Summary
Much of the genome is transcribed into long non-coding RNAs (ncRNAs). Previous data suggested that bithoraxoid (bxd) ncRNAs of the Drosophila bithorax complex prevent silencing of Ultrabithorax (Ubx), and recruit activating proteins of the trithorax group to their maintenance elements. We found that, surprisingly, Ubx and several bxd ncRNAs are expressed in non-overlapping patterns in both embryos and imaginal discs, suggesting that transcription of these ncRNAs is associated with repression, not activation, of Ubx. Our data rule out siRNA or miRNA-based mechanisms for repression by bxd ncRNAs. Rather, ncRNA transcription itself, acting in cis, represses Ubx. The Trithorax complex TAC1 binds the Ubx coding region in nuclei expressing Ubx, and the bxd region in nuclei not expressing Ubx. We propose that TAC1 promotes the mosaic pattern of Ubx expression by facilitating transcriptional elongation of bxd ncRNAs, which represses Ubx transcription.
Introduction
The Hox genes of the bithorax complex (BX-C) have spatially restricted expression patterns that vary within and between segments and tissues. Transcription factors encoded by segmentation genes (Carroll et al., 1988; Irish et al., 1989) establish the patterns of the Hox genes Ubx, abd-A, and Abd-B of the BX-C in embryos. After the segmentation proteins decay, Hox expression patterns are maintained epigenetically by proteins of the trithorax group (trxG) and the Polycomb group (PcG) (Grimaud et al., 2006b). PcG genes maintain the silent state, whereas trxG genes maintain the active state of Hox genes. PcG and trxG proteins act through partially overlapping sets of response elements known as maintenance elements (MEs, as in Figure 1A) (Hodgson et al., 2001; Pirrotta et al., 1995; Tillib et al., 1999).
Figure 1. Ubx and ncRNAs are expressed in complementary embryonic domains.
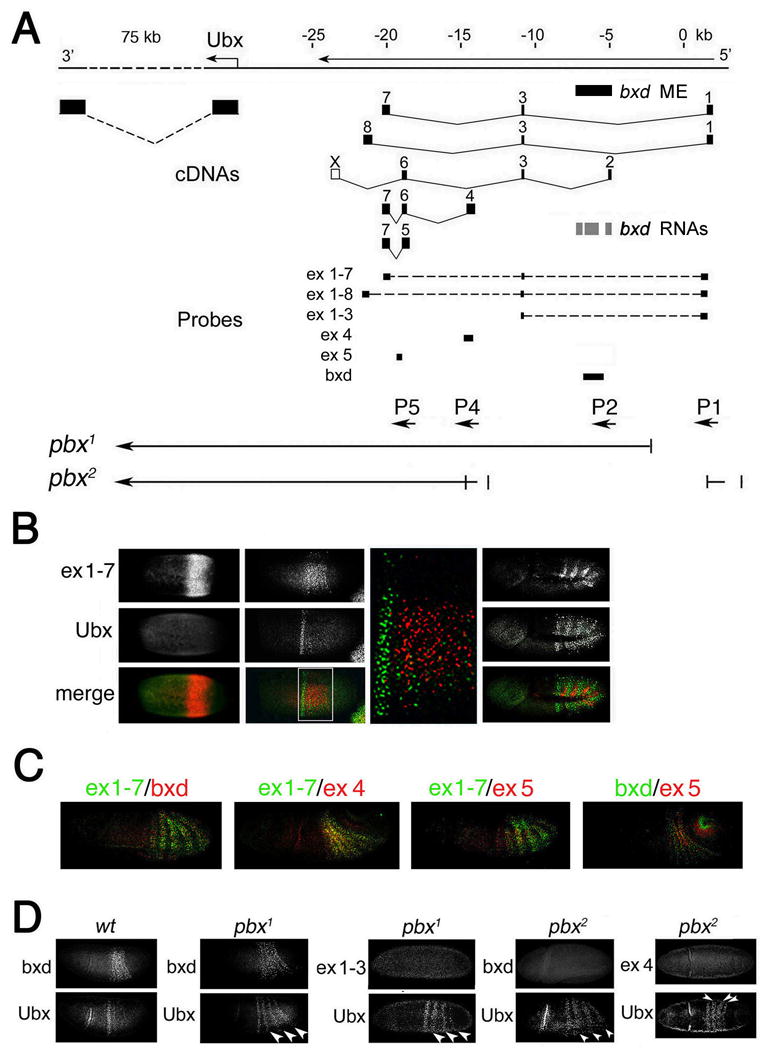
(A) Map of Ubx and intergenic transcripts. The exons of Ubx and intergenic transcripts (Lipshitz et al., 1987) are shown in black. bxd ME (Tillib et al., 1999) is shown by the black bar below DNA line. Described locations of three bxd transcripts (Sanchez-Elsner et al., 2006) are shown by grey bars. Probes used for in situ hybridization are diagrammed below the cDNAs as black bars. The bottom map shows the extent of deleted regions in pbx1 and pbx2 mutants. Promoters of ncRNAs are indicated by arrows below the probes.
(B) Time-course of nascent Ubx and ncRNA transcription. Nascent ncRNA was detected with probes indicated in A. At syncytial blastoderm, RNA 1–7 is expressed earlier than Ubx (left panels). The initial expression domain of Ubx is anterior to that of RNA 1–7 (middle panels, including blow-up of merge). During germband elongation, expression domains of RNA 1–7 in each parasegment are anterior to those of Ubx (right panels). Ubx-expressing cells do not express RNA 1–7, or any ncRNAs detected by probes shown in Figure 1A at any stage (some data not shown).
(C) RNA containing exons 1–7, 4, 5, and bxd RNA are expressed within the same intersegmental domains, although their expression differs in different germ layers.
(D) Effects of pbx1 and pbx2 deletions on expression of the indicated ncRNAs and endogenous Ubx.
One of the most startling discoveries of the genomic era has been that much of the genome is transcribed into non-coding RNAs (ncRNAs) (Eddy, 2002; Gottesman, 2002; Mattick and Makunin, 2006). Recent attention has focused on small interfering RNAs (siRNAs) and micro-RNAs (miRNAs) that modulate gene activity by an antisense mechanism termed RNA interference (RNAi), which interferes with mRNA stability or translation (Carthew, 2006; Massirer and Pasquinelli, 2006; Sen and Blau, 2006). However, the most abundant and least characterized class of ncRNAs are long and have mostly unknown functions (Goodrich and McClure, 1991; Mattick and Makunin, 2006).
The intergenic regions of the Hox genes in D. melanogaster produce many long ncRNAs that may regulate Hox gene coding sequences. Increasing attention has been directed to the role of transcription of MEs in the regulation of BX-C genes. Several ncRNAs are transcribed through a well-studied ME in the bxd regulatory region that lies between the Ubx and abd-A transcription units (Cumberledge et al., 1990; Lipshitz et al., 1987; Sanchez-Herrero and Akam, 1989). The bxd ME regulates Ubx (Chan et al., 1994; Muller and Bienz, 1991; Simon et al., 1993) (Figure 1A). Transcription through bxd precedes activation of Ubx coding RNAs (hereafter referred to as “Ubx RNA”, or simply as “Ubx”), suggesting that ncRNAs might regulate Ubx (Rank et al., 2002). Transcription patterns of ncRNAs appear similar to those of the neighboring Hox genes and are collinear with regulatory domains along the chromosome (Bae et al., 2002). A synthesis of genetic (Bender and Fitzgerald, 2002; Hogga and Karch, 2002; Rank et al., 2002) and transgenic studies (Schmitt et al., 2005) led to the idea that transcription of ncRNAs through MEs interferes with PcG-mediated silencing, perhaps by preventing recruitment of PcG proteins. A recent study suggests that transcription of MEs may recruit trxG proteins to maintenance elements (Sanchez-Elsner et al., 2006). If indeed transcription of MEs simultaneously prevents PcG binding and establishes trxG binding, this could be a key element of Hox regulation. Clearly, this model requires that intergenic bxd RNAs are expressed in the same cells as Ubx. However double-labeling of intergenic and coding RNAs at high resolution has not been performed, so this attractive model has not been rigorously tested.
The trxG protein complex TAC1 plays important roles in maintaining expression of homeotic genes throughout embryogenesis (Petruk et al., 2001). Recent attention has focused on the role of trxG proteins in histone modifications and in altering nucleosome positioning (Beisel et al., 2002; Smith et al., 2004). TAC1 contains three proteins, Trx, Sbf1 and dCBP, and thus acetylates histones and methylates histone H3 at Lys-4 (H3-K4), due to the enzymatic activities of dCBP and the SET domain of Trx, respectively (Petruk et al., 2001; Smith et al., 2004). How binding of Trx to MEs regulates expression of Hox genes is unclear. Because embryos have a mixture of cells expressing and not expressing each Hox gene, it has not been possible to determine precisely how trxG protein binding correlates with transcription of Ubx and bxd ncRNAs.
In this study, we show that Ubx is repressed by bxd transcription. The bxd ncRNAs do not act by siRNA or miRNA-based mechanisms, but repress Ubx in cis through a transcription-dependent mechanism. Alternative association of TAC1 with either Ubx or bxd correlates with their transcription. TAC1 appears to be part of an interdependent network of general elongation factors that associate with active genes. We suggest that a key role of TAC1 in establishing the mosaic pattern of Ubx expression is to promote elongation of bxd ncRNAs, which in turn represses expression of Ubx.
Results
Ubx and intergenic ncRNAs are expressed in different cells in embryos
We concentrated on Ubx, and the upstream bxd region because multiple ncRNAs are transcribed through the bxd region (Figure 1A). Two promoters of bxd ncRNAs (P1 and P2 in Figures 1A and 2C) are localized upstream of the bxd ME, and two others lie downstream (P3, P4). The bxd regulatory region contains several trx-responsive elements as separable regions within the ME (Tillib et al., 1999).
Figure 2. Intergenic transcription represses Ubx in cis.
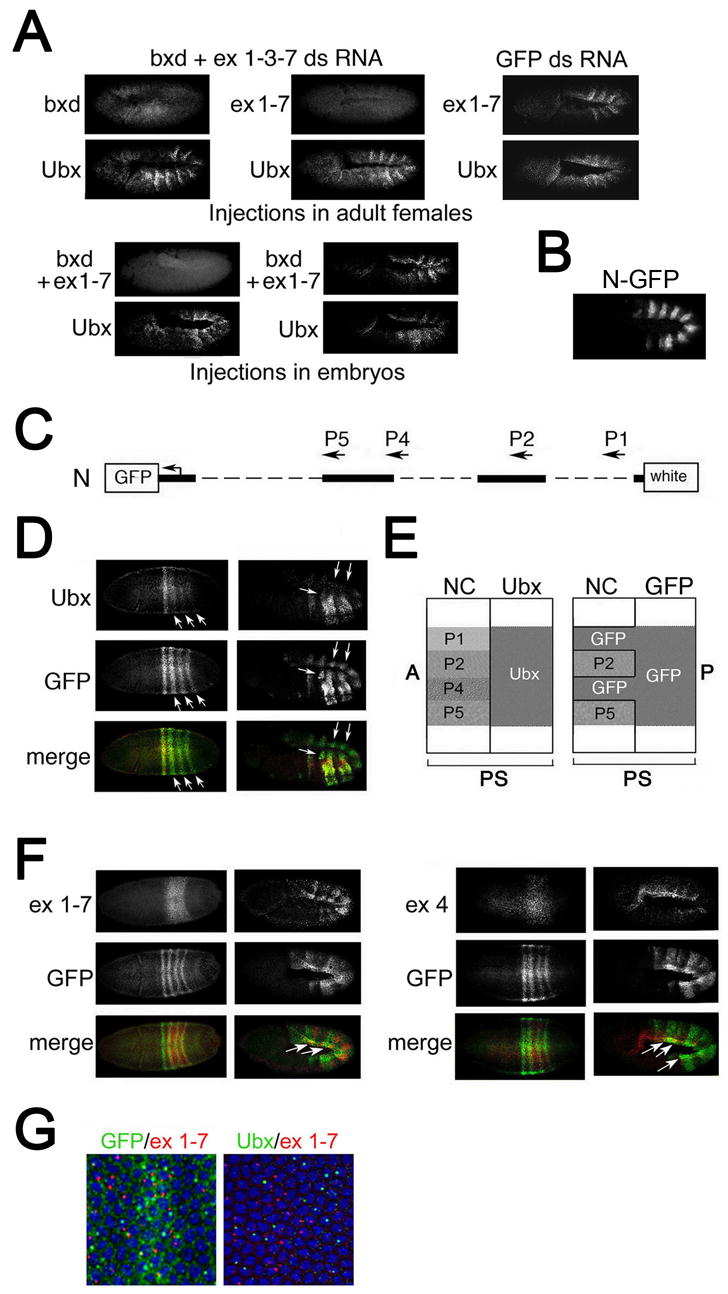
(A) Products of ncRNAs do not directly repress Ubx transcription. A mixture of dsRNAs specific to exons 1, 3, 7, and part of the bxd region, and control dsRNA specific to GFP were injected either into adult females (top panels) or into preblastoderm embryos. Embryos were analyzed by in situ hybridization with probes to Ubx, exons 1–7 and the mixture of probes to exons 1–7 and bxd, as indicated.
(B) Immunostaining of embryo carrying the Ubx-GFP transgenic construct N (see map in C) with GFP antibody.
(C) Map of the Ubx-GFP transgenic construct N. The transgene contains ncRNA promoters P2 and P5 but lacks promoters P1 and P4.
(D) The GFP reporter gene is ectopically expressed (arrows) at blastoderm and germband extension relative to endogenous Ubx.
(E) A diagram of predicted expression patterns of Ubx and GFP, assuming repression occurs only in cis. In the absence of the P1 and P4 promoters, the GFP transgene is expected to be expressed in cells where the corresponding endogenous RNAs are expressed. A, P, anterior and posterior of the embryo.
(F) Expression of the GFP reporter gene overlaps with expression of ncRNAs that are produced from the P1 (probe ex 1–7 left panels) and P4 (probe ex 4, right panels) promoters.
(G) Comparison of the expression of endogenous Ubx and the GFP reporter gene with RNA 1–7 at the blastoderm stage. Expression of GFP is seen in both cytoplasm and nuclei, and RNA 1–7 is detected in the nuclei of the same cells (left).
The development of a high-resolution, multiplex RNA in situ hybridization technique (Kosman et al. 2004) allows one to test the hypothesis that bxd ncRNAs are necessary for activation of Ubx. The model requires that Ubx and bxd ncRNAs be expressed in the same cells. We compared expression patterns of Ubx with those of bxd ncRNAs using 4 probes that are specific to several exons described previously (Lipshitz et al., 1987), as well as a probe that includes three transcripts (Sanchez-Elsner et al., 2006) (“bxd” in Figure 1A) described previously as giving a Ubx-like pattern (Rank et al., 2002). Expression of all tested bxd ncRNAs begins at syncytial blastoderm, and precedes that of Ubx (probes 1–7 and bxd in Figure 1B; bxd ncRNAs 1–8, 4 and 5: data not shown). At cellular blastoderm, the initial domain of Ubx expression is just anterior to the primary domains of ncRNA expression. During germband elongation, Ubx and bxd ncRNAs are expressed within the same restricted portion of the embryo, as reported previously. At all embryonic stages, no bxd ncRNA is expressed in cells that express Ubx (Figure 1B and data not shown). Ubx is expressed in the posterior, while bxd ncRNAs are expressed in the anterior, of each parasegment 6–13. This expression of Ubx and bxd ncRNAs in alternative sets of cells is confirmed by results described below using sorted nuclei.
The bxd ncRNA 1–7 shown in Figure 1B is expressed in a region that is more restricted dorsoventrally than that of Ubx. Overall, different bxd ncRNAs show significant overlap in their expression domains, which have the same anterior-posterior boundary. This overlap may be because these RNAs share alternatively spliced small exons. Strikingly, bxd ncRNAs are expressed in different germ layers (Figure 1C), suggesting that expression of individual bxd ncRNAs may be driven by different tissue-specific regulatory elements.
Elimination of intergenic transcripts leads to ectopic expression of Ubx
Our findings argue strongly that bxd ncRNAs cannot activate, but may instead repress, Ubx. To test this idea, we analyzed expression of Ubx in mutants that carry deletions in the transcription units of ncRNAs. The pbx1 mutation that causes homeotic defects in adults, deletes promoter P1 and the first exon of two ncRNAs, 1–7 and 1–8, while the pbx2 mutation deletes promoter regions P1, P2 and P4, and the 5’-exons of these and several other ncRNAs (Irvine et al., 1991) (Figures 1A and S2). Consistent with these molecular lesions, the corresponding bxd ncRNAs are not expressed in pbx1 and pbx2 mutant embryos (Figure 1D). Importantly, absence of bxd ncRNA is accompanied by clear ectopic expression of Ubx in the posterior region of the embryo where these bxd RNAs are expressed in wildtype embryos (Figure 1D), consistent with a role for bxd ncRNAs in Ubx repression. This ectopic expression of Ubx is very unlikely to be caused by deletion of an unmapped PcG response element, because mis-expression of Ubx in PcG maternal and zygotic homozygous mutant embryos is not yet detectable at this stage of embryogenesis (Soto et al., 1995; Struhl and Akam, 1985).
ncRNAs do not repress Ubx by RNAi-based mechanisms
Grimaud et al. (Grimaud et al., 2006a) report that mutations in genes required for RNA interference (RNAi) have no homeotic phenotypes, suggesting that RNAi-based mechanisms are not essential to the regulation of Ubx. To test directly whether bxd ncRNAs repress Ubx by RNAi, we prepared a mixture of dsRNAs that are specific to bxd RNA exons 1, 3 and 7, and to other bxd RNAs mapped previously (Sanchez-Elsner et al., 2006) (grey bars in Figure 1A) and introduced these into embryos using two strategies. First, we injected dsRNA into adult females, where it is taken up by oocytes (Dzitoyeva et al., 2003), and second, we injected the dsRNAs into preblastoderm embryos. These dsRNAs diffuse through the oocyte or embryo, and will eliminate any RNAs containing homologous sequences by RNAi.
As shown in Figure 2A, introduction of dsRNA by either method leads to an almost complete elimination of the products of the corresponding ncRNAs. In all tested embryos, elimination of bxd ncRNAs had no effect on Ubx expression (Figure 2A). This shows that an RNAi-based degradation mechanism (i.e. siRNA) cannot be responsible for repression of Ubx. Note that because our injected dsRNA did not cause Ubx degradation, we have also eliminated the possibility that our probes contain sequences that are normally responsible for degradation of Ubx. We also carefully examined expression of a reporter transgene, that mimics the Ubx pattern, but does not contain the Ubx 3’ UTR, a common target of miRNA-based translational repression (Figure 2C). There is no apparent ectopic expression of this reporter in regions of bxd transcription, arguing against miRNA-dependent translational repression (Figure 2B). These experiments together with the lack of homeotic phenotypes in RNAi mutant flies argue that ncRNAs do not act in trans, and suggest that repression is due to a cis-acting mechanism associated with bxd transcription per se.
Ubx expression is repressed by transcription from the promoters of ncRNAs
Transcription-based mechanisms of repression, namely promoter competition or transcriptional interference (Martens et al., 2004), offer an attractive possibility for cis-repression of Ubx by bxd ncRNAs. These cis-repression models predict that deletion of the promoter of an ncRNA will lead to expression of the repressed promoter in cells where these RNAs are normally transcribed. To test this, we used a Ubx transgene that closely mimics expression of endogenous Ubx in mid-to-late stage embryos (Tillib et al., 1999). This construct lacks promoters P1 and P4 which drive several ncRNAs, but contains promoters P2 (in the bxd region) and P5 (Figure 2C). The absence of these two promoters in the transgene leads to ectopic expression of the GFP reporter gene in the posterior region of blastoderm embryos (Figure 2D). During germband elongation, GFP is clearly expressed in some mesodermal regions where endogenous Ubx is not expressed, suggesting that the absence of transcription from the P1 and P4 promoters causes a loss of cis-repression of Ubx transcription in some mesodermal cells.
The cis-repression model of bxd ncRNA function predicts that the expanded domain of GFP expression from the transgene lacking P1 and P4 promoters should correspond to the domains of bxd ncRNAs normally transcribed from the endogenous promoters (see Figure 2E). As predicted, GFP expression significantly overlaps with that of the endogenous bxd ncRNAs 1–7 and 4, which are produced from the P1 and P4 promoters deleted in our transgene, in the posterior region of the blastoderm embryo (Figure 2F). Moreover, this overlap of GFP with ncRNAs in the mesodermal regions continues into germband extension (compare Figures. 2D and 2F, right panels). In contrast, endogenous Ubx does not overlap with any bxd ncRNA in embryos at either stage (Figures 1C and 2G). These results strongly corroborate a mechanism of Ubx repression by bxd transcription that acts in cis.
Ubx may be repressed by a transcriptional interference mechanism
Promoter competition occurs when nearby promoters, like those of bxd ncRNAs and Ubx, compete for rate-limiting transcription factors. The alternative cis-acting mechanism of transcriptional interference occurs when Pol II does not terminate at the 3’-exon of an upstream RNA, but proceeds through a promoter or enhancer, disrupting essential protein interactions with these regulatory elements. The results above cannot distinguish between these two mechanisms.
To address this issue, we asked whether transcription of bxd ncRNAs proceeds to the vicinity of the Ubx promoter. First, we tested by RT-PCR for the presence of transcripts in the 8kb region between the bxd ncRNA 3’-exons 7 and 8 and the Ubx start site (Figure 3A). As Figure 3B shows, cDNA synthesized from the primer located just upstream of the Ubx promoter contains sequences both from the vicinity of the Ubx promoter and exons 7 and 8 of bxd ncRNAs, as well as from intervening sequences. This suggests that Pol II transcribes bxd ncRNAs and continues to the vicinity of the Ubx promoter. To test whether these read-through RNAs are expressed in the cells where Ubx is repressed, we used a probe to the upstream regulatory region of Ubx (P5 in Figure 3A) for in situ hybridization. Figure 3C shows that there is almost complete overlap of RNA from the vicinity of the Ubx promoter with the 3’-exon 7 of ncRNAs. Importantly, as for the bxd ncRNAs in general, synthesis of these ncRNAs precedes synthesis of Ubx, and occurs in cells not expressing Ubx (Figure 3D), confirming that these read-through RNA products correlate with Ubx repression.
Figure 3. Ubx is repressed by read-through transcription from intergenic transcription units.
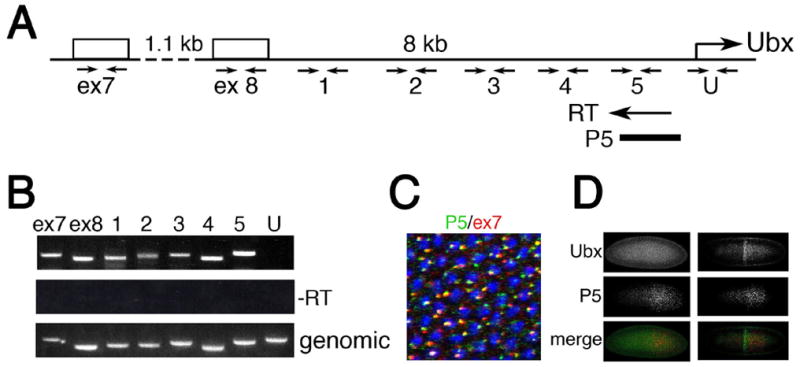
(A) Map of the intergenic region between the 3’-exons 7 and 8 of ncRNAs and the Ubx start site. cDNA was synthesized from the primer close to the Ubx start site (RT). Primer sets for PCR amplification are indicated below the map. Probe P5 for in situ hybridization is shown at the bottom. For coordinates of primers and in situ probes see Supplementary material.
(B) RT-PCR detection of transcripts in the region proximal to the Ubx promoter. Primer sets are those shown in (A).
(C) Transcripts from the region proximal to the Ubx start site are expressed in the same cells as transcripts of the ncRNAs containing exon 7.
(D) Transcripts from the region proximal to the Ubx start site are expressed earlier than Ubx in blastoderm embryos (left panels), and they are expressed in cells that do not express Ubx (right panels).
ncRNAs are not required for activation of Ubx
Our data suggest that expression of bxd ncRNAs represses expression of Ubx in embryos, in contrast to a report that expression of bxd ncRNAs (Figure 1A) are required for activation of Ubx in larval imaginal discs and in S2 cells by specific recruitment of the trxG protein Ash1 (Sanchez-Elsner et al., 2006). Therefore we re-examined the proposed activating role of ncRNAs in larval discs.
Our RT-PCR and in situ hybridization results (Figures 4A,B and S3) show that Ubx is expressed at low levels in wing disks, in agreement with studies showing that Ubx is expressed in the peripodial membrane but not in the epithelium proper (Brower, 1987; Pallavi and Shashidhara, 2003), but in contrast to recent reports (Sanchez-Elsner et al., 2006). Importantly, bxd ncRNAs detected by exon 1, exon 5 and bxd probes are not expressed in any of the three tested larval discs at any significant levels (Figures 4A,B and S3), agreeing with previous data for exon 1 and exon 5 RNAs in larval imaginal discs (Lipshitz et al., 1987), but disagreeing with (Sanchez-Elsner et al., 2006).
Figure 4. NcRNAs are not required for activation of Ubx in larval imaginal discs.
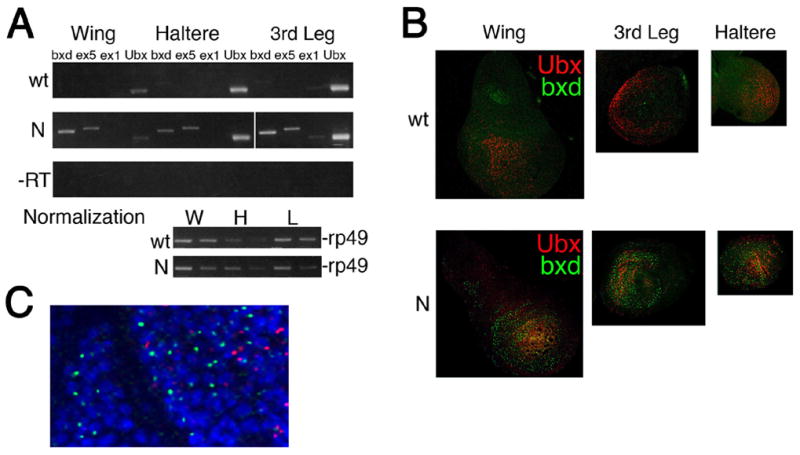
(A). RT-PCR analysis of the amounts of ex1, ex5, bxd, and Ubx RNAs in wing, haltere and 3rd leg imaginal discs in wildtype (wt) and transgenic (N) larvae. Controls without RT are shown in the third row. rp49 was used to normalize the amount of RNA in discs between the two strains (bottom). Primer sets are the same as in Figures 5B,E.
(B) In situ hybridization of Ubx and bxd probes to wing (W), haltere (H) and 3rd leg (L) larval imaginal discs in wildtype (wt) and transgenic (N) larvae. Probes are the same as in Figure 1B,C.
(C) High magnification detection of Ubx (red) and bxd (green) nascent RNAs by fluorescent in situ hybridization in 3rd leg disc of transgenic larvae. Probes are the same as in (B).
The Ubx-GFP transgene used above (Figure 2C) ectopically expresses exon 5 and the three ncRNAs (“bxd” in Figure 1A) described by Sanchez-Elsner et al. (2006) in wing, 3rd leg, and haltere discs (Figure 4A,B). Sanchez-Elsner (2006) report that overexpression of each of these transcripts induces expression of endogenous Ubx in wing discs. We show by RT-PCR that expression of bxd RNA from the transgene does not lead to an increase in Ubx expression, even in the wing disc which expresses low levels of Ubx endogenously (Figure 4A). Furthermore, bxd ncRNA transcribed from the transgene and endogenous Ubx RNA are expressed in different cells and in different regions in each of these discs (Figure 4B,C). The lack of overlap between bxd ncRNA and Ubx expression argues that bxd RNA does not activate Ubx in imaginal discs.
An approach to sorting nuclei based on Ubx expression
The previous experiments suggest that transcriptional elongation of bxd ncRNAs has a key role in Ubx regulation, but does not suggest how transcription of ncRNAs is regulated molecularly. To address this question, we investigated the role of the TAC1 complex. Embryos contain a mixture of cells expressing and not expressing Ubx and bxd ncRNAs respectively, so it is not possible to carry out informative biochemistry on whole embryos. As we have shown above, imaginal discs are not suitable for these studies because they do not express bxd ncRNAs significantly. Sorting embryonic cells based on expression of GFP-expressing transgenes has not worked for Drosophila embryos, mainly because of high levels of fluorescence from yolk proteins in the cytoplasm of embryonic gut cells (Figure 5A). To overcome this problem, we sorted nuclei rather than cells by flow cytometry, based on expression of a Ubx-GFP transgene. Our procedure for isolation of highly purified embryonic nuclei in large quantities is simple, reliable, and removes most of the material with non-specific fluorescence (see Experimental Procedures, Supplementary materials and Figure S1). It provides biochemically useful amounts of highly enriched nuclei without significant disruption of chromatin structure, which can be used to detect chromatin-associated proteins, to detect RNA by RT-PCR, and for expression profiling. This technique will be generally useful as nuclei expressing fluorescent proteins controlled by any regulatory region can be sorted from embryos.
Figure 5. Association of TAC1 complex with active and silenced Ubx in embryos.
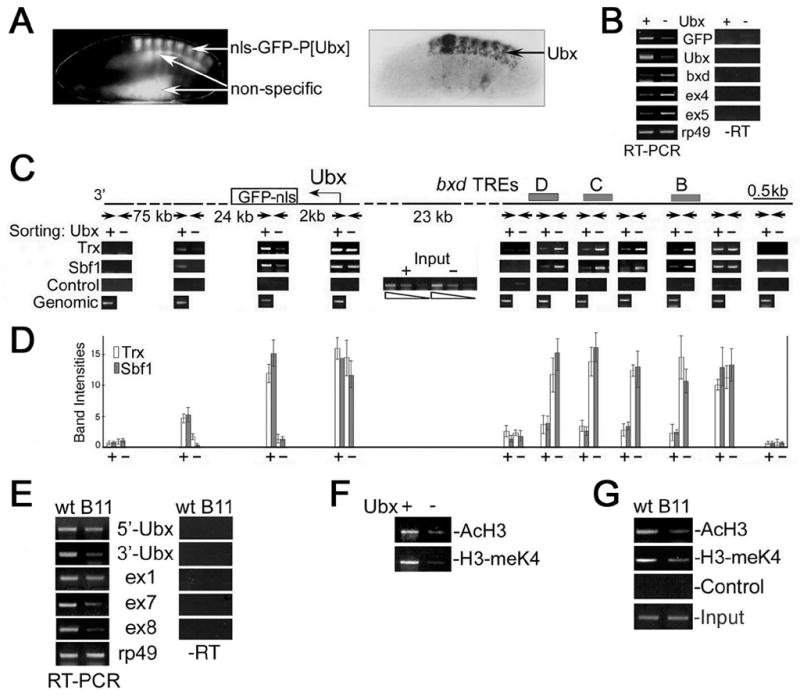
(A) Expression of GFP reporter (top) and endogenous Ubx (bottom) in mid-stage Drosophila embryos. Non-specific fluorescence is indicated.
(B) RT-PCR analysis of the amounts of GFP, Ubx, intergenic and rp49 transcripts in RNA from Ubx/GFP-positive (+) and Ubx/GFP-negative (-) nuclei from 7–13 hr embryos. Primers for exons 4 and 5 and the bxd region were designed according to (Lipshitz et al., 1987; Rank et al., 2002). rp49 was used to normalize the amount of RNA. Controls without RT are shown on the right.
(C) Top: map showing the regions of endogenous Ubx and the GFP-Ubx transgene that were tested for association of TAC1 components. Trx binding sites/response elements (TREs) in the bxd region (Tillib et al., 1999) are shown as grey bars, primer sets used for ChIP analysis are indicated by arrows, with distances between them. Bottom: ChIP analyses were performed with antibodies specific to Trx and Sbf1 (as shown on the left) from chromatin isolated from either Ubx/GFP-positive (+) or Ubx/GFP-negative (–) nuclei. Data shown is for the transcribed region 2 kb downstream of the endogenous Ubx start site; indistinguishable results were obtained with the corresponding primers for the GFP transgene. Control, no antibody. Input is shown in the middle.
(D) Graph of the relative levels of Trx and Sbf1 in the Ubx transcription unit. Background levels of cross-linking were determined by omitting the precipitating antibody and were then subtracted from the signals.
(E) RT-PCR analysis of the amounts of Ubx and ncRNAs in wildtype (wt) and trxB11 (B11) mutant embryos. rp49 was used to normalize the amount of RNA. For coordinates of primer sets for Ubx and exons 1, 7 and 8 see Supplementary material and Figure 1A. Controls without RT are shown on the right.
(F) ChIP analysis of histone modifications in the coding region of Ubx. Immunoprecipitations were performed with antibodies against acetylated H3 (AcH3) and H3 dimethylated at K4 (H3-meK4) from the chromatin isolated from Ubx/GFP+ and Ubx/GFP– nuclei. Immunoprecipitated material was PCR amplified using the primers for the region 2 kb downstream of the Ubx start site. Input is the same as in Figure 5C.
(G) Chromatin prepared from wildtype (wt) and trxB11 homozygous mutant embryos (B11) was immunoprecipitated with antibodies against AcH3 and H3-meK4. Primers were those for the sequence 2 kb downstream of the Ubx promoter. Control, without antibody.
The transgene expressing GFP under the regulation of 14 kb of DNA from the bxd regulatory region illustrated in Figures 2C and S1A was used to sort nuclei into those expressing GFP (Ubx+) and not expressing GFP (Ubx−). As shown in Figure 5A, the patterns of GFP and Ubx expression are very similar, showing that the transgene faithfully reproduces Ubx expression. Prior to use in these experiments, all batches of sorted nuclei were tested as shown in Figure S1C to ensure high levels of enrichment.
We first compared expression levels of GFP, endogenous Ubx and several bxd ncRNAs in our Ubx+ and Ubx− nuclei by RT-PCR. One pair of PCR primers was designed for the region that includes the “bxd” transcripts, and two other sets were designed for exons 4 and 5 of ncRNAs (Figure 1A; (Lipshitz et al., 1987). Figure 5B shows that in sharp contrast to endogenous Ubx and the GFP transgene, all three primer sets show the presence of ncRNA transcripts primarily in the Ubx− nuclei. Importantly, the results of our RT-PCR analysis of sorted nuclei are consistent with the patterns of expression (Figure 1B,C), and confirm that our sorting procedure allows efficient separation of cells that preferentially express either Ubx or bxd ncRNA.
TAC1 is involved in transcriptional elongation of Ubx
Using sorted nuclei from 7–13 hr embryos in the chromatin immunoprecipitation (ChIP) assay, we asked where TAC1 binds MEs, and at promoters or transcribed regions of either Ubx or bxd ncRNAs, in Ubx+ and Ubx− nuclei. In Ubx+ nuclei, the recruitment levels of Trx and Sbf1 are much lower in the bxd ME relative to that in Ubx− nuclei (Figure 5C,D). The data are simply explained if TAC1 has a role in transcription of both Ubx and bxd ncRNAs, which as we have shown, occurs in non-overlapping cell populations. As it is often assumed that trxG proteins binds the bxd ME only when Ubx is transcribed, this result suggests that the bxd ME is not the only element that TAC1 binds in the bxd region.
Recruitment levels of both Trx and Sbf1 are clearly higher in Ubx+ nuclei than in Ubx− nuclei in the region downstream of the transcription start site, at both endogenous Ubx (Figure 5C,D) and the GFP transgene (not shown), peaking at about 2 kb from the start site, suggesting that TAC1 binds downstream of actively transcribed promoters. TAC1 components were also detected, albeit at lower levels, in the middle of the Ubx gene (24 kb downstream of the start site), but were not significantly enriched at its 3’-end.
This binding pattern is consistent with a specific role for TAC1 in maintaining effective elongation. This idea is supported by RT-PCR analysis of trxB11 null mutants (Figure 5E), which shows that synthesis of the 3’-end both of the Ubx mRNA and of bxd ncRNAs is more strongly reduced than the 5’-end. Such a differential effect implicates trx function in the processivity of transcriptional elongation. These results are also consistent with our previous data showing that expression of Ubx is not completely abrogated in trxB11 mutant embryos (Mazo et al., 1990). Overall, our results show that alternative TAC1 (Trx and Sbf1) binding to Ubx and to the bxd region correlates with a function in transcriptional elongation in these complementary sets of cells.
TAC1 modifies histones in the coding region of Ubx
Since the TAC1 complex possesses HMT and HAT activities (Petruk et al., 2001; Smith et al., 2004), we tested whether association of this complex with the transcribed region of Ubx in sorted nuclei correlates with increased levels of modified histones. Consistent with the presence of active TAC1, the levels of both acetylated H3 and H3 dimethylated at K4 in the Ubx coding region are significantly greater in Ubx+ than in Ubx− nuclei (Figure 5F). Figure 5G shows that the amounts of methylated H3-K4 and acetylated H3 are significantly reduced in trxB11 embryos, demonstrating that modifications of nucleosomes in the coding region of activated Ubx are dependent on TAC1. This change in association of TAC1 may be a key determinant of whether Ubx expression is maintained in an active or a repressed state.
TAC1 recruitment to the coding regions of Ubx and bxd ncRNAs depends on elongation factors
If TAC1 is important for transcriptional elongation, then binding of TAC1 within the transcribed regions of Ubx and bxd ncRNAs and the associated H3-K4 methylation might be affected by mutations in elongation factors, such as Spt16 (a component of the FACT nucleosome assembly complex), Spt4 and Spt6. We examined Trx and Sbf1 binding in these mutants. Homozygous mutant embryos were selected using GFP-marked balancers as described previously (Smith et al., 2004). A mutation in Spt4 did not affect association of TAC1 or H3-meK4 within this region of Ubx (Figure 6A). However, binding of TAC1 was strongly decreased in both Spt6 and Spt16 mutant embryos (Figure 6A). Methylation of H3-K4 was also decreased in the same mutants. These data suggest that TAC1 may be associated with elongationally engaged Pol II.
Figure 6. Elongation factors are required for TAC1 recruitment to Ubx.
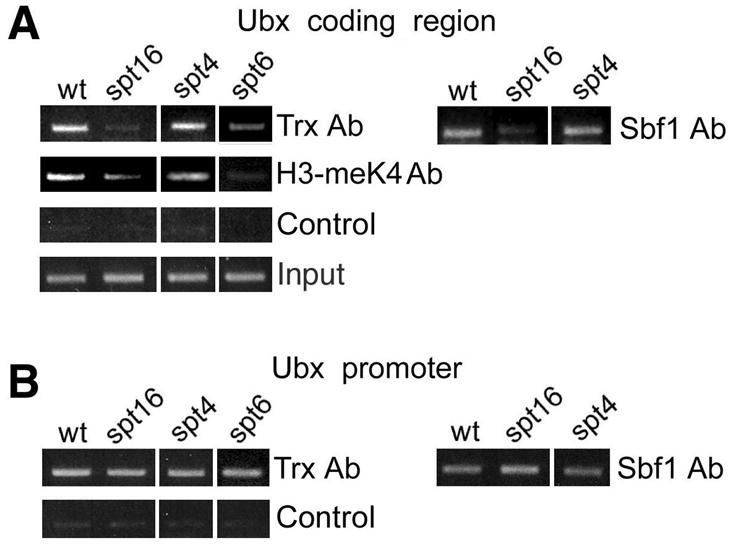
Chromatin was prepared from wildtype (wt) and the homozygous mutant embryos indicated above each set, and immunoprecipitated with antibodies against Trx, Sbf1 and histone H3-meK4 as indicated on the right. Immunoprecipitated material was PCR amplified with primers for the region 2 kb downstream of the start site (A) and to the promoter (B). Primer sets are the same as in Figure 5C. Control, no antibody.
TAC1 is essential for recruitment of Spt16
We then asked whether TAC1 is required for the recruitment of Spt16 to Ubx. Association of Spt16 with both the promoter and downstream regions of Ubx is significantly decreased in homozygous trxB11 null mutant embryos (Figure 7A), suggesting that Spt16 recruitment requires the presence of TAC1 during the initial phases of transcriptional elongation.
Figure 7. TAC1 is essential for association of FACT.
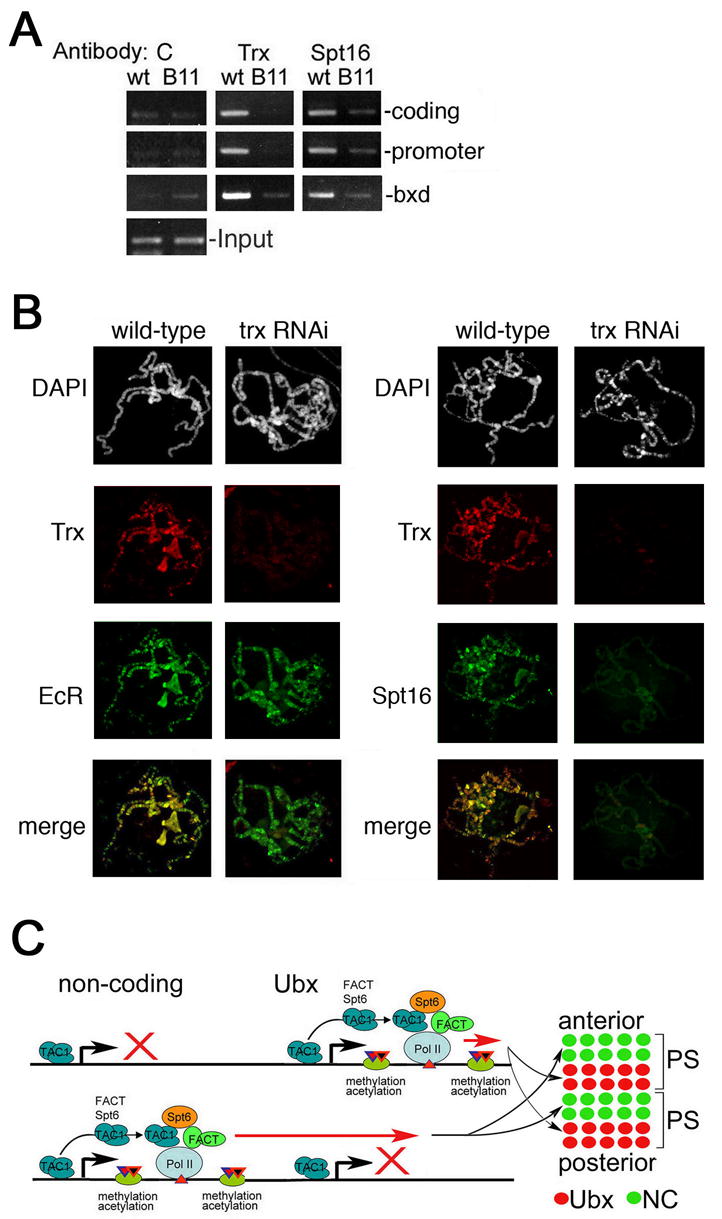
(A) Chromatin prepared from wildtype (wt) and trxB11 (B11) homozygous mutant embryos was immunoprecipitated with antibodies against Trx or Spt16. Immunoprecipitated material was PCR amplified in with primers for the region 2 kb downstream of the start site (top), for the Ubx promoter (middle), and for the central C bxd region (bottom) (Figure 5C). C, no antibody control. Input is shown at the bottom.
(B) Salivary gland polytene chromosomes were prepared from wildtype 3rd instar larvae and a line expressing trx RNAi. The overall structure of chromosomes is indistinguishable (first row). Binding by Trx (row 2) and Spt16 (right row 3) is almost completely abolished in the trx RNAi line, while Ecdysone Receptor (EcR) binding is unaffected (row 3 left). Merge is shown at the bottom.
(C) A model for the role of TAC1 in transcription. TAC1 recruitment along with elongation factors and histone modification is necessary for efficient transcriptional elongation of Ubx and ncRNAs in complementary cells in the posterior and anterior regions of each parasegment, respectively. Efficient elongation of Pol II from promoters of ncRNAs proceeds through the 5’-regulatory elements of Ubx, thus preventing its expression. This generates a mosaic pattern of Ubx expression within embryonic parasegments.
Components of the FACT elongation complex, Spt16 and SSRP1, were previously shown to be associated with the bxd region of Ubx (Shimojima et al., 2003). We find that Spt16 association with the bxd region is diminished in trxB11 embryos (Figure 7A). Therefore, association of FACT with the transcribed regions of both Ubx and bxd ncRNAs is TAC1-dependent, confirming that TAC1 is involved in transcriptional elongation of both Ubx and ncRNAs. Taken together, our results indicate that TAC1 and FACT are coordinately recruited to the elongating Pol II complex downstream of both the Ubx and bxd ncRNA promoters.
To extend this analysis, we asked whether TAC1 is required for FACT association with target genes on a broader scale by examining binding of Spt16 to salivary gland polytene chromosomes of third instar larvae from wildtype and trx RNAi mutant animals. We created a transgenic fly line that carries a Gal4-UAS-driven RNAi construct for the trx gene, in which expression of trx RNAi can be induced using the hsp70-Gal4 driver. Induction during the early third larval instar allows these animals to survive long enough to examine polytene chromosomes. In wild type, binding sites of Trx largely overlap with those of Spt16 (Figure 7B). The number of binding sites detected on polytene chromosomes with of anti-Trx antibody depend on the titer of antibody used. In these experiments, we used higher titer of Trx antibody to identify previously undetected minor sites, which are extensive (Figure 7B, compare to Smith et al. 2004). The structure of polytene chromosomes in trx RNAi larvae is indistinguishable from that of the wild type animals. In addition, binding of the control protein ecdysone receptor (EcR) to polytene chromosomes is unaffected (Figure 7B). However, binding of the Trx protein is strongly decreased in these mutants, especially at its characteristic strong binding sites. In the same larvae, binding of Spt16 is also strongly decreased (Figure 7B). Thus, Trx may be required for recruitment of the elongation factor Spt16 to most activated genes, suggesting a global role for Trx in transcriptional elongation.
TAC1 may also have a role in transcriptional initiation of Ubx and ncRNAs
Similar amounts of TAC1 are associated with the promoter region of Ubx in Ubx+ and Ubx− nuclei, and the same is true for the region distal to bxd ME B, which contains the P1 promoter of bxd ncRNAs (Figure 5C,D). We detected a slight but reproducible decrease in the levels of synthesis of the 5’-regions of both Ubx and ncRNAs in trxB11 mutants (Figure 5E). Interestingly, association of Trx and Sbf1 with the promoter region of Ubx is not affected in elongation factor mutants (Figure 6B). All of these results are consistent with the idea that TAC1 recruitment to the promoter is independent of the formation of the elongation complex. These results also show that overall expression of Trx and Sbf1 are not affected in elongation factor mutants. Taken together, our results are consistent with the notion that TAC1 is required primarily for efficient elongation of Ubx by Pol II, but that it may well play an additional role in initiation of transcription.
Discussion
An attractive notion has been that transcription of bxd ncRNAs, which precedes that of Ubx in embryos, facilitates correct spatial expression of Ubx. Previous studies showed that transcription through the ME could interfere with silencing (Bender and Fitzgerald, 2002; Hogga and Karch, 2002; Rank et al., 2002); (Schmitt et al., 2005), so it was proposed that bxd ncRNA transcription normally prevents recruitment of PcG proteins to the ME. However, our experiments unambiguously demonstrate that Ubx and bxd ncRNAs are transcribed in different cells in embryos. Our results also suggest that bxd ncRNAs do not facilitate Ubx expression in larval imaginal discs, as was recently proposed (Sanchez-Elsner et al., 2006). Instead, transcription of ncRNAs correlates with repression of Ubx. It is possible that the abnormal transcription induced in previous studies interfered with transcription of ncRNAs in the BX-C, rather than with ME function, a possibility that can be tested experimentally. It will be interesting to use our system of sorting Ubx+ and Ubx− nuclei to examine binding of PcG proteins in nuclei where bxd ncRNAs either are, or are not, transcribed.
Our experiments rule out trans-repression by bxd ncRNAs, and instead support repression of Ubx in cis by transcription of these RNAs per se. A likely mechanism of this repression is transcriptional interference, since we show that ncRNA transcription extends into the region just upstream of the Ubx initiation site, which may well disrupt protein-DNA interactions required for Ubx initiation. However, this does not rule out promoter competition, and both of these mechanisms may contribute to the observed effects. Previous genetic studies (Grimaud et al., 2006a) and the results presented here show that bxd ncRNAs do not work by RNAi. An RNAi-based repression mechanism has been described for the miRNA produced by the iab-4 transcript, which directly interacts with the 3’-untranslated region of Ubx and prevents translation (Ronshaugen et al., 2005). These authors show that ectopic expression of iab-4 leads to homeotic phenotypes in the haltere, but do not show that loss of RNAi prevents this effect, nor has the effect of loss of function mutations of the iab-4 transcript been tested, so it remains to be seen if the iab-4 transcript is a bona fide miRNA.
As we did not detect significant levels of bxd ncRNAs in imaginal discs, nor do they persist to late embryonic stages, they are unlikely be responsible for repression of Ubx throughout development. In fact, Papp and Muller (2006) report that Trx is bound to the bxd ME in both wing and haltere discs, which have low and high levels of Ubx expression, respectively. The difference between binding of Trx to the bxd ME in embryos and in discs (Papp and Muller, 2006) may be a consequence of the absence of transcription of bxd ncRNAs in discs and its presence in embryos, or to other uncharacterized differences between Ubx regulation in embryos and discs. Also, as we show that Trx binds constitutively in some areas of the ME, Papp and Muller may have detected such binding in imaginal discs.
Intergenic transcription also cannot explain repression of Ubx in the anterior of the embryo, where it is thought that hunchback and PcG genes set up and maintain the anterior boundary of Ubx expression. However, the pattern of bxd ncRNA transcription, which prefigures, in a complementary fashion, the mosaic pattern of Ubx expression within the parasegments of the embryonic trunk, appears to be essential for proper Ubx initiation. The Ubx pattern may then be maintained or modified at later embryonic stages through repression by other Hox proteins (i.e., abdA and AbdB) and by PcG genes. Thus, maintenance of Ubx expression likely requires multiple mechanisms that are employed at different developmental stages.
Our data support a role for Trx in transcriptional elongation as a mechanism for maintenance of a developmentally regulated gene. It has been argued that Trx does not have a direct role in activation of homeotic genes in Drosophila, but instead prevents repression of transcription by PcG proteins (Klymenko and Muller, 2004). However, our data suggest that trx is required for recruitment of elongation factors and for efficient completion of transcripts. Therefore, maintenance of transcriptional activity by Trx may be a consequence of its role in elongation, and a block in elongation might lead to the establishment of PcG-mediated repression. Alternatively, Trx may be required only for normal levels of Hox gene expression, and not for maintenance of low levels of expression, a possibility consistent with at least some aspects of the trx mutant phenotype.
This work strongly supports a general role for Trx and TAC1 in transcription, and agrees with our previous findings that TAC1 relocates from other genes to the transcribed region of hsp70 following induction of the cellular stress response (Smith et al., 2004). The histone methyltransferase activity of Set1, the SET domain protein homologous to Trx, has a role in transcription (Hampsey and Reinberg, 2003), and MLL was suggested to play a similar role in mammals (Guenther et al., 2005; Hughes et al., 2004; Milne et al., 2005; Yokoyama et al., 2004). We suggest that this role is in transcriptional elongation, because Trx and elongation factors are co-ordinately recruited, because Trx binds downstream of the promoter more strongly to the 5’ than the 3’ end, and because transcripts extending to the 3’ end are more strongly affected by trx mutations, for both Ubx and bxd ncRNAs.
TAC1 is also present at the promoter (Figure 5C,D), and this is unaffected by mutations in elongation factors (Figure 6B). Therefore, association of TAC1 with the promoter likely precedes the recruitment of elongation factors. Thus, TAC1 may play several distinct roles, one in initiation, another during the recruitment of the elongation complex and perhaps a third during subsequent elongation, where its ability to modify histones may be required for effective completion of long transcripts.
This work provides the first direct evidence of the involvement of long ncRNAs in regulation of homeotic genes of Drosophila. Repression of Ubx is apparently mediated by expression of several intergenic ncRNAs in different germ layers of Ubx-expressing parasegments. TAC1 may be required for efficient read-through by Pol II into the region upstream of the Ubx initiation site, and as a result, for efficient repression of Ubx (see model in Figure 7C). Therefore, we propose a direct link between elongation facilitated by the TAC1 epigenetic complex and repression of Ubx by intergenic transcription. A goal for the future will be to determine if other homeotic genes of Drosophila, and of other organisms, are also regulated by long ncRNAs whose expression is regulated by TAC1 proteins.
Experimental Procedures
Drosophila Genetics
Details on strains used and their construction can be found in the Supplementary material. All strains were maintained on standard medium at 25oC. Homozygous mutant embryos were collected from stocks carrying either the trxB11, spt4, spt6, or spt16 mutations over Kr-GAL4, UAS-GFP-carrying balancer chromosomes (Smith et al., 2004) based on the absence of GFP expression. The wildtype strain Oregon R was used as a control.
Isolation and sorting of embryonic nuclei
Nuclei were prepared using a procedure already described (Petruk et al. 2001), and details are given in Supplementary material. Nuclei were sorted on a Coulter ELITE ESP cell sorter at 40C. After sorting, an aliquot of 10,000 nuclei was used to prepare RNA, while the rest of the material was used for ChIP experiments (see Immunoprecipitation).
RNA preparation and RT-PCR
Each batch of sorted nuclei was analysed by RT-PCR for at least a 10-fold enrichment of Ubx RNA in GFP-positive nuclei using standard procedures. RNA from 25 wildtype or trxB11 mutant embryos or from 15 dissected discs was prepared using the High Pure RNA Isolation Kit (Roche), and RT was performed using random primers. For coordinates of primer sets, see Supplementary material.
Chromatin Immunoprecipitation
ChIP experiments were performed according to the Upstate Biotechnology protocol, using 150,000 to 200,000 sorted nuclei or 50 whole embryos per sample. Details of the procedure, antibodies used, and coordinates of primers are given in Supplementary Experimental Procedures.
In situ hybridization and immunostaining
Embryo and larvae fixation, preparation of labelled RNA probes, and nascent transcript RNA FISH were performed according to (Kosman et al., 2004; Kuzin et al., 1994). cDNA sequences were synthesized by PCR and subcloned into the pGEM-T vector (Promega). For coordinates of probes, see Supplementary material.
RNA probes were labelled with DIG-, and Biotin-conjugated UTP and were detected as follows: DIG: sheep anti-DIG (Roche), Alexa 555 donkey anti-sheep; Biotin: mouse anti-Biotin (Roche), Alexa 488 donkey anti-mouse. Images of embryos were obtained using a confocal microscope in the KCC imaging facility. Preparation and immunostaining of chromosome spreads were performed as described (Tillib at al., 1999).
Injections of dsRNAs
dsRNAs specific to exons 1-3-7 and bxd were synthesized from the same constructs that were used for in situ hybridization. Sense and antisense RNAs were synthesized using the Riboprobe in vitro Transcription System (Promega). Equal amounts of sense and antisense RNAs were annealed by heating at 900C for 3 min and cooling down slowly to room temperature. Equal amounts of the exon 1–7 and bxd dsRNAs were combined and used for injection at 10ng/ml either in preblastoderm embryos using standard procedures, or in adult females as described (Dzitoyeva et al., 2003). GFP (Stratagene) dsRNA was used as a control.
Supplementary Material
Acknowledgments
We thank S. Noselli, M. Bourouis and O. Beaudouin-Massiani for antibodies and mutant stocks, M. Fujioka for GFP-vector, S. Dzitoyeva for help with fly injections, and W. Jankowski for help in using the confocal microscope in the KCC Bioimaging Facility. This work was supported by the following grants: NIH 1R01GM075141 and 5P01CA50507 and March of Dimes 6-FY06-346 to A.M., 11671 from the Canadian Institute of Health Research to H.W.B., NIH T32-HL07780 to K.R., 17002018 from the MEXT of Japan to S.H. and NSF 0416760 and NIH 2R01GM50231 to J.B.J.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bae E, Calhoun VC, Levine M, Lewis EB, Drewell RA. Characterization of the intergenic RNA profile at abdominal-A and Abdominal-B in the Drosophila bithorax complex. Proc Natl Acad Sci U S A. 2002;99:16847–16852. doi: 10.1073/pnas.222671299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beisel C, Imhof A, Greene J, Kremmer E, Sauer F. Histone methylation by the Drosophila epigenetic transcriptional regulator Ash1. Nature. 2002;419:857–862. doi: 10.1038/nature01126. [DOI] [PubMed] [Google Scholar]
- Bender W, Fitzgerald DP. Transcription activates repressed domains in the Drosophila bithorax complex. Development. 2002;129:4923–4930. doi: 10.1242/dev.129.21.4923. [DOI] [PubMed] [Google Scholar]
- Brower DL. Ultrabithorax gene expression in Drosophila imaginal discs and larval nervous system. Development. 1987;101:83–92. doi: 10.1242/dev.101.1.83. [DOI] [PubMed] [Google Scholar]
- Carroll SB, DiNardo S, O'Farrell PH, White RA, Scott MP. Temporal and spatial relationships between segmentation and homeotic gene expression in Drosophila embryos: distributions of the fushi tarazu, engrailed, Sex combs reduced, Antennapedia, and Ultrabithorax proteins. Genes Dev. 1988;2:350–360. doi: 10.1101/gad.2.3.350. [DOI] [PubMed] [Google Scholar]
- Carthew RW. Gene regulation by microRNAs. Curr Opin Genet Dev. 2006;16:203–208. doi: 10.1016/j.gde.2006.02.012. [DOI] [PubMed] [Google Scholar]
- Chan CS, Rastelli L, Pirrotta V. A Polycomb response element in the Ubx gene that determines an epigenetically inherited state of repression. Embo J. 1994;13:2553–2564. doi: 10.1002/j.1460-2075.1994.tb06545.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cumberledge S, Zaratzian A, Sakonju S. Characterization of two RNAs transcribed from the cis-regulatory region of the abd-A domain within the Drosophila bithorax complex. Proc Natl Acad Sci U S A. 1990;87:3259–3263. doi: 10.1073/pnas.87.9.3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dzitoyeva S, Dimitrijevic N, Manev H. Identification of a novel Drosophila gene, beltless, using injectable embryonic and adult RNA interference (RNAi) BMC Genomics. 2003;4:33. doi: 10.1186/1471-2164-4-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy SR. Computational genomics of noncoding RNA genes. Cell. 2002;109:137–140. doi: 10.1016/s0092-8674(02)00727-4. [DOI] [PubMed] [Google Scholar]
- Goodrich JA, McClure WR. Competing promoters in prokaryotic transcription. Trends Biochem Sci. 1991;16:394–397. doi: 10.1016/0968-0004(91)90162-o. [DOI] [PubMed] [Google Scholar]
- Gottesman S. Stealth regulation: biological circuits with small RNA switches. Genes Dev. 2002;16:2829–2842. doi: 10.1101/gad.1030302. [DOI] [PubMed] [Google Scholar]
- Grimaud C, Bantignies F, Pal-Bhadra M, Ghana P, Bhadra U, Cavalli G. RNAi components are required for nuclear clustering of Polycomb group response elements. Cell. 2006a;124:957–971. doi: 10.1016/j.cell.2006.01.036. [DOI] [PubMed] [Google Scholar]
- Grimaud C, Negre N, Cavalli G. From genetics to epigenetics: the tale of Polycomb group and trithorax group genes. Chromosome Res. 2006b;14:363–375. doi: 10.1007/s10577-006-1069-y. [DOI] [PubMed] [Google Scholar]
- Guenther MG, Jenner RG, Chevalier B, Nakamura T, Croce CM, Canaani E, Young RA. Global and Hox-specific roles for the MLL1 methyltransferase. Proc Natl Acad Sci U S A. 2005;102:8603–8608. doi: 10.1073/pnas.0503072102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampsey M, Reinberg D. Tails of intrigue: phosphorylation of RNA polymerase II mediates histone methylation. Cell. 2003;113:429–432. doi: 10.1016/s0092-8674(03)00360-x. [DOI] [PubMed] [Google Scholar]
- Hodgson JW, Argiropoulos B, Brock HW. Site-specific recognition of a 70-base-pair element containing d(GA)(n) repeats mediates bithoraxoid polycomb group response element-dependent silencing. Mol Cell Biol. 2001;21:4528–4543. doi: 10.1128/MCB.21.14.4528-4543.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogga I, Karch F. Transcription through the iab-7 cis-regulatory domain of the bithorax complex interferes with maintenance of Polycomb-mediated silencing. Development. 2002;129:4915–4922. doi: 10.1242/dev.129.21.4915. [DOI] [PubMed] [Google Scholar]
- Hughes CM, Rozenblatt-Rosen O, Milne TA, Copeland TD, Levine SS, Lee JC, Hayes DN, Shanmugam KS, Bhattacharjee A, Biondi CA, et al. Menin associates with a trithorax family histone methyltransferase complex and with the hoxc8 locus. Mol Cell. 2004;13:587–597. doi: 10.1016/s1097-2765(04)00081-4. [DOI] [PubMed] [Google Scholar]
- Irish VF, Martinez-Arias A, Akam M. Spatial regulation of the Antennapedia and Ultrabithorax homeotic genes during Drosophila early development. Embo J. 1989;8:1527–1537. doi: 10.1002/j.1460-2075.1989.tb03537.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irvine KD, Helfand SL, Hogness DS. The large upstream control region of the Drosophila homeotic gene Ultrabithorax. Development. 1991;111:407–424. doi: 10.1242/dev.111.2.407. [DOI] [PubMed] [Google Scholar]
- Klymenko T, Muller J. The histone methyltransferases Trithorax and Ash1 prevent transcriptional silencing by Polycomb group proteins. EMBO Rep. 2004;5:373–377. doi: 10.1038/sj.embor.7400111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosman D, Mizutani CM, Lemons D, Cox WG, McGinnis W, Bier E. Multiplex detection of RNA expression in Drosophila embryos. Science. 2004;305:846. doi: 10.1126/science.1099247. [DOI] [PubMed] [Google Scholar]
- Kuzin B, Tillib S, Sedkov Y, Mizrokhi L, Mazo A. The Drosophila trithorax gene encodes a chromosomal protein and directly regulates the region-specific homeotic gene fork head. Genes Dev. 1994;8:2478–2490. doi: 10.1101/gad.8.20.2478. [DOI] [PubMed] [Google Scholar]
- Lipshitz HD, Peattie DA, Hogness DS. Novel transcripts from the Ultrabithorax domain of the bithorax complex. Genes Dev. 1987;1:307–322. doi: 10.1101/gad.1.3.307. [DOI] [PubMed] [Google Scholar]
- Martens JA, Laprade L, Winston F. Intergenic transcription is required to repress the Saccharomyces cerevisiae SER3 gene. Nature. 2004;429:571–574. doi: 10.1038/nature02538. [DOI] [PubMed] [Google Scholar]
- Massirer KB, Pasquinelli AE. The evolving role of microRNAs in animal gene expression. Bioessays. 2006;28:449–452. doi: 10.1002/bies.20406. [DOI] [PubMed] [Google Scholar]
- Mattick JS, Makunin IV. Non-coding RNA. Hum Mol Genet . 2006;15:R17–29. doi: 10.1093/hmg/ddl046. Spec No 1. [DOI] [PubMed] [Google Scholar]
- Mazo AM, Huang DH, Mozer BA, Dawid IB. The trithorax gene, a trans-acting regulator of the bithorax complex in Drosophila, encodes a protein with zinc-binding domains. Proc Natl Acad Sci U S A. 1990;87:2112–2116. doi: 10.1073/pnas.87.6.2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milne TA, Dou Y, Martin ME, Brock HW, Roeder RG, Hess JL. MLL associates specifically with a subset of transcriptionally active target genes. Proc Natl Acad Sci U S A. 2005;102:14765–14770. doi: 10.1073/pnas.0503630102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller J, Bienz M. Long range repression conferring boundaries of Ultrabithorax expression in the Drosophila embryo. Embo J. 1991;10:3147–3155. doi: 10.1002/j.1460-2075.1991.tb04876.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pallavi SK, Shashidhara LS. Egfr/Ras pathway mediates interactions between peripodial and disc proper cells in Drosophila wing discs. Development. 2003;130:4931–4941. doi: 10.1242/dev.00719. [DOI] [PubMed] [Google Scholar]
- Papp B, Muller J. Histone trimethylation and the maintenance of transcriptional ONand OFF states by trxG and PcG proteins. Genes Dev. 2006;20:2041–2054. doi: 10.1101/gad.388706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petruk S, Sedkov Y, Smith S, Tillib S, Kraevski V, Nakamura T, Canaani E, Croce CM, Mazo A. Trithorax and dCBP Acting in a Complex to Maintain Expression of a Homeotic Gene. Science. 2001;294:1331–1334. doi: 10.1126/science.1065683. [DOI] [PubMed] [Google Scholar]
- Pirrotta V, Chan CS, McCabe D, Qian S. Distinct parasegmental and imaginal enhancers and the establishment of the expression pattern of the Ubx gene. Genetics. 1995;141:1439–1450. doi: 10.1093/genetics/141.4.1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rank G, Prestel M, Paro R. Transcription through intergenic chromosomal memory elements of the Drosophila bithorax complex correlates with an epigenetic switch. Mol Cell Biol. 2002;22:8026–8034. doi: 10.1128/MCB.22.22.8026-8034.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronshaugen M, Biemar F, Piel J, Levine M, Lai EC. The Drosophila microRNA iab-4 causes a dominant homeotic transformation of halteres to wings. Genes Dev. 2005;19:2947–2952. doi: 10.1101/gad.1372505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Elsner T, Gou D, Kremmer E, Sauer F. Noncoding RNAs of trithorax response elements recruit Drosophila Ash1 to Ultrabithorax. Science. 2006;311:1118–1123. doi: 10.1126/science.1117705. [DOI] [PubMed] [Google Scholar]
- Sanchez-Herrero E, Akam M. Spatially ordered transcription of regulatory DNA in the bithorax complex of Drosophila. Development. 1989;107:321–329. doi: 10.1242/dev.107.2.321. [DOI] [PubMed] [Google Scholar]
- Schmitt S, Prestel M, Paro R. Intergenic transcription through a polycomb group response element counteracts silencing. Genes Dev. 2005;19:697–708. doi: 10.1101/gad.326205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen GL, Blau HM. A brief history of RNAi: the silence of the genes. Faseb J. 2006;20:1293–1299. doi: 10.1096/fj.06-6014rev. [DOI] [PubMed] [Google Scholar]
- Shimojima T, Okada M, Nakayama T, Ueda H, Okawa K, Iwamatsu A, Handa H, Hirose S. Drosophila FACT contributes to Hox gene expression through physical and functional interactions with GAGA factor. Genes Dev. 2003;17:1605–1616. doi: 10.1101/gad.1086803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon J, Chiang A, Bender W, Shimell MJ, O'Connor M. Elements of the Drosophila bithorax complex that mediate repression by Polycomb group products. Dev Biol. 1993;158:131–144. doi: 10.1006/dbio.1993.1174. [DOI] [PubMed] [Google Scholar]
- Smith ST, Petruk S, Sedkov Y, Cho E, Tillib S, Canaani E, Mazo A. Modulation of heat shock gene expression by the TAC1 chromatin-modifying complex. Nat Cell Biol. 2004;6:162–167. doi: 10.1038/ncb1088. [DOI] [PubMed] [Google Scholar]
- Soto MC, Chou TB, Bender W. Comparison of germline mosaics of genes in the Polycomb group of Drosophila melanogaster. Genetics. 1995;140:231–243. doi: 10.1093/genetics/140.1.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl G, Akam M. Altered distributions of Ultrabithorax transcripts in extra sex combs mutant embryos of Drosophila. Embo J. 1985;4:3259–3264. doi: 10.1002/j.1460-2075.1985.tb04075.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tillib S, Petruk S, Sedkov Y, Kuzin A, Fujioka M, Goto T, Mazo A. Trithorax- and Polycomb-group response elements within an Ultrabithorax transcription maintenance unit consist of closely situated but separable sequences. Mol Cell Biol. 1999;19:5189–5202. doi: 10.1128/mcb.19.7.5189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama A, Wang Z, Wysocka J, Sanyal M, Aufiero DJ, Kitabayashi I, Herr W, Cleary ML. Leukemia proto-oncoprotein MLL forms a SET1-like histone methyltransferase complex with menin to regulate Hox gene expression. Mol Cell Biol. 2004;24:5639–5649. doi: 10.1128/MCB.24.13.5639-5649.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


