Abstract
The structural maintenance of chromosome (SMC) proteins are required for a number of essential nuclear processes, including those of chromosome condensation, chromatid cohesion, and DNA repair. Eukaryotic SMC proteins form heterodimers capable of binding DNA and possess a DNA-stimulated ATPase activity. They have a characteristic structure of terminal globular domains with two internal arms that are predicted to form a coiled-coil structure interspaced with a globular “hinge” domain. We report here that the predicted coiled-coil arms are disrupted at conserved sites in SMC proteins. These disruptions, which vary in length and sequence identity, abolish the otherwise symmetrical secondary structure of antiparallel SMC heterodimers and provide the first evidence for a possible functional orientation of eukaryotic SMC complexes. The retention of these breaks between evolutionarily distant, yet related, SMC members indicates that they may have a fundamental role in SMC heterodimer function.
The class of proteins collectively known as structural maintenance of chromosomes (SMCs) have received considerable attention of late because of their involvement in functions critical to genome stability and maintenance—specifically chromosome condensation, chromatid cohesion, and DNA repair (for review, see Cobbe and Heck 2000).
Three distinct SMC-containing complexes have been identified in eukaryotes, which participate in different aspects of chromosome dynamics. The complexes contain specialized SMC heterodimer pairs and other complex-specific accessory proteins, which are thought to modulate SMC protein function. The SMC1-SMC3 and SMC2-SMC4 heterodimers are core components of the cohesin (Michaelis et al. 1997; Losada et al. 1998) and condensin (Hirano et al. 1997) complexes, respectively. The cohesin complex is required for the formation and maintenance of sister chromatid cohesion (Guacci et al. 1997; Michaelis et al. 1997; Uhlmann and Nasmyth 1998). The condensin complex is responsible for condensing the replicated genome (Saka et al. 1994; Hirano et al. 1997). The most recently recognized SMC heterodimer pair, denoted as Rad18-Spr18 in fission yeast and as SMC5-SMC6 in mammals, participates in DNA replication and repair (Lehmann et al. 1995; Verkade et al. 1999).
Phylogenetic analysis of co-aligned SMC sequences shows closest evolutionary relationships between the paralogous SMC1 and SMC4 subfamilies and, similarly, the SMC2 and SMC3 subfamilies (Cobbe and Heck 2000; Jones and Sgouros 2001). Primary sequence comparisons indicate that the Rad18-Spr18 heterodimer evolved from an SMC branch separate from that of a progenitor cohesin/condensin complex, which also includes the prokaryotic MukB and BsSMC proteins (Jones and Sgouros 2001). Archael and bacterial genomes encode solitary SMC proteins that presumably homodimerize (Hirano and Hirano 1998; Melby at al. 1998). Bacterial SMC mutants show decondensed chromosomes and impaired viability through chromosome loss during division (Niki et al. 1991, 1992; Yamanaka et al. 1994; Britton et al. 1998). This is consistent with SMC duplication and diversification from the ancestral type occurring early in the eukaryotic lineage, permitting specialization of the eukaryotic heterodimers.
SMC proteins are clearly identifiable by both sequence homology and their conserved secondary structure profiles. All SMCs possess a distinctive predicted secondary structure, with terminal globular domains linked by two coiled-coil regions of ∼300 amino acids interspaced with a globular “hinge” domain (Strunnikov et al. 1993). Electron microscopic (EM) studies of both prokaryotic and eukaryotic SMC proteins (Melby et al 1998, Hirano et al 2001; Anderson et al. 2002) are consistent with a dimeric antiparallel “head-to-tail” configuration, with an extended interaction surface between the coiled-coil domains of the two proteins. This antiparallel orientation, which is strongly supported by X-ray crystallographic studies (Löwe et al. 2001), readily explains how the N-terminal Walker A and C-terminal Walker B ATP hydrolysis motifs (Saitoh et al. 1994) interact to generate full ATPase activity. The elongated coiled-coil arms and flexible central hinge indicate a pivotal, scissors-type motion between the two termini (Melby et al. 1998; Hirano et al. 2001). This, in combination with the observation that the N- and C-terminal globular domains possess ATP- and DNA-binding activities (Akhmedov et al. 1998a), indicates that the SMC heterodimers play a mechanical role in chromosome cohesion, condensation, and repair (Strunnikov et al. 1993).
With the exception of the characteristic sequence differences within the conserved globular domains that classify the SMC subfamilies, there have been no reported structural characteristics distinguishing between the six eukaryotic SMC subfamilies. Thus, it remains unclear how the specialist SMC protein pairs and their associated accessory proteins interact to accomplish such disparate cellular roles.
In the present study, we describe an analysis of secondary structure predictions of eukaryotic SMC proteins. Our studies have identified disruptions in the predicted coiled-coil regions at conserved sites in the sequences of eukaryotic cohesion and condensin SMC members that display evolutionary, but not functional, conservation, namely, SMC1/SMC4 and SMC2/SMC3. Similar coil-breaking regions were also observed in the evolutionary divergent Rad18 (SMC6) and Spr18 (SMC5) proteins, indicating a new degree of structural relatedness between all eukaryotic SMC heterodimeric complexes and leading us to propose a revised eukaryotic SMC lineage founded on predicted structure rather than on primary sequence. The disruptions of the cohesin and condensin SMCs possess a consensus sequence that in most cases is notably conserved within the orthologs, with an increased degree of local homology amid the less well conserved flanking coiled-coil sequence. It is suggested that the disruptions in the predicted coiled-coil secondary structure may result in a weakened or broken α-helix, an imperfection that may be integral to SMC dimer function, as similar disruptions are observed in eubacterial SMC sequences. Furthermore, these disruptions abolish the symmetrical structure of SMCs, providing the first evidence that there may be a functional orientation to the eukaryotic SMC complexes.
RESULTS
Cohesins SMC1 and SMC3 Have Conserved Disruptions to Their Predicted Coiled-Coil Domains
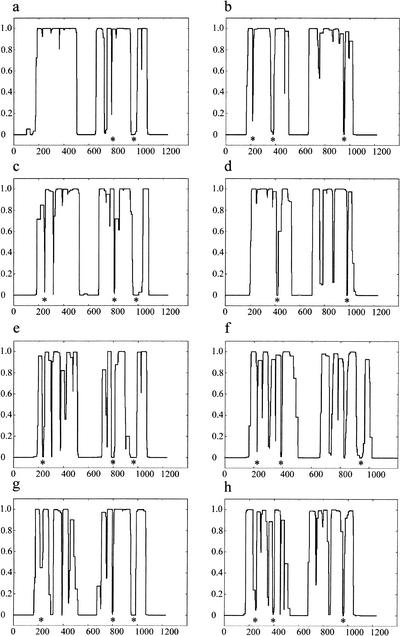
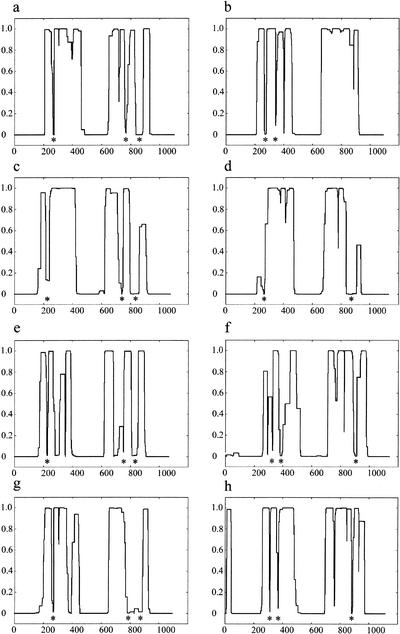
In the course of assessing the secondary structures of the mammalian cohesins, it was noticed by us and others (Stursberg et al. 1999) that the human SMC1 has a prominent disruption of ∼50 residues toward the end of its second (most C-terminal) coiled-coil domain (amino acids 935–984; Fig. 1a). In this region, the prediction of a coiled-coil secondary structure by the COILS program (Lupas et al. 1991) drops from near certainty to zero, regardless of whether scanning windows of 14, 21, or 28 amino acids (representing two, three, or four heptad turns of a coiled-coil helix) were used. Examination of SMC1 orthologs in Drosophila, Schizosaccharomyces pombe, and Saccharomyces cerevisiae revealed a break of similar size and position (Fig. 1c,e,g). A second, shorter break of ∼14 amino acids, located near the beginning of the C-terminal coiled-coil domain, was also evident in the human SMC1 sequence (at residue 779) and at analogous sites in other SMC1 orthologs. In addition, although not prominent in the 28-amino-acid window output for the human SMC1 sequence, a third conserved break was identified within 50 residues of the beginning of the first (more N-terminal) coiled-coil domain in the remaining SMC1 orthologs.
Figure 1.
COILS prediction of SMC1 and SMC3 orthologs. The COILS output for Homo sapiens SMC1 (a), H. sapiens SMC3 (b) , Drosophila melanogaster SMC1 (c), D. melanogaster SMC3 (d), fission yeast (Schizosaccharomyces pombe) SMC1 (e), fission yeast SMC3 (f), budding yeast (Saccharomyces cerevisiae) SMC1 (g), and budding yeast SMC3 (h) . The vertical axis denotes the probability of the amino acid sequence adopting a coiled-coil structure based on a scanning window of 28 residues. Although the original COILS output included windows of 14 and 21 residues, these have not been displayed for clarity. Conserved disruptions in the coiled-coil structure prediction to the cohesin SMC sequences are indicated beneath the COILS profile with an asterisk.
The heterodimeric partner of human SMC1, human SMC3, displayed disruptions within both the first (amino acids 213– 228 and 376–394) and second (amino acids 947–961) coiled-coil domains (Fig. 1b). These, too, are positionally conserved in Drosophila and yeast SMC3 orthologs (Fig. 1d,f,h). For clarity, we will henceforth denote these disruptions as SMC1 loop I (amino acid 222; 13 residues), SMC1 loop II (amino acid 779; 14 residues), SMC1 loop III (amino acid 935; 50 residues), SMC3 loop I (amino acid 213; 16 residues), SMC3 loop II (amino acid 376; 20 residues), and SMC3 loop III (amino acid 947; 15 residues).
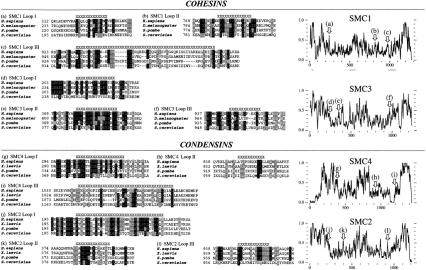
Alignment of the regions disrupting the predicted coiled-coil structures of the cohesin SMCs revealed conserved motifs (Fig. 2a–f) that are notable for their high local homology amid less conserved flanking coiled-coil sequences, as illustrated in the alignment of human and S. pombe SMC3 N-terminal coiled-coil (Fig. 3). In the shorter coil-breaking motifs (SMC1 loop I, SMC1 loop II, SMC3 loops I through III), residues that might be expected to exert important structural determinants owing to their unique properties influencing helical turn (G, P) or having a rigid, planar, side-chain (F, Y, and W) were conserved across all phyla in almost every instance. The larger disruption of ∼50 amino acids in SMC1 loop III appears less well conserved, in both length and composition, yet its primary sequence is nonetheless preserved across significant evolutionary distances. Several other coiled-coil disruptions predicted in the SMC1 and SMC3 proteins were also examined; however, they did not appear to be as conserved in location between the orthologs or as conserved in primary sequence as those addressed above.
Figure 2.
Alignments of the disruptions in the predicted coiled-coil arms of cohesin and condensin SMC proteins. Disruptions to the first coiled-coil (a) and second coiled-coil (b, c) domains of SMC1, and first coiled-coil (d, e) and second coiled-coil (f) domains of SMC3 orthologs. Disruptions to the first coiled-coil (g) and second coiled-coil (h, i) domains of SMC4, and first coiled-coil (j, k) and second coiled-coil (l) domains of SMC2 orthologs. Regions with poor coiled-coil structure prediction are indicated by X's above the alignment. Right panels indicate the location of the sequence alignments presented in a–l on amino acid identity plots (window size, 8) generated from global co-alignment of the full-length SMC orthologous sequences from which alignments a–l are derived.
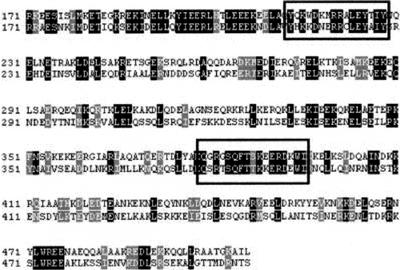
Figure 3.
Disruptions to the coiled-coil arms display notable sequence conservation amid poorly conserved flanking coiled-coil regions. An alignment of the first coiled-coil arms of the human (top) and fission yeast (Schizosaccharomyces pombe; bottom) SMC3 orthologs. The disruptions (SMC3 loop I and SMC3 loop II) to the predicted coiled-coil domain are boxed. These disruptions are present within regions of high local homology relative to the remainder of the sequence.
Condensins SMC2 and SMC4 Have Analogous Disruptions to Their Coiled-Coil Domains That Display Positional, but Not Sequence, Conservation to Their Cohesin SMC Counterparts
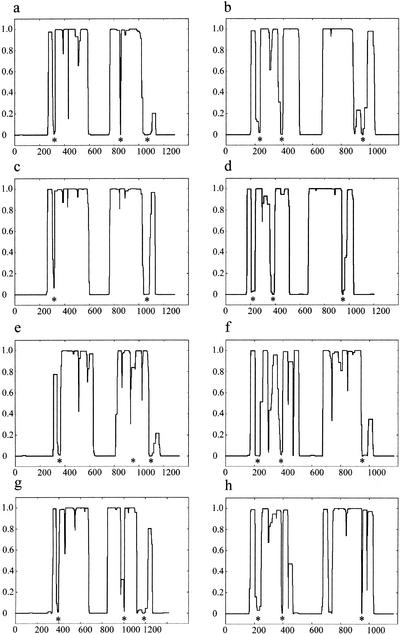
Examination of the condensin SMC (SMC2 and SMC4) sequences using an identical approach to that above identified a disruption in the second (C-terminal) predicted coiled-coil human SMC4 of a similar size (∼40 residues) and position to that of SMC1 loop III. This disruption was conserved in SMC4 orthologs from a diverse range of phyla (SMC4 loop III, amino acids 1041–1079; Fig. 4a,c,e,g). Likewise, two smaller disruptions, corresponding in position to SMC1 loop I and SMC1 loop II, were noted at amino acids 306–325 (SMC4 loop I) and 848–858 (SMC4 loop II), respectively, in the human SMC4 sequence. These disruptions are also not restricted to human SMC4 but are evident in the Xenopus, S. pombe, and S. cerevisiae orthologs, although the second disruption appeared to be absent from the COILS prediction of the Xenopus SMC4 protein (Fig. 4c).
Figure 4.
COILS prediction of SMC2 and SMC4 orthologs. The COILS output for Homo sapiens SMC4 (a), H. sapiens SMC2 (b), Xenopus laevi SMC4 (c), X. laevi SMC2 (d), fission yeast (Schizosaccharomyces pombe) SMC4 (e), fission yeast SMC2 (f), budding yeast (Saccharomyces cerevisiae) SMC4 (g), and budding yeast SMC2 (h). The vertical axis denotes the probability of the amino acid sequence adopting a coiled-coil structure based on a scanning window of 28 residues. Conserved disruptions in the coiled-coil structure prediction to the condensin SMC sequences are indicated beneath the COILS profile with an asterisk.
Analysis of SMC2 orthologs also displayed breaks in both coiled-coil domains reminiscent of SMC3 (SMC2 loop I, amino acids 205–241; SMC2 loop II, amino acids 384–397; SMC2 loop III, amino acids 948–960; Fig. 4b,d,f,h). When aligned, these breaks were also found to be conserved in sequence (Fig. 2g–l), although, significantly, the consensus differs completely from those of the cohesins.
The structural relationship of SMC1 to SMC4 and of SMC2 to SMC3 is supported by the SMC phylogenetic tree compiled from their primary sequences (Cobbe and Heck 2000). Both methods of analysis are consistent with the notion that cohesin and condensin subfamily members evolved from a progenitor SMC heterodimer, which was later duplicated and diverged to create the individual subfamilies. That these structural features should remain conserved through the evolution of SMC complexes specific for chromatid cohesion and chromosomal condensation strongly indicates that they play a common, critical role in SMC function.
The Coiled-Coil Disruptions of the SMC5 and SMC6 Heterodimer Appear Structurally Related to the Cohesin and Condensin SMCs
The Rad18-Spr18 SMC heterodimer, involved in DNA replication and repair in S. pombe, has metazoan orthologs called SMC6 and SMC5 (Taylor et al. 2001), respectively. The phylogeny of the entire SMC family assembled from the primary sequences of eukaryotic and bacterial SMCs indicated that the SMC5 and SMC6 subfamilies are among the most removed of the eukaryotic SMC members (Cobbe and Heck 2000; Jones and Sgouros 2001; Taylor et al. 2001). By this method, the radiation of SMC5 and SMC6 from the evolutionary branch, which leads to the heterodimeric cohesin and condensin complexes, appears to have been much earlier than their divergence from each other. The inference of the primary sequence analyses conducted was that by virtue of their closer relatedness to the eubacterial SMCs and to each other, the origin of the SMC5-SMC6 heterodimer was independent from that of the progenitor cohesin/condensin heterodimer. However, contrary to expectations of unique disruptions in their coiled-coil arms, it was observed that SMC5 and SMC6 orthologs possessed disruptions characteristic of the SMC1/SMC4 and SMC2/SMC3 subfamilies, respectively (Fig. 5).
Figure 5.
COILS prediction of SMC5 and SMC6 orthologs. The COILS output for Homo sapiens SMC5 (a), H. sapiens SMC6 (b), Drosophila melanogaster SMC5 (c), C. elegans SMC6 (d), fission yeast (Schizosaccharomyces pombe) Spr18 (e), fission yeast Rad18 (f), budding yeast (Saccharomyces cerevisiae) Spr18 ortholog (g), and the budding yeast Rhc18 (h). The vertical axis denotes the probability of the amino acid sequence adopting a coiled-coil structure based on a scanning window of 28 residues. Conserved disruptions in the coiled-coil structure prediction to the SMC5/SMC6 SMC sequences are indicated beneath the COILS profile with an asterisk.
S. pombe Rad18 (Fig. 5f), the seminal SMC6 member, has three disruptions, which were conserved among orthologous sequences. Loop I and loop II reside in the first (N-terminal) coiled-coil arm at positions 286–306 and 369–403 amino acids, respectively. Loop III is in the second (C-terminal) coiled-coil arm at positions 886–912 amino acids. Equivalent disruptions are observed in the coiled-coil predictions of S. cerevisiae Rhc18 (Fig. 5h) and of C. elegans (Fig. 5d) and human SMC6 (Fig. 5b), although loop II appears to be absent from C. elegans SMC6, and loop III, from the human homolog.
S. pombe Spr18, the heterodimeric partner of Rad18, appears to have a reversal of the distribution of disruptions to the coiled-coil arms. Loop I appears in the N-terminal arm between amino acids 220–233, and loop II and loop III can be found in the C-terminal arm at amino acids 731–751 and 804–854, respectively (Fig. 5e). Disruptions corresponding in size and position to the Spr18 loops I, II, and III appear in the human, C. elegans, and S. cerevisiae orthologs (Fig. 5a,c,g, respectively). Significantly, unlike the other SMCs analyzed, the Rad18/SMC6 and Spr18/SMC5 disruptions were not conserved in sequence between orthologs (data not shown).
Disruptions to the Coiled-Coil Arms of Heterodimeric SMC Complexes Are Mirrored in an Antiparallel Orientation
If the positions of the coiled-coil disruptions are plotted onto an antiparallel alignment of the secondary structures of each of the eukaryotic cohesin, condensin, and Rad18/Spr18 orthologs, a striking symmetry is observed (Fig. 6a). The juxtaposition of each disruption with those identified in their corresponding heterodimeric partner is very close. For example, in the human cohesin proteins, the distance from the start of the first coiled-coil arm to SMC1 loop I is ∼45 residues, which corresponds to the distance from the end of SMC3 loop III to the end of the second arm (∼50 residues). Likewise, the interstitial regions between the loops are of a similar length. For example, the region between human SMC1 loop II and loop III is ∼145 residues in length, and the corresponding interval between human SMC3 loop I and loop II is ∼148 residues. The absolute correspondence in the alignment of these coil-breaking motifs when aligned in an antiparallel orientation cannot be readily attributed to chance alone. These data are supported by recent EM studies of human and Xenopus cohesin and condensin complexes, which are consistent with these SMCs adopting an antiparallel head-to-tail dimeric configuration (Anderson et al. 2002). This study also revealed distinct bends in the SMC coiled-coil arms in some forms of these complexes. The most notable of these bends occurs approximately one third of the distance between the hinge and globular domains of both complexes, which strikingly matches the position of the postulated loop II in these proteins.
Figure 6.
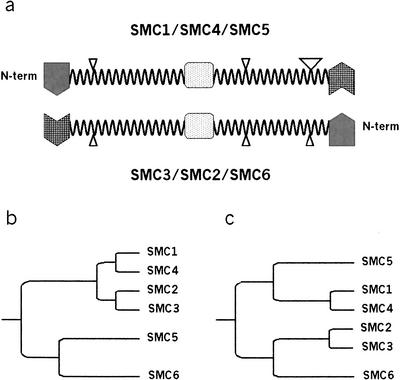
The conserved disruptions to the predicted coiled-coil arm domains of eukaryotic SMC heterodimers abolish SMC structural symmetry and indicate a common ancestral complex for all eukaryotic SMC heterodimers. (a) Schematic representation of the domain structure of cohesin, condensin, and SMC5-SMC6 SMC heterodimer. The globular N-terminal (shaded) and C-terminal (hatched) domains are separated by two helical coiled-coil arms and a central hinge domain (dots). Coiled-coil disruptions to the arm domains (loops) are indicated by triangles with size that is proportional to the length of the loop. Because of the conserved domain architecture of SMC proteins and the antiparallel association of the heterodimers, SMC complexes lack secondary structure orientation and can be freely rotated around the hinge domain and about a lateral axis along the arm lengths. However, with an appreciation of the observed disruptions to the coiled-coil domains as structural features of eukaryotic SMCs, the heterodimers gain a structural directionality that may translate into a functional orientation of the complexes. Not to scale. (b) A dendrogram of eukaryotic SMC evolution derived from the primary sequences of the cohesin, condensin, and SMC5-SMC6 heterodimers as presented by Cobbe and Heck (2000) and Jones and Sgouros (2001). It differs from a phylogeny that incorporates the structural similarities between all eukaryotic heterodimeric SMC complexes (c), which have a conserved distribution of disruptions to their coiled-coil domains. Arm distances are only illustrative of evolutionary distances of SMC subfamilies.
DISCUSSION
Comparative Analysis of Coiled-Coil Predictions Reveals a New Structural Feature of SMC Proteins
Here we report the identification of a new structural feature of SMC proteins—that of disruptions to the predicted coiled-coil domains of eukaryotic heterodimeric SMC members. These disruptions have been retained during the evolutionary divergence of an antecedent SMC dimer into the separate functions of chromosome condensation and cohesion and of DNA replication and repair, indicating that they play a role fundamental to all these activities. Additionally, orthologous cohesin and condensin SMC members have disruptions that form a consensus sequence that is especially notable within the context of the poorly conserved primary sequence of the surrounding coiled-coil. This indicates that the structural characteristics of these loops are essential for SMC function and contrast with the more substitution-tolerant coiled-coil regions within which they are embedded.
The COILS program uses a database of parallel two-stranded coiled-coils to calculate the likelihood of an input sequence adopting a similar structure (Lupas et al. 1991). The extended, antiparallel orientation of MukB coiled-coil arms and, by analogy, other SMC complexes is a unique feature adopted by the SMC family (Melby et al. 1998). Clearly, the structural requirements for the antiparallel conformation must be suitably similar to that of a parallel helix for the high predictive indices of most regions along SMC “arms.” When the COILS prediction lapses in the middle of an SMC arm, it could be owing to a region of sequence with poor facility of the COILS program to predict because of the antiparallel nature of the structure. We believe that this is highly unlikely, because we observe high predictive scores with SMC sequences from a diverse range of phyla that display low primary sequence identity. The consistently low coiled-coil probability calculated within these postulated loop regions argues strongly that these predicted disruptions do indeed have poor potential for a coiled-coil structure. Additionally, there is strong evidence that the predicted disruptions to the second arm of MukB did not contribute to the overall length of the arm visible by EM (Melby et al. 1998).
Predicted Secondary Structure Analysis Reveals Basal SMC Protein Phylogeny
The SMC5 and SMC6 orthologs appeared identical to the cohesin or condensin SMCs with respect to the distribution of their coiled-coil disruptions. This is incongruent with the phylogeny of the broader SMC family, inferred from the primary sequences of both eukaryotic and bacterial SMC members, in which the Rad18/SMC6 and Spr18/SMC5 subfamilies are representatives of one of the three major branches of SMC proteins (Cobbe and Heck 2000; Jones and Sgouros 2001). The other two branches later diverge into the individual subunits of the cohesin and condensin SMC members. The inference of these primary sequence analyses is that by virtue of their closer relatedness to the eubacterial SMCs and to each other, the origin of the SMC5-SMC6 heterodimer was independent from that of the progenitor cohesin/condensin heterodimer (Fig. 6b). Our observations indicate a further structural relatedness between the eukaryotic cohesin and condensin complexes than might be supposed from an analysis of the primary sequences. This leads us to propose a new phylogeny based on the secondary structure of SMCs. Although it is not possible to estimate the evolutionary divergence from the proposed structural similarities, Figure 6c illustrates the basic proposition in which all eukaryotic heterodimeric SMC complexes are composed of representatives from two broader families: (1) members of the SMC1, SMC4, and SMC6 subfamilies and (2) members of SMC2, SMC3, and SMC5. Our proposed lineage postulates that the last ancestral SMC protein in common to all of the eukaryotic heterodimeric SMC complexes was a heterodimer containing symmetrical disruptions in the coiled-coil domains instead of a homodimer as indicated by primary sequence alignments.
Disruptions to the Coiled-Coil Arms May Form Nonhelical Loops and Abolish the Structural Symmetry of SMC Heterodimers
Until now, the structural homogeneity of SMC proteins and the antiparallel association of the heterodimeric complex have not provided any indication of a functional orientation between the two ends of the complex. Furthermore, the lengths of the predicted coiled-coil arms display notable, and possibly significant, symmetry about the hinge domain. This generalization holds true for the bacterial homodimers and the eukaryotic SMCs (Melby et al. 1998). However, with a classification based on the helix disruptions, it becomes apparent that the heterodimers may no longer be freely rotated along either longitudinal (rotation around the hinge domain) or latitudinal axes (rotation about the arm interface) without changing the structural orientation of the complex (Fig. 6a). In addition, the antiparallel alignment displays a reversal in the positioning and spacing of each of the loops between the two component SMCs, resulting in a heterodimer with “mirrored” disruptions at facing sides of the paired antiparallel coiled-coil arms. This nonrandom distribution supports the hypothesis that eukaryotic heterodimeric SMC complexes, as with the bacterial homodimeric SMCs, adopt an antiparallel configuration.
The E. coli MukB and B. subtilis BsSMC have both been purified and visualized using EM (Melby et al. 1998; Hirano et al. 2001). Both proteins adopted a symmetrical structure with three globular domains separated by long rods of equal length, that is, exactly the appearance expected from the coiled-coil profile widely judged as characteristic of SMC proteins. However, closer examination of the COILS output for both MukB and BsSMC reveals significant drops in coiled-coil probability for lengths of up to ∼90 residues and ∼40 residues, respectively (data not shown). This was also noted by Melby et al. (1998). These investigators concluded that the disruptions to the predicted second coiled-coil arm were not contributing to the observed coiled-coil length of the arm. This was also the conclusion of another EM study of an SMC member written by Anderson et al. (2001), who analyzed the structure of the S. cerevisiae Rad50-Mre11 complex. Although this latter study presents compelling evidence to indicate that Rad50 does not form antiparallel heterodimers as predicted for the other eukaryotic SMCs, only by omission of the disruptions to the predicted coiled-coil arms of Rad50 could the investigators reconcile the disparity between the observed and predicted arm lengths.
The irregular distribution of the disruptions to the predicted coiled-coil arms of SMC1, SMC4, and SMC5 (Fig. 6a), with both loop II and loop III located in the latter half of the second arm (and, conversely, loops I and II in the first arm of SMC2, SMC3, and SMC6), may imply that one extremity of the heterodimer possesses added flexibility to an otherwise presupposed rigid arm domain. This flexibility of the coiled-coil arms may be essential for basic SMC function. EM studies of immobilized BsSMC and MukB homodimers (Melby et al. 1998; Hirano et al. 2001) and of human and Xenopus cohesin and condensin complexes (Anderson et al. 2002) show pronounced kinks in the arms of some forms of these complexes. The direct observation of kinks in the coiled-coil arms of these SMC proteins, the most notable in the cohesin/condensin complexes closely correlating with the location of loop II, strongly supports the contention that these coil-breaking motifs contribute to SMC arm flexibility in vivo.
What function might the coil-breaking disruptions have, and what form might they take? A disruption to the prediction of a coiled-coil domain might not necessarily result in an extended discontinuity to the helical structure of the SMC dimer in all cases. Such an insertion could form a loop from the surface of the helix, as indicated by the EM studies cited above, with the regular periodicity of the partner coiled-coil continuing. However, there would be some torsional twist created that might act to destabilize the coiled-coil in the region of the loop, especially as the disruptions appear to coincide at approximately equivalent positions in the arms of antiparallel heterodimers. Insertions of three or four residues have been observed in coiled-coils and have the effect of inducing local underwinding or overwinding of the helix (Brown et al. 1996; Burkhard et al. 2001). These relatively short disruptions have been “corrected,” in the case of the intermediate filament protein vimentin, by inserting residues to complete the heptad turn or by substituting residues to create a continuous heptad repeat between two coiled-coil domains (Herrmann et al. 1999). However, in both instances, changing the nature of the disruptions to the coiled-coils also impaired the ability of the protein to correctly form higher order filaments. The larger loops in all SMC proteins examined may therefore serve to modulate the pairing and rigidity of the arms, possibly transmitting a signal to or from accessory proteins with which they interact. This might also explain our observation that although the loops remain positionally conserved between the functionally divergent SMC1/SMC4 and SMC2/SMC3 proteins, the consensus sequence for each loop is different for each subfamily. The architecture of the cohesin and condensin complexes is built around a SMC heterodimer; however, the associated proteins in both complexes are unrelated (Losada et al. 1998), and although it remains uncharacterized, the Rad18-Spr18 heterodimer is also an element of a larger complex (Fousteri and Lehmann 2000). If the loops were to have a role in binding other proteins, then as the composition of the complex changed, so too would the sequence of the interacting loops. Alternatively, a study by Akhmedov et al. (1998b) showed that the coiled-coil domain of SMC1/3 dimer was capable of binding DNA directly, and consequently, disruptions to the coiled-coil may be crucial in modulating such interactions.
Current attempts at furthering the understanding of SMC function have focused on dissecting those domains already identified, namely, to make mutants of the active terminal ATPase (Verkade et al. 1999; Hirano et al. 2001) and of the central hinge domain (Hirano et al. 2001). Until now, the interstitial coiled-coil arms have been treated as an ancillary feature of the structure. However, a thorough understanding of the mechanism of SMC activity will require study of this newly recognized feature of the SMC proteins, that of a conserved disruption to the predicted coiled-coils. It seems likely that these disruptions will prove important to SMC function, as they are a conserved feature of all classes of eukaryotic SMC heterodimers, as well as appearing in the eubacterial SMC proteins. Hopefully, these studies will provide some insight into the how and why of the important roles of SMC proteins in chromosomal dynamics.
METHODS
Coiled-Coil Structure Prediction
Coiled-coil secondary structure was predicted using the COILS program (Lupas et al. 1991) at the Swiss node of the EMBnetserver using Web access to the site http://www.ch.embnet.org/software/COILS_form.html. All predictions were performed using the MTIDK matrix, which was derived from the sequences of myosins, paramyosins, tropomyosins, intermediate filaments type I–IV, desmosomal proteins, and kinesins (Lupas et al. 1991).
SMC sequences were obtained from GenBank entries (protein accession codes) as follows: C. elegans from SMC5, AAK31464; and SMC6 (hypothetical protein F54D5.14), T20288; Drosophila melanogaster from SMC1, CAB76376; and SMC3, S70553; Homo sapiens from SMC1, NP 006297; HCAP-E (SMC2), AAC72360; SMC3/Bamacan, NP 005436; HCAP-C (SMC4), BAA73535; SMC5, CAC39247; and SMC6, CAC39248; Saccharomyces cerevisiae from SMC1, BAA09230; SMC2, NP_116687; SMC3, NP_012461; SMC4, NP_013187; Rhc18 (Rad18 homolog), Q12749; and ORF YOL034w (Spr18 homolog), CAA99034; and Schizosaccharomyces pombe from Smc1, T40059; cut14 (Smc2), S51623; Smc3, CAA15722; cut3 (Smc4), S51622; Rad18, P53692; and Spr18, O13710; and Xenopus laevis -XCAP-E (SMC2), B55094; and XCAP-C (SMC4), A55094.
WEB SITE REFERENCES
http://www.ch.embnet.org/software/COILS_form.html; Location of the COILS program (the EMBnetserver) used to predict Coiled-coil secondary structure.
Acknowledgments
We gratefully acknowledge Jamie Rossjohn for assistance with the manuscript preparation and Roland Kanaar for comments on the manuscript.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL m.mckay@pmci.unimelb.edu.au; FAX 61-3-9656-1411.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr107302.
REFERENCES
- Akhmedov AT, Frei C, Tsai-Pflugfelder M, Kemper B, Gasser SM, Jessberger R. Structural maintenance of chromosomes protein C-terminal domains bind preferentially to DNA with secondary structure. J Biol Chem. 1998a;273:24088–24094. doi: 10.1074/jbc.273.37.24088. [DOI] [PubMed] [Google Scholar]
- Akhmedov AT, Gross B, Jessberger R. Mammalian SMC3 C-terminal and coiled-coil protein domains specifically bind palindromic DNA, do not block DNA ends, and prevent DNA bending. J Biol Chem. 1998b;274:38216–38224. doi: 10.1074/jbc.274.53.38216. [DOI] [PubMed] [Google Scholar]
- Anderson DE, Trujillo KM, Sung P, Erickson HP. Structure of the Rad50 × Mre11 DNA repair complex from Saccharomyces cerevisiaeby electron microscopy. J Biol Chem. 2001;276:37027–37033. doi: 10.1074/jbc.M106179200. [DOI] [PubMed] [Google Scholar]
- Anderson DE, Losada A, Erickson HP, Hirano T. Codensin and cohesin display different arm conformations with characteristic hinge angles. J Cell Biol. 2002;156:419–424. doi: 10.1083/jcb.200111002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britton RA, Lin DC, Grossman AD. Characterization of a prokaryotic SMC protein involved in chromosome partitioning. Genes Dev. 1998;12:1254–1259. doi: 10.1101/gad.12.9.1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JH, Cohen C, Parry DA. Heptad breaks in α-helical coiled coils: Stutters and stammers. Proteins. 1996;26:134–145. doi: 10.1002/(SICI)1097-0134(199610)26:2<134::AID-PROT3>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- Burkhard P, Strelkov SV, Stetefeld J. Coiled coils: A highly versatile protein folding motif. Trends Cell Biol. 2001;11:82–88. doi: 10.1016/s0962-8924(00)01898-5. [DOI] [PubMed] [Google Scholar]
- Cobbe N, Heck MM. SMCs in the world of chromosome biology: From prokaryotes to higher eukaryotes. J Struct Biol. 2000;129:123–143. doi: 10.1006/jsbi.2000.4255. [DOI] [PubMed] [Google Scholar]
- Fousteri MI, Lehmann AR. A novel SMC protein complex in Schizosaccharomyces pombecontains the Rad18 DNA repair protein. EMBO J. 2000;19:1691–1702. doi: 10.1093/emboj/19.7.1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guacci V, Koshland D, Strunnikov A. A direct link between sister chromatid cohesion and chromosome condensation revealed through the analysis of MCD1 in S. cerevisiae. Cell. 1997;91:47–57. doi: 10.1016/s0092-8674(01)80008-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann H, Haner M, Brettel M, Ku NO, Aebi U. Characterization of distinct early assembly units of different intermediate filament proteins. J Mol Biol. 1999;286:1403–1420. doi: 10.1006/jmbi.1999.2528. [DOI] [PubMed] [Google Scholar]
- Hirano M, Hirano T. ATP-dependent aggregation of single-stranded DNA by a bacterial SMC homodimer. EMBO J. 1998;17:7139–7148. doi: 10.1093/emboj/17.23.7139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano M, Anderson DE, Erickson HP, Hirano T. Bimodal activation of SMC ATPase by intra- and inter-molecular interactions. EMBO J. 2001;20:3238–3250. doi: 10.1093/emboj/20.12.3238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano T, Kobayashi R, Hirano M. Condensins, chromosome condensation protein complexes containing XCAP- C, XCAP-E and a Xenopus homolog of the DrosophilaBarren protein. Cell. 1997;89:511–521. doi: 10.1016/s0092-8674(00)80233-0. [DOI] [PubMed] [Google Scholar]
- Jones S, Sgouros J. The cohesin complex: Sequence homologies, interaction networks and shared motifs. Genome Biol. 2001;2:RESEARCH0009. doi: 10.1186/gb-2001-2-3-research0009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann AR, Walicka M, Griffiths DJ, Murray JM, Watts FZ, McCready S, Carr AM. The rad18 gene of Schizosaccharomyces pombedefines a new subgroup of the SMC superfamily involved in DNA repair. Mol Cell Biol. 1995;15:7067–7080. doi: 10.1128/mcb.15.12.7067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losada A, Hirano M, Hirano T. Identification of XenopusSMC protein complexes required for sister chromatid cohesion. Genes Dev. 1998;12:1986–1997. doi: 10.1101/gad.12.13.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Löwe J, Cordell SC, van den Ent F. Crystal structure of the SMC head domain: An ABC ATPase with 900 residues antiparallel coiled-coil inserted. J Mol Biol. 2001;306:25–35. doi: 10.1006/jmbi.2000.4379. [DOI] [PubMed] [Google Scholar]
- Lupas A, Van Dyke M, Stock J. Predicting coiled coils from protein sequences. Science. 1991;252:1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- Melby TE, Ciampaglio CN, Briscoe G, Erickson HP. The symmetrical structure of structural maintenance of chromosomes (SMC) and MukB proteins: Long, antiparallel coiled coils, folded at a flexible hinge. J Cell Biol. 1998;142:1595–1604. doi: 10.1083/jcb.142.6.1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaelis C, Ciosk R, Nasmyth K. Cohesins: Chromosomal proteins that prevent premature separation of sister chromatids. Cell. 1997;91:35–45. doi: 10.1016/s0092-8674(01)80007-6. [DOI] [PubMed] [Google Scholar]
- Niki H, Jaffe A, Imamura R, Ogura T, Hiraga S. The new gene mukB codes for a 177 kd protein with coiled-coil domains involved in chromosome partitioning of E. coli. EMBO J. 1991;10:183–193. doi: 10.1002/j.1460-2075.1991.tb07935.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niki H, Imamura R, Kitaoka M, Yamanaka K, Ogura T, Hiraga S. E. coliMukB protein involved in chromosome partition forms a homodimer with a rod-and-hinge structure having DNA binding and ATP/GTP binding activities. EMBO J. 1992;11:5101–5109. doi: 10.1002/j.1460-2075.1992.tb05617.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitoh N, Goldberg IG, Wood ER, Earnshaw W. ScII: An abundant chromosome scaffold protein is a member of a family of putative ATPases with an unusual predicted tertiary structure. J Cell Biol. 1994;127:303–318. doi: 10.1083/jcb.127.2.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saka Y, Sutani T, Yamashita Y, Saitoh S, Takeuchi M, Nakaseko Y, Yanagida M. Fission yeast cut3 and cut14, members of a ubiquitous protein family, are required for chromosome condensation and segregation in mitosis. EMBO J. 1994;13:4938–4952. doi: 10.1002/j.1460-2075.1994.tb06821.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strunnikov AV, Larionov VL, Koshland D. SMC1: An essential yeast gene encoding a putative head-rod-tail protein is required for nuclear division and defines a new ubiquitous protein family. J Cell Biol. 1993;123:1635–1648. doi: 10.1083/jcb.123.6.1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stursberg S, Riwar B, Jessberger J. Cloning and characterization of mammalian SMC1 and SMC3 genes and proteins, components of the DNA recombination complexes RC-1. Gene. 1999;228:1–12. doi: 10.1016/s0378-1119(99)00021-9. [DOI] [PubMed] [Google Scholar]
- Taylor EM, Moghraby JS, Lees JH, Smit B, Moens PB, Lehmann AR. Characterization of a novel human SMC heterodimer homologous to the Schizosaccharomyces pombeRad18/Spr18 complex. Mol Biol Cell. 2001;12:1583–1594. doi: 10.1091/mbc.12.6.1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlmann F, Nasmyth K. Cohesion between sister chromatids must be established during DNA replication. Curr Biol. 1998;8:1095–1101. doi: 10.1016/s0960-9822(98)70463-4. [DOI] [PubMed] [Google Scholar]
- Verkade HM, Bugg SJ, Lindsay HD, Carr AM, O'Connell MJ. Rad18 is required for DNA repair and checkpoint responses in fission yeast. Mol Biol Cell. 1999;10:2905–2918. doi: 10.1091/mbc.10.9.2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanaka K, Mitani T, Feng J, Ogura T, Niki H, Hiraga S. Two mutant alleles of mukB, a gene essential for chromosome partition in Escherichia coli. FEMS Microbiol Lett. 1994;123:27–31. doi: 10.1111/j.1574-6968.1994.tb07196.x. [DOI] [PubMed] [Google Scholar]