenies.
Figure 2.
Supertrees of 45 species constructed with 730 trees. (A) Supertree based on trees made by BIONJ and a gamma distribution estimation of evolutionary rate heterogeneity. (B) Supertree made with ML trees. Only bootstrap values over 50% are shown.
Comparison of Gene Trees
As noted above, it is difficult to study lateral transfers using phylogeny because the extent to which the rRNA tree, or any other reference, represents something more than the phylogeny of a gene is unknown. To bypass this problem, we made all of the possible comparisons between gene phylogenies by using principal coordinates analysis (PCO). If a group of genes tends to have similar phylogenies, it may be representative of a common history.
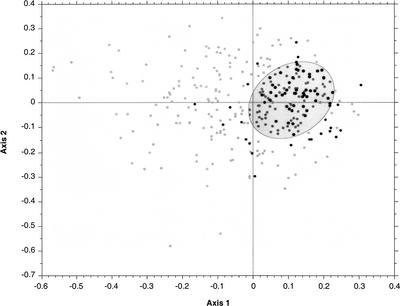
We used the Robinson-Foulds (RF) topological distance (Robinson and Foulds 1981) to compare trees with each other. It was not possible to consider every domain (Archaea, Bacteria, and Eukarya) at the same time since too many pairs of trees were not comparable due to lack of common species. We therefore computed topological distances between all 310 trees containing at least ten bacterial species. Only results based on distance-based trees are shown, because ML-based trees gave very similar results. The result of this analysis of 310 trees is particularly interesting: the representation of the two first axes of PCO (Fig. 3) shows a cloud that is very dense on the right with a tail on the left. This structure is mainly due to the first axis, the other axes displaying a distribution that is centered on the origin. The structuring on the first axis suggests that genes gathered in the densest region of the cloud share, at least partially, a common phylogenetic signal, while trees present in the tail are perturbed by lateral transfers, hidden paralogies, or reconstruction artifacts. When considering the position of informational and operational genes in the cloud, it is very striking that informational genes are almost all grouped in the densest region, while the tail is formed only by operational genes. This result is consistent with previous studies (Rivera et al. 1998; Jain et al. 1999) that present evidence of a better conservation of phylogenetic information in informational genes. However, since operational genes are also well represented in the dense region, this result suggests that, contrarily to informational genes, this definition refers to a heterogeneous group, which contain genes that may be as constrained as informational genes through evolution.
Figure 3.
Plot of the two first axes of the PCO made from 310 BIONJ trees compared with RF distance. The trees chosen contained at least 10 bacterial species. The same experiment with ML-trees gave very similar results. Black dots correspond to informational genes, and gray dots correspond to operational genes. The ellipse contains the 121 trees retained for supertree reconstruction (see Table 1).
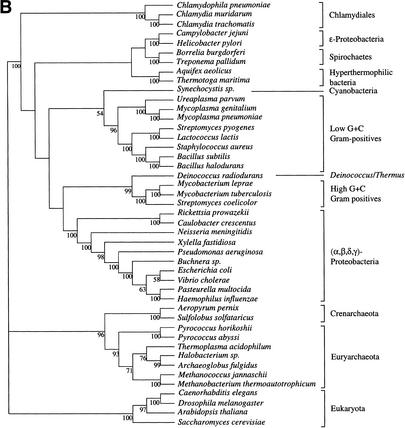
Supertree from the Core of Genes
PCO analysis of the 310 genes allowed identification of a pool sharing similar topologies. It seems thus parsimonious to suppose that this grouping relies on common history rather than on artifacts acting in the same way on different genes. We therefore selected the genes present in the densest region of the cloud, as shown in Figure 3. This left 121 trees for supertree reconstruction for the gamma-corrected distance experiment and 118 for the ML experiment. Slight variations of the limits of this region gave exactly the same topology, although with variations in bootstrap values. The supertrees obtained are shown in Figure 4. As in the 730-gene supertree, the three domains of life are monophyletic. Low resolution of the Archaeal part of the tree is due to the fact that genes present only in Archaea or shared only by Archaea and Eukaryotes were removed from the gene sample to allow PCO computation. The eukaryotic part of the tree has the same topology as in Figure 2. As might be expected, the bacterial part presents higher bootstrap values and appears thus more resolved, especially using the distance-based trees. The groups cited earlier remain monophyletic. However, ɛ-Proteobacteria are here grouped with other Proteobacteria with a significant bootstrap value. The remainder of the tree shows substantial differences with rRNA phylogenies which place hyperthermophilic (Aquifex, Thermotoga) and radioresistant (Deinococcus) bacteria close to the root (Woese 1987). The supertree gives no evidence for such early emergence of these groups and tends to give them positions close to mesophilic bacteria and particularly Proteobacteria, although with relatively low bootstrap support. Instead, the basal position in the bacterial tree is occupied by Spirochaetes and Chlamydiales with significant bootstrap values in the distance-based tree.
Figure 4.
Supertrees of 45 species built with the trees selected using the PCO results. (A) Supertree based on 121 trees made by BIONJ and a gamma distribution estimation of evolutionary rate heterogeneity. (B) Supertree based on 118 ML trees.
Discussion
Defining Orthologs
We selected orthologous gene families (see Methods) with the intent of removing lateral transfers and paralogy as often as possible. Only families containing one gene per species were kept. Therefore, only orthologous replacement and hidden paralogies (i.e., differential loss of the two copies in two lineages) can occur in selected families. These two types of events are expected to be comparatively rare. This stringent criterion led us to exclude certain genes that are considered good tools for phylogeny. For example, Synechocystis (strain PCC 6803), Vibrio cholerae, and Streptomyces coelicolor have been found to possess several genes from the EF-G family (HOBACGEN family number HBG000251), which may result from either lateral transfers or hidden paralogies.
As several transfers between domains have been described, we removed or corrected (by dismissing the transferred sequences when the transfer was evident) families in which Bacteria were not monophyletic (Brown et al. 2001) or containing only Archaea and hyperthermophilic bacteria (Logsdon and Faguy 1999; Nesbo et al. 2001). The assumption of monophyly of Bacteria can be criticized, in light of the proposal by Gupta (1998) that Archaea derive from Gram-positive bacteria. The families that were removed necessitated hypothesizing several events of lateral transfers between bacteria and other domains. All of the corrections made on families were due to probable unannotated eukaryotic genes of mitochondrial or chloroplastic origin (i.e., with a branching of eukaryotes within Proteobacteria or with the branching of Arabidopsis with Synechocystis). Overall, very few families were involved.
Supertree Compared to Other Genome Trees
Many methods using information from complete genomes to infer phylogenetic relationships between prokaryotes have been proposed. Among them, we mention here those based on concatenation of genes and those using gene content. Regarding the former method, one of the most remarkable works is that of Brown et al. (2001). Those investigators used a set of 23 ubiquitous and well-conserved genes to infer the phylogeny of 45 organisms. Their tree supports, with high bootstrap values, the basal position of Spirochaetes and Chlamydiales. However, they found that nine of these genes (which represented about 40% of their data set) had been subject to interdomain lateral transfers. Notably, some of them were identified as transfers involving Archaea and Spirochaetes, which could be responsible for the basal position of these bacteria. After removal of those genes from the set, a phylogeny that is sensitive to reconstruction methods and with low support for several deep branches was obtained. This topology is however in general agreement with the rRNA-based topology, notably for the position of hyperthermophilic bacteria that occupy the most basal position. Although this method enables one to obtain alignments of respectable length, it remains limited by the number of genes it can take into account. Moreover, Brown et al. (2001) showed that the presence of laterally transferred genes radically changes the topology of the tree in the concatenation method. Thus, if 40% of the genes retained have undergone interdomain lateral transfers, what is the rate of lateral transfers among bacteria and what is their impact on the final phylogeny?
Another objection to the concatenation approach is the weighting accorded to a gene. For example, in the 14-protein alignment of Brown et al. (2001), only four proteins represent more than half of all sites. Thus, if a gene family has undergone a lateral transfer, it may impose its topology if the protein is long enough. A solution to these problems could be the addition of a large number of genes, since a common phylogenetic signal may emerge through discordant information due to lateral transfers (Eernisse and Kluge 1993). However, other approaches must be developed, because ubiquitous genes are rare.
The methods based on gene content may be summarized as follows: if one considers that events of gain and loss of genes are relatively rare, then the presence or absence of a gene in a genome can be considered an informative binary character. Hence, a phylogeny minimizing these events can be reconstructed and may represent the phylogeny of the genomes. Several authors have proposed schemes derived from this idea. Though these methods can give very interesting results (Snel et al. 1999), the hypothesis on which this model is based could be discussed at length, since many investigators consider gene loss and lateral transfers the main driving force of bacterial evolution. For example, Ochman et al. (2000) estimated that prokaryotic genomes may contain 0%–16.6% genes (with a mean of ∼6%) acquired recently enough to conserve an atypical nucleotide composition. Moreover, Mira et al. (2001) proposed a model of genome size maintenance in which gain and loss of genes play the most important role. Thus, gene content-based methods may encounter problems due to convergence.
The strength of the supertree method is that it allows a large amount of data to be considered. As discussed above, this property should allow the recovery of a phylogenetic signal in the presence of lateral transfers. Moreover, each gene tree brings a comparable amount of information, whatever its length. However, the topology of the supertree based on 730 genes, and particularly its bacterial part, suggests that it is necessary to remove trees containing long branch artifacts, lateral transfers, or hidden paralogy. It is worth noting that the ML trees seem to be more subject to reconstruction problems, because the grouping of ɛ-Proteobacteria, hyperthermophilic bacteria, and Spirochaetes is clearly artefactual. The PCO analysis made of trees containing comparable sets of species (see Methods) revealed that a group of genes possess similar topologies for the bacterial part of the tree. This group contains almost all informational genes contained in the data set. This result is in agreement with the vision of a core of genes that remains associated for long periods in prokaryotes. As proposed earlier, informational genes seem to be an essential component of this core, but it appears that this is also the case for several operational genes. However, operational genes undoubtedly display a larger range of topology, which highlights the fact that this functional class regroups genes having very different evolutionary patterns. Though it may be due to lateral transfers, it is worth noting that the genes present in the tail of the cloud shown in Figure 3 tend to contain fewer species. Hence, they may also be subject to reconstruction problems due to low number of taxa (Lecointre et al. 1993) or may contain hidden paralogies, which are more difficult to detect in gene families containing few species (Salzberg et al. 2001).
Horizontal Transfers: “Genome Space” or Core of Genes?
Although some deep nodes have low bootstrap support, the level of resolution of the supertree reported here is in strong disagreement with the “genome space” (Bellgard et al. 1999) vision of the prokaryotic world predicting a “star phylogeny.” One could argue that grouping of species in the supertree would only reflect the frequency of gene exchanges between these species. This interpretation can be excluded because the supertree method would then not be expected to give a tree topology radically different from gene content-based trees (Snel et al. 1999; Tekaia et al. 1999; Lin and Gerstein 2000), which are predicted to be very sensitive to this problem. It is worth noting that a particularly stringent selection of protein families was exercised for building the supertree. In particular, a phylogenetic definition of orthology rather than a definition based on reciprocal best BLAST hits was used (Eisen 2000a; Koski and Golding 2001), as is often the case for practical reasons. Thus, all gene trees where a species was represented more than once were excluded from analysis. This selection allowed us to make absolutely no a priori assumptions about the topology of the trees, except for the monophyly of Bacteria, and to reduce the probability of taking hidden paralogies into account. Although the PCO analysis led to a strong reduction of the length of the supermatrix (e.g., from 5382 sites in Fig. 2A to 1891 sites in Fig. 4A), bootstrap values increased for most bacterial nodes. This increase of bootstrap values in supertrees reveals that the group of genes selected after the PCO analysis contains congruent information on the phylogeny of Bacteria. This suggests a vision of bacterial evolution where a “core” of genes tends to remain stable through evolution (Snel et al. 1999; Eisen 2000b).
The HOBACGEN-CG annotations of the gene families present in the dense region of the PCO data with BIONJ trees are shown in Table 1. As noted above, this set of genes is strongly enriched in informational genes compared to the complete data set. However, about half of the genes have operational functions. A substantial fraction of these genes have no known function. Their presence in the inferred core of genes suggests that they may have an important function.
Table 1.
Protein Families Found in the Dense Region of the Cloud in Figure 3
| Family | Dom. | Class | Spec. | Definition |
|---|---|---|---|---|
| HBG000224 | 1 | info | 31 | NAD-dependent DNA ligase family |
| HBG000226 | 1 | info | 28 | DNA polymerase III α chain |
| HBG000253 | 1 | info | 31 | EF-Ts family |
| HBG000387 | 2 | info | 19 | Glutamyl-tRNA reductase family |
| HBG000436 | 1 | info | 28 | Translation initiation factor IF-1 |
| HBG000440 | 3 | info | 39 | Translation initiation factor IF-2 |
| HBG000445 | 1 | info | 31 | Translation initiation factor IF-3 |
| HBG000531 | 1 | info | 23 | Transcriptional-repair coupling factor |
| HBG000910 | 1 | info | 30 | S5P family of ribosomal proteins |
| HBG003694 | 1 | info | 29 | Excinuclease ABC subunit C UvrC family |
| HBG005458 | 3 | info | 42 | 30S ribosomal protein S10P |
| HBG006229 | 1 | info | 28 | tRNA δ(2)-isopentenylpyrophosphate transferase |
| HBG008973 | 2 | info | 41 | Alanyl-tRNA synthetase |
| HBG008994 | 1 | info | 30 | Leucyl-tRNA synthetase |
| HBG009221 | 2 | info | 33 | Threonyl-tRNA synthetase |
| HBG010120 | 1 | info | 25 | DNA repair protein RadA |
| HBG011587 | 3 | info | 45 | S7P family of ribosomal proteins |
| HBG011828 | 3 | info | 44 | 30S ribosomal protein S3P |
| HBG011949 | 1 | info | 28 | 50S ribosomal protein L17 |
| HBG012008 | 1 | info | 30 | S15P family of ribosomal proteins |
| HBG012054 | 1 | info | 32 | Peptidyl-tRNA hydrolase Pth family |
| HBG012248 | 1 | info | 32 | L20P family of ribosomal proteins |
| HBG012249 | 1 | info | 29 | Phenylalanyl-tRNA synthetase α chain |
| HBG012416 | 2 | info | 39 | Valyl-tRNA synthetase |
| HBG012497 | 3 | info | 43 | L6P family of ribosomal proteins |
| HBG012522 | 1 | info | 26 | Phenylalanyl-tRNA synthetase β chain |
| HBG012593 | 3 | info | 43 | 50S ribosomal protein L1P |
| HBG012594 | 1 | info | 30 | DNA-directed RNA polymerase β chain |
| HBG012618 | 3 | info | 44 | S2P family of ribosomal proteins |
| HBG012650 | 1 | info | 15 | Ribosomal protein L11 methyltransferase |
| HBG013445 | 1 | info | 31 | L19P family of ribosomal proteins |
| HBG013652 | 1 | info | 31 | Ribosome releasing factor RRF family |
| HBG013954 | 1 | info | 22 | DNA repair and genetic recombination protein RecN |
| HBG014295 | 1 | info | 30 | Holliday junction DNA helicase RuvB |
| HBG014467 | 1 | info | 28 | DNA-directed RNA polymerase β′ chain |
| HBG014469 | 3 | info | 42 | L11P family of ribosomal proteins |
| HBG014584 | 1 | info | 29 | DNA-directed RNA polymerase α chain |
| HBG014585 | 3 | info | 44 | 50S ribosomal protein 15P |
| HBG014588 | 1 | info | 31 | 50S ribosomal protein L16 |
| HBG014596 | 1 | info | 32 | Aspartyl-tRNA synthetase |
| HBG014727 | 3 | info | 42 | Seryl-tRNA synthetase |
| HBG015438 | 1 | info | 14 | EF-1 for elongation factor 1-α |
| HBG016715 | 1 | info | 25 | RecG subfamily of helicases |
| HBG016735 | 1 | info | 31 | L18P family of ribosomal proteins |
| HBG016737 | 1 | info | 30 | L22P family of ribosomal proteins |
| HBG016768 | 1 | info | 31 | L4P family of ribosomal proteins |
| HBG016876 | 1 | info | 31 | tRNA methyltransferase |
| HBG018696 | 1 | info | 25 | DNA polymase type-A family PolA |
| HBG019433 | 2 | info | 18 | DNA mismatch repair MutL/HexB family |
| HBG020375 | 1 | info | 19 | A/G-specific adenine glycosylase MutY |
| HBG057220 | 1 | info | 25 | Prolyl-tRNA synthetase |
| HBG000042 | 2 | oper | 24 | Adenine phosphoribosyltransferase |
| HBG000063 | 1 | oper | 24 | ATP synthase α chain |
| HBG000069 | 1 | oper | 24 | ATPase γ chain family |
| HBG000092 | 2 | oper | 20 | Biotin and lipoic acid synthetases family |
| HBG000137 | 1 | oper | 28 | ATP-dependent Clp protease |
| HBG000315 | 2 | oper | 13 | Ferritin-like protein |
| HBG000337 | 1 | oper | 21 | Riboflavin biosynthesis protein RibA |
| HBG000348 | 1 | oper | 14 | Glucose inhibited division protein B homolog GidB |
| HBG000379 | 1 | oper | 23 | GMP synthetase |
| HBG000380 | 2 | oper | 35 | GTP-binding protein Guf1 |
| HBG000388 | 3 | oper | 30 | Alanine dehydrogenase family |
| HBG000421 | 1 | oper | 10 | Hpr serine/threonine protein kinase PtsK family |
| HBG000471 | 2 | oper | 32 | Guanylate kinase Gmk protein |
| HBG000529 | 2 | oper | 31 | Adenosylmethionine synthetase family |
| HBG000560 | 1 | oper | 24 | UDP-N-acetylmuramoylalanine-D-glutamate ligase MutD |
| HBG000564 | 1 | oper | 25 | MurCDEF family |
| HBG000595 | 1 | oper | 28 | N utilization substance protein A homolog |
| HBG001251 | 1 | oper | 32 | UPF0117 family unknown function |
| HBG001398 | 1 | oper | 24 | Menaquinone biosynthesis methyltransferase |
| HBG001475 | 1 | oper | 21 | GTP-binding protein TypA/BipA homolog |
| HBG001546 | 1 | oper | 12 | Flagella basal body Rod proteins family |
| HBG001832 | 1 | oper | 21 | Unknown function |
| HBG002257 | 2 | oper | 20 | Stationary-phase survival protein SurE |
| HBG002555 | 1 | oper | 14 | CysE/LacA/LpxA/NodL family of acetyltransferases |
| HBG002569 | 1 | oper | 15 | 3-deoxy-manno-octulosonate cytidylyltransferase |
| HBG002592 | 1 | oper | 22 | Exodeoxyribonuclease VII large subunit |
| HBG002674 | 1 | oper | 20 | HemK protein homolog |
| HBG002766 | 1 | oper | 17 | Heat shock protein HsIV |
| HBG002767 | 1 | oper | 17 | ATP-dependent Hsl protease |
| HBG002854 | 2 | oper | 32 | GatB family |
| HBG002953 | 2 | oper | 28 | UPF0038 family unknown function |
| HBG003029 | 2 | oper | 29 | Glycerol-3-phosphate dehydrogenase [NAD+] |
| HBG003087 | 1 | oper | 16 | UDP-3-O-[3-hydroxymyristoyl] N-acetylglucosamine deacetylase |
| HBG003095 | 2 | oper | 22 | Scc-independent protein translocase protein TalC |
| HBG003169 | 1 | oper | 27 | Guanosine pentaphosphate synthetase |
| HBG003178 | 1 | oper | 26 | Unknown function |
| HBG003219 | 1 | oper | 31 | Preprotein translocase SecY subunit |
| HBG003373 | 1 | oper | 14 | Penicillin-binding protein Pbp2 |
| HBG004350 | 2 | oper | 27 | Phosphoribosylamine—glycine ligase |
| HBG005358 | 2 | oper | 34 | Protease Qri7 |
| HBG005857 | 2 | oper | 16 | Unknown function |
| HBG006494 | 1 | oper | 15 | Unknown function |
| HBG006702 | 1 | oper | 12 | Bacteriocin gene regulator Hfq protein |
| HBG007038 | 1 | oper | 23 | Unknown function |
| HBG007596 | 1 | oper | 10 | Flagellar protein FliS |
| HBG007971 | 1 | oper | 19 | Fpg family |
| HBG008259 | 1 | oper | 16 | UPF0040 family unknown function |
| HBG008387 | 1 | oper | 16 | UPF0088 family unknown function |
| HBG008920 | 1 | oper | 27 | Cytidylate kinase family |
| HBG008981 | 3 | oper | 44 | GTP-binding protein Gtp1/Obg family |
| HBG009096 | 1 | oper | 15 | Unknown function |
| HBG009213 | 1 | oper | 16 | Unknown function |
| HBG009256 | 1 | oper | 16 | General stress protein Ctc |
| HBG009692 | 1 | oper | 18 | Unknown function |
| HBG011361 | 2 | oper | 25 | GTP-binding protein HflX |
| HBG012031 | 1 | oper | 32 | SmpB protein family |
| HBG012163 | 1 | oper | 12 | Flagellar protein FLiG |
| HBG012209 | 1 | oper | 23 | MurCDEF family |
| HBG012397 | 1 | oper | 19 | PhoH family |
| HBG013158 | 1 | oper | 27 | RNAse III family |
| HBG014707 | 2 | oper | 39 | Phosphoglycerate kinase |
| HBG016560 | 1 | oper | 13 | Phosphoglyceromutase |
| HBG016999 | 1 | oper | 29 | UPF0011 family unknown function |
| HBG017494 | 1 | oper | 15 | Unknown function |
| HBG024417 | 1 | oper | 10 | Phosphate acetyltransferase |
| HBG033861 | 2 | oper | 16 | UPF0044 family unknown function |
| HBG042814 | 1 | oper | 26 | CDP-alcohol phosphatidyltransferase class-I family |
| HBG057111 | 2 | oper | 13 | GTP-binding protein Gtp1/Obg family |
| HBG057805 | 1 | oper | 25 | MurCDEF family |
| HBG070402 | 1 | oper | 27 | GTP-binding protein Era |
Family, HOBACGEN-CC family number; Dom., number of domains (i.e., Bateria, Archaea, and Eukarya) represented in the family; Spec., number of species represented in the family; Definition, simplified HOBACGEN-CG description of the family.
Which Artifacts May Affect the Supertree?
The sample of completely sequenced bacterial genomes is currently strongly biased toward species of medical interest. Thus, the supertree contains many parasites that display peculiar evolutionary patterns. Based only on topology and statistical support, our supertree method is expected to be sensitive to systematic artifacts of reconstruction. Nevertheless, although systematic bias exists, artifacts are not likely to systematically gather the same species, depending on the species sampling, which may differ between gene families in a supertree approach. In this case, even weak congruent information due to phylogenetic signal would be stronger than conflicting artefactual information. For instance, Mycoplasma species have a very low genomic G+C content (25% for Ureaplasma parvum and 32% for Mycoplasma pneumoniae), and are known to have a very reduced genome and fast evolutionary rate (Ochman et al. 1999). This is probably why these species tend to have a very basal position in several single (Gupta 1998; Klenk et al. 1999) and multiple gene phylogenies (Teichmann and Mitchison 1999; Hansmann and Martin 2000; Lin and Gerstein 2000). Therefore, the fact that Mycoplasma species are unambiguously grouped with Bacillus in the supertrees suggests that our approach is robust against biases related to G+C content and evolutionary rates. The same remarks can be made for Helicobacter pylori, which shows a high level of genetic variation between strains (Wang et al. 1999) and tends to have an aberrant position in many phylogenies (Gupta 2000), probably due to its high evolutionary rate.
The Supertree of Life: Questions About Bacterial History
The topology of the supertrees (Fig. 4) strongly supports the monophyly of each of the three domains of life (Bacteria, Archaea, and Eukarya). The phylogeny of Proteobacteria appears to be relatively well resolved at this level and is in agreement with the rRNA phylogeny and protein-based works (for review, see Gupta 2000). Their monophyly (including H. pylori and Campilobacter jejunii) is well supported, and this last result is particularly valuable because it has rarely been found with genome tree methods (Teichmann and Mitchison 1999; Tekaia et al. 1999; Lin and Gerstein 2000). Equally interesting is the position of the thermophilic bacteria, Aquifex aeolicus and Thermotoga maritima, which are strongly grouped. First, the monophyly of these bacteria contradicts small subunit ribosomal RNA analysis, which branch them successively at the base of the Bacteria and thus supports a thermophilic origin of Bacteria (Woese 1987; Barns et al. 1996, Bocchetta et al. 2000). If thermophilic bacteria are shown to be monophyletic, even with a basal position, the hypotheses of a thermophilic or mesophilic bacterial ancestor become at least equally parsimonious. However, since proteins of thermophilic bacteria and Archaea have been shown to possess a very peculiar amino acid composition (Kreil and Ouzounis 2001), it remains possible that the grouping of Thermotoga and Aquifex rests on a systematic artifact present in the majority of the trees. Second, Aquifex and Thermotoga are significantly excluded from the basal position in the gamma distance-based supertree. Thus, the genomic supertree brings no evidence for an early divergence of thermophilic lineages and is more consistent with a mesophilic last universal common ancestor (LUCA; Forterre 1996; Galtier et al. 1999). This view interprets the early emergence of these lineages in rRNA trees as a reconstruction artifact (Forterre 1996; Klenk et al. 1999) due to a bias of rRNA toward high-G+C content in hyperthermophiles (Galtier et al. 1999). Our result rather confirms that Thermotoga and Aquifex were secondarily adapted to high temperature (Miller and Lazcano 1995; Forterre 1996). Several studies have already reported a clustering of Aquifex with Proteobacteria (Klenk et al. 1999; Gupta 2000) or of Thermotoga with Gram-positives (Tiboni et al. 1993; Gribaldo et al. 1999). Thus, though our results cast a shadow on the basal position of thermophilic bacteria, their exact position remains an open question.
The basal position of Spirochaetes and Chlamydiales seems to have some level of support. The deep nodes of the supertree based on gamma-corrected distances are indeed supported by bootstrap values over 70%. The fact that these bacteria are vertebrate parasites does not preclude their basal position, because they possess close free-living relatives (Paster and Dewhirst 2000). Remarkably, Brown et al. (2001) using a set of 23 concatenated proteins found a very similar topology and interpreted this result as an artifact due to lateral transfers between Bacteria and Archaea in some of these proteins. However, such an explanation could not be proposed in the present case, since only families compatible with monophyletic Bacteria were selected. Noticeably, among the 121 trees retained to build the supertree, only 35 contain information for the position of the root of Bacteria by spanning two or more domains (see Table 1). Few studies have inferred the position of the root of Bacteria with so much data, but this number is still relatively low. Thus, this result must be confirmed by adding species, and particularly species close to Spirochaetes and Chlamydiales.
The monophyly of low G+C Gram-positives (including Bacillus and Mycoplasma) on one side and of high G+C Gram-positives on the other side appears to be very robust, but the significant support for the position of Deinococcus radiodurans suggests polyphyly of the Gram-positives. This position is very striking because Deinococcus is usually considered to have a much more basal position among bacteria (Woese 1987). Huang and Ito (1999) noted such a position, close to Gram-positives, with a DNA polymerase C phylogeny. The Brown et al. (2001) study also gives strong support to this position. These results suggest that two independent losses of the external membrane occurred in high-G+C and low-G+C Gram-positive bacteria. Nevertheless, it is interesting to note that the bootstrap value supporting this grouping is the only one that decreases after the PCO analysis. Thus, it remains possible that this position is due to the high G+C content of the genome of Deinococcus. Indeed, Deinococcus is a close relative of Thermus aquaticus, which is a Gram-negative thermophilic bacterium. Though Deinococcus is positive to the Gram coloration, it has been shown to possess an external membrane, unlike Gram-positives (Murray 1986). Thus, though this position of Deinococcus seems to have some degree of support in several studies (including our present work), it still needs to be confirmed, in particular by the addition of Thermus in the supertree.
The Archaeal part of the tree shows rather low bootstrap values in Figure 4. This may be due to the fact that all genes present only in Archaea were removed from the PCO analysis. This part of the tree appears to be rather well resolved when considering the 730 trees, especially with ML-based trees (Fig. 2B). This supertree shows strong support for both Archaea monophyly and their division in two groups, that is, Crenarchaeota and Euryarchaeota. ML-based trees and gamma distance-based trees support a different position for the species Thermoplasma acidophilum. Hence, the topology of the Archaeal part of the tree should be considered with caution. Our experience of supertrees suggests that such problems will be resolved when more Archaeal genome sequences become available.
The eukaryotic part of the tree supports a clade gathering plants and animals, which is in contradiction with more precise studies (Baldauf et al. 2000). However, since this work was not aimed at eukaryotic phylogeny, genes that were specific to eukaryotes were not retained. Thus, the topology of this part of the tree is based on only a few of the available genes. Moreover, it is difficult to infer relations of orthology when considering so few species (Salzberg et al. 2001), especially among eukaryotes, where the frequency of multigene families is high. Indeed, it is well known that reconstruction methods often fail to find the true phylogeny with small taxa samples (Lecointre et al. 1993). A supertree study of relationships among eukaryotes should use a completely different method of selecting gene families than the one proposed here.
Conclusion
Resolving the question of whether a prokaryotic phylogeny can be reconstructed encounters two major obstacles: first, the frequency at which lateral transfers and hidden paralogy occur; second, the loss of phylogenetic signal for deep branches. To bypass these obstacles, several strategies have been used. Some ignore the information contained in sequences, because it may be misleading, and consider the presence of a gene as a character in itself. These methods are predicted to be very sensitive to lateral transfers and gene loss. Other methods try to increase phylogenetic signal by concatenating genes. For them, lateral transfers raise severe problems comparable to those encountered when reconstructing phylogeny with genes that have recombined (Schierup and Hein 2000). The present supertree method appears to be a good tool to infer phylogeny because it does take into account molecular phylogenetic information of hundreds of genes and provides a way to cumulate all of the phylogenetic signal while considering its statistical significance. However, such an approach is meaningful only if a core of genes remaining stable during evolution exists. We obtained evidence of such a core of genes using topological comparisons of trees, and we used these genes to build a supertree. Although the supertree provides good support for several well known lineages, some internal branches remain unresolved, and some groupings may be due to systematic reconstruction artifacts. Because the number of completely sequenced genomes, and simultaneously, that of large gene families is increasing very quickly, this method can be expected to increase in efficiency. Moreover, the probability of identifying hidden paralogy increases with the number of orthologs (Salzberg et al. 2001). Although the results presented here must be confirmed by experiments gathering more complete genomes, they already suggest a tree of life that has some level of support, and show that it may be possible to extract the information concerning deep nodes of the bacterial phylogeny.
METHODS
Family Selection
A special release of the HOBACGEN database (Perrière et al. 2000) called HOBACGEN-CG was made, gathering all protein sequences into families of homologous genes from the completely or almost completely sequenced genomes of 41 prokaryotes and four eukaryotes. We retained as orthologous gene families those containing only one gene per species, or several genes more similar within species than between. We considered in this second case only one of the paralogs. Although it may miss some hidden paralogy, especially in domains for which few species are available such as eukaryotes, this definition of orthology has been shown to be much more accurate than a reciprocal BLAST hit-based one (Koski and Golding 2001). Eukaryote sequences known to encode proteins with a mitochondrial or chloroplastic location were removed, to reduce the problems due to horizontal transfers between mitochondrial, chloroplastic, and nuclear genomes. Protein sequences from hyperthermophilic bacteria with orthologs only in Archaea were removed from the family they belong to because these genes are suspected to have been acquired by lateral transfers (Nelson et al. 1999; Logsdon and Faguy 1999; Nesbo et al. 2001). Only families containing at least seven species were used for further analysis.
Alignments and Tree Construction
The sequences of each family were aligned using CLUSTAL W (Higgins et al. 1996), with all default parameters. To select parts of the alignments for which homology between sites can be assumed with good confidence, we used the GBLOCKS program (Castresana 2000). This program identifies blocks in an alignment for which homology of sites can be assumed with good confidence and regions that contain reliable phylogenetic information. It has been shown to give alignments that are almost independent of the different options of CLUSTAL W. We retained for tree construction only the alignments having conserved at least twice more sites than species.
An ML tree was computed for each family with the protML program (Kishino et al. 1990) (options: JTT model of substitution, quick add OTUs search, 300 trees retained) which gives an approximate bootstrap probability for each node. For each family, a BIONJ (Gascuel 1997) tree was also constructed using the distance matrix provided by PUZZLE (Strimmer and von Haeseler 1996) under a gamma law-based model of substitution (alpha parameter estimated by PUZZLE, eight gamma rate categories) and bootstrapped using SEQBOOT and CONSENSE from the PHYLIP package (Felsenstein 1989). To reduce the impact of interdomain lateral transfers, we applied the same criteria used by Brown et al. (2001); that is, we screened the trees where bacteria were not monophyletic and we removed these families from the data set or corrected them by removing the transferred sequences from the alignment when it was evident. We made no assumption about the monophyly of the Archaeal domain, as this problem has already been discussed (Martin and Muller 1998). Thus, 730 families containing at least seven of the 45 species available in HOBACGEN-CG were selected. Informational and operational genes were identified using annotations from HOBACGEN-CG.
Comparisons Between Trees
Trees were compared using a C program performing the following steps: (1) For each pair of trees, trees are reduced to the species they have in common. (2) The Robinson-Foulds (RF) topological distance (Robinson and Foulds 1981) is then computed for the pair. (3) On the n × n distance matrix obtained (n is the number of trees), a PCO is computed using ADE-4 (Thioulouse et al. 1997). PCO is a multivariate ordination method based on distance matrices, and it allowed us to embed our n trees in a space of up to n − 1 dimensions (Gower 1966). By taking the most significant two first dimensions and plotting the objects (the trees) along these, the major trends and groupings in the data can be determined by visual inspection.
Processing of the complete data set was difficult because several pairs of trees have nonoverlapping sets of species, producing a matrix with many holes. To reduce this problem, we performed the PCO analysis on trees containing at least ten bacterial species. This reduced the number of holes present in the matrix to less than 10% of the total, which allowed us to substitute remaining holes by the mean of the distances present in the matrix (D. Chessel, pers. comm.).
Supertree Computation
Trees chosen for the supertree computation were coded into a binary matrix using the coding scheme of Baum (1992) and Ragan (1992): each tree obtained for a set of species from a single gene family is coded into a binary matrix of informative sites with respect to bootstrap values, as shown in Figure 1. The matrices obtained are concatenated into a supermatrix in which species absent from a gene family are encoded as unknown state. The supertree is calculated on the supermatrix using program DNAPARS (default options) from the PHYLIP package. Bootstrap values on the supermatrix are obtained using SEQBOOT and CONSENSE.
All of the data used to build the trees as well as all supertrees mentioned here are available at ftp://pbil.univ-lyon1.fr/pub/datasets/GR2002. The HOBACGEN-CG database can be accessed on the PBIL server through the FamFetch interface (http://pbil.univ-lyon1.fr/databases/hobacgen.html).
WEB SITE REFERENCES
http://pbil.univ-lyon1.fr/databases/hobacgen.html; The HOBACGEN-CG database can be accessed on the PBIL server through the FamFetch interface.
Acknowledgments
We thank G. Marais and E. Lerat for daily discussions, D. Chessel for advice in PCO computation, and L. Duret for critical reading of the paper. This work was supported by Centre National de la Recherche Scientifique and Ministére de la Recherche.The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL daubin@biomserv.univ-lyon1.fr; FAX +33 478-89-27-19.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.187002.
REFERENCES
- Baldauf SL, Roger AJ, Wenk-Siefert I, Doolittle WF. A kingdom-level phylogeny of eukaryotes based on combined protein data. Science. 2000;290:972–977. doi: 10.1126/science.290.5493.972. [DOI] [PubMed] [Google Scholar]
- Barns SM, Delwiche CF, Palmer JD, Pace NR. Perspectives on archaeal diversity, thermophily and monophyly from environmental rRNA sequences. Proc Natl Acad Sci. 1996;93:9188–9193. doi: 10.1073/pnas.93.17.9188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum BR. Combining trees as a way of combining data sets for phylogenetic inference, and the desirability of combining gene trees. Taxon. 1992;41:3–10. [Google Scholar]
- Bellgard MI, Itoh T, Watanabe H, Imanishi T, Gojobori T. Dynamic evolution of genomes and the concept of genome space. Ann NY Acad Sci. 1999;870:293–300. doi: 10.1111/j.1749-6632.1999.tb08891.x. [DOI] [PubMed] [Google Scholar]
- Bocchetta M, Gribaldo S, Sanangelantoni A, Cammarano P. Phylogenetic depth of the bacterial genera Aquifex and Thermotoga inferred from analysis of ribosomal protein, elongation factor, and RNA polymerase subunit sequences. J Mol Evol. 2000;50:366–380. doi: 10.1007/s002399910040. [DOI] [PubMed] [Google Scholar]
- Brochier C, Philippe H, Moreira D. The evolutionary history of ribosomal protein Rps14: Horizontal gene transfer at the heart of the ribosome. Trends Genet. 2000;16:529–533. doi: 10.1016/s0168-9525(00)02142-9. [DOI] [PubMed] [Google Scholar]
- Brown JR, Douady CJ, Italia MJ, Marshall WE, Stanhope MJ. Universal trees based on large combined protein sequence data sets. Nat Genet. 2001;28:281–285. doi: 10.1038/90129. [DOI] [PubMed] [Google Scholar]
- Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 2000;17:540–552. doi: 10.1093/oxfordjournals.molbev.a026334. [DOI] [PubMed] [Google Scholar]
- Eernisse DJ, Kluge AG. Taxonomic congruence versus total evidence, and amniote phylogeny inferred from fossils, molecules, and morphology. Mol Biol Evol. 1993;10:1170–1195. doi: 10.1093/oxfordjournals.molbev.a040071. [DOI] [PubMed] [Google Scholar]
- Eisen JA. Assessing evolutionary relationships among microbes from whole-genome analysis. Curr Opin Microbiol. 2000a;3:475–480. doi: 10.1016/s1369-5274(00)00125-9. [DOI] [PubMed] [Google Scholar]
- Eisen JA. Horizontal gene transfer among microbial genomes: New insights from complete genome analysis. Curr Opin Genet Dev. 2000b;10:606–611. doi: 10.1016/s0959-437x(00)00143-x. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. PHYLIP – Phylogeny inference package (Version 3.2) Cladistics. 1989;5:164–166. [Google Scholar]
- Forterre P. A hot topic: The origin of hyperthermophiles. Cell. 1996;85:789–792. doi: 10.1016/s0092-8674(00)81262-3. [DOI] [PubMed] [Google Scholar]
- Galtier N, Tourasse N, Gouy M. A nonhyperthermophilic common ancestor to extant life forms. Science. 1999;283:220–221. doi: 10.1126/science.283.5399.220. [DOI] [PubMed] [Google Scholar]
- Gascuel O. BIONJ: An improved version of the NJ algorithm based on a simple model of sequence data. Mol Biol Evol. 1997;14:685–695. doi: 10.1093/oxfordjournals.molbev.a025808. [DOI] [PubMed] [Google Scholar]
- Glansdorff N. About the last common ancestor, the universal life-tree and lateral gene transfer: A reappraisal. Mol Microbiol. 2000;38:177–185. doi: 10.1046/j.1365-2958.2000.02126.x. [DOI] [PubMed] [Google Scholar]
- Gower JC. Some distance properties of latent root and vector methods used in multivariate analysis. Biometrika. 1966;53:325–328. [Google Scholar]
- Gribaldo S, Lumia V, Creti R, de Macario EC, Sanangelantoni A, Cammarano P. Discontinuous occurrence of the hsp70 (dnaK) gene among Archaea and sequence features of Hsp70 suggest a novel outlook on phylogenies inferred from this protein. J Bacteriol. 1999;181:434–443. doi: 10.1128/jb.181.2.434-443.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S, Perriere G. Intragenomic base content variation is a potential source of biases when searching for horizontally transferred genes. Mol Biol Evol. 2001;18:1838–1840. doi: 10.1093/oxfordjournals.molbev.a003972. [DOI] [PubMed] [Google Scholar]
- Gupta RS. Protein phylogenies and signature sequences: A reappraisal of evolutionary relationships among archaebacteria, eubacteria, and eukaryotes. Microbiol Mol Biol Rev. 1998;62:1435–1491. doi: 10.1128/mmbr.62.4.1435-1491.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— The phylogeny of proteobacteria: Relationships to other eubacterial phyla and eukaryotes. FEMS Microbiol Rev. 2000;24:367–402. doi: 10.1111/j.1574-6976.2000.tb00547.x. [DOI] [PubMed] [Google Scholar]
- Hansmann S, Martin W. Phylogeny of 33 ribosomal and six other proteins encoded in an ancient gene cluster that is conserved across prokaryotic genomes: Influence of excluding poorly alignable sites from analysis. Int J Syst Evol Microbiol. 2000;50:1655–1663. doi: 10.1099/00207713-50-4-1655. [DOI] [PubMed] [Google Scholar]
- Higgins DG, Thompson JD, Gibson TJ. Using CLUSTAL for multiple sequence alignments. Methods Enzymol. 1996;266:383–402. doi: 10.1016/s0076-6879(96)66024-8. [DOI] [PubMed] [Google Scholar]
- Huang Y-P, Ito J. DNA polymerase C of the thermophilic bacterium Thermus aquaticus: Classification and phylogenetic analysis of the family C DNA polymerases. J Mol Evol. 1999;48:756–769. doi: 10.1007/pl00006520. [DOI] [PubMed] [Google Scholar]
- Jain R, Rivera MC, Lake JA. Horizontal gene transfer among genomes: The complexity hypothesis. Proc Natl Acad Sci. 1999;96:3801–3806. doi: 10.1073/pnas.96.7.3801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishino H, Miyata T, Hasegawa M. Maximum likelihood inference of protein phylogeny and the origin of chloroplasts. J Mol Evol. 1990;30:151–160. [Google Scholar]
- Klenk HP, Meier TD, Durovic P, Schwass V, Lottspeich F, Dennis PP, Zillig W. RNA polymerase of Aquifex pyrophilus: Implications for the evolution of the bacterial rpoBC operon and extremely thermophilic bacteria. J Mol Evol. 1999;48:528–541. doi: 10.1007/pl00006496. [DOI] [PubMed] [Google Scholar]
- Koski LB, Golding GB. The closest BLAST hit is often not the nearest neighbor. J Mol Evol. 2001;52:540–542. doi: 10.1007/s002390010184. [DOI] [PubMed] [Google Scholar]
- Koski LB, Morton RA, Golding GB. Codon bias and base composition are poor indicators of horizontally transferred. Mol Biol Evol. 2001;18:404–412. doi: 10.1093/oxfordjournals.molbev.a003816. [DOI] [PubMed] [Google Scholar]
- Kreil DP, Ouzounis CA. Identification of thermophilic species by the amino acid compositions deduced from their genomes. Nucleic Acids Res. 2001;29:1608–1615. doi: 10.1093/nar/29.7.1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecointre G, Philippe H, Van Le HL, Le Guyader H. Species sampling has a major impact on phylogenetic inference. Mol Phyl Evol. 1993;2:205–224. doi: 10.1006/mpev.1993.1021. [DOI] [PubMed] [Google Scholar]
- Lin J, Gerstein M. Whole-genome trees based on the occurrence of folds and orthologs: Implications for comparing genomes on different levels. Genome Res. 2000;10:808–818. doi: 10.1101/gr.10.6.808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu FG, Miyamoto MM, Freire NP, Ong PQ, Tennant MR, Young TS, Gugel KF. Molecular and morphological supertrees for eutherian (placental) mammals. Science. 2001;291:1786–1789. doi: 10.1126/science.1056346. [DOI] [PubMed] [Google Scholar]
- Logsdon JM, Faguy DM. Thermotoga heats up lateral gene transfer. Curr Biol. 1999;9:R747–R751. doi: 10.1016/s0960-9822(99)80474-6. [DOI] [PubMed] [Google Scholar]
- Martin W, Muller M. The hydrogen hypothesis for the first eukaryote. Nature. 1998;392:37–41. doi: 10.1038/32096. [DOI] [PubMed] [Google Scholar]
- Miller SL, Lazcano A. The origin of life – did it occur at high temperatures? J Mol Evol. 1995;41:689–692. doi: 10.1007/BF00173146. [DOI] [PubMed] [Google Scholar]
- Mira A, Ochman H, Moran NA. Deletional bias and the evolution of bacterial genomes. Trends Genet. 2001;17:589–596. doi: 10.1016/s0168-9525(01)02447-7. [DOI] [PubMed] [Google Scholar]
- Murray RGE. Family II. Deinococcaceae Brooks and Murray 1981. 356VP. In: Sneath PHA, editor. Bergey's manual of systematic bacteriology. Baltimore: Williams and Wilkins; 1986. pp. 1035–1043. [Google Scholar]
- Nelson KE, Clayton RA, Gill SR, Gwinn ML, Dodson RJ, Haft DH, Hickey EK, Peterson JD, Nelson WC, Ketchum KA, et al. Evidence for lateral gene transfer between Archaea and Bacteria from genome sequence of Thermotoga maritima. Nature. 1999;399:323–329. doi: 10.1038/20601. [DOI] [PubMed] [Google Scholar]
- Nesbo CL, L'Haridon S, Stetter KO, Doolittle WF. Phylogenetic analyses of two “archaeal” genes in Thermotoga maritima reveal multiple transfers between archaea and bacteria. Mol Biol Evol. 2001;18:362–375. doi: 10.1093/oxfordjournals.molbev.a003812. [DOI] [PubMed] [Google Scholar]
- Ochman H, Elwyn S, Moran NA. Calibrating bacterial evolution. Proc Natl Acad Sci. 1999;96:12638–12643. doi: 10.1073/pnas.96.22.12638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochman H, Lawrence JG, Groisman EA. Lateral gene transfer and the nature of bacterial innovation. Nature. 2000;405:299–304. doi: 10.1038/35012500. [DOI] [PubMed] [Google Scholar]
- Paster BJ, Dewhirst FE. Phylogenetic foundation of spirochetes. J Mol Microbiol Biotechnology. 2000;2:341–344. [PubMed] [Google Scholar]
- Perrière G, Duret L, Gouy M. HOBACGEN: Database system for comparative genomics in bacteria. Genome Res. 2000;10:379–385. doi: 10.1101/gr.10.3.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippe H, Laurent J. How good are deep phylogenetic trees? Curr Opin Genet Dev. 1998;8:616–623. doi: 10.1016/s0959-437x(98)80028-2. [DOI] [PubMed] [Google Scholar]
- Ragan MA. Phylogenetic inference based on matrix representation of trees. Mol Phylogenet Evol. 1992;1:53–58. doi: 10.1016/1055-7903(92)90035-f. [DOI] [PubMed] [Google Scholar]
- Ragan MA. On surrogate methods for detecting lateral gene transfer. FEMS Microbiol Lett. 2001;201:187–191. doi: 10.1111/j.1574-6968.2001.tb10755.x. [DOI] [PubMed] [Google Scholar]
- Rivera MC, Jain R, Moore JE, Lake JA. Genomic evidence for two functionally distinct gene classes. Proc Natl Acad Sci. 1998;95:6239–6244. doi: 10.1073/pnas.95.11.6239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson DF, Foulds LR. Comparison of phylogenetic trees. Math Biosci. 1981;53:131–147. [Google Scholar]
- Salzberg SL, White O, Peterson J, Eisen JA. Microbial genes in the human genome: Lateral transfer or gene loss? Science. 2001;292:1903–1906. doi: 10.1126/science.1061036. [DOI] [PubMed] [Google Scholar]
- Schierup MH, Hein J. Consequences of recombination on traditional phylogenetic analysis. Genetics. 2000;156:879–891. doi: 10.1093/genetics/156.2.879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snel B, Bork P, Huynen MA. Genome phylogeny based on gene content. Nat Genet. 1999;21:108–110. doi: 10.1038/5052. [DOI] [PubMed] [Google Scholar]
- Strimmer K, von Haeseler A. Quartet puzzling: A quartet maximum likelihood method for reconstructing tree topologies. Mol Biol Evol. 1996;13:964–969. [Google Scholar]
- Teichmann SA, Mitchison G. Is there a phylogenetic signal in prokaryote proteins? J Mol Evol. 1999;49:98–107. doi: 10.1007/pl00006538. [DOI] [PubMed] [Google Scholar]
- Tekaia F, Lazcano A, Dujon B. The genomic tree as revealed from whole proteome comparisons. Genome Res. 1999;9:550–557. [PMC free article] [PubMed] [Google Scholar]
- Thioulouse J, Chessel D, Dolédec S, Olivier JM. ADE-4: A multivariate analysis and graphical display software. Stat and Comput. 1997;7:75–83. [Google Scholar]
- Tiboni O, Cammarano P, Sanangelantoni AM. Cloning and sequencing of the gene encoding glutamine synthetase I from the archaeum Pyrococcus woesei: Anomalous phylogenies inferred from analysis of archaeal and bacterial glutamine synthetase I sequences. J Bacteriol. 1993;175:2961–2969. doi: 10.1128/jb.175.10.2961-2969.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B. Limitations of compositional approach to identifying horizontally transferred genes. J Mol Evol. 2001;53:244–250. doi: 10.1007/s002390010214. [DOI] [PubMed] [Google Scholar]
- Wang G, Humayun MZ, Taylor DE. Mutation as an origin of genetic variability in Helicobacter pylori. Trends Microbiol. 1999;7:488–493. doi: 10.1016/s0966-842x(99)01632-7. [DOI] [PubMed] [Google Scholar]
- Woese C. Bacterial evolution. Microbiol Rev. 1987;51:221–271. doi: 10.1128/mr.51.2.221-271.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]