Abstract
The developmentally important Hedgehog (Hh) signaling pathway has recently been implicated in several forms of solid cancer. Current drug development programs focus on targeting the protooncogene Smoothened, a key transmembrane pathway member. These drug candidates, albeit promising, do not address the scenario in which pathway activation occurs downstream of Smoothened, as observed in cases of medulloblastoma, glioma, pericytoma, breast cancer, and prostate cancer. A cellular screen for small-molecule antagonists of GLI-mediated transcription, which constitutes the final step in the Hh pathway, revealed two molecules that are able to selectively inhibit GLI-mediated gene transactivation. We provide genetic evidence of downstream pathway blockade by these compounds and demonstrate the ineffectiveness of upstream antagonists such as cyclopamine in such situations. Mechanistically, both inhibitors act in the nucleus to block GLI function, and one of them interferes with GLI1 DNA binding in living cells. Importantly, the discovered compounds efficiently inhibited in vitro tumor cell proliferation in a GLI-dependent manner and successfully blocked cell growth in an in vivo xenograft model using human prostate cancer cells harboring downstream activation of the Hh pathway.
Keywords: GLI inhibitors, Hedgehog signaling, cancer
Ectopic activation of the Hedgehog (Hh) signaling pathway has recently been shown to be involved in several malignancies, such as basal cell carcinoma of the skin, cerebellar medulloblastoma, rhabdomyosarcoma, or cancers of the pancreas, colon, stomach, lung, and prostate (1–8).
Vertebrate organisms possess three Hh proteins [Sonic Hh (Shh), Indiana Hh (Ihh), and Desert Hh (Dhh)], which all bind to the same receptor, Ptch1. Without ligand stimulation, Ptch1 restrains signaling of the seven-pass transmembrane protein and protooncogene Smoothened (Smo). Upon Hh binding, this inhibition is relieved and Smo transduces the signal to the ultimate effectors of the pathway, the zinc-finger transcription factors Gli1, Gli2, and Gli3. With respect to tumor formation, Gli1 and Gli2 are the prime transcriptional effectors involved, and constitutive activation of at least one of them is of critical importance for cancer development (9–13). Additional pathway members that act between Smo and Gli include negative components such as Suppressor of Fused (Sufu), Rab23, or Ren, as well as proteins exerting a positive effect on Hh signaling, such as IFT proteins, Tectonic, or MIM/BEG4 (14–19).
In several cases, Smo-independent activation mechanisms have been documented, such as mutations in or loss of heterozygosity of the SUFU gene, which was found in medulloblastoma (20, 21), prostate cancer (22), rhabdomyosarcoma (7), and a human lung cancer cell line (23). In addition, other downstream pathway activation mechanisms have been revealed, such as REN deletions (15), GLI gene amplifications (24–26), GLI1 chromosomal translocations (27), or GLI2 protein stabilization (28). Moreover, reports on frequent cyclopamine-insensitivity of GLI1-positive cell lines derived from pancreatic adenocarcinoma can be found in the literature (6). Taken together, this result demonstrates diversity in Smo-independent activating mechanisms and shows that, apart from the autocrine stimulation loops and receptor mutations documented in several cancers (2, 4, 5), a significant fraction of tumors possesses downstream activation of the pathway.
At present, there is no clinically available treatment that specifically targets the Hh signaling pathway. Cyclopamine, a plant steroidal alkaloid that inhibits Smo (29), along with other Smo-targeting compounds (Curis, Cambridge, MA), is currently entering phase I.
To address the need for broadly active downstream inhibitors of Hh signaling, we set up a cell-based screen to identify small-molecule antagonists of GLI function, which constitutes the final step in the Hh pathway. Such molecules should be of greater applicability than selective upstream inhibitors in a broad spectrum of GLI-dependent cancers because they act irrespective of the mode of pathway activation.
Results
A Cell-Based Screen for Inhibitors of GLI1-Mediated Transcription.
To block downstream Hh signaling, we decided to target the GLI proteins because these members of the family of zinc-finger transcription factors constitute the essential and ultimate effectors of the pathway. The screen we performed was carried out by using GLI1 because the importance of GLI1 in tumor development and progression is well documented in mouse models and human cell culture systems (5, 8, 10, 12). However, given the high degree of homology between GLI1 and GLI2, we reasoned that a potential GLI1 inhibitor would also target GLI2.
Two compounds that were capable of reducing GLI1-mediated transcription emerged from a screen using HEK293 cells transiently expressing GLI1 and a GLI-dependent luciferase reporter (Fig. 1 A and Materials and Methods). We termed these molecules GANT61 (for Gli-ANTagonist) and GANT58 (NSC 136476 and NSC 75503, respectively). Fig. 1A shows the structure of these compounds and a scheme of the small-molecule screen. Chemically, the two screening hits represent different classes, with GANT61 being a hexahydropyrimidine derivative and GANT58 possessing a thiophene core with four pyridine rings. The identity and purity of the compounds were verified by liquid chromatography-coupled mass spectrometry. Despite their structural differences, both molecules were capable of interfering with GLI1 as well as GLI2-mediated transcription in a dose-dependent manner, as shown in Fig. 1 B and C. In particular, GANT61 was able to efficiently block GLI1 as well as GLI2-induced transcription. The GLI2 variant used was a constitutively active version containing an N-terminal deletion of its repressor domain (ΔN-GLI2, GLI2β) (30, 31). In these assays, Suppressor of Fused (SUFU) was cotransfected as a positive control. SUFU directly binds GLI proteins and efficiently represses transcription. It should be noted that the artificial overexpression of GLI1/GLI2 likely required higher concentrations of antagonist than would normally be needed at physiological expression levels.
Fig. 1.
Inhibition of GLI-induced transcription in transfected HEK293 cells. (A) Schematic illustration of the compound screen to identify small-molecule GLI antagonists. GliBS, Gli binding site; Luc, firefly luciferase. The structures of two hits, GANT61 and GANT58, are given in the upper right. (B) GLI1 inhibition. (C) GLI2 (ΔN-GLI2) inhibition. SUFU (SF) was cotransfected as a positive control. To achieve equal transfection efficiencies in all wells, transfections were done on large plates and then split on smaller wells and treated. All values were normalized to total protein amount. Treatment time was 24 h, and control cells were treated with DMSO only. Shown is the mean of three independent experiments. Error bars indicate SD.
Inhibition of Endogenous Hh Signaling by the GANT Compounds.
To investigate the inhibitory properties of the Gli antagonists under more physiological conditions, we turned to systems using induction of the endogenous Gli1 gene, such as the Hh signaling competent murine NIH 3T3 cell line. We treated a clonal NIH 3T3 cell line, in which a Gli reporter gene was stably incorporated [Shh-LIGHT2 (Shh-L2) cells], with the synthetic Smo agonist SAG (32) and confirmed the ability of GANT61 and GANT58 to suppress signaling. The IC50 in this assay was ≈5 μM for both compounds (Fig. 2A). Cyclopamine, a steroidal alkaloid from the corn lily that binds to and inactivates Smo (29), was included as a positive control. None of the compounds directly inhibited the reporter enzyme, as assayed by the expression of luciferase in a Gli-independent manner in NIH 3T3 (data not shown). In addition, no toxicity or reduction in cell viability [which would have resulted in subconfluency and subsequent pathway reduction (29)] could be observed for several cell lines tested [supporting information (SI) Fig. 7].
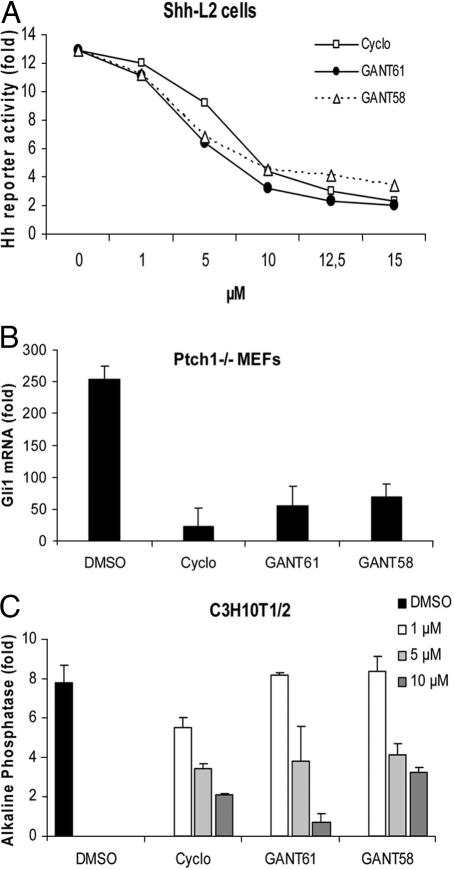
Fig. 2.
Inhibition of endogenous Hh signaling. (A) Dose–response curve of GANT61 and GANT58 in comparison to cyclopamine (Cyclo) in SAG-treated Shh-L2 cells. All three compounds are capable of inhibiting Hh signaling. Treatment time was 48 h, and normalization was against Renilla luciferase. Shown is the fold increase of Hh reporter activity compared with cells not treated with SAG. (B) Determination of Gli1 mRNA levels by quantitative PCR in Ptch1−/− MEFs (fold induction compared with wild-type MEFs). Confluent cells were treated with 10 μM compound for 2–3 days. Data were normalized to Gapdh expression. (C) Inhibition of SAG-induced alkaline phosphatase expression in C3H10T1/2 cells after treatment with compounds and SAG for 4 days. Values were normalized to total protein amount. Shown is the fold alkaline phosphatase induction compared with cells not treated with SAG. All experiments were repeated at least three times. Error bars depict SD.
Next, we analyzed endogenous Hh target gene activation in Ptch1−/− mouse embryonic fibroblasts (MEFs). Because Ptch1 is the repressive Hh receptor, loss of this gene results in ligand-independent, constitutive activation of the pathway with concomitant expression of Hh target genes such as Gli1. The latter can thus serve as a readout for pathway activity. As can be seen in Fig. 2B, both GANTs significantly reduced Gli1 mRNA levels at a con-centration of 10 μM, which is indicative of strong pathway inhibition. Similar findings were obtained by a luminometric assay measuring endogenous Ptch1 expression (SI Fig. 8) and by measuring inhibition of alkaline phosphatase induction in C3H10T1/2 cells (Fig. 2C).
GANT61 and GANT58 Are Downstream Inhibitors of Hh Signaling.
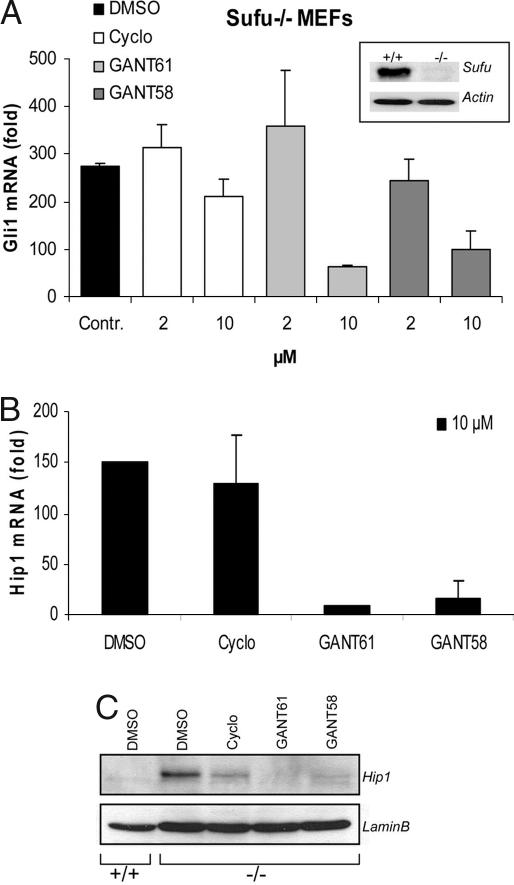
To further elucidate the concept of downstream pathway inhibition by GANT61 and GANT58, we made use of Sufu-null MEFs. These cells have unrestricted, cell-autonomous activation of high levels of Hh signaling due to genetic ablation of the downstream negative factor Sufu (14).
Treatment of Sufu−/− cells with 10 μM GANTs led to a significant reduction of the high expression levels of the Hh target genes, Gli1 and Hip1, as shown by quantitative PCR (Fig. 3 A and B). As expected, cyclopamine was unable to repress signaling in this system because pathway activation occurs downstream of Smo. When Hip1 protein levels were analyzed by Western blotting, we observed complete abrogation of the signal upon GANT exposure (Fig. 3C). Some reduction was also seen in samples from cyclopamine-treated cells, although no reduction was apparent on the mRNA level. Taken together, these findings confirm that GANT61 and GANT58 are indeed inhibitors of Hh signaling downstream of Smo and Sufu.
Fig. 3.
Downstream inhibition of the Hh pathway. (A and B) Quantitative RT-PCR for Gli1 mRNA (A) and Hip1 mRNA (B) in Sufu−/− MEFs treated with the indicated compounds for 3 days. Results were normalized against Gapdh mRNA levels. Shown is the fold increase in comparison to Sufu+/+ MEFs (n = at least three, error bars depict SD). (A Inset) Immunoblot verifying the absence of Sufu protein in Sufu−/− cells. (C) Hip1 Western blot of Sufu−/− MEFs treated with 10 μM compound.
The GANT Compounds Display Selectivity for the Hh Pathway.
To evaluate the specificity for Hh signaling, we investigated the influence of GANT61 and GANT58 on several unrelated signal transduction pathways: TNF signaling/NFκB activation, glucocorticoid receptor gene transactivation, and the Ras–Raf–Mek–Mapk cascade. As can be seen in SI Fig. 9, none of these pathways was influenced on treatment with up to 10 μM GANT61/GANT58, demonstrating a high degree of selectivity for Hh/Gli signaling. Furthermore, these results show that general and common cellular events like correct protein folding, nuclear transport, or basal transcriptional machinery assembly are not disturbed. More data on specificity can be found on the National Cancer Institute's web site (www.dtp.nci.nih.gov), where it is shown that GANT61 and GANT58 are inactive in several other screens for inhibitors of HGF/Met, C/EBPα, or HIF-1. Also, an mRNA microarray experiment on GANT61-treated Ptch1−/− MEFs revealed no marked inhibition pattern implicating additional cellular signaling cascades (SI Appendix 1 and later sections).
Inhibition of Tumor Cell Growth.
NIH 3T3 cells form colonies in soft agar when transfected with dominant-active Smo (29). In addition, fibroblast transformation was shown to be mediated by Gli1 and, to a lesser extent, Gli2, but not Gli3 (12). To evaluate whether GANT61/GANT58 could suppress Hh/Gli-mediated transformation in culture, we determined colony formation in NIH 3T3 cells transfected with an expression plasmid for SHH. In line with Gli inhibition by the GANTs, both molecules were capable of inhibiting cellular transformation, whereas numerous dense colonies formed in solvent-treated samples (SI Fig. 10).
Several recent reports confirm that proliferation of HH/GLI-positive human tumor cells can be halted by Hh-neutralizing antibodies, Smo antagonists, or siRNA against Gli1 (5, 6, 8). To elucidate the potential use of the GANT molecules in putative future cancer therapy, we investigated whether GLI-dependent growth of human cancer cells could be blocked. We included a panel of cells with low (HepG2 hepatocellular carcinoma cells, Jurkat T cell leukemia cells) and elevated (PANC1 pancreatic adenocarcinoma cells, 22Rv1 prostate carcinoma cells) GLI1 levels (SI Fig. 11).
Incubation of PANC1 or 22Rv1 cells with 5 μM GANT61 or GANT58 for 48 h led to a reduction in GLI1 and PTCH expression, consistent with significant GLI inhibition (Fig. 4A). In contrast, treatment with the same concentration of cyclopamine resulted in minor changes in GLI1/PTCH expression, suggesting that in these two cell lines Hh pathway activation has occurred downstream of SMO. It is interesting to note that 22Rv1 cells have also been shown to possess stabilized levels of GLI2 protein, and that growth of PANC1 cells has been previously reported to be cyclopamine-insensitive (6, 28).
Fig. 4.
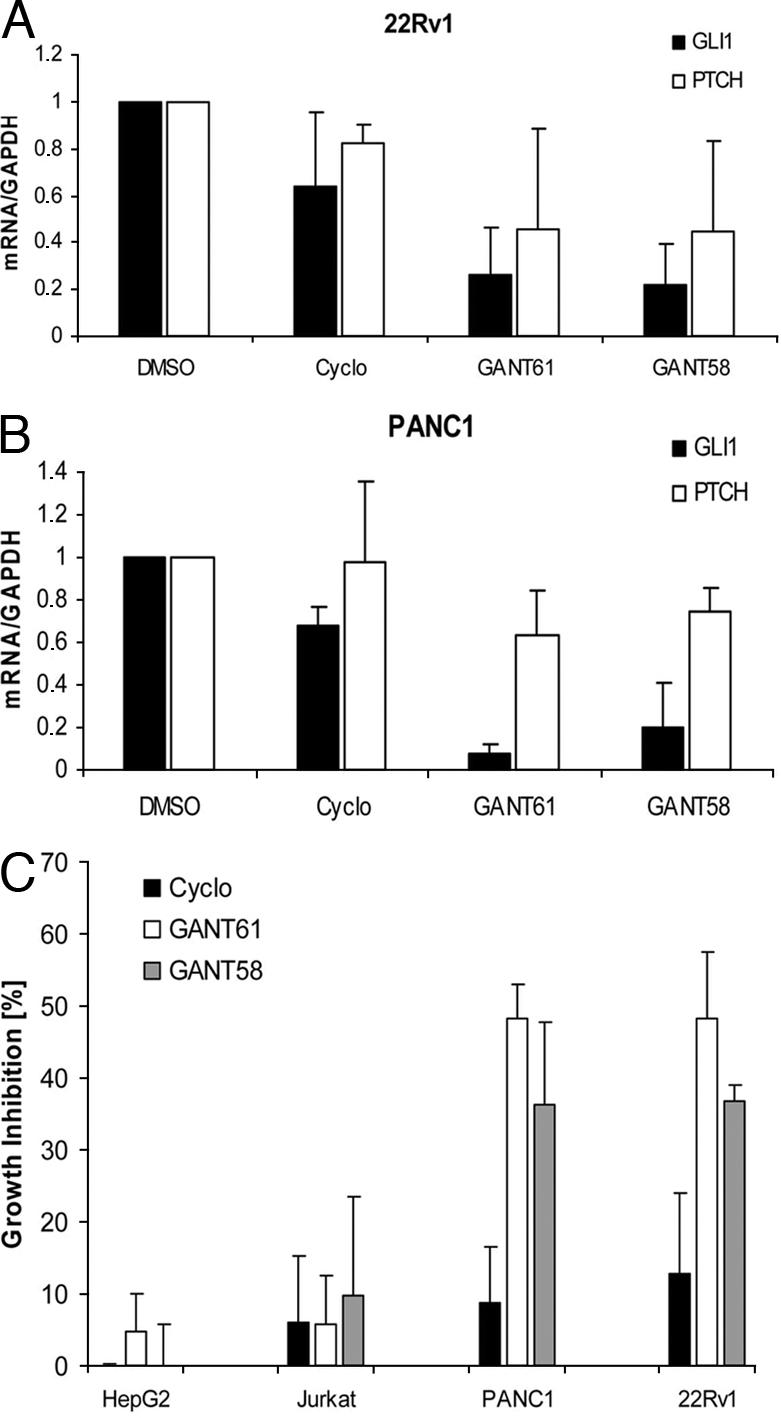
Inhibition of GLI-dependent human tumor cell growth. (A) Change of GAPDH-normalized GLI1 and PTCH expression in 22Rv1 and PANC1 cells on treatment with 5 μM compound for 48 h. (B) Inhibition of cell proliferation as measured by BrdU incorporation. Shown is the percentage of inhibition of BrdU incorporation in comparison to DMSO-treated samples (n = ≥4, error bars = indicate SD).
We then asked whether the reduction in GLI-mediated gene activation was translated into diminished growth. Quantification of BrdU incorporation in cells treated with 5-μM compound for 48 h revealed a GLI-dependent pattern of inhibition of cell proliferation: The GLI1-positive cell lines PANC1 and 22Rv1 were much more susceptible to GANT61/GANT58 (≈40–50% inhibition) than the low-GLI1 cell lines HepG2 and Jurkat (≈0–10% inhibition) (Fig. 4B). Inhibition of Smo activity by cyclopamine treatment had only marginal effects on cell proliferation (≈0–12% inhibition), which is in line with the quantitative PCR data presented in Fig. 4A (results from treatment of additional cell lines are summarized in SI Fig. 12).
To verify the influence of GLI transcription factors on cell proliferation, we knocked down GLI1 and GLI2 expression by siRNAs in 22Rv1 and PANC1 cells, which resulted in a reduction of ≥60% in mRNA levels 48 h after transfection (SI Fig. 13A). Interestingly, PTCH mRNA was only down-regulated on GLI2 knockdown, supporting findings in murine fibroblasts showing that Ptch1 is primarily a target gene of Gli2 rather than Gli1 (33).
As reported previously for other cancer cell lines (8), knocking down GLI1 levels by siRNA led to an almost 20% reduction in proliferation in 22Rv1 and PANC1 cells (SI Fig. 13B). Because GLI2 mRNA levels in 22Rv1 cells are very low, the effects of GLI2 knockdown were not as pronounced in this cell line (≈7–10% inhibition) as they were in PANC1 (≈15% inhibition), where GLI2 levels are higher (≈30-fold higher, data not shown). This result might explain why a combination of siRNAs against GLI1 and GLI2 resulted in an additional growth inhibition only in PANC1, but not in 22Rv1 cells (SI Fig. 13B). It is noteworthy that the true growth-inhibition rates upon interference with GLI function are likely to be higher because siRNA transfections efficiencies are <100%.
To circumvent the problem of suboptimal transfection efficiencies and to investigate the GLI-dependency of the observed antiproliferative effect in more detail, we turned to Ptch1−/− MEFs. Proliferation of these cells has previously been shown to be reduced upon pathway inhibition by cyclopamine (29). As can be seen in SI Fig. 14, treatment of Ptch1−/− cells with cyclopamine or GANTs led to a dose-dependent 40–60% reduction in BrdU incorporation. If these cells were pretreated with 10 μM cyclopamine for 24 h to block upstream Hh pathway initiation and were then cultured in the presence of cyclopamine plus GANT61 or GANT58, the antiproliferative effect of GLI antagonism was strongly reduced. In contrast, the strong antiproliferative agent mitomycin C blocked cell growth irrespective of the Gli status. These results strongly suggest that the antiproliferative effects of GANT61 and GANT58 are mediated by Gli inhibition.
Besides reduced growth, inhibition of the Hh signaling pathway has been reported to result in increased apoptosis (2, 6, 34). However, using concentrations sufficient for GLI inhibition, no significant induction of apoptosis with cyclopamine, GANT61, or GANT58 was detected (SI Fig. 15). This finding suggests that the reduction in BrdU incorporation was not caused by apoptosis, but rather by cytostatic mechanisms.
GANT Treatment Suppresses Human Tumor Cell Growth in Vivo.
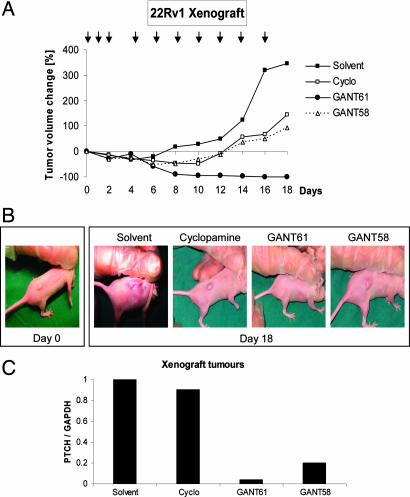
To address the ability of the two GANT molecules to block GLI function and subsequent cell growth also in vivo, we turned to a human xenograft model. Nude mice were injected s.c. with GLI1-positive 22Rv1 prostate cancer cells, and tumors were established (median size ≈250 mm3). We began treatment with daily s.c. injections at a concentration of 50 mg/kg of cyclopamine, GANT61, GANT58, or solvent only (n = 4–5 for each group). However, after 3 days, cyclopamine-treated animals presented with severe ulcerations at the injection sites (data not shown). Therefore, we decided to change the treatment regimen to injections only every second day. To be able to compare all compounds, this protocol was also introduced for the GANTs, although mice treated with these compounds showed no such signs of toxicity. All injections were done 2–3 cm away from the tumors. During an 18-day treatment period, suppression of tumor cell growth was observed for all compounds (Fig. 5 A and B). Treatment with cyclopamine or GANT58 resulted in the inhibition of additional xenograft growth and limited the increase in tumor size. Importantly, GANT61 induced growth regression until no tumor was palpable. During the 18-day treatment time, no adverse side effects, such as weight loss, ulcerations, or general non-well being of the animals, were observed with either of the GANT compounds.
Fig. 5.
Human prostate cancer xenograft. (A) Change of tumor volume during treatment period. Time points of injections are given as arrows above the curve. (B) Macroscopic appearance of xenografts at the beginning of treatment (day 0) and at the end of treatment (day 18). No tumor could be seen in GANT61-treated animals (the bulge in the picture is part of the rib cage). (C) Quantification of PTCH mRNA by quantitative PCR in treated 22Rv1 tumors. Values were normalized against GAPDH. Shown is the mean of the analysis of two tumors for each treatment.
To analyze the tumors in more detail, two animals of each group were killed and the tumors were removed. Because no tumor mass was left in the GANT61-treated cohort, we took two solvent-treated mice on day 18 and injected them for 5 consecutive days with 50 mg/kg GANT61 before removing tumors for further analysis (SI Fig. 16). Staining for BrdU incorporation revealed a clear inhibition of cell proliferation for all three compounds in comparison with control, where the number of proliferating cells was high. Staining for cleaved caspase 3 as a marker of apoptosis resulted in a weak staining in solvent and cyclopamine-treated tumors, which was significantly increased in GANT61- and GANT58-treated samples. This result might indicate that GLI inhibition favors induction of apoptosis in an in vivo situation because no cell death could be found in vitro using the same cell line (SI Fig. 15).
Quantitative PCR analysis of the Hh pathway status in the xenografted cells revealed that, in line with in vitro data, GANT61 as well as GANT58, but not cyclopamine, strongly reduced the expression levels of the target gene PTCH (technical difficulties precluded analysis of GLI1 in this case) (Fig. 5C).
To elucidate whether the tumor regression seen with GANT61 was complete or whether some cancer cells had survived, we stopped treatment of four mice on day 18 and left them untreated for 16 days. Tumors reappeared in only two of these four animals, suggesting that in the other two mice all cancer cells had been completely eradicated.
To address the issue of Gli specificity in more detail and in an attempt to identify putative GANT61 targets unrelated to the Hh pathway, which might underlie the strong antiproliferative effect, we performed microarray-based expression profiling of Ptch1−/− MEFs treated with 10 μM GANT61. SI Appendix 1 gives a summary of signaling pathways and physiological networks implicated in cell proliferation. However, the changes in gene expression seen in the microarray experiment appeared to be quite weak and could not be linked to a consistent inhibition of any of these pathways, again suggesting a high selectivity of GANT61 for Hh/Gli signaling.
GANT61 Interferes with Cellular DNA Binding of GLI1.
To address the molecular mechanism of Gli inhibition in more detail, we performed several experiments showing that (i) primary cilia, which are indispensable for proper Hh signaling (17), are not disturbed in morphology and frequency as determined in GANT-exposed NIH 3T3 cells stained for acetylated tubulin (data not shown). This result is in line with the finding that both GANTs block GLI1, a protein shown to be independent of primary cilia (17). Furthermore, GLI inhibition was observed in HEK293 cells (Fig. 1 B and C), which do not possess primary cilia (data not shown). (ii) The GANT compounds seem to act on nuclear Gli because they could inhibit a nuclear-accumulated Gli mutant (35) as well as wild-type Gli (SI Fig. 17 A and B). (iii) Unexpectedly, GANT61 treatment led to a strong accumulation of transfected GLI1 in the nucleus of transfected cells (SI Fig. 17 C and D), further supporting the strong inhibition of nuclear GLI1. (iv) Neither GANT61 nor GANT58 induced phosphorylation of Creb, a protein target of protein kinase A (PKA) (SI Fig. 18A). Activated PKA is a known inhibitor of Gli function. This result suggests that none of the GANTs inhibits GLI via PKA activation, as does forskolin. This conclusion was supported by results from luciferase assays (SI Fig. 18B). (v) As verified by EMSAs, in vitro DNA binding of GLI1 was not altered by either of the GANT molecules. However, when cells were pretreated with GANT61 and lysates were analyzed for GLI DNA binding, GANT61 strongly reduced the intensity of the shifted band (Fig. 6 and SI Fig. 19), indicating that GANT61 induces a modification of GLI1 that compromises proper DNA binding.
Fig. 6.
Inhibition of GLI1 DNA binding. (Upper Left) EMSA. Compounds were added to whole-cell lysates of GLI1-transfected HEK293 cells (in vitro). (Lower Left) Mean band intensities from two independent EMSA experiments. (Upper Right) EMSA. Compounds were added to Flag-GLI1-transfected HEK293 cells in culture (in vivo), and whole-cell lysates were prepared 24 h later. Lysate input was normalized to equal GLI1 loading. (Inset) Western blot (WB) using α-Flag. (Lower Right) Mean band intensities from two independent lysate preparations and EMSA experiments. GliBS, Gli binding site.
Discussion
In this study, we identify two low-molecular-weight compounds that, based on genetic evidence, inhibit Hh signaling downstream of Smo and Sufu at the level of Gli with an IC50 of ≈5 μM in cellular assays. Exploring the mechanism of inhibition, we found that both GANT61 and GANT58 mainly act at the nuclear level because transcription induced by GLI1 with a mutated nuclear export signal was still blocked. Moreover, we discovered that treatment with GANT61, representing the most promising inhibitor, surprisingly led to an accumulation of GLI1 in the nucleus, but that at the same time GLI1 transcriptional activity was potently inhibited. Consistent with this observation, DNA binding of GLI1 in GANT61-treated cells was markedly reduced, suggesting that GANT61 may induce posttranslational modifications of GLI1 that either prevent DNA binding or destabilize the GLI1–DNA complex. In Western blots, we noted a shift in GLI1 band mobility in GANT61-treated samples, which might point toward an increase in phosphorylation. The precise nature of this potential GLI1 modification is currently under investigation.
Surprisingly, we found some discrepancies between our in vitro and in vivo cell proliferation data. For instance, cyclopamine showed no clear inhibition of cell growth of the prostate carcinoma cell line 22Rv1 in vitro, but prohibited additional tumor growth in the xenograft assay. This finding might indicate that cyclopamine does not directly act on the xenografted tumor cells, but on the surrounding stroma. It has been suggested previously that in prostate cancer tumor-derived SHH activates Gli expression in the stroma with subsequent provision of growth support for the tumor (36). Moreover, 22Rv1 cells express IHH (data not shown), making it conceivable that cyclopamine inhibits IHH-mediated signaling from tumor cells to surrounding stromal cells and, as a result, eliminates the growth-promoting influence of the stroma. The hypothesis of such a tumor–stroma cross-talk is supported by the finding that mouse cells surrounding the human xenograft have reduced Gli1 levels if treated with cyclopamine, GANT58, or GANT61 (SI Fig. 20).
The lack of a stromal compartment might explain the inefficacy of cyclopamine in our in vitro experiments. The involvement of a stromal component might also explain the in vitro vs. in vivo difference in apoptosis induction that we observed. In the xenograft experiment, GANT61 induced strong growth regression until no tumor mass was visible. After discontinuation of the treatment, tumors did not reappear in 50% of the animals (two of four), demonstrating the feasibility of complete tumor cell eradication by treatment with GANT61. The high efficiency of GANT61 may be explained by the fact that GLI inhibition affects the tumor cells as well as the stromal cells. Differences in the observed in vivo efficacies between GANT61 and GANT58 could, at least in part, be caused by the treatment protocol (injections only every second day and for 18 days in total) because it would favor compounds with more advantageous pharmacokinetic properties.
With respect to the numerous potential mutational targets within the Hh pathway downstream of Smo already discovered, the group of tumors for which direct GLI inhibition is beneficial is substantial and likely to increase. Hence, the small-molecule inhibitors of GLI-mediated transcription described here will be valuable research tools in elucidating the involvement of GLI in normal embryonic development and tumor growth. In addition, data presented in this work might encourage further medicinal chemistry efforts to explore the full potential of these compound classes. Having the latter in mind, it is interesting to note that both GANT molecules meet the criteria concerning effective absorption and distribution of potential drug molecules (37).
Materials and Methods
Compound Screen.
HEK293 cells were transfected with GLI1 expression plasmid, together with the reporter plasmids 12xGliBS-Luc and R-Luc on 10-cm plates (day 0). Twenty-four hours later, cells were seeded in white 96-well plates with clear bottom at a density of 15,000 cells per well. Cells were allowed to attach, and compounds (Diversity Set, 1990 compounds; National Cancer Institute, Bethesda, MD) were added at a final concentration of 10 μM in DMSO (0.5% final DMSO concentration) (day 1.5). Cells were grown for another 24 h, subsequently lysed, and then analyzed by using the Dual Luciferase kit (Promega, Madison, WI). Plates were read on a Berthold Technologies (Bad Wildbad, Germany) microplate luminometer.
BrdU Incorporation Assay.
Subconfluent cells were grown in reduced FBS (2.5%) for 48 h (8) in the presence of 5 μM test compound (or DMSO) on white 96-well plates with clear bottom. Subsequently, cells were labeled for 2 h with BrdU, fixed, and analyzed (luminometric BrdU cell proliferation ELISA; Roche Diagnostics, Indianapolis, IN). Samples were read on a Molecular Devices (Sunnyvale, CA) SpectraMax Gemini EM.
RNA Interference.
Predesigned, double-stranded siRNAs targeting human GLI1 and GLI2 were purchased from Dharmacon (SMARTpool reagents; Dharmacon RNA Technologies, Lafayette, CO). As controls, nontargeting siRNA and siRNA-targeting GAPDH were used (Dharmacon RNA Technologies). Transfections were performed with Dharmafect1 (Dharmacon Technologies) according to the manufacturer's instructions.
Xenograft Experiment.
5 × 106 22Rv1 cells were suspended in a total volume of 100 μl of a 1:1 mixture of RPMI medium 1640:Matrigel (E1270, Sigma–Aldrich, St. Louis, MO). The cell suspension was injected s.c. at the posterior flank of female BALB/c nude mice (nu/nu). Tumors were grown until they reached a median size of ≈250 mm3 (5–6 days). Animals were randomly divided into four groups (n = 4–5) and treated with solvent only (corn oil:ethanol, 4:1) or compounds in solvent (50 mg/kg) for 16 days (treatment scheme according to Fig. 5). s.c. injections of compounds were performed several centimeters away from the tumor. Tumor volumes were calculated by the formula length × width × 0.5 × (length + width) (3). At the end of the treatment period, animals were given a BrdU pulse (50 mg/kg) for 30 min, and tumors were removed. All animal experiments were approved by local ethics authorities. Cyclopamine was purchased from LC Laboratories (Woburn, MA).
Further information is available in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank the National Cancer Institute (Developmental Therapeutics Program), P. Beachy (Johns Hopkins University School of Medicine, Baltimore, MD), F. Aberger (University of Salzburg, Salzburg, Austria), and J. Bergman (Karolinska Institute, Huddinge, Sweden) for kind provision of materials; J. Westman for mass spectrometry analysis and discussions; S. Strömblad and B. Rozell for advice on xenograft experiments; and S. Teglund for helpful comments. The use of animal facilities within the Clinical Research Center, Karolinska University Hospital, is gratefully acknowledged. This work was supported by stipends from the Swedish Cancer Society, the Wenner-Gren Foundation, and the Karolinska Institute (to M.L.); a Marie Curie International Incoming Fellowship (to T.S.); and grants from the Swedish Cancer Society and Swiss Bridge (to R.T.).
Abbreviations
- Hh
Hedgehog
- GANT
Gli antagonist
- Gli
glioma-associated oncogene
- MEF
mouse embryonic fibroblast
- Sufu
Suppressor of Fused.
Footnotes
Conflict of interest statement: The inhibitors are the subject of a filed patent application.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0609699104/DC1.
References
- 1.Beachy PA, Karhadkar SS, Berman DM. Nature. 2004;432:324–331. doi: 10.1038/nature03100. [DOI] [PubMed] [Google Scholar]
- 2.Watkins DN, Berman DM, Burkholder SG, Wang B, Beachy PA, Baylin SB. Nature. 2003;422:313–317. doi: 10.1038/nature01493. [DOI] [PubMed] [Google Scholar]
- 3.Berman DM, Karhadkar SS, Hallahan AR, Pritchard JI, Eberhart CG, Watkins DN, Chen JK, Cooper MK, Taipale J, Olson JM, et al. Science. 2002;297:1559–1561. doi: 10.1126/science.1073733. [DOI] [PubMed] [Google Scholar]
- 4.Berman DM, Karhadkar SS, Maitra A, Montes DO, Gerstenblith MR, Briggs K, Parker AR, Shimada Y, Eshleman JR, Watkins DN, et al. Nature. 2003;425:846–851. doi: 10.1038/nature01972. [DOI] [PubMed] [Google Scholar]
- 5.Karhadkar SS, Bova GS, Abdallah N, Dhara S, Gardner D, Maitra A, Isaacs JT, Berman DM, Beachy PA. Nature. 2004;431:707–712. doi: 10.1038/nature02962. [DOI] [PubMed] [Google Scholar]
- 6.Thayer SP, di Magliano MP, Heiser PW, Nielsen CM, Roberts DJ, Lauwers GY, Qi YP, Gysin S, Fernandez-del Castillo C, et al. Nature. 2003;425:851–856. doi: 10.1038/nature02009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tostar U, Malm CJ, Meis-Kindblom JM, Kindblom LG, Toftgard R, Unden AB. J Pathol. 2006;208:17–25. doi: 10.1002/path.1882. [DOI] [PubMed] [Google Scholar]
- 8.Sanchez P, Hernandez AM, Stecca B, Kahler AJ, DeGueme AM, Barrett A, Beyna M, Datta MW, Datta S, Altaba A. Proc Natl Acad Sci USA. 2004;101:12561–12566. doi: 10.1073/pnas.0404956101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dahmane N, Lee J, Robins P, Heller P, Altaba A. Nature. 1997;389:876–881. doi: 10.1038/39918. [DOI] [PubMed] [Google Scholar]
- 10.Nilsson M, Unden AB, Krause D, Malmqwist U, Raza K, Zaphiropoulos PG, Toftgard R. Proc Natl Acad Sci USA. 2000;97:3438–3443. doi: 10.1073/pnas.050467397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grachtchouk M, Mo R, Yu S, Zhang X, Sasaki H, Hui CC, Dlugosz AA. Nat Genet. 2000;24:216–217. doi: 10.1038/73417. [DOI] [PubMed] [Google Scholar]
- 12.Kimura H, Stephen D, Joyner A, Curran T. Oncogene. 2005;24:4026–4036. doi: 10.1038/sj.onc.1208567. [DOI] [PubMed] [Google Scholar]
- 13.Sheng H, Goich S, Wang A, Grachtchouk M, Lowe L, Mo R, Lin K, de Sauvage FJ, Sasaki H, Hui CC, et al. Cancer Res. 2002;62:5308–5316. [PubMed] [Google Scholar]
- 14.Svard J, Heby-Henricson K, Persson-Lek M, Rozell B, Lauth M, Bergstrom A, Ericson J, Toftgard R, Teglund S. Dev Cell. 2006;10:187–197. doi: 10.1016/j.devcel.2005.12.013. [DOI] [PubMed] [Google Scholar]
- 15.Di Marcotullio L, Ferretti E, De Smaele E, Argenti B, Mincione C, Zazzeroni F, Gallo R, Masuelli L, Napolitano M, Maroder M, et al. Proc Natl Acad Sci USA. 2004;101:10833–10838. doi: 10.1073/pnas.0400690101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eggenschwiler JT, Bulgakov OV, Qin J, Li T, Anderson KV. Dev Biol. 2006;290:1–12. doi: 10.1016/j.ydbio.2005.09.022. [DOI] [PubMed] [Google Scholar]
- 17.Haycraft CJ, Banizs B, Aydin-Son Y, Zhang Q, Michaud EJ, Yoder BK. PLoS Genet. 2005;1:e53. doi: 10.1371/journal.pgen.0010053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reiter JF, Skarnes WC. Genes Dev. 2006;20:22–27. doi: 10.1101/gad.1363606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Callahan CA, Ofstad T, Horng L, Wang JK, Zhen HH, Coulombe PA, Oro AE. Genes Dev. 2004;18:2724–2729. doi: 10.1101/gad.1221804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Taylor MD, Liu L, Raffel C, Hui CC, Mainprize TG, Zhang X, Agatep R, Chiappa S, Gao L, Lowrance A, et al. Nat Genet. 2002;31:306–310. doi: 10.1038/ng916. [DOI] [PubMed] [Google Scholar]
- 21.Thompson MC, Fuller C, Hogg TL, Dalton J, Finkelstein D, Lau CC, Chintagumpala M, Adesina A, Ashley DM, Kellie SJ, et al. J Clin Oncol. 2006;24:1924–1931. doi: 10.1200/JCO.2005.04.4974. [DOI] [PubMed] [Google Scholar]
- 22.Sheng T, Li C, Zhang X, Chi S, He N, Chen K, McCormick F, Gatalica Z, Xie J. Mol Cancer. 2004;3:29. doi: 10.1186/1476-4598-3-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chi S, Huang S, Li C, Zhang X, He N, Bhutani MS, Jones D, Castro CY, Logrono R, Haque A, et al. Cancer Lett. 2006;244:53–60. doi: 10.1016/j.canlet.2005.11.036. [DOI] [PubMed] [Google Scholar]
- 24.Kinzler KW, Bigner SH, Bigner DD, Trent JM, Law ML, O'Brien SJ, Wong AJ, Vogelstein B. Science. 1987;236:70–73. doi: 10.1126/science.3563490. [DOI] [PubMed] [Google Scholar]
- 25.Nessling M, Richter K, Schwaenen C, Roerig P, Wrobel G, Wessendorf S, Fritz B, Bentz M, Sinn HP, Radlwimmer B, et al. Cancer Res. 2005;65:439–447. [PubMed] [Google Scholar]
- 26.Sasai K, Romer JT, Lee Y, Finkelstein D, Fuller C, McKinnon PJ, Curran T. Cancer Res. 2006;66:4215–4222. doi: 10.1158/0008-5472.CAN-05-4505. [DOI] [PubMed] [Google Scholar]
- 27.Dahlen A, Fletcher CD, Mertens F, Fletcher JA, Perez-Atayde AR, Hicks MJ, Debiec-Rychter M, Sciot R, Wejde J, Wedin R, et al. Am J Pathol. 2004;164:1645–1653. doi: 10.1016/s0002-9440(10)63723-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bhatia N, Thiyagarajan S, Elcheva I, Saleem M, Dlugosz A, Mukhtar H, Spiegelman VS. J Biol Chem. 2006;281:19320–19326. doi: 10.1074/jbc.M513203200. [DOI] [PubMed] [Google Scholar]
- 29.Taipale J, Chen JK, Cooper MK, Wang B, Mann RK, Milenkovic L, Scott MP, Beachy PA. Nature. 2000;406:1005–1009. doi: 10.1038/35023008. [DOI] [PubMed] [Google Scholar]
- 30.Regl G, Neill GW, Eichberger T, Kasper M, Ikram MS, Koller J, Hintner H, Quinn AG, Frischauf AM, Aberger F. Oncogene. 2002;21:5529–5539. doi: 10.1038/sj.onc.1205748. [DOI] [PubMed] [Google Scholar]
- 31.Roessler E, Ermilov AN, Grange DK, Wang A, Grachtchouk M, Dlugosz AA, Muenke M. Hum Mol Genet. 2005;14:2181–2188. doi: 10.1093/hmg/ddi222. [DOI] [PubMed] [Google Scholar]
- 32.Chen JK, Taipale J, Young KE, Maiti T, Beachy PA. Proc Natl Acad Sci USA. 2002;99:14071–14076. doi: 10.1073/pnas.182542899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lipinski RJ, Gipp JJ, Zhang J, Doles JD, Bushman W. Exp Cell Res. 2006;312:1925–1938. doi: 10.1016/j.yexcr.2006.02.019. [DOI] [PubMed] [Google Scholar]
- 34.Ma X, Sheng T, Zhang Y, Zhang X, He J, Huang S, Chen K, Sultz J, Adegboyega PA, Zhang H, et al. Int J Cancer. 2006;118:139–148. doi: 10.1002/ijc.21295. [DOI] [PubMed] [Google Scholar]
- 35.Mao J, Maye P, Kogerman P, Tejedor FJ, Toftgard R, Xie W, Wu G, Wu D. J Biol Chem. 2002;277:35156–35161. doi: 10.1074/jbc.M206743200. [DOI] [PubMed] [Google Scholar]
- 36.Fan L, Pepicelli CV, Dibble CC, Catbagan W, Zarycki JL, Laciak R, Gipp J, Shaw A, Lamm ML, Munoz A, et al. Endocrinology. 2004;145:3961–3970. doi: 10.1210/en.2004-0079. [DOI] [PubMed] [Google Scholar]
- 37.Neidle S, Thurston DE. Nat Rev Cancer. 2005;5:285–296. doi: 10.1038/nrc1587. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.