Abstract
By applying graph representations to biochemical pathways, a new computational pipeline is proposed to find potential operons in microbial genomes. The algorithm relies on the fact that enzyme genes in operons tend to catalyze successive reactions in metabolic pathways. We applied this algorithm to 42 microbial genomes to identify putative operon structures. The predicted operons from Escherichia coli were compared with a selected metabolism-related operon dataset from the RegulonDB database, yielding a prediction sensitivity (89%) and specificity (87%) relative to this dataset. Several examples of detected operons are given and analyzed. Modular gene cluster transfer and operon fusion are observed. A further use of predicted operon data to assign function to putative genes was suggested and, as an example, a previous putative gene (MJ1604) from Methanococcus jannaschii is now annotated as a phosphofructokinase, which was regarded previously as a missing enzyme in this organism. GC content changes in the operon region and nonoperon region were examined. The results reveal a clear GC content transition at the boundaries of putative operons. We looked further into the conservation of operons across genomes. A trp operon alignment is analyzed in depth to show gene loss and rearrangement in different organisms during operon evolution.
The increasing availability of sequenced microbial genomes enables us to perform high-throughput computational analysis with increasing predictive accuracy. It has been observed both experimentally and computationally that genes in microbial genomes tend to form modular functional units that are conserved during evolution (Tamames et al. 1997; Overbeek et al. 1999; Ettema et al. 2001). Operon structures are known to be an important family among these conserved functionally related genomic units. Moreover, these units often appear in multiple genomes and perform highly compartmentalized activity in biochemical pathways. In this work, we will describe a method to automatically detect neighboring enzyme clusters in the genome that catalyze successive chemical reactions in the metabolic pathways. These neighboring enzyme clusters have been shown to be candidate operons (Ogata et al. 2000). Because the proximity of functionally related genes gains efficiency by coordinating activities and regulation, it is encouraged by selection. By group regulation of certain highly expressed metabolic genes needed under specific growth conditions, microorganisms can minimize their energy expenditure. In most bacterial genomes, functionally coupled gene clusters are often regulated under the same upstream promoter, forming a polycistronic transcribed operon unit with associated regulatory sites. Loss or disruption of such proximity may decrease metabolic flux and have deleterious consequences in individual survival. Experimental detection or confirmation of operons is time-consuming (Walters et al. 2001) and relatively difficult to implement in the laboratory as a high-throughput process. Computational methods to reconstruct metabolic mechanisms of newly sequenced microbial organisms have gained increased attention in recent years (Selkov et al. 1997a; Bono et al. 1998). Computational identification of operons using the currently available sequence data and computerized knowledge representation of biochemical rules is thus helpful in extracting important compartmentalized features of metabolic pathways in different organisms.
Several computational algorithms for operon modeling and prediction have been suggested recently, mostly based on the model organism Escherichia coli (Yada et al. 1999; Salgado et al. 2000; Ermolaeva et al. 2001), in which many promoters and terminators are well known. Many of them rely on modeling of sequence motifs in promoter and terminator sites. These methods are less effective when promoter or terminator sequences are not well conserved. It has been pointed out that these sequences are not necessarily fully conserved during evolution among microbial genomes (Itoh et al. 1999). Other methods combine the observation that operons have much shorter intergenic distances than genes at the borders of transcription units with the functional category assignments (Selkov et al. 1997a). Another natural approach for this problem is through the mechanism of validating functional clusters by the frequency of their appearance in multiple genomes (Ermolaeva et al. 2001).
Alternatively, genes in operons interact with each other either by physical assembly of their products or through participating networked reactions in cells, so that they are likely to be involved in closely related biological processes. The knowledge of metabolic biochemical pathways offers us an appealing universal methodology for inferring such functional coupling. Computationally, the problem is reduced to developing an effective algorithm for detecting a correspondence between a possibly large metabolic pathway and a gene cluster. More precisely, we need to detect a subgraph in a pathway map in which the genes encoding the major players appear in a gene cluster on the genome. Fortunately, several groups have generated easily accessible pathway network data, which enables us to compute functional clusters by using pathway databases and graph algorithms (Karp et al 1996; Selkov et al. 1997b; Kanehisa and Goto 2000). In a seminal effort led by M. Kanehisa, a heuristic graph comparison method has been proposed by Ogata et al. (2000) for identifying functional clusters by detecting correspondences in the genome graph and the pathway graph.
In this work, we report a conceptually simple computational method that could be used to detect metabolism-related operons with high accuracy. We provide a summary of our results with predicted operons for many of the currently annotated organisms. The complete list of putative operons for microorganisms that we analyzed is available over the Internet (http://genomics4.bu.edu/operons). In addition, we also documented the applicability of this method for functional annotation of sandwich or gap genes that occur in the middle of putative operons (see our website http://genomics4.bu.edu/operons/gappedgene).
For theoretical completeness, we report on the computational complexity of detecting functionally related gene clusters. In particular, we provide a formal proof that several versions of the problem of detecting functionally related clusters are NP-complete. Finally, we discuss the applicability of these and more general methods for functional annotation of genes and gene clusters.
RESULTS
Analysis of Operon Data in E. coli
E. coli is a well-studied organism, which we can use to evaluate the performance of our method. The RegulonDB database by Huerta et al. (1998) integrates knowledge of transcription regulatory signals of E. coli. It has been reported that the total number of currently known operons in E. coli is 237 (Salgado et al. 2000), from which we compiled a set of 128 (54%) metabolism-related operons as our test set (91 of them are experimentally verified). Our method predicted 124 operons, which includes 114 of the 128 real operons (sensitivity = 89%). Of the 124 predicted operons, 108 (specificity = 87%) share at least two enzymes with operons in the test set. The number of exact correspondences is 55, ∼44% of all predictions.
Compared with the FREC (functionally related enzyme clusters) results by Ogata et al.(2000), our method detects more possible operons, including the example shown in Figure 1. Another example that we found, the Men operon in E. coli, is shown in Figure 2. Five contiguous genes catalyze successive reactions in the ubiquinone biosynthetic pathway. The graph comparison method (Ogata et al. 2000) only detects two genes (b2264c and b2265c) from the whole operon.
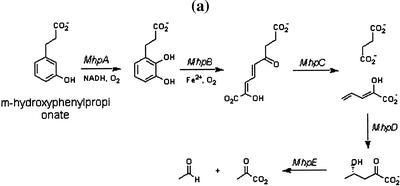
Figure 1.
(a) Phenylpropionate catabolic pathway, mhpABCDE catalyzes successive reactions. (b) (Subsets of genes in the mhp operon involved in different pathways left) and the actual reaction chains in the pathways catalyzed by these genes (right). This figure gives an example where computing transitive closure of smaller operons on the chromosome gives a larger operon. All genes run in the same direction.
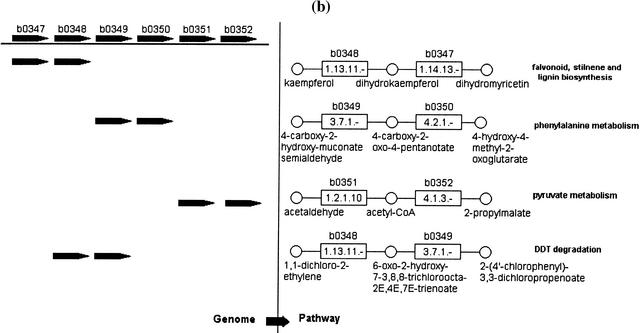
Figure 2.
Men operon in E. coli. Part of the ubiquinone biosynthetic pathway is shown on the left. The genomic region containing this operon is shown on the right. Inside the enzyme nodes (rectangle) of the pathway, the names of the matched genes are shown in brackets, e.g., b2264 encodes a bifunctional protein with two enzymatic activities (Palaniappan et al. 1992). The gene filled with gray (b2263) encodes a product that is currently annotated as a hypothetical protein.
The size distribution of predicted E. coli operons is shown in Figure 3. According to a previous study (Overbeek et al. 1999), the number of predicted operons diminishes with the length of the operons roughly according to a Poisson distribution. Prediction accuracy, however, increases with the length of the operons (data not shown). The larger a predicted gene cluster is, the more likely it is a real operon. Most predicted operons from randomly shuffled genomes are of size 2.
Figure 3.
Distribution of operon length in E. coli. The solid line shows the distribution of operon length in the E. coli genome. The broken line shows the distribution in the randomly shuffled E. coli genome. (inset) A normalized histogram of operon length distribution in E. coli.
Operons in 42 Microbial Organisms
The results from 42 microbial organisms are shown in Table 1. The entire predicted operon list for each organism can be accessed on http://genomics4.bu.edu/operons. The average number of operons obtained from random shuffling experiments for each genome is also shown in Table 1. The number of operons and their composition partly reflects the current status of genomic annotation. In E. coli and Bacillus subtilis, the two best-studied bacteria, the number of predicted operons is naturally high. The quality of our detection procedure depends on our current knowledge of metabolic pathways, and our results are necessarily biased toward the organisms with well-documented pathways.
Table 1.
Summary of Results for 42 Organisms
| Species | No. of predicted operons | Genome size (Mb) | No. of enzymes in operons/No. of total enzymes* | Ratio of enzymes of operons | Average length of operons | Number of operons in a shuffled genome | P-value |
|---|---|---|---|---|---|---|---|
| E. coli | 124 | 4.7 | 374/562 | 0.66 | 3.0 | 22 | 0.001 |
| H. influenzae | 52 | 1.8 | 160/293 | 0.54 | 3.1 | 11 | 0.006 |
| X. fastidiosa | 42 | 2.7 | 148/365 | 0.39 | 3.5 | 5 | 0.005 |
| V. cholerae | 80 | 4.0 | 237/562 | 0.42 | 3.0 | 11 | 0.001 |
| P. aeruginosa | 101 | 6.4 | 290/715 | 0.41 | 2.9 | 14 | 0.002 |
| Buchnera sp APS | 30 | 0.7 | 122/224 | 0.54 | 4.1 | 8 | 0.02 |
| P. multocida | 57 | 2.3 | 169/418 | 0.40 | 3.0 | 12 | 0.004 |
| N. meningitidis B | 38 | 2.3 | 134/404 | 0.33 | 3.5 | 8 | 0.007 |
| H. pylori | 25 | 1.7 | 81/280 | 0.29 | 3.2 | 6 | 0.02 |
| C. jejuni | 33 | 1.7 | 112/335 | 0.33 | 3.4 | 8 | 0.009 |
| R. prowazekii | 25 | 1.1 | 71/182 | 0.39 | 2.8 | 6 | 0.007 |
| M. lot̂i | 88 | 7.1 | 260/729 | 0.36 | 3.0 | 8 | 0.003 |
| C. crescentus | 48 | 4.1 | 135/372 | 0.36 | 2.8 | 5 | 0.004 |
| B. subtilis | 105 | 4.3 | 323/510 | 0.63 | 3.1 | 13 | 0.001 |
| B. halodurans | 98 | 4.3 | 308/563 | 0.55 | 3.1 | 13 | 0.001 |
| M. genitalium | 13 | 0.6 | 40/86 | 0.47 | 3.1 | 5 | 0.10 |
| M. pneumoniae | 17 | 0.8 | 50/117 | 0.43 | 2.9 | 6 | 0.03 |
| M. pulmonis | 19 | 1.0 | 60/116 | 0.52 | 3.2 | 4 | 0.01 |
| U. urealyticum | 11 | 0.8 | 33/101 | 0.33 | 3.0 | 3 | 0.04 |
| L. lactis | 62 | 2.4 | 203/367 | 0.55 | 3.3 | 9 | 0.002 |
| S. pyogenes | 46 | 1.9 | 152/283 | 0.54 | 3.3 | 10 | 0.001 |
| S. aureus Mu50 | 6 | 2.9 | 21/43 | 0.49 | 3.5 | 0 | 0.005 |
| M. tuberculosis | 89 | 4.5 | 266/591 | 0.45 | 3.0 | 16 | 0.003 |
| M. leprae | 47 | 3.3 | 134/326 | 0.41 | 2.9 | 4 | 0.002 |
| C. trachomatis | 27 | 1.0 | 81/187 | 0.43 | 3.0 | 5 | 0.007 |
| C. pneumoniae | 24 | 1.2 | 75/190 | 0.39 | 3.1 | 4 | 0.007 |
| B. burgdorferi | 20 | 1.5 | 50/138 | 0.36 | 2.5 | 2 | 0.001 |
| T. pallidum | 15 | 1.2 | 44/152 | 0.29 | 2.9 | 5 | 0.03 |
| Synechocystis | 24 | 3.6 | 59/453 | 0.13 | 2.5 | 8 | 0.02 |
| D. radiodurans | 46 | 2.7 | 136/434 | 0.31 | 3.0 | 7 | 0.002 |
| A. aeolicus | 31 | 1.6 | 90/387 | 0.23 | 2.9 | 10 | 0.01 |
| T. maritima | 42 | 1.9 | 172/368 | 0.47 | 4.1 | 7 | 0.005 |
| M. jannaschii | 28 | 1.7 | 96/274 | 0.35 | 3.4 | 9 | 0.01 |
| M. thermoautotrophicum | 46 | 1.8 | 164/349 | 0.47 | 3.6 | 10 | 0.006 |
| A. fulgidus | 59 | 2.2 | 178/406 | 0.44 | 3.0 | 13 | 0.003 |
| T. acidophilum | 34 | 1.6 | 107/271 | 0.39 | 3.2 | 8 | 0.03 |
| T. volcanium | 35 | 1.6 | 116/277 | 0.42 | 3.3 | 7 | 0.006 |
| P. horikoshii | 23 | 1.8 | 80/231 | 0.35 | 3.5 | 4 | 0.01 |
| P. abyssi | 30 | 1.8 | 125/280 | 0.45 | 4.2 | 6 | 0.009 |
| A. pernix | 33 | 1.7 | 95/274 | 0.37 | 2.9 | 4 | 0.004 |
| S. solfataricus | 59 | 3.0 | 190/458 | 0.41 | 3.2 | 11 | 0.007 |
| S. cerevisiae | 33 | 13 | 74/692 | 0.11 | 2.2 | 16 | 0.047 |
The total number of enzymes that are involved in the metabolic pathways.
We analyzed the GC content changes between genes both inside the operons and at the boundaries of the operons. The results for B. subtilis and E. coli are shown below in Figure 4. There are extended tails (marked in the ellipses in the figure) in the histogram of boundary GC content changes for both organisms, which reveal jumps of GC content at the boundaries of the operons. At the same time, GC content between genes inside operons does not change much (mostly <0.05). There are several possible explanations for the higher GC content changes at the boundaries of operons, for example, possible horizontal transfer of operons from other organisms (Lawrence 1997), or the larger intergenic regions between operons other than inside operons.
Figure 4.
GC content change inside operons and at operon boundaries. Histogram of GC content change (x-axis, 0 ∼1) in operon and boundary regions in E. coli (a) and B. subtilis (b). GC content change was computed from two genes next to each other. If both of them are inside an operon, it is counted as inside operons. If either one is outside of the operon, it is counted as at the boundaries. GC content change is calculated for each gene. Ellipses mark the high GC transitions at the boundaries of the operons.
Interestingly, a small number of predicted operons are reported with gene gaps inside, which we named sandwich genes or gap genes. Analysis of these gap genes suggests that they might fall into a similar broad functional category with their gene neighbors. In cases in which a gap gene could not be annotated by a conventional sequence comparison method, identification of an operon around it may help us unravel its functional role. An example of this methodology is the functional interpretation of MJ1604 in the Methanococcus jannaschi genome. The current annotation status for MJ1604 is hypothetical protein. From our results, we found that it is located inside of the detected operon (MJ1603, MJ1605). These two genes encode two key enzymes in the pentose phosphate pathway, ribose 5-phosphate isomerase (MJ1603) and glucose-6-phosphate isomerase (MJ1605). We suspected that MJ1604 might also be involved in the pentose phosphate pathway. In addition, we also knew that another key enzyme in the pentose phosphate pathway, phosphofructokinase, was reported missing in the M. jannaschi genome by Bult et al. (1996). With these observations in mind, we performed a BLASTP search (Altschul et al. 1997). The top three hits with bitscore >400 and E-value <1E-100 are ADP-dependent phosphofructokinase in Thermococcus zilligii (Genbank AAF97356), ADP-dependent phosphofructokinase in Thermococcous litoralis (Genbank BAB69952), and phosphofructokinase in Pyrococcus furiosus (Genbank AAD48400). This immediately indicates that MJ1604 is likely to be the M. jannaschii phosphofructokinase. Thus, the gene cluster MJ1603, MJ1604, and MJ1605 almost certainly encodes three key enzymes and catalyzes successive reactions in the pentose phosphate pathway. The homologous sequences reported by BLAST are new sequences that were not available at the time of the original annotation. The annotation based on gene location in operons agrees with the annotation obtained by sequence comparison.
There are also completely unknown genes that appear inside operons. This provides clues leading to the possible annotations of these unknown genes. One such example is an operon reported in Archaeoglobus fulgidus encoding H+-transporting ATP synthetase (AF1159, AF1160, AF1161, AF1162, AF1163, AF1164, AF1165, AF1166, AF1167, AF1168, AF1169). Except for the unknown gene AF1161 (pid, 2649430), every other gene encodes a subunit of this H+-transporting ATP synthetase complex. AF1161 does not have any homologous sequences based on a BLAST search in the current nonredundant database, whereas the location of this gene suggests the possibility that it encodes another subunit of this complex if it is not the result of a sequencing error or gene-finding error. Further analysis shows that although this long gene cluster is highly conserved in other microorganisms, gene AF1161 is not conserved with the other genes, which, in turn, suggests that AF1161 might encode a subunit that is only present in A. fulgidus.
We observed that the average length of operons (= number of enzymes that participate in operons/number of operons) remains a constant around 3 in most of the genomes. The average length of operons can be taken as a measure of the degree of modularity of biochemical pathways in the genomes. For the randomly shuffled genome of E. coli, the average length is close to 2.0, because it is the shortest the algorithm could identify. For bacteria with high average length, for example, E. coli, B. subtilis, and Buchnera, it suggests that their genomes are highly organized into operons compared with other bacteria. However, other bacteria, for example, B. burgdorferi and Synechocystis have smaller average operon lengths, suggesting that their genomes have undergone more frequent gene translocations or that they contain many enzymes in operons that are currently unannotated.
Conservation of Operons Across Organisms
We then studied the conservation of operons across genomes. Operon alignment provides us with an informative tool to identify orthologous relationships between genes. Additionally, operon alignment may help us understand the evolution of operon structure and evolutionary transformations between microorganisms. To show the utility of this approach, we constructed a trp operon alignment using the operon database we built. The trp operon controls the biosynthesis of tryptophan in the cell from the initial precursor chorismic acid. TrpE and trpD produce anthranilate synthetase, an enzyme catalyzing the first two reactions in the trypotophan pathway. TrpC produces indole glycerolphosphate synthetase, which is responsible for catalyzing the next two steps in the pathway. TrpA and trpB produce tryptophan synthetase, which catalyzes the final step in the pathway.
We examined 42 microorganisms and succeeded in identifying the trp operon in 26 of them. In these 26 organisms that contain the trp operon enzymes, the degree of conservation varies. Figure 5 shows an alignment of operons in several organisms. There are several interesting phenomena in this operon alignment. In E. coli, B. subtilis, and A. fulgidus, all of the trpABCDE genes lie in a single operon. Gene fusions were observed as follows: Anthranilate isomerase (EC:5.3.1.24) and indole-3-glycerol phosphate synthase (EC:4.1.1.48) are fused into one gene in Helicobacter pylori, Haemophilus influenzae, and Buchnera, but are separated in other organisms; anthranilate phosphoribosyltransferase (EC:2.4.2.18) are fused with anthranilate synthase (EC:4.1.3.27) in E. coli and indole-3-glycerol phosphate synthase (EC:4.1.1.48) in A. fulgidus. Alhough fusions are inferred from Enzyme Commission (EC) number comparison, subsequent sequence analysis using BLAST (Altschul et al. 1997) gave supportive evidence (data not shown) that they are real. Genes inside the trp operons often have overlapping open reading frames (ORFs), which provide a genetic basis for protein fusions. In E. coli, an attenuation sequence (trpL) is present, encoding a leader peptide. However, we did not find a similar sequence in any of the other organisms, suggesting the absence of the attenuation mechanism in trypotophan synthesis (Shigenobu et al. 2000). Aeropyrum pernix has a special trp gene cluster in which genes encoding trp-related enzymes are found on both strands. However, it is noteworthy that these genes all appear together on the genome. In H. influenzae, the entire trp operon is broken into two parts, which appear as two separate operons in the genome. This provides an example of proposed operon fusion, in which small operons can fuse into a larger operon. Unlike most gene fusions between protein domains, operon fusions are between gene clusters.
Figure 5.
trp operon alignment. Genes are drawn with directions (sharp end is a transcription stop) and are labeled with all the enzymatic activities they have. Genes colored gray are nonenzymes and are not inside the operon. The single line represents a DNA strand (5′ to 3′). Double lines represent both strands.
DISCUSSION
Automatic representation and computation of biochemical pathway knowledge has become a key research area in computational genomics in recent years. In particular, metabolic pathways in bacteria provide an example of how to abstract biochemical knowledge into common templates, namely, graph representations. Here, we report the application of an efficient graph algorithm to predict operons using metabolic pathways. This approach has been shown to be high-throughput and highly specific. The computational pipeline used in this work requires a genome enzyme catalog and documented pathway information. For a newly sequenced genome, the enzyme catalog can be acquired by identifying homologous relations with known genes via sequence comparison methodology. Moreover, the pipeline can provide a nonhomology-based tool to identify genes within operons that are candidates to encode missing enzyme of the pathway that cannot be annotated by the conventional homology method.
It is important to realize the crucial role of knowledge representation of biochemical rules (Fukuda et al. 2001), especially when one considers the plasticity of certain metabolic pathways in microorganisms (Dandekar et al. 1999). For highly divergent regulatory pathways or more complex biological processes in higher organisms, they may differ from one organism to another. Then, it becomes a challenge to make a generalization of pathways to perform high-throughput computation. More research needs to be done toward building a consistent infrastructure.
Operon identification helps us understand how genes are regulated as a group in bacterial genomes. Metabolic genes are commonly highly expressed in bacterial cells. By turning on and off a set of functionally related genes together instead of regulating them individually, operon structures help the cell cope with variations of environmental conditions and enhance the chances of cell survival. Our method helps identify putative operons related to metabolic processes in each microorganism. It opens a gate for investigating the process of gaining and losing of operons across genomes during evolution. In pathogens, this approach could create opportunities for identifying possible drug targets.
We provide a formal proof that identifying gene clusters by matching to a subgraph in a pathway is NP complete. Our result suggests (at least in theory) that the most general statement of the problem might be computationally difficult. In practice, however, we expect that we can take advantage of several constraints that make the cluster identification problem much easier. The most significant constraints are as follows: (1) the length of most functionally related gene clusters appears to be bounded by a small constant; (2) it is relatively rare that all genes appearing in a gene cluster have multiple matches in a pathway graph. If any of the genes in a cluster has a unique occurrence in a pathway graph, it immediately provides a strong constraint on the number of matching subgraphs that must be considered by the algorithm.
Consequently, a number of simple ideas can be used to solve the general problem optimally. For example, we could build a dictionary by hashing all possible connected subgraphs and find all possible partial matches during a linear time scan of the genome. Then, we can easily extend these partial matches to identify all maximally sized functionally related clusters (in a BLAST-like fashion).
Compared with other algorithms suggested earlier (Ogata et al. 2000), our algorithm is simpler to implement and appears to have at least comparable sensitivity and specificity. The running time of our algorithm is more or less linear with the total size of the pathway graph. This, of course, assumes that the genomes have been preprocessed previously and each position of a gene has been indexed. We took a heuristic approach to reduce the computational complexity by making several biologically meaningful assumptions. The matching scheme can be revised and generalized to capture different biological contexts and different definitions of functionally related gene clusters that can include protein–protein interactions or other events.
METHODS
Data
All sequence data is taken from the GENES database in KEGG. (http://www.genome.ad.jp/kegg/kegg2.html). In addition to the original annotations given by each sequencing group, KEGG keeps a well-maintained enzyme catalog for each organism. The organisms analyzed include bacteria, archaea, and budding yeast (see Table 1 for a full list of organisms analyzed). Metabolic pathways data are taken from the PATHWAY (http://www.genome.ad.jp/kegg/metabolism.html) and the BRITE databases (http://www.genome.ad.jp/brite/) in KEGG. The pathway data includes about 90 reference metabolic pathways now in KEGG.
Graph Construction
Pathway data retrieved from KEGG are in a binary relation format representing pairwise interactions in pathway reactions (Goto et al. 1997). Using this binary relational data, we constructed a graph representation for each pathway. All metabolic pathways are represented as labeled undirected graphs, G (V,E), in which V is a set of vertices in a pathway graph and E is a set of edges connecting two vertices. There are two types of vertices in the graphs, as used in KEGG pathway diagrams, a compound type, which corresponds to either reactants or products in a particular pathway reaction, and an enzyme type, which is needed to catalyze this reaction. They differ in their labels, in which the compound vertex uses a compound's ID from the KEGG database as labels, and the enzyme vertex uses an EC number as a label. In the pathway graph, there is at least one enzyme vertex between two different compound vertices, representing a biochemical conversion catalyzed by this enzyme. In cases in which a reaction can be catalyzed by multiple enzymes, each enzyme vertex is present between compound vertices. The edges of the graph are weighted (all weights are equal to one for simplicity). The edge distance between two compound vertices, or metabolic distance (Ettema et al. 2001), is a measure of how many reaction steps are needed to accomplish this chemical conversion in a metabolic process. Graphs are built and analyzed using variants of algorithms in the LEDA package (Library of Efficient Data types and Algorithm, http://www.mpi-sub.mpg.de/LEDA/leda.html).
Algorithm
The specific computational problem we addressed is to identify a subgraph in which vertices in this subgraph appear in close proximity on the genome. Naturally, an important special case of this problem is identification of a genomic cluster that appears in the same pathway. A solution to this problem requires an efficient way of traversing the pathway graph and at the same time keeping track of the metabolic distance between vertices. Several graph traversal algorithms such as breadth-first search and depth-first search could be used. In this effort, we use a variant of breadth-first search.
We start the traversal from a chosen vertex (root) and visit vertices in stages; vertices connected to the root are reached first and placed in the second layer. We iteratively visit all vertices that are reachable in one step from vertices in layer I and have not been visited before, place them in layer I + 1 and proceed to the next layer. The most distant vertices from the root are reached last. By setting a depth parameter, we can control how far in the pathway graph a traversal can reach. As a special case, we can traverse the entire pathway by setting the depth larger than the diameter of the graph. The traversal will return a tree of vertices, in which the root is the start vertex and all other vertices are ordered in layers by their distances from the root. After we have the traversal tree, we then look into the genome, examining whether the vertex set in the tree falls into a neighborhood on the genome, as illustrated in Figure 6. A breadth-first search is started from each enzyme vertex in the pathway graph. As a result, the total number of searches is linearly proportional to the size of the pathway graph.
Figure 6.
A graphical interpretation of breadth-first search (BFS) graph traversal. The black vertex is the start vertex for BFS traversal in a metabolic pathway. In this example, the depth parameter is set to 2; the first layer is filled with dark gray and the second layer is filled with light gray. After a tree is returned from traversal, we locate the gene in the genome with the same EC number as the start vertex and extend a window on each side of it. We then compare genes in this window and in the traversal tree by EC numbers. If there is more than one match, this gene cluster window is marked for further pruning.
Enzyme vertices in the reference metabolic pathways can be matched with genes by comparing EC numbers. EC numbers are abstracted in four levels (e.g., EC:1.1.1.1). Not all enzymes have clear assignments in all four levels. The lower the level to which it has been assigned, the more specifically we know about its biochemical activity. When EC numbers are not available in all four levels, a loose matching scheme could be used assuming enzymes with unknown activity at a certain level could function as any type of enzyme at that level. However, to avoid overprediction, the results presented here do not utilize the loosely matched genes, that is, we only rely on exact matching of EC numbers.
Because this work focuses on inference of operons rather than general functionally related clusters, a conservative depth parameter of 3 is currently used, which means the breadth-first search can go as deep as three reactions away from the starting vertex. In follow-up studies, we will report the results for other much looser depth settings of parameters and additional statistical results from comparative genomics of functionally related gene clusters (Y. Zheng, in prep.). Genes are considered to be in close proximity if they are separated by fewer than three ORFs. Naturally, other settings of these gap parameters (both in the pathway graph and in the genome) will yield slightly different results.
To improve the specificity of our algorithm, a pruning step is added after an initial putative operon is detected. The idea of pruning is illustrated in Figure 7. With the depth parameter equal to three, one of the genes in a cluster could appear somewhat separated from other genes that form a more closely connected subgraph. We found that such remotely occurring genes are less likely to be part of an operon. The purpose of the pruning procedure is to eliminate such genes and thereby improve the overall specificity of the detection algorithm.
Figure 7.
Graphical illustration of pruning procedure. Nodes with the labels A,B,C,D in the pathway graph and the genome (line) are matched enzymes. The black vertex is the remote gene, which is three reaction steps away from the nearest gene (A) in the graph and three open reading frames (ORFs) away from the nearest gene (A) on the chromosome (gray genes are genes that were not matched). Consequently, the black vertex gets pruned from the cluster. The idea of pruning is implemented by computing the shortest distance in the graph from each matched vertex to the nearest matched vertex. A special case occurs when only two genes are reported as a possible operon. If their metabolic distance is equal to 3, they are pruned out.
For each genome, this procedure is applied using all reference metabolic pathways independently. A set of potential operons is reported for each of 90 metabolic pathway maps. Operons reported from each different pathway may overlap with each other on the genome. Accordingly, a final clustering step is added to perform a transitive closure, in which all potential operons are projected back onto the genome according to their relative positions, and overlapped operons are merged into larger putative operons. Park and Kim (2001) proposed that functional units can be assembled into a larger operon by a modular type gene transfer (Park and Kim 2001), which partly justifies the final clustering step (Fig. 1). A simplified pipeline flowchart is shown (Fig. 8). All source codes are written in C++ and available upon request.
Figure 8.
Flowchart of our computational pipeline.
We applied this procedure to each genome using all reference metabolic pathways independently. This is a single operon(mhp) in E. coli with six genes as follows: mhpA(b0347), mhpB(b0348), mhpC(b0349), mhpD(b0350), mhpF(b0351), and mhpE(b0352). The Mhp operon catalyzes successive reactions in the phenylpropionate catabolic pathway (Fig. 1a) (Burlingame et al. 1986). The KEGG pathway database does not have this pathway, so our algorithm cannot detect it from a single pathway. Instead, it first detects this operon in pieces from different pathways, which are later assembled into the whole operon by a clustering step. The mhp operon in E. coli is not found intact in any other microorganism genomes analyzed in this work. However, in Mycobacterium tuberculosis, a smaller gene cluster similar to b0350, b0351, and b0352 is identified (Rv3534c, Rv3535c, Rv3536c) with EC numbers matched and gene order conserved. We later found that the transcriptional activator of the mhp operon is encoded by b0346, which is also part of the mhp operon. However, because gene b0346 does not have an EC number entry, the algorithm ignores it, which partly shows a potential limitation of this method. The above example suggests that a large operon can be divided into smaller conserved gene clusters and subsets of a large operon can participate in different pathways.
Statistical Significance
The statistical significance of the number of predicted operons is tested against the expected number of predicted operons in a randomly shuffled genome. The shuffling process simulates genomic rearrangements by randomly picking two genes and exchanging their positions in the genome. For each shuffling experiment, the gene exchange step is repeated for a sufficient number of times to generate a random genome. Then, we can calculate the number of putative operons by applying the operon identification pipeline to shuffled genomes and averaging the results. Such shuffling experiments are repeated ten times for each genome. The P-value of the number of operons in a genome is given by Chebyshev's inequality: P<[(N-μ)/σ]−2 (reported in Table 1), in which N is the number of operons predicted in a genome, μ is the mean of the number of operons from the shuffling experiments, and σ is the standard deviation.
WEB SITE REFERENCES
http://genomics4.bu.edu/operons; operon list for each organism included in this study.
http://genomics4.bu.edu/operons/gappedgene; gap gene list.
http://www.genome.ad.jp/kegg/kegg2.html; KEGG database.
http://www.genome.ad.jp/kegg/metabolism.html; KEGG PATHWAY database.
http://www.genome.ad.jp/brite; KEGG BRITE database.
http://www.mpi-sub.mpg.de/LEDA/leda.html; LEDA package.
Acknowledgments
We thank the KEGG group, for their invaluable efforts in organizing the metabolic pathways and making them publicly accessible. This research has been supported in part by NSF-KDI0196227.
Computational Complexity
The problem of computing functional clusters in bacterial genomes appears to be relatively simple. However, at least in theory we can provide evidence that a general algorithm for extracting functional clusters by graph matching is likely to require exponential time. In other words, it is NP-complete.
We briefly describe two simple variants of this problem and sketch out a proof of the intractability result:
Variant 1: Given a labeled line graph (genome) G and a labeled pathway graph P, is there a contiguous cluster of genes of length K that appears as a connected reaction chain in the subgraph of the pathway graph P? This problem is clearly NP-complete since it is equivalent to the Hamiltonian cycle problem. Simply, consider the case where all the nodes are not labeled (or labeled with the same tag). We need to locate a path in the pathway graph of length K where each node is visited only once in order to include every gene in the cluster.
Variant 2: Here we define a more natural (i.e., more relevant to the detection of biologically plausible clusters) variant of the problem, which is still provably intractable. Given a labeled line graph G and a labeled pathway P, where all the nodes in the genome (or a large subset of it) are uniquely labeled, is there a connected component of P of size K that occurs as a cluster in G?
This problem is also NP-complete. We will use reduction from the best-known NP-complete problem that requires finding a satisfying Boolean assignment to the variables of a formula to make the formula TRUE. The formula is assumed to be in 3-CNF form with m clauses and n variables. That is, given a logical formula of this form we will show an efficient reduction to the genome-pathway cluster detection problem. This will establish that if we can solve the pathway problem we can also solve the satisfiability problem for a logical formula.
Given a formula F, we create the following graph. For each variable xi we create two nodes labeled xi and xi (xi means not xi). We then create a node for each clause and one “supernode”. The supernode is connected to all of the xi nodes and xi nodes. The clause node is connected to the variables in its clause. The nodes xi and xi are labeled with the same functional label i. The clauses and the supernode all have different labels. Then it's easy to see that we have a connected component of m+n+1 nodes of different functional labels iff the original formula is satisfied.
These two natural variants of the cluster identification problem suggest that the general problem is as computationally difficult as many other problems in computational biology, such as multiple alignment. Therefore the heuristics used in our paper as well as the previous papers are better justified in view of the computational intractability of the general problem.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL kasif@bu.edu; FAX (617) 353-6766.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.200602.
REFERENCES
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: A new generation of protein database search algorithms. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bono H, Ogata H, Goto S, Kanehisa M. Reconstruction of amino acid biosynthesis pathways from the complete genome sequence. Genome Res. 1998;8:203–210. doi: 10.1101/gr.8.3.203. [DOI] [PubMed] [Google Scholar]
- Bult CJ, White O, Olsen GJ, Zhou L, Fleischmann RD, Suttonh GG, Blake JA, Fitzgerald LM, Clauton RA, Gocayne JD, et al. Complete genome sequence of the Methanogenic archaeon, Methanococcus jannaschi. Science. 1996;273:1058–1073. doi: 10.1126/science.273.5278.1058. [DOI] [PubMed] [Google Scholar]
- Burlingame RP, Wyman L, Chapman PJ. Isolation and characterization of Escherichia colimutants defective for phenylpropionate degradation. J Bacteriol. 1986;168:55–64. doi: 10.1128/jb.168.1.55-64.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dandekar T, Schuster S, Snel B, Huynen M, Bork P. Pathway alignment: Application to the comparative analysis of glycolytic enzymes. Biochem J. 1999;343:115–124. [PMC free article] [PubMed] [Google Scholar]
- Ermolaeva MD, White O, Salzberg SL. Prediction of operons in microbial genomes. Nucleic Acids Res. 2001;29:1216–1221. doi: 10.1093/nar/29.5.1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ettema T, van der Oost J, Huynen M. Modularity in the gain and loss of genes: Application for function prediction. Trends Genet. 2001;17:485–487. doi: 10.1016/s0168-9525(01)02384-8. [DOI] [PubMed] [Google Scholar]
- Fukuda K, Takagi T. Knowledge representation of signal transduction pathways. Bioinformatics. 2001;17:829–837. doi: 10.1093/bioinformatics/17.9.829. [DOI] [PubMed] [Google Scholar]
- Goto, S., Bono, H., Ogata, H., Fujibuchi, W., Nishioka, T., Sato, K., and Kanehisa, M. 1997. Organizing and computing metabolic pathway data in terms of binary relations. Pacific Symp. Biocomput. 175–186. [PubMed]
- Huerta AM, Salgado H, Thieffry D, Collado-Vides J. RegulonDB: A database on transcriptional regulation in Escherichia coli. Nucleic Acids Res. 1998;26:55–59. doi: 10.1093/nar/26.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh T, Takemoto K, Mori H, Gojobori T. Evolutionary instability of operon structures disclosed by sequence comparison of complete microbial genomes. Mol Biol Evol. 1999;16:332–346. doi: 10.1093/oxfordjournals.molbev.a026114. [DOI] [PubMed] [Google Scholar]
- Kanehisa M, Goto S. KEGG: Kyoto Encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karp PD, Riley M, Paley SM, Pelligrini-Toole A. EcoCyc: An encyclopedia of Escherichia coligenes and metabolism. Nucleic Acids Res. 1996;24:32–39. doi: 10.1093/nar/24.1.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence JG. Selfish operons and speciation by gene transfer. Trends Microbiol. 1997;5:355–359. doi: 10.1016/S0966-842X(97)01110-4. [DOI] [PubMed] [Google Scholar]
- Ogata H, Fujibuchi W, Goto S, Kanehisa M. A heuristic graph comparison algorithm and its application to detect functionally related enzyme clusters. Nucleic Acids Res. 2000;28:4021–4028. doi: 10.1093/nar/28.20.4021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overbeek R, Fonstein M, D'souza M, Pusch GD, Maltsev N. The use of gene clusters to infer functional coupling. Proc Natl Acad Sci. 1999;96:2896–2901. doi: 10.1073/pnas.96.6.2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palaniappan C, Sharma V, Hudspeth ME, Meganathan R. Menaquinone (vitamin K2) biosynthesis: Evidence that the Escherichia coli menDgene encodes both 2-succinyl-6-hydroxy-2,4-cyclohexadiene-1-carboxylic acid synthase and alpha-ketoglutarate decarboxylase activities. J Bacteriol. 1992;174:8111–8118. doi: 10.1128/jb.174.24.8111-8118.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park H, Kim H. Genetic and structural organization of the aminophenol catabolic operon and its implication for evolutionary process. J Bacteriol. 2001;183:5074–5081. doi: 10.1128/JB.183.17.5074-5081.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salgado H, Moreno-Hagelsieb G, Smith TF, Collado-Vides J. Operons in Escherichia coli: Genomic analysis and predictions. Proc Natl Acad Sci. 2000;97:6652–6657. doi: 10.1073/pnas.110147297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selkov E, Maltsev N, Olsen GJ, Overbeek R, Whitman WB. A reconstruction of the metabolism of Methanococcus jannaschiifrom sequence data. Gene. 1997a;197:11–26. doi: 10.1016/s0378-1119(97)00307-7. [DOI] [PubMed] [Google Scholar]
- Selkov E, Galimova M, Goryanin I, Gretchkin Y, Ivanova N, Komarov Y, Maltsev N, Mikhailova N, Nenashev V, Overbeek R, et al. The metabolic pathway collection: An update. Nucleic Acids Res. 1997b;25:37–38. doi: 10.1093/nar/25.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shigenobu S, Watanabe H, Hattori M, Sakaki Y, Ishikawa H. Genome sequence of the endocellular bacterial symbiont of aphids Buchnera sp. APS Nature. 2000;407:81–86. doi: 10.1038/35024074. [DOI] [PubMed] [Google Scholar]
- Tamames J, Casari G, Ouzounis C, Valencia A. Conserved clusters of functionally related genes in two bacterial genomes. J Mol Evol. 1997;44:66–73. doi: 10.1007/pl00006122. [DOI] [PubMed] [Google Scholar]
- Walters DM, Russ R, Knackmuss H-J, Rouviere PE. High-density sampling of a bacterial operon using mRNA differential display. Gene. 2001;273:305–315. doi: 10.1016/s0378-1119(01)00597-2. [DOI] [PubMed] [Google Scholar]
- Yada T, Nakao M, Totoki Y, Nakai K. Modeling and predicting transcriptional units of Escherichia coligenes using hidden Markov models. Bioinformatics. 1999;15:987–993. doi: 10.1093/bioinformatics/15.12.987. [DOI] [PubMed] [Google Scholar]