Abstract
Suppression subtractive hybridization was used to clone genes associated with the activation of hepatic stellate cells and 13 genes were found to be dominantly expressed in activated stellate cells. Among them, one was identical to the 421-837th base pairs of cDNA sequence reported for rat prion-related protein (PrP). In cultured stellate cells, PrP mRNA expression increased in a time-dependent manner in parallel with smooth muscle (SM) α-actin mRNA expression. In situ hybridization demonstrated that PrP mRNA was localized in and around the fibrous septa of carbon tetrachloride (CCl4)-treated liver. Cellular PrP (PrPc) was produced by culture-activated stellate cells, and immunohistochemically detected in the fibrous septa of CCl4-damaged liver and sinusoidal linings of common bile duct-ligated liver, consistent with the localization of SM α-actin. Immunoelectron microscopy revealed that PrPc resided on the plasma membrane of stellate cells. These results indicate that PrP expression is closely related to stellate cell activation associated with fibrogenic stimuli.
Hepatic stellate cells constitute the sinusoidal walls as liver-specific pericytes and store vitamin A-associated lipid droplets. 1 When the liver suffers an injury, these cells undergo activation and proliferation in response to various growth factors and oxidative stress. 2-4 In the course of activation, stellate cells are dramatically altered in the metabolism of extracellular matrix materials, 5,6 smooth muscle (SM) α-actin, 7 and nerve-related materials such as glial fibrillary acidic protein (GFAP) 8,9 and neural cell adhesion molecule (N-CAM). 10,11 Although the proteins characteristic of activated stellate cells have been thus extensively studied, data on genes related to stellate cell activation remain very limited. In fact, a novel immediate early gene Zf-9 has been very recently cloned from rat stellate cells. 12 Stellate cell activation is an important event in the pathology of liver diseases because it is believed to play roles in the development of fibrosis. 2,5,6 The analysis of genes associated with stellate cell activation may thus provide a clue to the molecular mechanism and gene therapy of fibrotic liver diseases.
In the present study, we employed suppression subtractive hybridization to clone the genes which were dominantly expressed in activated phenotype of stellate cells and demonstrated for the first time that prion-related protein (PrP) gene expression and protein (cellular PrP, PrPc) production were induced in the course of stellate cell activation both in vitro and in vivo, comparing to the expression of SM α-actin, a commonly used marker for stellate cell activation.
Materials and Methods
Animals
Male Wistar rats and male Syrian hamsters were purchased from Japan SLC, Inc. (Shizuoka, Japan), housed under a specific pathogen-free condition at a constant temperature, and fed standard chow pellets and water ad libitum. All experiments were carried out according to the standard guidelines of Osaka City University for animal experimentation.
Isolation and Culture of Stellate Cells
Stellate cells were isolated from the liver of rats or hamsters as previously reported. 13 In brief, after the liver was perfusion-digested with pronase E (Merck, Darmstadt, Germany) and collagenase (Wako Pure Chemical, Osaka, Japan), it was taken out, minced, and incubated in a Krebs Ringer solution containing 0.05% pronase E, 0.05% collagenase, and 20 μg/ml DNase (Boehringer Mannheim, Mannheim, Germany) for 30 minutes at pH 7.3. After removing undigested materials, a stellate cell-enriched fraction was obtained by centrifugation of the mixture in an 8.2% Nycodenz (Nycomed Pharma AS, Oslo, Norway) at 1400 g and 4°C for 20 minutes. The cells were washed and suspended in RPMI 1640 supplemented with 10% fetal bovine serum (FBS, GIBCO BRL, Gaithersburg, MD) and antibiotics (10 5 units/L penicillin G and 100 mg/L streptomycin). The purity of stellate cells was always more than 95% as assessed by the presence of yellow-colored droplets. They were cultured on uncoated plastic dishes for 14 days after plating.
Suppression Subtractive Hybridization
Poly A+ RNA was extracted from 14-day-cultured or freshly isolated rat stellate cells using a Micro-Fast Track kit (Invitrogen, Carlsbad, CA). Each 2 μg of poly A+ RNA was used to make the tester and the driver cDNA. Suppression subtractive hybridization was performed using a PCR-select cDNA subtraction kit (Clontech, Palo Alto, CA) according to the manufacturer’s protocol. Briefly, the tester and the driver cDNA were digested with Rsal, and the tester cDNA was ligated to the adaptor DNA. After twice repeated hybridization with the tester and the driver cDNA, the resulting mixture was amplified by PCR using flanking and nested primers which anneal the adaptor DNA to produce subtracted PCR fragments. They were subcloned into pGEM-T vector (Promega, Madison, WI). Amplified cDNA fragments were sequenced using a Rhodamine Terminator Cycle Sequencing Kit (Perkin-Elmer, Foster City, CA) and ABI PRISM 310 automated fluorescent DNA sequencer (Perkin-Elmer). DNA database searches were performed using the FASTA program by directly accessing the database of DNA Data Bank of Japan.
Northern Blot Analysis
Total RNA was extracted from stellate cells using Isogene (Nippon Gene, Tokyo). Each 20 μg of the extracted RNA was denatured and then subjected to electrophoresis in 1% agarose gel containing 7.4% formaldehyde for 3 hours at 100 V. The electrophoresed RNA was transferred to a nylon membrane (Hybond-N, Amersham, Buckinghamshire, UK). After fixation, the membrane was incubated for 2 hours at 42°C in a prehybridization buffer containing 5× saline sodium citrate (SSC, 3 mol/L NaCl and 0.3 mol/L sodium citrate), 5× Denhardt’s solution, 50% formamide, 0.5% sodium dodecyl sulfate (SDS), and 20 μg/ml salmon sperm DNA. The membrane was then incubated in the same buffer supplemented with cDNA for PrP, SM α-actin, or β-actin which was labeled with [32P]dCTP by using a multiprime labeling system (Amersham). After overnight incubation at 42°C, the membrane was washed twice with 2× SSC containing 0.1% SDS and then with 0.1× SSC containing 0.1% SDS. Autoradiography was performed using a Fujix BAS 2000 bio-imaging analyzer (Fuji Photo-Film, Tokyo, Japan).
Western Blot Analysis
Hamster stellate cells cultured for indicated periods were homogenized in 50 mmol/L of ice-cold Tris buffer containing 1 mmol/L EDTA (pH 7.4). The homogenates (10 μg protein) were solubilized in SDS sample buffer, subjected to 12% SDS-polyacrylamide gel electrophoresis (PAGE) and then transferred onto Immobilon-P (Millipore, Bedford, MA). After washing, the membranes were treated with 1% BSA at 42°C overnight and incubated with either anti-SM α-actin monoclonal antibody (1:1000, Boehringer Mannheim) or anti-human PrP monoclonal antibody (1:1000, Chemicon International, Temecula, CA) at room temperature for 2 hours. After washing, the membranes were incubated with peroxidase-conjugated rabbit anti-mouse IgG (1:1000, Dako, Glostrup, Denmark) at room temperature for 1 hour. After washing, immunoreactive bands were visualized on x-ray film (Kodak XAR 5) using ECL Western blotting detection reagents (Amersham).
Induction of Hepatic Fibrosis
Hepatic fibrosis was induced in rats and hamsters either by subcutaneous administration of 100 μl of carbon tetrachloride (CCl4)/100 g body weight twice a week for 9 weeks or by ligation of common bile duct for a week.
Fixation
Under ether anesthesia, the liver was taken out. A part of the liver was fixed with 4% formaldehyde and paraffin sections were stained with Azan Mallory staining. Another part of the liver was fixed in 4% paraformaldehyde and used for in situ hybridization, immunohistochemistry, and electron microscopy.
In Situ Hybridization
Sense and antisense RNA probes corresponding to rat PrP cDNA fragment were inserted into the pGEM-T vector and labeled with digoxigenin-labeled UTP (Boehringer Mannheim) by using T7 or SP6 RNA polymerase, respectively. Cryosections of paraformaldehyde-fixed liver specimens 5 μm thick were mounted on glass slides coated with TESTA (3-amino propyltriethoxysilane, Sigma, St. Louis, MO) and air-dried. The sections were rinsed twice in 0.1 mol/L phosphate buffer (PB) and then treated with 10 μg/ml proteinase K in 50 mmol/L Tris-HCl and 5 mmol/L EDTA for 5 minutes. They were refixed in 4% paraformaldehyde, rinsed once in DEPC water, acetylated in 0.25% acetic anhydride in 0.1 mol/L triethanolamine, and then rinsed once in 0.1 mol/L PB prior to dehydration. Hybridization was performed at 55°C for 16 hours in a solution (pH 8.0) containing 50% formamide, 20 mmol/L Tris-HCl, 5 mmol/L EDTA (pH 8.0), 0.3 mol/L NaCl, 10 mmol/L PB, 1× Denhardt’s solution, 10% dextran sulfate, 0.2% sarcosyl, 500 μg/ml yeast tRNA, 200 μg/ml salmon sperm DNA and 2.5 μg/ml of digoxigenin-labeled riboprobe. After washing at 65°C in a high-stringency solution containing 50% formamide, 2× SSC and 10% mercaptoethanol for 30 minutes, sections were treated with RNase A (1 μg/ml) in 10 mmol/L Tris-HCl, 1 mmol/L EDTA, and 0.5 mol/L NaCl for 10 minutes at 37°C. They were washed at 65°C in the high-stringency solution for 30 minutes in 2× SSC for 10 minutes, and in 0.1× SSC for 10 minutes. Immunological detection was performed using the Genius nonradioactive DNA labeling and detection kit (Boehringer Mannheim) according to the manufacturer’s protocol. After color development, sections were counterstained with methyl green and mounted.
Immunohistochemistry
Paraformaldehyde-fixed hamster liver specimens were cut by cryostat into sections 5 μm thick, mounted on glass slides coated with TESTA, and air-dried. Samples were incubated with 0.3% hydrogen peroxide in methanol for 30 minutes. After washing, they were treated with normal animal serum for 1 hour at room temperature and further incubated with anti-SM α-actin monoclonal antibody (1:500) or anti-human PrP monoclonal antibody (1:500) overnight at 4°C. After washing, they were incubated with biotinylated secondary antibody (1:500, Dako) for 1 hour at room temperature, followed by incubation with avidin-biotin-peroxidase complex (Vectastain, Burlingame, CA). Reaction products were visualized by incubating with 0.025% diaminobenzidine (DAB) and 0.003% hydrogen peroxide and counterstaining with methyl green.
Immunoelectron Microscopy
Liver specimens fixed with 4% paraformaldehyde were cut into 50-μm-thick sections by a Microslicer (Dosaka, Kyoto, Japan). Sections were immunostained for PrP as described above except that the endogenous peroxidase blocking step was omitted. After colorization with DAB and hydrogen peroxide, sections were postfixed in 1% osmium tetraoxide, dehydrated in ethanol, and embedded in polybed (Polyscience, Warrington, PA). Thin sections were stained with saturated lead citrate for 5 minutes and observed under a JEM-1200 EX electron microscope (JEOL, Tokyo, Japan) at 100 kV.
Results
Expression of PrP Gene in Cultured Rat Stellate Cells
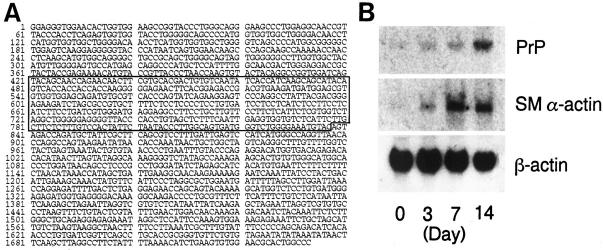
We used freshly isolated and 14-day-cultured rat stellate cells for quiescent and activated, respectively, cells as demonstrated by the negative and positive, respectively, immunocytochemical stain for SM α-actin (data not shown), and isolated 13 genes exclusively expressed in activated stellate cells by using suppression subtractive hybridization. DNA sequence analysis revealed that four were novel genes, four were genes already reported to express in stellate cells (ie, SM α-actin, 7 laminin β1, 14 entactin, 15 and oxidized low-density lipoprotein receptor) 16 and the remaining five were known genes but their expression in stellate cells has not been reported so far. Of great interest, one gene from the third group was a PrP gene, indicated by the fact that the sequence of cDNA fragment was 100% identical to the 421–837th base pairs of cDNA sequence reported for rat PrP (Genbank, M20313) 17 (Figure 1A) ▶ . Time-course Northern blot analysis showed that, while PrP mRNA expression was scarce in the stellate cells at day 0 after culture, it increased in a time-dependent manner by day 14, accompanied by the increase in SM α-actin mRNA expression (Figure 1B) ▶ .
Figure 1.
A: Nucleotide sequence of rat PrP cDNA as previously reported. 17 The enclosed part represents the cDNA fragment obtained here from activated stellate cells by suppression subtractive hybridization. B: Northern blot analysis of mRNA expression for PrP, SM α-actin and β-actin in rat stellate cells cultured for indicated periods. Note that PrP mRNA expression increased by culture in a time-dependent manner in parallel with SM α-actin mRNA expression, indicative of close relation of PrP mRNA expression to stellate cell activation.
Expression of PrP mRNA in Rat Livers
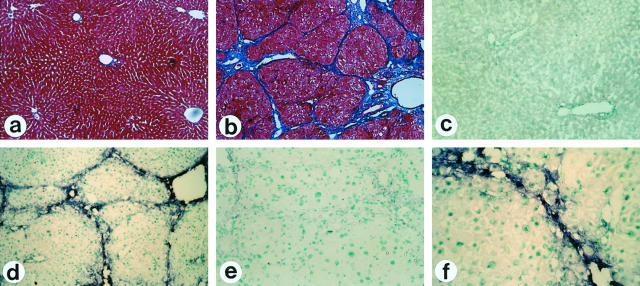
To demonstrate PrP mRNA in liver tissues, in situ hybridization was performed in untreated (Figure 2a) ▶ and CCl4-induced fibrotic rat livers (Figure 2b) ▶ . While PrP mRNA expression was negligible in untreated livers (Figure 2c) ▶ , it was prominent in and around the fibrous septa of fibrotic livers (Figure 2d, f) ▶ . There was no staining with sense riboprobe in fibrotic livers (Figure 2e) ▶ .
Figure 2.
Detection of PrP mRNA expression in rat liver tissue. Untreated ((a, c) ) and CCl4-treated ((b, d–f)) rat livers were stained with Azan-Mallory stain ((a, b) ) and in situ hybridization for PrP mRNA expression with antisense riboprobe ((c, d, f)) and sense probe ((e)). Note that PrP mRNA is expressed in the cells located in and around the septa of fibrotic liver. Magnification, ×25 ((a, b)), ×50 ((c, d)), ×100 ((e)), ×125 ((f)).
Expression of PrPc in Cultured Hamster Stellate Cells
Because anti-human PrPc monoclonal antibody cross-reacts with hamster PrPc but not with rat PrPc, we used cultured hamster stellate cells for the analysis of PrPc expression. Western blot analysis demonstrated that the bands indicating PrPc with molecular weight of 30–35 kDa were absent at day 0, appeared at day 3, and significantly increased at days 7 and 14 (Figure 3A) ▶ . The time course of SM α-actin expression was very similar to that of PrPc expression; SM α-actin was negligible at day 0 and increased in a time-dependent manner, reaching the maximum at days 7 and 14 (Figure 3A) ▶ .
Figure 3.
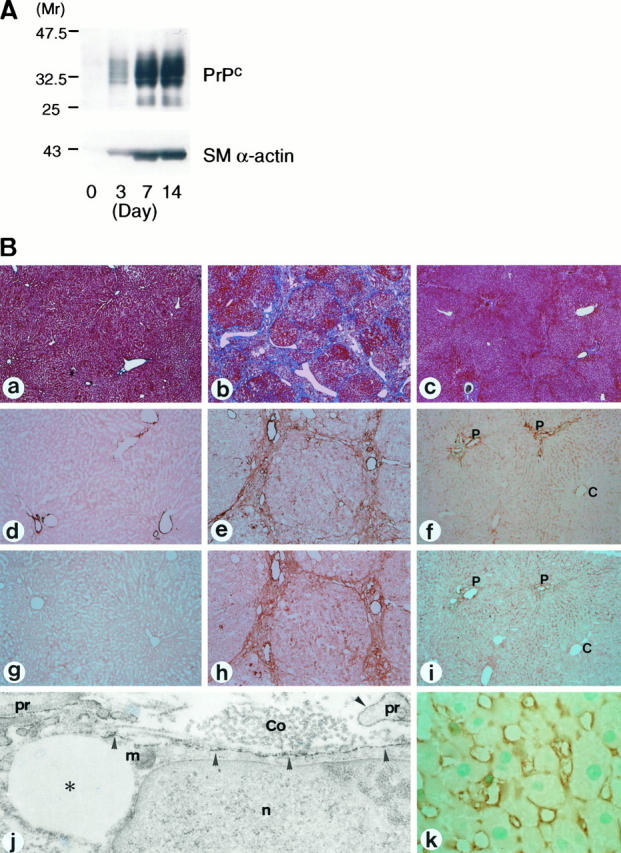
A: Western blot analysis for PrPc and SM α-actin in hamster stellate cells cultured for indicated periods. Note that PrPc expression increased in a time-dependent manner in parallel with SM α-actin expression. B: Detection of PrPc expression in hamster liver tissue. Untreated ((a, d, g)), CCl4-treated ((b, e, h, j) ), and common bile duct-ligated hamster livers ((c, f, i, k) ) were stained with Azan-Mallory stain ((a–c)) and immunohistochemical stain for SM α-actin ((d–f)) and PrP ((g–k)). In bile duct-ligated livers, positive cells for SM α-actin ((f)) and PrP ((i)) were distributed preferentially in the periportal and intermediate zones of liver lobule and scarce in the central zone. Immunoelectron microscopy has demonstrated that positive cells for PrP in the septa are stellate cells containing lipid droplets ((asterisks)); the reaction products ((arrowheads)) are located on the plasma membrane of the cell body and processes (pr). Collagen fibers (Co) are closely associated with the stellate cell. P, portal veins; C, central veins; n, nucleus; m, mitochondria. Magnification, ×20 ((a–c)), ×50 ((d–i)), ×22,000 ((j)), ×200 ((k)).
Expression of PrPc in Hamster Livers
To demonstrate PrPc expression in liver tissues, immunohistochemical detection of the protein was done in untreated (Figure 3B ▶ -a), CCl4-treated (Figure 3B ▶ -b), and common bile duct-ligated (Figure 3B ▶ -c) hamster livers. In untreated livers, SM α-actin was expressed in perivascular smooth muscles (Figure 3B ▶ -d), whereas PrPc was not detected (Figure 3B ▶ -g) except in lymphocytes and basement membrane-like structures of the portal vein (data not shown). In and around the fibrous septa of CCl4-induced fibrotic livers, numerous cells stained positively for SM α-actin (Figure 3B ▶ -e) and PrPc (Figure 3B ▶ -h). Immunoelectron microscopy of CCl4-treated livers revealed that reaction deposits for PrPc were located along the plasma membrane of stellate cells containing lipid droplets (Figure 3B ▶ -j). In common bile duct-ligated livers, PrPc-positive cells were found along the sinusoids preferentially in the periportal and intermediate zones of liver lobule (Figure 3B ▶ -i, k). SM α-actin-positive cells exhibited similar intralobular distribution as PrPc-positive ones (Figure 3B ▶ -f).
Discussion
The molecular mechanisms of stellate cell activation and regulation have been extensively investigated to elucidate the pathogenesis of liver fibrosis and to develop potential therapeutic strategies to prevent liver cirrhosis. 4,18,19 The present study has for the first time demonstrated that PrP is expressed in activated stellate cells at both the mRNA and protein levels. Expression of PrP was well correlated with that of SM α-actin, a well known marker for stellate cell activation.
Prions are causative agents for diseases termed spongiform encephalopathies, which are not only infectious in livestock and human beings but also inheritable in the latter. They are devoid of nucleic acids and consist of a single protein termed scrapie-type PrP (PrPSc). There is no difference in amino acid sequence between pathogenic PrPSc and its normal counterpart PrPc. It is reported that PrPSc converts its host-encoded isoform PrPc into insoluble aggregates of PrPSc concomitantly with pathologic modifications. 20-23 As a result, PrPSc causes neural cell degeneration. However, the pathophysiological function of PrPc is poorly understood.
PrPc is a constituent glycoprotein present at a high concentration in the brain. It is also found in the heart, kidney, and lung at intermediate levels, but is not detectable in the liver. 24,25 This study demonstrated that expression of PrP mRNA and its protein was negligible in normal liver tissue, a finding consistent with previous studies. However, it was found to increase dramatically in diseased livers associated with stellate cell activation. It is reported that activated stellate cells undergo metabolic alterations which induce increased or decreased the production of nerve-related proteins such as N-CAM 11 or GFAP, 8 respectively, in rats. The present finding of PrP expression in activated stellate cells may provide further evidence of a metabolic relation of activated stellate cells with nerve cells. It has been also suggested that PrPc may function as a neural cell receptor or a cell adhesion molecule, directing and/or maintaining the architecture of the nervous system. 26,27 The demonstration by immunoelectron microscopy that PrPc localizes on the surface of activated stellate cells is consistent with this view.
It was recently reported that PrPc may function as a copper-binding protein and modulate copper-dependent enzyme activities. 28 Thus, it is conceivable that Cu2+/Zn2+-superoxide dismutase activity could be regulated by PrPc. Because stellate cell activation is accelerated by oxidative stress 4,29 and down-regulated by antioxidants, 13 PrPc might be involved in the response to oxidative stimuli. Although the association of stellate cell activation with other reported functions of PrPc needs to be studied in detail, the present finding of the restricted expression of PrP mRNA and protein in activated stellate cells both in vitro and in vivo suggests that PrP expression could be involved in the development of liver fibrosis.
Acknowledgments
We thank Prof. Hiroshi Takagi and Dr. Takunari Yoneda, Department of Anatomy, Osaka City University Medical School, for their valuable advice for this study.
Footnotes
Address correspondence to: Kazuo Ikeda, M.D., Second Department of Anatomy, Osaka City University Medical School, 1-4-3, Asahimachi, Abeno, Osaka 545-8585, Japan. E-mail: ikeda\@med.osaka-cu.ac.jp.
Supported by grants-in-aid from the Ministry of Education, Science and Culture of Japan (No. 09770014 to KI, No. 09770372 to NK) KI was supported by Osaka City University Medical Research Foundation for Medical Research (1997).
References
- 1.Wake K: Perisinusoidal stellate cells (fat-storing cells, interstitial cells, lipocytes), their related structure in and around the liver sinusoids, and vitamin A-storing cells in extrahepatic organ. Int Rev Cytol 1980, 66:303-353 [DOI] [PubMed] [Google Scholar]
- 2.Friedman SL: The cellular basis of hepatic fibrosis. N Engl J Med 1993, 328:1828-1835 [DOI] [PubMed] [Google Scholar]
- 3.Pinzani M: Novel insights into the biology and physiology of the Ito cell. Pharmacol Ther 1995, 66:387-412 [DOI] [PubMed] [Google Scholar]
- 4.Lee KS, Buck M, Houglum K, Chojkier M: Activation of hepatic stellate cells by TGFα and collagen type I is mediated by oxidative stress through c-myb expression. J Clin Invest 1995, 96:2461-2468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Maher JJ, Bissell DM, Friedman SL, Roll FJ: Collagen measured in primary cultures of normal rat hepatocytes derived from lipocytes within the monolayer. J Clin Invest 1988, 82:450-459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gressner AM, Bachem MG: Cellular sources of noncollagenous matrix proteins: role of fat-storing cells in fibrogenesis. Smin Liver Dis 1990, 10:30-46 [DOI] [PubMed] [Google Scholar]
- 7.Radamori G: The stellate cell (Ito cell, fat-storing cell, lipocyte, perisinusoidal cell) of the liver. New insights into pathophysiology of an intriguing cell. Virchows Arch B Cell Pathol 1991, 61:147-158 [DOI] [PubMed] [Google Scholar]
- 8.Niki T, de Bleser PJ, Xu G, von den Berg K, Wisse E, Geerts A: Comparison of glial fibrillary acidic protein and desmin staining in normal and CCl4-induced fibrotic rat livers. Hepatology 1996, 23:1538-1545 [DOI] [PubMed] [Google Scholar]
- 9.Grad A, White F, Dutton G: Extra-neural glial fibrillary acidic protein (GFAP) immunoreactivity in perisinusoidal stellate cells of rat liver. J Neuroimmunol 1985, 8:359-375 [DOI] [PubMed] [Google Scholar]
- 10.Nakatani K, Seki S, Kawada N, Kobayashi K, Kaneda K: Expression of neural cell adhesion molecule (N-CAM) in perisinusoidal stellate cells of the human liver. Cell Tissue Res 1996, 283:159-165 [DOI] [PubMed] [Google Scholar]
- 11.Knittel T, Aurisch S, Neubauer K, Eichhorst S, Ramadori G: Cell-type-specific expression of neural cell adhesion molecule (N-CAM) in Ito cells of rat liver: up-regulation during in vitro activation and in hepatic tissue repair. Am J Pathol 1996, 149:449-462 [PMC free article] [PubMed] [Google Scholar]
- 12.Lalazar A, Wong L, Yamasaki G, Friedman SL: Early genes induced in hepatic stellate cells during wound healing. Gene 1997, 22:235-243 [DOI] [PubMed] [Google Scholar]
- 13.Kawada N, Seki S, Inoue M, Kuroki T: Effect of antioxidants, resveratrol, quercetin, and N-acetylcysteine, on the functions of cultured rat hepatic stellate cells and Kupffer cells. Hepatology 1998, 27:1265-1274 [DOI] [PubMed] [Google Scholar]
- 14.Geerts A, Greenwel P, Cunningham M, DeBleser P, Rogiers V, Wisse E, Rojkind M: Identification of connective tissue gene transcripts in freshly isolated parenchymal, endothelial, Kupffer and fat-storing cells by northern hybridization analysis. J Hepatol 1993, 19:148-153 [DOI] [PubMed] [Google Scholar]
- 15.Loreal O, Levavasseur F, Fromaget C, Gros D, Guillouzo A, Clement B: Cooperation of Ito cells and hepatocytes in the deposition of extracellular matrix in vitro. Am J Pathol 1993, 143:538-544 [PMC free article] [PubMed] [Google Scholar]
- 16.Bachem MG, Scneiderhan W, Scmid-Kotsas A, Grab H, Zorn U, Celik E, Menke A, Weidenbach H, Adler G, Grunert A: Human hepatic stellate cells express the scavenger receptor CD36 to which oxidized low density lipoproteins bind and stimulate extracellular matrix synthesis. Hepatology 1997, 26:184A. [DOI] [PubMed] [Google Scholar]
- 17.Liao YC, Tokes Z, Lim E, Lackey A, Woo CH, Button JD, Clawson GA: Cloning of rat “prion-related protein” cDNA. Lab Invest 1987, 57:370-374 [PubMed] [Google Scholar]
- 18.Wong L, Yamasaki G, Johnson RJ, Friedman SL: Induction of β-platelet-derived growth factor receptor in rat hepatic lipocytes during cellular activation in vivo and in culture. J Clin Invest 1994, 94:1563-1569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rockey DC, Maher JJ, Jarnagin WR, Gabbiani G, Friedman SL: Inhibition of rat lipocyte activation in culture by interferon-γ. Hepatology 1992, 16:776-784 [DOI] [PubMed] [Google Scholar]
- 20.Prusiner SB: Molecular biology of prion diseases. Science 1991, 252:1515-1522 [DOI] [PubMed] [Google Scholar]
- 21.Weissmann C: Molecular biology of prion diseases. Trends Cell Biol 1994, 4:10-14 [DOI] [PubMed] [Google Scholar]
- 22.Edenhofer F, Weiss S, Winnacker EL, Famulok M: Chemistry and molecular biology of transmissible spongiform encephalopathies. Angew Chem Int Ed Engl 1997, 36:1674-1694 [Google Scholar]
- 23.Prusiner SB, Scott MR, DeArmond SJ, Cohen FE: Prion protein biology. Cell 1998, 93:337-348 [DOI] [PubMed] [Google Scholar]
- 24.Bendheim RE, Brown HR, Rudelli RD, Scala LJ, Goller NL, Wen GY, Kascsak RJ, Cashman NR, Bolton DC: Nearly ubiquitous tissue distribution of the scrapie agent precursor protein. Neurology 1992, 42:149-156 [DOI] [PubMed] [Google Scholar]
- 25.Horiuchi M, Yamazaki N, Ikeda T, Ishiguro N, Shinagawa M: A cellular form of prion protein (PrPc) exists in many non-neuronal tissue of sheep. J Gen Virol 1995, 76:2583-2587 [DOI] [PubMed] [Google Scholar]
- 26.Manson J, West JD, Thomson V, McBridge P, Kaufman MH, Hope J: The prion protein gene: a role in mouse embryogenesis? Development 1992, 115:117-122 [DOI] [PubMed] [Google Scholar]
- 27.Rieger R, Edenhofer F, Lasmezas CI, Weiss S: The human 37-KDa laminin receptor precursor interacts with the prion protein in eukaryotic cells. Nat Med 1997, 3:1383-1388 [DOI] [PubMed] [Google Scholar]
- 28.Brown DR, Qin K, Herms JW, Madlung A, Manson J, Strome R, Fraser PE, Kruck T, von Bohlens A, Schultz-Schaeffer W, Giese A, Westaway D, Kretzschmar H: The cellular prion protein binds copper in vivo. Nature 1997, 390:684-687 [DOI] [PubMed] [Google Scholar]
- 29.Casini A, Ceni E, Salzano R, Milani S, Schuppan D, Surrenti C: Acetaldehyde regulates the gene expression of matrix metalloproteinase-1 and -2 in human fat-storing cells. Life Sci 1994, 55:1311-1316 [DOI] [PubMed] [Google Scholar]