Abstract
A novel set of polymerase chain reaction (PCR) primers, designated SPF1 and SPF2 and located in the L1 region, was developed for universal detection of human papillomavirus (HPV). A short PCR fragment (SPF) of only 65 pb was synthesized. SPF amplimers were detected in a microtiter-based hybridization system, using a mixture of oligonucleotide probes. The SPF system allowed detection of at least 43 different HPV genotypes. The clinical performance of the novel SPF system was assessed in three different patient groups. 1) Analysis of 534 cervical scrapes, obtained from treated patients, showed that the detection rate in 447 (83.7%) scrapes with normal cytology was significantly higher using the SPF system as compared with the universal primer set GP5+/6+ (P < 0.001). 2) The SPF assay detected HPV DNA in 299 (98.4%) of 304 scrapes with cytological dyskaryosis. 3) The SPF system detected HPV DNA in 100% of 184 formalin-fixed, paraffin-embedded cervical carcinoma specimens. In conclusion, the novel SPF system permitted universal and highly sensitive detection of HPV DNA in diverse clinical materials and may improve the molecular diagnosis and epidemiology of this important virus.
Cervical cancer is the second most important cause of cancer-related mortality in women after breast cancer, 1 annually affecting an estimated 500,000 women worldwide. 2 Smoking, age of first sexual contact, number of sexual partners, and immunological status have been identified as significant risk factors for cervical carcinoma. 3-6 Infection with human papillomavirus (HPV) also is considered to be an important risk factor. 7,8 HPV is an epitheliotropic virus and contains a circular double-stranded DNA genome of approximately 7900 bp. To date, at least 77 distinct HPV genotypes have been described that can be classified into a cutaneous and a mucosal group, as established by their associated clinical manifestations. 8 Among the mucosal genotypes that infect the anogenital tract, potentially high-risk HPV genotypes 16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 66, and 69 have been defined based on their prevalence in cervical intraepithelial neoplasia (CIN) lesions and cervical carcinomas. 8 The HPV genotypes 6, 11, 34, 40, 42, 43, 44, 53, 54, 55, 59, 61, 62, 67, 68, 71, and 74 are classified as putative low-risk factors. By definition, infection with high-risk HPV genotypes favors an increased risk for cancer. 9-11 However, the distinction between high-risk and low-risk genotypes is a moot point as low-risk HPV genotypes have also been found in carcinomas. 9
As HPV cannot be cultured in vitro and serological assays are still ineffective, diagnosis of HPV injury is based on the use of molecular tools. Direct dot-spot detection and in situ hybridization assays have been described, 12,13 but these methods are tedious and appear to lack sensitivity and specificity. DNA amplification methods, such as the polymerase chain reaction (PCR), permit more sensitive detection of the viral DNA. Besides type-specific PCR primers for individual HPV genotypes, 14,15 several universal PCR primer sets have been developed, including My11/My09, 16 OBI/II, 17 CPI/CPII, 18 and GP5+/6+. 19 At present, these HPV primer sets are widely used for routine diagnosis of HPV infections. However, it has been shown that none of these general primer sets permits adequate detection of the still expanding spectrum of anogenital HPV genotypes. 20,21 Moreover, as the sensitivity of these universal primer sets is limited, their use will underestimate the true prevalence of HPV. 14,20,21,22
The aim of the present study was to develop a set of HPV PCR primers for highly sensitive detection of all known mucosal HPV types. The assay comprised a novel set of primers for broad-spectrum detection of HPV DNA by short-fragment PCR and a microtiter-format assay for detection of HPV-derived PCR products by a mixture of probes. The sensitivity and specificity of this novel HPV detection system were analyzed and compared with type-specific as well as with other general HPV PCR primer sets. Finally, the performance of the method was evaluated by testing clinical samples from various patient populations, including 1) cervical scrapes, mainly classified as normal cytology from patients previously treated for CIN lesions, 2) mild, moderate, and severe dyskaryotic smears, and 3) formalin-fixed, paraffin-embedded cervical carcinoma samples.
Materials and Methods
Patients
The present study comprised three different groups of patients. Group 1 comprised 534 cervical scrapes obtained from consecutive women that were treated in the gynecology outpatient clinic of a community hospital in Delft, The Netherlands. Patients had a history of treatment for dysplasia by loop excision of the transformation zone (LETZ). Cervical scrapes were obtained with a Cervex brush (Rovers, Oss, The Netherlands) and were resuspended and transported in 1.5 ml of phosphate-buffered saline (PBS), pH 7.2, at room temperature. Smear preparations were examined and classified.
Group 2 consisted of 304 cervical scrapes of a total of 320 scrapes from women that were tested for the presence of HPV during a community screening program between 1988 and 1993 in The Netherlands. 23 The surveyed patients had either one cervical scrape showing severe dyskaryosis (n = 153) or two scrapes showing mild or moderate dyskaryosis (n = 151) with a maximum interval of 1 year. Of the unavailable scrapes, 10 samples were severely dyskaryotic and 6 were mild or moderate dyskaryotic.
Group 3 comprised 184 patients with cervical carcinoma. The cervical biopsies were obtained from women visiting the Russian Cancer Center in Moscow between 1988 and 1994. 24 The specimens had been fixed in formalin, embedded in paraffin, histologically examined after hematoxylin and eosin staining, and classified as squamous cell carcinoma (n = 132), adenocarcinoma (n = 31), or adenosquamous carcinoma (n = 21).
DNA Isolation
Cervical scrapes were resuspended in 1.5 ml of PBS pH 7.2. To prepare DNA for PCR, the cell suspension was vigorously shaken, and 120 μl was treated by adding 40 μl of proteinase K (200 μg/ml) in 3% Triton X-100 for 1 hour at 37°C. The proteinase was inactivated by incubation at 95°C for 10 minutes. Subsequently, 10 μl of the solution was used in a PCR reaction.
In group 3, DNA was isolated from formalin-fixed, paraffin-embedded tissue sections by a modified version of the method as described by Claas et al. 25 Briefly, a 10-μm section was collected in a 1.5-ml tube and deparaffinized by adding 500 μl of xylol. After gentle shaking for 2 minutes and centrifuging for 5 minutes at 12,000 rpm, the pellet was retreated with 500 μl of xylol. The pellet was then washed twice with 500 μl of 96% ethanol and once with 500 μl of acetone. Subsequently, the pellet was air dried, dissolved in 200 μl of 5 mmol/L TrisHCl, pH 9.0, containing 1 mg/ml proteinase K, and incubated overnight at 37°C. Finally, the proteinase K was activated by incubation at 95°C for 10 minutes, and the supernatant was used directly for PCR.
Plasmids
Plasmids containing HPV genomic DNA were kindly provided by Dr. E.-M. de Villiers, Heidelberg, Germany (HPV genotypes 6b, 11, 13, 16, 18, 40, 45, 51, and 53), Dr. R. Ostrow, Minneapolis, MN (HPV genotype 26), Dr. A. Lorincz, Silver Springs, MD (HPV genotypes 31, 35, 43, 44, 56, 61, and 64), Dr. T. Matsukura, Tokyo, Japan (HPV genotypes 58, 59, 62, 67, and 69), and Dr. G. Orth, Paris, France (HPV genotypes 30, 33, 34, 39, 42, 52, 54, 55, 66, 68, 70, and 74). HPV genotypes were classified according to the Los Alamos Database (available at http://hpv-web.lanl.gov).
PCR
The SPF primers developed in this study are located in the L1 open reading frame and listed in Table 1 ▶ . SPF was performed in a final reaction volume of 100 μl, containing 10 μl of the isolated DNA, 10 mmol/L TrisHCl, pH 9.0, 50 mmol/L KCl, 2.5 mmol/L MgCl2, 0.1% Triton X-100, 0.01% gelatin, 200 mmol/L each deoxynucleoside triphosphate, 20 pmol of forward and reverse primers, and 0.25 U of SuperTaq (Sphaero Q, Cambridge, UK). PCR conditions were as follows: preheating for 1 minute at 94°C was followed by 40 cycles of 1 minute at 94°C, 1 minute at 45°C, and 1 minute at 72°C and a final extension of 5 minutes at 72°C. Each PCR experiment was performed with positive and several negative PCR controls.
Table 1.
SPF Primers and Probes
| Primer designation | Sequence 5′ → 3′ | Position* |
|---|---|---|
| SPF primers | ||
| SPF1A | GCiCAGGGiCACAATAATGG | 6582–6601 |
| SPF1B | GCiCAGGGiCATAACAATGG | 6582–6601 |
| SPF1C | GCiCAGGGiCATAATAATGG | 6582–6601 |
| SPF1D | GCiCAAGGiCATAATAATGG | 6582–6601 |
| SPF2B-bio | GTiGTATCiACAACAGTAACAAA | 6624–6646 |
| SPF2D-bio | GTiGTATCiACTACAGTAACAAA | 6624–6646 |
| SPF probes | ||
| HPVuni1A | CAiAATAATGGCATiTGTTGGC | 6591–6612 |
| HPVuni1B | CAiAACAATGGCATiTGTTGGC | 6591–6612 |
| HPVuni1C | CACAATAATGGCATTTGTTGGGG | 6591–6613 |
| HPVuni2 | CAiAATAATGGTATiTGTTGGG | 6591–6612 |
| HPVuni3 | CAiAACAATGGTATiTGTTGGC | 6591–6612 |
| HPVuni4 | AACAATGGTATiTGCTGG | 6594–6611 |
| HPVuni5 | AACAATGGTGTTTGCTGG | 6594–6611 |
| HPVuni6 | AATAATGGCATiTGCTGG | 6594–6611 |
| HPVuni7 | AACAATGGCATiTGCTGG | 6594–6611 |
i, inosine.
*Sequence positions according to HPV genotype 16 sequence PPH16; Genbank accession number K02718.
Several universal primer sequences were published earlier. Primer set GP5+/6+ was described by De Roda Husman et al, 19 and My09/11 was reported by Manos et al. 16 Type-specific PCR primers for HPV genotypes 16, 18, 31, and 33 have been described by Baay et al. 14 All of these PCRs were performed as described. 14,16,19
HPV DNA Detection in Microtiter Plates
Amplimers, synthesized by biotinylated PCR primers, were detected by hybridization to a mixture of HPV-specific probes (Table 1) ▶ . Ten microliters of PCR product was diluted in 100 μl of hybridization buffer (150 mmol/L NaCl, 15 mmol/L sodium citrate, pH 7.0, 0.1% Tween 20) and incubated at 42°C for 30 minutes in streptavidin-coated microtiter plates. Noncaptured materials were removed by three washes with hybridization buffer. The double-stranded captured PCR products were denatured by addition of 100 μl of denaturation solution (100 mmol/L NaOH) and incubated for 5 minutes at room temperature, followed by three washes with hybridization buffer. A mixture of digoxigenin (DIG)-labeled HPV-specific probes (Table 1) ▶ were diluted in hybridization buffer and added to the well and incubated at 42°C for 45 minutes. Wells were washed three times, and anti-DIG alkaline phosphatase conjugate was added and incubated at 42°C for 15 minutes. After five washes, substrate was added and incubated at room temperature for 15 minutes. The reaction was stopped by adding 100 μl of 0.5 mmol/L H2SO4. Optical densities (OD) were determined at 450 nm in a microtiter plate reader. Samples were considered positive if the OD450 was 2.5 times higher than the negative PCR control. In each run, negative controls as well as positive and borderline positive controls were tested together with the clinical samples.
Southern Blot Hybridization
Southern blot hybridization was performed according to standard procedures. 26 Briefly, 20 μl of the PCR product (GP5+/6+ and My11/09) was electrophoresed on a 2% Tris-borate/ethylenediaminetetra-acetate (TBE) agarose gel. The small (65-bp) SPF1/2 amplimers were separated on a 3% agarose gel. Subsequently, amplimers were transferred onto a nylon membrane (Hybond N+, Amersham, Little Chalfont, UK) by vacuum blotting in the presence of 0.4 N NaOH. The Southern blots were hybridized with a 32P-labeled oligonucleotide probe for 16 hours at 42°C in a solution containing 5X SSC (1X SSC contains 150 mmol/L NaCl and 15 mmol/L sodium citrate), 5X Denhardt’s (1X Denhardt’s contains 0.02% bovine serum albumin, 0.02% polyvinyl pyrrolidone, and 0.02% Ficoll), 0.5% SDS, 75 mmol/L EDTA, and 0.1 mg/ml denatured herring sperm DNA. Subsequently, the blots were washed twice in 2X SSC/0.1% SDS at 42°C for 15 minutes. Autoradiography was performed with intensifying screens for 3.5 hours at −70°C, using Kodak X-Omat AR film.
Sequence Analysis
SPF1/2 amplimers were excised from a 2% low-melting-point Tris-acetate/ethylenediaminetetra-acetate (TAE)-agarose gel and directly analyzed by the AmpliCycle sequencing kit (Perkin Elmer, Norwalk, CT) using one of the SPF primers. The concentration of dideoxyribonucleosides in the termination mixtures was increased to permit adequate sequencing of the short 65-bp PCR products. Sequence products were separated and detected by an automated sequencing system (Alf-Express, Pharmacia, Uppsala, Sweden). As the software of the sequencing unit was not suitable for interpretation of very short sequences, the profiles were read manually. Sequences were analyzed with the PC-Gene software (Intelligenetics, Mountain View, CA) and compared with HPV sequences of known types using the computer program basic local alignment search tool (BLAST). 27
Statistical Analyses
For statistical analyses χ 2 and Fisher’s exact test were used to compare the prevalence of a dichotomous variable between two groups. P values <0.05 were considered to be statistically significant.
Results
Development of Novel Universal Primers and Probes
The L1 region sequences from 39 HPV genotypes were obtained from Genbank. Alignment of these sequences identified a relatively well conserved part in the L1 region, located between nucleotides 6582 and 6646, as shown in Figure 1 ▶ (all numbers are according to the prototype PPH16 sequence of HPV type 16; Genbank accession number K02718). Based on this sequence alignment, target regions for forward and reverse primers were chosen that were relatively well conserved and designated as SPF1 (nt 6582-6601) and SPF2 (nt 6624-6646), respectively. Four primers (SPF1A to -D) were designed as forward PCR primers, aimed at the SPF1 region. Similarly, two reverse primers (SPF2B and -D) were aimed at the SPF2 region. These six primers (Table 1) ▶ were used in equimolar quantities for universal HPV DNA amplification. The expected amplimer size was only 65 bp (nt 6582-6646), including a 22 bp interprimer fragment, flanked by the SPF1 and SPF2 priming regions. The SPF primer target regions are similar to the ones used by the sense primers My11 and GP5+. 16,19 In the SPF system, the later target region is used for antisense priming.
Figure 1.
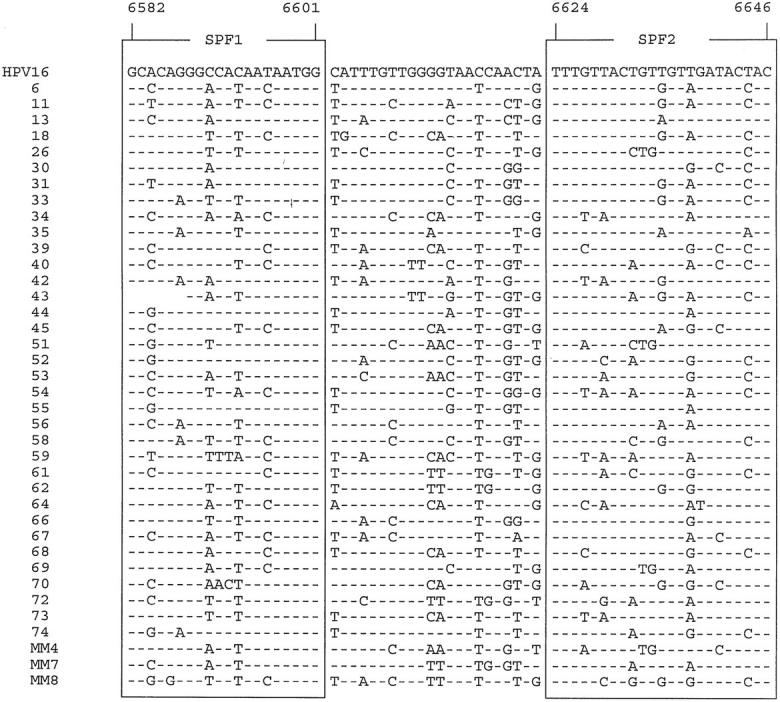
Nucleotide sequence alignments, positions 6582 to 6646 (numbered according to HPV-16 sequence PPH16, Genbank accession number K02718), of 39 anogenital HPV genotypes. HPV genotypes are identified by numbers. Hyphens indicate the presence of nucleotides identical to the top sequence (HPV16). Primer regions for SPF1 and SPF2 are shown by boxes.
As shown in Figure 1 ▶ , the sequences of the interprimer region, located between SPF1 and SPF2, are heterogeneous. This sequence variation is sufficient to permit recognition of each of the different HPV genotypes, except for types 68 and 73, which have identical interprimer regions.
For universal detection of HPV DNA amplimers from a broad range of genotypes, a total of nine probes (Table 1) ▶ were designed that permitted hybridization to all anogenital HPV genotypes. These probes were used in a microtiter-based amplimer detection assay to determine the presence of amplified HPV DNA after SPF-mediated PCR.
Specificity and Sensitivity of the SPF1/2 Primers
To assess the efficacy of the novel broad-spectrum primer set, plasmids containing partial or complete HPV genomic sequences from 34 different genotypes (as listed in Materials and Methods) were analyzed by the SPF system. Amplimers of the expected size of 65 bp were obtained from all plasmids, as determined by gel electrophoresis (data not shown). Sequences of the 22-bp interprimer regions were completely concordant with the published L1 region sequence of each HPV genotype as shown in Figure 1 ▶ . Subsequently, the SPF1/2 amplimers were tested in the microtiter plate hybridization assay. To test the detection of HPV genotypes 73, MM4, and MM7, clinical material was used. Hybridization with the mixture of the nine universal probes yielded positive signals for all of the 37 amplimers. These results showed that the SPF1/2-mediated PCR products of all tested HPV genotypes could be detected by the mixture of HPV DNA detection probes.
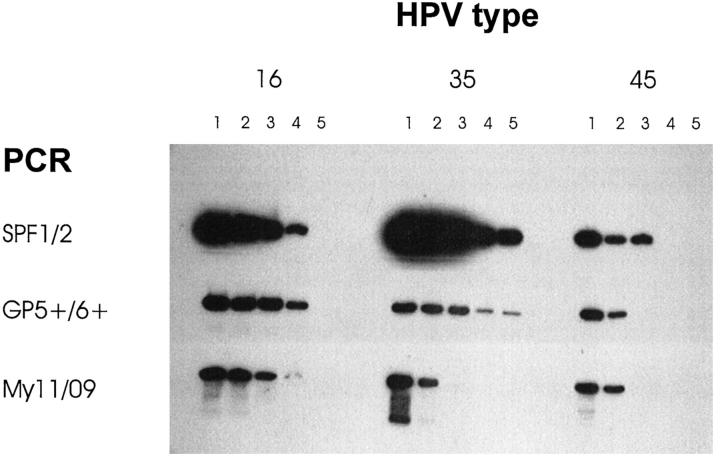
To determine the sensitivity of the novel primer set, series of 10-fold dilutions of plasmids containing genomic DNA from HPV types 16, 35, and 45 were tested by PCR and Southern blot hybridization. The sensitivity of the SPF1/2 primers was compared with those of the primer sets My11/My09 and GP5+/GP6+ (Figure 2) ▶ . For HPV type 16, the three PCR primer sets demonstrated the same sensitivity. For HPV type 35, the My11/09 primer set was at least 100-fold less sensitive than SPF1/2 and GP5+/6+. Primer sets GP5+/6+ and My11/09 reached equal sensitivity for type 45, whereas SPF1/2 was 10 times more sensitive. These results showed the high sensitivity of the novel SPF1/2 primers for these HPV genotypes.
Figure 2.
Southern blot hybridization of PCR products, amplified by SPF1/2 (top row), GP5+/6+ (middle row), and My11/09 (bottom row). PCR products from 10-fold dilutions of plasmids, containing genomic sequences of HPV genotypes 16, 35, and 45 were analyzed. Lanes 1 to 5 represent PCR products starting with 100 fg, 10 fg, 1 fg, 100 ag, and 10 ag of HPV DNA, respectively.
Clinical Evaluation of the Novel SPF Detection System
The clinical performance of the novel HPV DNA detection system was investigated by analysis of materials from three different patient groups. Group 1 comprised a total of 534 cervical scrapes that were obtained during follow-up from women after treatment for cervical intraepithelial neoplasia (CIN). Results of the cytological examinations are summarized in Table 2 ▶ . The great majority (83.7%) of the follow-up smears did not show cytological abnormalities. All cervical scrapes were tested for the presence of HPV DNA with the primer sets SPF1/2 and GP5+/6+. The PCR results are also shown in Table 2 ▶ . Overall, a total of 160 (30.0%) of the 534 samples were found to be positive for the SPF1/2 primers, which is significantly higher than the 113 (21.2%) positive samples detected by GP5+/6+ PCR (P < 0.001). All 113 GP5+/6+-positive samples were also positive with the SPF1/2 primer set. PCR results of the 47 cases that were initially positive by the SPF1/2 primers, but negative by the GP5+/6+ primer set, were all confirmed by repeating the PCR with both systems. The HPV DNA detection rate in scrapes with a normal cytology (n = 447) was significantly higher by SPF1/2-mediated PCR as compared with GP5+/6+ PCR (P < 0.001). The detection rates among scrapes with atypical squamous cells of undetermined significance (ASCUS) or mild to moderate dyskaryotic lesions were not significantly different. The numbers of cases where no cytological analysis was possible or with severe dyskaryosis or carcinoma were too low to compare the efficacy of both primer sets in this group.
Table 2.
Morphology and HPV DNA Detection among 534 Cervical Scrapes from Patient Group 1
| Cytological classification | Number of scrapes | Number of SPF1/2 positive (%) | Number of GP5+/6+ positive (%) |
|---|---|---|---|
| No diagnosis possible | 4 | 2 | 2 |
| Normal cytology | 447 | 102 (22.8) | 63 (14.1)* |
| ASCUS | 41 | 15 (36.6) | 12 (29.3)† |
| Mild/moderate dyskaryosis | 35 | 34 (97.1) | 30 (85.7)† |
| Severe dyskaryosis | 6 | 6 | 6 |
| Carcinoma | 1 | 1 | 1 |
| Total | 534 | 160 (30.0) | 113 (21.2) |
ASCUS, atypical squamous cells of undetermined significance.
*P < 0.001.
†Not significant.
SPF1/2 amplimers from the 160 positive cases were subjected to direct sequence analysis, and the presence of HPV-specific sequences corresponding to the L1 region sequences of various HPV genotypes was confirmed in all cases (data not shown). Based on the 22-bp interprimer sequences, 70 (59.8%) and 23 (19.7%) of the 117 HPV-positive cases with normal cytology (n = 102) or ASCUS (n = 15) were classified as high-risk and low-risk HPV genotypes, respectively (Table 3) ▶ . The distinction between high-risk and low-risk HPV genotypes was according to zur Hausen. 8 Among the 34 SPF-positive cases in group 1 with mild or moderate dyskaryotic lesions, 79.4% and 17.6% were classified as high risk and low risk, respectively. The prevalence of sequences that did not exactly match any of the known HPV genotype sequences (designated as unknown in Table 3 ▶ ) among cases with normal cytology or ASCUS (20.5%) was significantly higher than among the mild or moderate dyskaryotic cases (3.0%). In this analysis, cases with no cytological diagnosis possible, severe dyskaryosis, or carcinoma were excluded due to low numbers.
Table 3.
Distribution of HPV Genotypes in Three Different Patient Groups
| HPV type | Patient group 1 (n = 534) | Patient group 2 (n = 304) | Patient group 3 (n = 184) | ||
|---|---|---|---|---|---|
| Normal cytology ASCUS* | Mild or moderate dyskaryosis | Mild or moderate dyskaryosis | Severe dyskaryosis | Carcinoma | |
| High risk | 70 (59.8%) | 27 (79.4%) | 140 (95.2%) | 150 (98.7%) | 177 (96.2%) |
| Low risk | 23 (19.7%) | 6 (19.7%) | 5 (3.4%) | 0 | 1 (0.5%)† |
| Unknown‡ | 24 (20.5%) | 1 (3.0%) | 2 (1.4%) | 2 (1.3%) | 6 (3.3%) |
| Total | 117 | 34 | 147 | 152 | 184 |
HPV genotypes were recognized in HPV-positive cases by sequence analysis of the SPF1/2 22-bp interprimer region and compared with published L1 region sequences of defined HPV genotypes. HPV genotypes 16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 66, and 69 were considered as high-risk genotypes. HPV genotypes 6, 11, 34, 40, 42, 43, 44, 52, 54, 55, 59, 61, 62, 67, 68, 71, and 74 were considered as low-risk genotypes. 8
*Of the 160 SPF-positive cases in group 1, 9 samples, classified as no diagnosis possible, severe dyskaryosis, and carcinoma were excluded.
†Sequence of the 22-bp interprimer region from this case was identical to HPV genotype 55.
‡Sequences of the 22-bp interprimer region not exactly matching any published L1 sequence of a defined HPV genotype.
As patient group 1 contained only a limited number of cases with dyskaryosis, a second patient group was analyzed, comprising a total of 304 cases that were cytologically classified as mild or moderate dyskaryosis (n = 151) or severe dyskaryosis (n = 153). Cervical scrapes were tested for the presence of HPV DNA with the SPF1/2 primer set, and the results are shown in Table 3 ▶ . A total of 299 (98.4%) of the 304 cases were positive by SPF-mediated PCR, and only five scrapes (four times mild or moderate dyskaryosis and one with severe dyskaryosis) were found to be HPV DNA negative. Again, the amplimers were directly sequenced, and the presence of HPV-specific sequences from various HPV genotypes was confirmed in all cases. A total of 140 (95.2%) and 5 (3.4%) of the 147 SPF-positive mild or moderate dyskaryotic cases were classified as high-risk and low-risk HPV genotypes, respectively. Among the 152 SPF-positive severe dyskaryotic cases, the prevalence of high-risk genotypes was 98.7% (Table 3) ▶ .
The presence of HPV in cervical carcinoma was assessed by SPF analysis of the third group, comprising 184 formalin-fixed, paraffin-embedded cervical cancer biopsies obtained from Russian women. The SPF1/2 primer set yielded positive results in 100% of cases (184/184). Amplimers were analyzed by direct sequencing, and the presence of HPV-specific sequences was confirmed in all cases. Among the 184 carcinomas, 177 (96.2%) and 1 (0.5%) were classified into high-risk and low-risk groups, respectively. The low-risk type sequence was associated with HPV genotype 55. 8 The remaining six yielded sequences that did not exactly match with known HPV genotypes (Table 3) ▶ .
Taken together, a total of 634 sequences of the 22-bp SPF interprimer regions, obtained from patient groups 1, 2, and 3 were compared with corresponding L1-region sequences of the different HPV genotypes, as shown in Table 3 ▶ . A total of 35 (5.5%) of the 634 sequences did not exactly match with any of the known HPV genotype L1 sequences. These were classified as unknown and require more extensive sequence analysis. The great majority of these 35 unrecognized sequences were found among scrapes with normal cytology or ASCUS in group 1 patients. Also, sequences, matching low-risk HPV genotypes were found almost exclusively among scrapes with normal cytology or ASCUS and appeared to be virtually absent in the dyskaryotic or carcinoma cases from groups 2 and 3.
Discussion
There is increasing evidence that the presence of HPV is associated with the development of cervical cancer. If true, early detection of HPV, followed by treatment of the dysplasia, may reduce the risk for the development of cervical cancer considerably. 28
As HPV cannot be cultured in vitro and serological assays are still not sensitive enough, diagnosis of HPV infection is currently based on detection of the viral DNA genome. The existence of various HPV genotypes with heterogeneous DNA genomes requires either type-specific or broad-spectrum detection of HPV DNA. Type-specific PCR detection would involve a large number of separate PCR assays for each individual clinical sample and is therefore impractical for routine diagnosis. Several broad-spectrum PCR primer sets have been described for detection of multiple HPV types by a single PCR assay. 16-18,29 However, none of the currently available universal primer sets is able to effectively detect all anogenital HPV genotypes, 21,30 and recent studies have shown that the overall prevalence of HPV can be underestimated considerably if only a single DNA detection method is used. 14,20,30,31
Development of a Novel HPV DNA Detection System
Alignment of a large number of L1-region sequences permitted the design of a novel set of broad-spectrum primers. The amplimer size of SPF1/2 was only 65 bp, whereas the amplimer sizes of GP5+/6+ and My11/09 were 150 and 450 bp, respectively. In general, the sensitivity of DNA detection by PCR is inversely related to the size of the amplimer, as the kinetics of the PCR reaction favor smaller amplimers. The importance of small amplimers has also been shown for the detection of the hepatitis C virus. 32 Moreover, as the efficacy of PCR also depends on the quality of the DNA extracted from the clinical specimen, detection with small amplimers will be advantageous, especially when using formalin-fixed, paraffin-embedded materials. 14,33 Taken together, the small amplimer size may explain the high sensitivity of the SPF assay.
The other universal primer sets located in the L1 open reading frame either use degenerated primers (My11/09) 16 or consensus primers (GP5+/6+). 19 The novel SPF system employs a mixture of defined primers for the SPF1 and SPF2 target regions. These regions are also used for My11 and GP5+ priming, respectively. As the heterogeneity of the primer target sites is limited, this mixture of primers may be more effective than consensus or degenerated primers. The 3′ terminus of a primer target region is crucial for the efficacy of the PCR primer. From the sequence alignment (Figure 1) ▶ , it is clear that the 3′ ends of SPF1 and SPF2 primers are both highly conserved among all HPV genotypes.
The SPF1/2 primers permitted effective detection of genomic DNA of 34 different anogenital HPV genotypes from plasmid clones. In addition, the SPF1/2 primers also allowed detection of cutaneous HPV genotypes 3, 4, 5, 8, 27, 32, 37, 65, and 71 (data not shown). Sequences of the 22-bp interprimer region were completely concordant with the published sequence for each of the HPV genotypes. This confirms the effective detection of a broad spectrum of HPV genotypes by the novel SPF assay.
For several HPV genotypes, the sensitivity of the SPF primer set was investigated. Results indicated that the SPF-mediated PCR was more or equally sensitive than GP5+/6+ and My11/09 for HPV 16, 35, and 45. My11/09 is less sensitive for HPV types 35 and 45, whereas GP5+/6+ is less sensitive for type 45 as compared with SPF1/2. These results indicated the high sensitivity of SPF1/2, whereas other general primer sets may be less sensitive for specific genotypes, as has been reported earlier. 21
Clinical Evaluation of the SPF System
The performance of the SPF1/2 primers was evaluated by testing a large number of clinical specimens, representing clinically different patient categories.
Among group 1 patients, with the majority of cases classified as normal cytology, the HPV detection rate using the SPF1/2 primers was significantly higher than with the GP5+/6+ primers. Semiquantitative studies have shown that the viral load is associated with the cytological grade of abnormality, with the lowest level of HPV DNA in scrapes with normal cytology and ASCUS. 30 These results suggest that the SPF assay is able to detect low concentrations of HPV DNA in clinical samples and confirms the higher sensitivity of the SPF primer set as compared with GP5+/6+ primers for a broad range of HPV genotypes.
The detection rate of the SPF1/2 primer set among the second group of patients with scrapes classified as dyskaryotic was close to 100%. Materials from this group had been analyzed by the GP5/6 primer set earlier, 23 with a detection rate of only 63.6% and 82.6% among mild or moderate dyskaryotic and severe dyskaryotic cases, respectively. Again this confirmed the high sensitivity of the SPF1/2 primers. These samples have not been tested with the adapted GP5+/6+ primers, which should have a higher sensitivity than GP5/6. 19 Among scrapes classified as mild or moderate dyskaryotic, the HPV DNA detection rate, as determined by various universal PCR primer sets, ranged from 71% to 76%. 30,34,35 Similarly, among scrapes classified as severe dyskaryotic, the HPV DNA detection rate ranged from 86% to 94%. 30,35 The HPV DNA detection rate of the SPF1/2 primer set was considerably higher in both groups, indicating the high sensitivity of this novel method.
Among cervical carcinoma specimens, HPV DNA has been generally found in approximately 70% of the cases using direct DNA detection by Southern blot analysis or type-specific PCR. 36,37 Using a combination of multiple universal PCR primers, HPV DNA can be detected in up to 90% of the carcinomas. 20,29 There are several explanations for the failure to detect HPV DNA in all cervical carcinomas. It has been postulated that truly HPV-DNA-negative cervical carcinomas exist, but the prevalence is considered to be very low. 38 Linear integration of the circular HPV DNA genome into the chromosomal DNA of the host cell may affect the PCR target region, although the preferred cleavage integration site is located in the E2 region. 39 Uneven distribution of HPV in the carcinoma tissue may lead to sampling errors, yielding false negative PCR results. Most likely, however, the sensitivity of the current PCR systems may be too limited to detect very low concentrations of HPV DNA in 100% of the 184 formalin-fixed, paraffin-embedded cervical cancer biopsies presented in patient group 3. The high efficacy of the SPF primer set was also shown by comparison with results of other general HPV primer sets in these materials. Among 30 samples that remained negative by HPV-16, -18, -31, and -33 type-specific PCR and positive by the SPF assay, HPV DNA was detected in only three cases by the general primer sets My11/My09 and/or GP5+/6+ (data not shown). This confirmed the high HPV DNA detection rate of the SPF primer set in formalin-fixed, paraffin-embedded cervical carcinomas, most likely due to the very small amplimer size.
Association between HPV Genotypes and Severity of Morphological Changes
The relative risk association of the different HPV types is still disputed. Based on the observed distribution of genotypes in low- and high-grade lesions and carcinomas, HPV genotypes have been classified into high-risk, intermediate-risk, or low-risk groups. 40 However, detection and identification of HPV types in various clinical materials strongly depends on the methods used. Direct hybridization methods have a relatively low detection level whereas universal PCR primer sets might have differential sensitivity for different HPV DNA genotypes. 21 Therefore, the data obtained so far from epidemiological studies may be incomplete and not completely accurate. The general primer sets My11/09 and GP5+/6+ are both aimed at the L1 region, which is one of the more conserved parts of the HPV genome. Classification of HPV genotypes as well as identification of novel genotypes is based on phylogenetic analysis of the 450-bp PCR fragment generated by My11/My09 primers. 8 Direct sequencing analysis of SPF amplimers obtained from reference HPV genomic clones confirmed the distinct heterogeneity of the 22-bp interprimer sequence. Consistent sequence variation in this region can be used to recognize different HPV genotypes. However, formal genotyping analysis of HPV DNA, based exclusively on sequence variation of the SPF1/2 amplimer, needs to be confirmed by phylogenetic analysis. Preliminary data from more extensive sequence analysis of the L1 region indicate that genotype recognition in the SPF1/2 fragment is highly reliable (data not shown). Comparison of the HPV genotype distribution, based on analysis of the 22-bp SPF interprimer sequences, revealed some interesting differences between patient groups with different degrees of cytological abnormalities. Among group 1 patients with normal cytology or ASCUS, the prevalence of low-risk HPV genotypes was approximately 24%. By contrast, in patients belonging to groups 2 and 3 with dyskaryosis or carcinoma, these low-risk genotypes were virtually absent, and all cases contained high-risk genotypes. A single carcinoma case appeared to contain HPV genotype 55. Detection of this potentially low-risk type by direct sequencing could be caused by a mixed infection with a high-risk type, but HPV genotype 55 has been observed in carcinoma earlier. 9 Also, the number of 22-bp interprimer sequences that did not exactly match any of the known HPV genotypes, was significantly higher among patients with normal cytology or ASCUS than among patients with at least dyskaryosis. These sequences may either represent novel HPV genotypes or variants of known HPV genotypes. This will be the subject of additional, more extensive phylogenetic studies. Overall, the frequency distribution of HPV genotypes appeared to be consistent with the hypothesis that patients can be initially infected with a broad range of HPV genotypes, but only those infected with potentially high-risk HPV genotypes are at risk for carcinoma.
In conclusion, the present study describes a novel PCR primer set for the detection of a broad spectrum of HPV genotypes. At present, the SPF1/2 primers have been shown to efficiently amplify sequences from at least 43 different HPV genotypes. These results indicated that the SPF1/2 primer set permits highly sensitive detection of a broad range of HPV genotypes. The sequence variation in the amplified fragment probably permits consistent identification of HPV genotypes. PCR mediated by the SPF1/2 primer set seems to be highly sensitive and may facilitate future studies on the clinical importance of HPV infections.
Acknowledgments
We thank Nathalie Nouhan for technical assistance, Jannie Baars, Miranda van Asselt, Cora Buis, and Nel Francka for providing data on the cytological analysis, Dr. Frank Smedts for histological analysis, Dr. Semyon Petrov for providing the carcinoma biopsies, Dr. Ron Berkhout for scientific discussions, and Fred Shapiro for manuscript editing.
Footnotes
Address reprint requests to Dr. Bernhard Kleter, Delft Diagnostic Laboratory, R. de Graafweg 7, PO Box 5100, 2600 GA Delft, The Netherlands.
References
- 1.Parkin DM, Laara E, Muir CS: Estimates of the worldwide frequency of sixteen major cancers in 1980. Int J Cancer 1988, 41:184-197 [DOI] [PubMed] [Google Scholar]
- 2.Parkin DM, Pisani P, Ferlay J: Estimates of the worldwide incidence of eighteen major cancers in 1985. Int J Cancer 1993, 54:606. [DOI] [PubMed] [Google Scholar]
- 3.Burger MP, Hollema H, Gouw AS, Pieters WJ, Quint WG: Cigarette smoking and human papillomavirus in patients with reported cervical cytological abnormality. Br Med J 1993, 306:749-752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carson LF, Twiggs LB, Fukushima M, Ostrow RS, Faras AJ, Okagaki T: Human genital papilloma infections: an evaluation of immunologic competence in the genital neoplasia-papilloma syndrome. Am J Obstet Gynecol 1986, 155:784-789 [DOI] [PubMed] [Google Scholar]
- 5.Gopalkrishna V, Murthy NS, Sharma JK, Roy M, Das DK, Luthra UK, Das BC: Increased human papillomavirus infection with the increasing number of pregnancies in Indian women. J Infect Dis 1995, 171:254-255 [DOI] [PubMed] [Google Scholar]
- 6.Herrero R, Brinton LA, Reeves WC, Brenes MM, Tenorio F, de BR, Gaitan E, Garcia M, Rawls WE: Sexual behavior, venereal diseases, hygiene practices, and invasive cervical cancer in a high-risk population. Cancer 1990, 65:380-386 [DOI] [PubMed] [Google Scholar]
- 7.zur Hausen H: Condylomata acuminata and human genital cancer. Cancer Res 1976, 36:794. [PubMed] [Google Scholar]
- 8.zur Hausen H: Papillomavirus infections: a major cause of human cancers. Biochim Biophys Acta 1996, 1288:F55-F78 [DOI] [PubMed] [Google Scholar]
- 9.Bosch FX, Manos MM, Munoz N, Sherman M, Jansen AM, Peto J, Schiffman MH, Moreno V, Kurman R, Shah KV: Prevalence of human papillomavirus in cervical cancer: a worldwide perspective. International Biological Study on Cervical Cancer (IBSCC) Study Group. J Natl Cancer Inst 1995, 87:796-802 [DOI] [PubMed] [Google Scholar]
- 10.Cuzick J, Sasieni P, Singer A: Risk factors for invasive cervix cancer in young women. Eur J Cancer 1996, 32A:836-841 [DOI] [PubMed] [Google Scholar]
- 11.Cuzick J, Szarewski A, Terry G, Ho L, Hanby A, Maddox P, Anderson M, Kocjan G, Steele ST, Guillebaud J: Human papillomavirus testing in primary cervical screening. Lancet 1995, 345:1533-1536 [DOI] [PubMed] [Google Scholar]
- 12.Melchers WJ, Herbrink P, Quint WG, Walboomers JM, Meijer CJ, Lindeman J: Prevalence of genital HPV infections in a regularly screened population in The Netherlands in relation to cervical cytology. J Med Virol 1988, 25:11-16 [DOI] [PubMed] [Google Scholar]
- 13.Melchers WJ, Herbrink P, Walboomers JM, Meijer CJ, van der Drift H, Lindeman J, Quint WG: Optimization of human papillomavirus genotype detection in cervical scrapes by a modified filter in situ hybridization test. J Clin Microbiol 1989, 27:106-110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Baay MF, Quint WG, Koudstaal J, Hollema H, Duk JM, Burger MP, Stolz E, Herbrink P: Comprehensive study of several general and type-specific primer pairs for detection of human papillomavirus DNA by PCR in paraffin-embedded cervical carcinomas. J Clin Microbiol 1996, 34:745-747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van den Brule AJ, Claas EC, Du Maine M, Melchers WJ, Helmerhorst T, Quint WG, Lindeman J, Meijer CJ, Walboomers JM: Use of anticontamination primers in the polymerase chain reaction for the detection of human papilloma virus genotypes in cervical scrapes and biopsies. J Med Virol 1989, 29:20-27 [DOI] [PubMed] [Google Scholar]
- 16.Manos MM, Ting Y, Wright DK, Lewis AJ, Broker TR, Wolinsky SM: Use of polymerase chain reaction amplification for detection of genital human papillomavirus. Cancer Cells 1989, 7:209-214 [Google Scholar]
- 17.Jenkins A, Kristiansen BE, Ask E, Oskarsen B, Kristiansen E, Lindqvist B, Trope C, Kjorstad K: Detection of genital papillomavirus types by polymerase chain reaction using common primers. APMIS 1991, 99:667-673 [DOI] [PubMed] [Google Scholar]
- 18.Tieben LM, ter Schegget J, Minnaar RP, Bouwes Bavinck J, Berkhout RJ, Vermeer BJ, Jebbink MF, Smits HL: Detection of cutaneous and genital HPV types in clinical samples by PCR using consensus primers. J Virol Methods 1993, 42:265-279 [DOI] [PubMed] [Google Scholar]
- 19.de Roda Husman AM, Walboomers JM, van den Brule AJ, Meijer CJ, Snijders PJ: The use of general primers GP5 and GP6 elongated at their 3′ ends with adjacent highly conserved sequences improves human papillomavirus detection by PCR. J Gen Virol 1995, 76:1057-1062 [DOI] [PubMed] [Google Scholar]
- 20.Karlsen F, Kalantari M, Jenkins A, Pettersen E, Kristensen G, Holm R, Johansson B, Hagmar B: Use of multiple PCR primer sets for optimal detection of human papillomavirus. J Clin Microbiol 1996, 34:2095-2100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Qu W, Jiang G, Cruz Y, Chang CJ, Ho GY, Klein RS, Burk RD: PCR detection of human papillomavirus: comparison between MY09/My11 and GP5+/6+ primer systems. J Clin Microbiol 1997, 35:1304-1310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zehbe I, Wilander E: Two consensus primer systems and nested polymerase chain reaction for human papillomavirus detection in cervical biopsies: a study of sensitivity. Hum Pathol 1996, 27:812-815 [DOI] [PubMed] [Google Scholar]
- 23.Burger MP, Hollema H, Pieters WJ, Quint WG: Predictive value of human papillomavirus type for histological diagnosis of women with cervical cytological abnormalities. Br Med J 1995, 310:94-95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.ter Harmsel B, van Muyden R, Smedts F, Hermans J, Kuijpers J, Raikhlin N, Petrov S, Ledebev A, Ramaekers F, Trimbos B: The significance of cell type and tumor growth markers in the prognosis of unscreened cervical cancer patients. Int J Gyn Cancer 1998, 8:336-344 [Google Scholar]
- 25.Claas EC, Melchers WJ, van der Linden H, Lindeman J, Quint WG: Human papillomavirus detection in paraffin-embedded cervical carcinomas and metastases of the carcinomas by the polymerase chain reaction. Am J Pathol 1989, 135:703-709 [PMC free article] [PubMed] [Google Scholar]
- 26.Sambrook J, Fritsch IF, Maniatis T: Molecular Cloning: A Laboratory Manual, 2. 1989. NY, Cold Spring Harbor Laboratory Press, Cold Spring Harbor
- 27.Altschul SF, Gish W, Miller W, Meyers EW, Lipman DJ: Basic local alignment search tool. J Mol Biol 1990, 215:403-410 [DOI] [PubMed] [Google Scholar]
- 28.Koss LG: The Papanicolaou test for cervical cancer detection: a triumph or a tragedy. J Am Med Assoc 1989, 261:737-743 [PubMed] [Google Scholar]
- 29.van den Brule AJ, Snijders PJ, Gordijn RL, Bleker OP, Meijer CJ, Walboomers JM: General primer-mediated polymerase chain reaction permits the detection of sequenced and still unsequenced human papillomavirus genotypes in cervical scrapes and carcinomas. Int J Cancer 1990, 45:644-649 [DOI] [PubMed] [Google Scholar]
- 30.Smits HL, Bollen LJ, Tjong-A-Hung SP, Vonk J, van der Velden J, ten Kate FJ, Kaan JA, Mol BW, ter Schegget J: Intermethod variation in detection of human papillomavirus DNA in cervical smears. J Clin Microbiol 1995, 33:2631-2636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smits HL: The use of consensus PCR and direct sequence analysis for the identification of HPV. Methods Mol Biol 1996, 50:109-126 [DOI] [PubMed] [Google Scholar]
- 32.Garson JA, Ring CJA, Tuke PW: Improvement of HCV genome detection with “short” PCR products. Lancet 1991, 338:1466-1467 [DOI] [PubMed] [Google Scholar]
- 33.Ohara Y, Honma M, Iwasaki Y: Sensitivity of the polymerase chain reaction for detecting human T-cell leukemia virus type I sequences in paraffin-embedded tissue: effect of unbuffered formalin fixation. J Virol Methods 1992, 37:83-88 [DOI] [PubMed] [Google Scholar]
- 34.Bollen LJ, Tjong-A-Hung SP, van der Velden P, Brouwer K, Mol BW, ten Kate FJ, ter Schegget J: Human papillomavirus deoxyribonucleic acid detection in mildly or moderately dysplastic smears: a possible method for selecting patients for colposcopy. Am J Obstet Gynecol 1997, 177:548-553 [DOI] [PubMed] [Google Scholar]
- 35.Jacobs MV, Snijders PJ, van den Brule AJ, Helmerhorst TJ, Meijer CJ, Walboomers JM: A general primer GP5+/6+(+)-mediated PCR-enzyme immunoassay method for rapid detection of 14 high-risk and 6 low-risk human papillomavirus genotypes in cervical scrapings. J Clin Microbiol 1997, 35:791-795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ikenberg H, Sauerbrei W, Schottmuller U, Spitz C, Pfleiderer A: Human papillomavirus DNA in cervical carcinoma: correlation with clinical data and influence on prognosis. Int J Cancer 1994, 59:322-326 [DOI] [PubMed] [Google Scholar]
- 37.Kristiansen E, Jenkins A, Holm R: Coexistence of episomal and integrated HPV16 DNA in squamous cell carcinoma of the cervix. J Clin Pathol 1994, 47:253-256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.zur Hausen H, De Barahona O: Human papillomaviruses. Annu Rev Microbiol 1994, 48:427-447 [DOI] [PubMed] [Google Scholar]
- 39.Schwarz E, Freese UK, Gissmann L, Mayer W, Roggenbuck B, Stremlau A, zur Hausen H: Structure and transcription of human papillomavirus sequences in cervical carcinoma cells. Nature 1985, 314:111-114 [DOI] [PubMed] [Google Scholar]
- 40.Lorincz AT, Reid R, Jenson AB, Greenberg MD, Lancaster W, Kurman RJ: Human papillomavirus infection of the cervix: relative risk associations of 15 common anogenital types. Obstet Gynecol 1992, 79:328-337 [DOI] [PubMed] [Google Scholar]