Abstract
Consumption of ethanol solutions by rodents in two-bottle choice tests is a model to study human alcohol intake. Mice of the C57BL/6ByJ strain have higher ethanol preferences and intakes than do mice of the 129P3/J strain. F2 hybrids between these two strains were phenotyped using two-bottle tests involving a choice between water and either 3% or 10% ethanol. High ethanol preferences and intakes of the B6 mice were inherited as additive or dominant traits in the F2 generation. A genome screen using these F2 mice identified three significant linkages. Two loci, on distal chromosome 4 (Ap3q) and proximal chromosome 7 (Ap7q), strongly affected 10% ethanol intake and weakly affected 3% ethanol intake. A male-specific locus on proximal chromosome 8 (Ap8q) affected 3% ethanol preference, but not indexes of 10% ethanol consumption. In addition, six suggestive linkages (on chromosomes 2, 9, 12, 13, 17, and 18) affecting indexes of 3% and/or 10% ethanol consumption were detected. The loci with significant and suggestive linkages accounted for 35–44% of the genetic variation in ethanol consumption phenotypes. No additive-by-additive epistatic interactions were detected for the primary loci with significant and suggestive linkages. However, there were a few modifiers of the primary linkages and a number of interactions among unlinked loci. This demonstrates a significant role of the genetic background in the variation of ethanol consumption.
[Supplementary material is available online at www.genome.org.]
Human alcohol consumption is a complex trait regulated by multiple mechanisms. Consistent with this, the inheritance of ethanol consumption by humans and animals is multigenic. Studies in humans have detected several candidate genes associated with human alcoholism, although the evidence is still somewhat controversial (for review, see Foroud and Li 1999). Another approach, using genome-wide linkage analyses in humans, also yielded several loci contributing to human alcoholism (Long et al. 1998; Foroud et al. 2000). Human studies have had limited statistical power and reproducibility, but genetic analyses using animal models complement them and help to overcome their limitations (Foroud and Li 1999). Several ethanol-related phenotypes in experimental animals reproduce different aspects of the complex effects of ethanol (Crabbe et al. 1994, 1999). One of the phenotypes, ethanol preference (also referred to as “voluntary consumption”), is assessed using two-bottle choice tests with an ethanol solution presented in one bottle and water presented in the second bottle. Although, like any model, mouse ethanol preference does not replicate the full spectrum of mechanisms involved in human alcohol consumption, it is considered relevant to human alcohol drinking (e.g., Lester and Freed 1973; Li et al. 1979; Dole and Gentry 1984; Dole 1986; Crabbe et al. 1994; Erickson 1996; Myers 1996; Arola et al. 1997) and is widely used in experimental studies on alcoholism.
Several laboratories have mapped quantitative trait loci (QTL) affecting ethanol preference in animal models using parental strains with contrasting phenotypes (for review, see Crabbe et al. 1999; Foroud and Li 1999; Belknap and Atkins 2001). In mice, the C57BL/6 strain has been most commonly used as one parent because of its high ethanol consumption. In most studies, the low-preferring strain used has been DBA/2 (e.g., Phillips et al. 1994, 1998; Melo et al. 1996; Belknap et al. 1997; Tarantino et al. 1998; Whatley et al. 1999), with a few exceptions (Gill et al. 1998; Vadasz et al., 2000). In this study, we produced an F2 intercross of the high ethanol-preferring C57BL/6ByJ (B6) strain with the 129P3/J (129) strain, which has low ethanol consumption (Belknap et al. 1993; Bachmanov et al. 1996b, 2000; Logue et al. 1998) and had not been used previously in ethanol-related mapping studies.
In the previous studies on mapping ethanol preference, a single 10% ethanol concentration was used. Although strain differences in ethanol consumption extend over a range of concentrations (Belknap et al. 1993), it is possible that responses to different concentrations involve different mechanisms (e.g., sensory, metabolic, or pharmacological) and thus may have a different genetic architecture. To investigate this, we tested the F2 mice sequentially with 3% and 10% ethanol solutions paired with water.
To determine chromosomal positions of genetic loci affecting ethanol consumption, a genome screen was conducted by genotyping polymorphic markers in the F2 mice. In addition to mapping primary QTL, we conducted a genome-wide search of additive-by-additive epistatic interactions. These interactions between loci in the homozygous condition are present in inbred, recombinant inbred, and congenic strains. Thus, the additive-by-additive interactions detected in F2 should be of practical significance for selection of congenic strains, which we plan to conduct. Using the B6 × 129 cross, two different ethanol concentrations, and genome-wide analysis of epistasis allowed us to discover novel ethanol preference loci.
RESULTS
Analyses of Phenotypes
Parental Strains
All indexes of ethanol consumption (intakes and preference scores for 3% and 10% ethanol) were overall higher in the B6 mice than in the 129 mice [effect of strain, F(1,35) >32.6, P<0.000003; two-way general analysis of variance (ANOVA)]. Females had higher 3% ethanol intakes and preferences than did males [effect of gender, F(1,35) > 5.5, P<0.03], but there were no significant gender differences for indexes of 10% ethanol consumption. The only significant strain × gender interaction by ANOVA was found for 3% ethanol preferences [F(1,35) = 9.2, P<0.005]; the strain differences were larger in males than in females.
F2 Hybrids
The F2 mice from three types of reciprocal crosses were compared using two-way ANOVA with gender and cross as between-group factors. Indexes of ethanol consumption for both 3% and 10% concentrations did not differ between the reciprocal crosses. Therefore, the data for all F2 mice were combined in subsequent analyses.
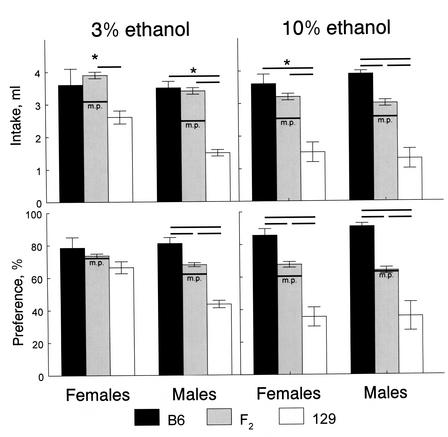
To assess the mode of inheritance (additive and dominant effects characterizing allelic interactions), the indexes of ethanol consumption of the F2 mice were compared with those of the B6 and 129 mice, and with corresponding midparental values, using two-way ANOVA with genotype (B6, 129, or F2) and gender as between-group factors. For all indexes of 3% and 10% ethanol consumption, there were significant differences among the three genotypes [F(2,483) > 10, P<0.001; Fig. 1]. Gender differences were found only for 3% ethanol preferences [they were higher in females than in males; F(1,483) = 5.1, P<0.05]. With the exception of 10% ethanol intake by males, ethanol intakes of the F2 mice were significantly higher than corresponding midparental and 129 values, and they did not differ from the B6 values, which indicates directional dominance of the B6 alleles. Ethanol preferences (and 10% ethanol intake by males) of the F2 mice did not differ significantly from midparental values, which indicates additive inheritance of these traits. These modes of inheritance suggest that the F2 intercross is appropriate for linkage analyses.
Figure 1.
Average daily ethanol intakes and preferences of B6, 129, and F2 mice (means ± standard errors). Horizontal lines above the bars represent significant differences between groups (P<0.05, planned comparisons, ANOVA). Horizontal lines labeled “m.p.” on the F2 bars show midparental values (average of B6 and 129 means). *Significant difference between F2 and midparental values (P<0.05, planned comparisons between the F2 and a collapsed value of the two parental strains).
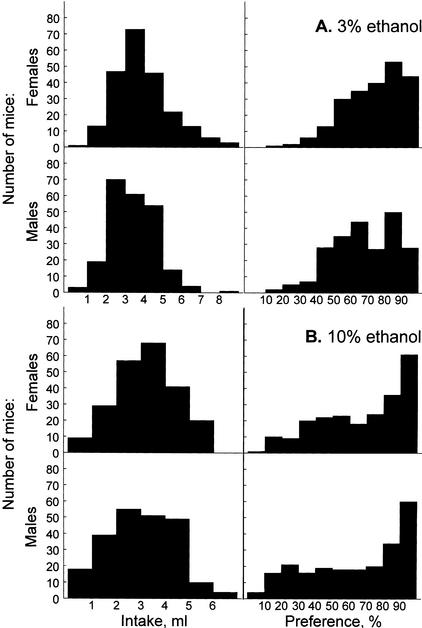
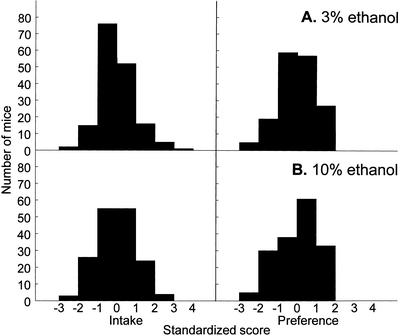
Distributions of ethanol intakes by the F2 mice did not differ significantly from the normal distribution (Fig. 2, left panels; P>0.2, Kolmogorov-Smirnov test, except for 3% ethanol intakes by females, P<0.05). Distributions for preference scores differed significantly from normal (Fig. 2, right panels; P<0.05), with a large proportion of mice showing preferences close to the upper limit of this index (100%). Square-root, decimal, and natural logarithm transformations did not alleviate the departure of the preference scores from the normal distribution. We therefore used untransformed preference scores in linkage analyses. Distributions of the same indexes were similar in males and females; any shifts between the distributions for the two genders (when their means differed) were offset by standardizing data within each gender (see Methods). Distributions of these standardized scores of the 169 F2 mice that were randomly selected for genotyping are shown in Figure 3; these distributions did not differ significantly from normal (P>0.1, Kolmogorov-Smirnov test), except for 10% ethanol preference scores (P<0.01).
Figure 2.
Distributions of 3% (A) and 10% (B) ethanol intakes (left) and preferences (right) in F2 mice. Top rows, females (n = 224); bottom rows, males (n = 226).
Figure 3.
Distributions of standardized 3% (A) and 10% (B) ethanol intakes (left) and preferences (right) in a subset of F2 mice of both genders used for genotyping (n = 169).
Heritability was estimated based on variances in the segregating (F2) and nonsegregating (B6 and 129) populations. The heritability indexes in males for 3% ethanol were 84% (intake) and 73% (preference), and for 10% ethanol they were 68% (intake) and 50% (preference). In females, the heritabilities were 56, 17, 55, and 60%, respectively. Thus, with the exception of 3% ethanol preference by females, at least half of the variance in the F2 hybrids was the result of genetic factors.
Correlations among indexes of ethanol consumption were all positive and significant, with strong correlations for intakes and preferences for the same ethanol concentration, and only moderate correlations between the indexes for different (3 and 10%) ethanol concentrations (Table 1).
Table 1.
Correlations Among Indexes of Ethanol Consumption in F2 Females (Top) and Males (Bottom)
| 3% Ethanol preference | 10% Ethanol intake | 10% Ethanol preference | |
|---|---|---|---|
| Females (n = 224) | |||
| 3% Ethanol intake | +0.73** | +0.36** | +0.15* |
| 3% Ethanol preference | – | +0.43** | +0.44** |
| 10% Ethanol intake | – | – | +0.88** |
| Males (n = 226) | |||
| 3% Ethanol intake | +0.75** | +0.53** | +0.34** |
| 3% Ethanol preference | – | +0.51** | +0.57** |
| 10% Ethanol intake | – | – | +0.89** |
P < 0.05; **P < 0.0042 (Bonferroni correction for 12 comparisons, 0.05/12 ≈ 0.0042).
Linkage Analyses
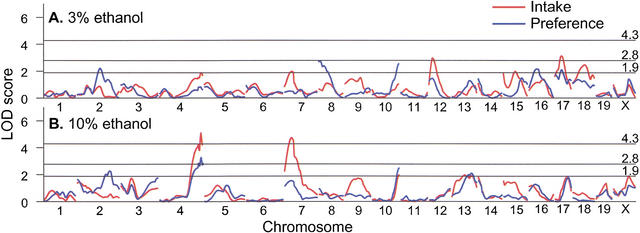
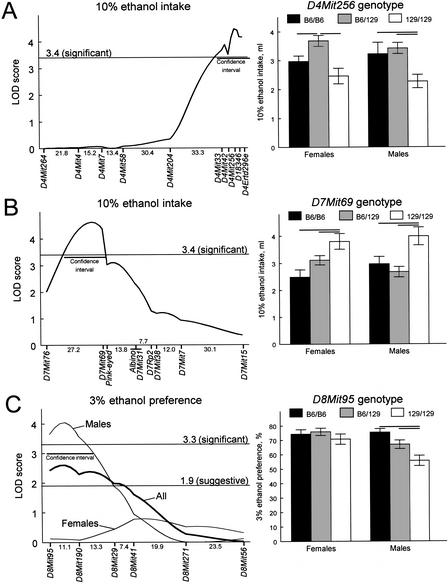
The results of the genome scan for indexes of ethanol consumption are shown in Figure 4 (unconstrained model) and Table 2 (locus-specific modes of inheritance). Loci on chromosomes 4 and 7 (Fig. 5A,B) significantly affected 10% ethanol intake and also had a weaker effect on 10% ethanol preference and 3% ethanol intake (Fig. 4). Effects of these loci on 10% ethanol intake were similar in males and females. A male-specific locus on chromosome 8 affected 3% ethanol preference (significant linkage, Fig. 5C) and intake (suggestive linkage, Table 2), but not indexes of 10% ethanol consumption. The linkage on distal chromosome 4 most likely corresponds to the Ap3q (alcohol preference 3 QTL) locus (Tarantino et al. 1998). The loci on chromosomes 7 and 8 were novel, and they were assigned symbols Ap7q and Ap8q (alcohol preference 7 and 8 QTL), respectively.
Figure 4.
Genome-wide scan for linkages with indexes of 3% (A) and 10% (B) ethanol consumption. Distances between markers were estimated using MAPMAKER/EXP. Curves trace the logarithm of the odds ratio (LOD) scores calculated under an unconstrained (free) model using MAPMAKER/QTL. Horizontal lines show thresholds for significant (LOD 4.3) and suggestive (LOD 2.8) linkages under the unconstrained model (Lander and Kruglyak 1995), and a more relaxed threshold (LOD 1.9) used to identify regions of potential linkages for additional genotyping.
Table 2.
Summary of Suggestive and Significant Linkages
| Markera | Position | Populationd | Mode of inheritancee | LODf | % of Phenotypical variance explainedg | Allele increasing phenotypical score | |
|---|---|---|---|---|---|---|---|
| MITb | MGDc | ||||||
| 3% Ethanol intake | |||||||
| D4Mit42 | 76.5 | 81.0 | Males | B6-dominant | 2.65* | 14.7 | B6 |
| D8Mit95 | 6.6 | 8.0 | Males | Additive | 1.92* | 11.1 | B6 |
| D9Mit25 | 20.8 | 26.0 | Males | 129-dominant | 2.61* | 17.6 | B6 |
| D12Mit46 | 13.1 | 17.0 | Both genders | B6-dominant | 2.75* | 8.5 | 129 |
| D17Mit6 | 27.3 | 31.0 | Both genders | 129-dominant | 3.13* | 8.3 | 129 |
| D18Mit55 | 16.4 | 25.0 | Both genders | Additive | 2.24* | 7.0 | B6 |
| 3% Ethanol preference | |||||||
| D2Mit12 | 50.3 | 50.3 | Both genders | 129-dominant | 2.22* | 6.0 | 129 |
| D8Mit95 | 6.6 | 8.0 | Males | Additive | 3.65** | 19.3 | B6 |
| D17Mit6 | 27.3 | 31.0 | Both genders | 129-dominant | 2.09* | 5.6 | 129 |
| 10% Ethanol intake | |||||||
| D4Mit256 | 82.0 | 82.7 | Both genders | B6-dominant | 4.47** | 13.3 | B6 |
| D7Mit69 | 20.8 | 24.5 | Both genders | B6-dominant | 4.37** | 11.8 | 129 |
| 10% Ethanol preference | |||||||
| D2Mit168h | 66.7 | 81.7 | Both genders | Additive | 1.94* | 5.4 | 129 |
| D4Mit256 | 82.0 | 82.7 | Both genders | B6-dominant | 2.92* | 8.7 | B6 |
| D13Mit147 | 37.2 | 51.0 | Both genders | Additive | 1.93* | 5.3 | B6 |
Marker showing the strongest linkage.
MIT—MIT/Whitehead Institute database (http://www-genome.wi.mit.edu/).
MGD—Mouse Genome Database at The Jackson Laboratory (http://www.informatics.jax.org/).
Only one gender is shown when linkage was above significant or suggestive threshold for this gender, but not for the other gender or for the combined sample of both genders. Note that linkage for distal chromosome 4 is present in both genders for 10% ethanol intake and preference (D4Mit256) and is male-specific for 3% ethanol intake (D4Mit42).
Estimated based on LOD scores calculated using MAPMAKER/QTL under unconstrained, additive, recessive, and dominant models.
LOD, logarithm of the odds ratio. Estimated for the most likely mode of inheritance.
Estimated for the unconstrained model.
For 10% ethanol preference, peak LOD score on chromosome 2 was at the agouti coat color locus (LOD 2.2, explained 6.6% of phenotypical variance, unconstrained model), but because this is a dominant marker (genotypes AA and Aa cannot be distinguished), mode of inheritance for this QTL was assessed based on genotype of another linked marker, D2Mit168.
Suggestive linkage (LOD thresholds 1.9 for additive and 2.0 for dominant/recessive models).
Significant linkage (logarithm of the odds ratio (LOD) thresholds 3.3 for additive and 3.4 for dominant/recessive models).
Figure 5.
Significant linkages. Left panels show results of interval mapping. Distances between markers in cM were estimated using MAPMAKER/EXP and are shown below the X-axis. Curves trace the logarithm of the odds ratio (LOD) scores calculated under locus-specific models using MAPMAKER/QTL. Horizontal lines show thresholds for suggestive and/or significant linkages under the locus-specific models (Lander and Kruglyak 1995). Confidence intervals (LOD drops of 1.0 from the peak) are shown by horizontal bars. Right panels show ethanol intakes or preferences by mice with different genotypes at a maker with the strongest linkage (means ± standard errors). Horizontal bars represent significant differences between groups (P<0.05, planned comparisons, two-way ANOVA). (A) A locus on distal chromosome 4 (Ap3q) affecting 10% ethanol intake. Left: B6-dominant model; the confidence interval spans region from 4 cM proximal to D4Mit33 to the telomeric end. Right: effects of D4Mit256 genotype, F(2,154) = 13.14, P = 0.000005, gender, F(1,154) = 0.11, P = 0.74, and their interaction, F(2,154) = 0.61, P = 0.55. (B) A locus on proximal chromosome 7 (Ap7q) affecting 10% ethanol intake. Left: B6-dominant model; the confidence interval spans region from 8 cM distally to D7Mit76 to 1 cM distally to D7Mit69. Right: effects of D7Mit69 genotype, F(2,160) = 11.08, P = 0.000031, gender, F(1,160) = 0.14, P = 0.71, and their interaction, F(2,160) = 2.19, P = 0.12. (C) A male-specific locus on proximal chromosome 8 (Ap8q) affecting 3% ethanol preference. Left: additive model; the confidence interval spans region from the centromeric end to 6 cM distally to D8Mit190. Right: effects of D8Mit95 genotype, F(2,161) = 6.40, P = 0.0021, gender, F(1,161) = 8.69, P = 0.0037, and their interaction, F(2,161) = 2.90, P = 0.058.
Suggestive linkages were found on chromosomes 2 (3% and 10% ethanol preferences), 9 (a male-specific locus affecting 3% ethanol intake), 12 (3% ethanol intake), 13 (10% ethanol preference), 17 (3% ethanol intake and preference), and 18 (3% ethanol intake; Table 2). Several potential linkages that did not reach the threshold of suggestive linkage (with logarithm of the odds ratio [LOD] scores above 1.9) were detected on chromosomes 10 (3% and 10% ethanol preference), 15 (3% ethanol intake), 16 (3% ethanol preference), and X (10% ethanol intake; Fig. 4).
In addition to interval mapping using MAPMAKER, linkages to the X chromosome markers were analyzed separately using one-way ANOVA with five genotypes (three for females and two for males). The marginally significant effect of DXMit69 genotypes on 10% ethanol intake [F(4,147) = 2.34, P = 0.054] was the only effect found, which is consistent with the weak X-chromosome linkage detected using interval mapping. In females, intakes of 10% ethanol were 3.3 ± 0.3 mL (B6/B6 homozygotes at DXMit69), 3.2 ± 0.2 mL (B6/129 heterozygotes), and 2.7 ± 0.3 mL (129/129 homozygotes; P>0.1, planned comparison tests). Males with the B6 allele of this marker consumed more ethanol than did males with the 129 allele (3.4 ± 0.2 and 2.7 ± 0.2 mL, respectively; P = 0.016, planned comparison test).
Of the nine significant and suggestive loci affecting indexes of 3% and/or 10% ethanol consumption, five had B6 alleles increasing ethanol consumption (chromosomes 4, 8, 9, 13, and 18) and four had B6 alleles decreasing it (chromosomes 2, 7, 12, and 17). The B6 alleles were dominant or partially dominant at three loci (chromosomes 4, 7, and 12), they were recessive or partially recessive at two loci (chromosomes 9 and 17), and allelic interactions for the loci on chromosomes 8, 13, and 18 were additive. The locus on chromosome 2 had a 129-dominant mode of inheritance for 3% ethanol preference, and an additive mode of inheritance for 10% ethanol preference; LOD peaks on chromosome 2 for these two phenotypes were slightly offset.
The combined effect of the significant and suggestive QTL was determined using MAPMAKER/QTL by fitting all primary QTL (listed in Table 2) simultaneously under the unconstrained (free) model. The significant and suggestive linkages explained 31% (3% ethanol intake), 17% (3% ethanol preference), 25% (10% ethanol intake), and 19% (10% ethanol preference) of phenotypical variance, and they explained respectively 44, 38, 40, and 35% of genetic variance (calculated as “% of phenotypical variance explained by QTLs”/“average heritability for males and females”×100). The remaining unexplained genetic variance must be attributed to the other loci that did not reach levels of suggestive linkage, and to epistatic interactions described below.
Analysis of Epistatic Interactions
Sequential analyses by Epistat and ANOVA (see details in Methods) detected 5–11 significant additive-by-additive epistatic interactions for each trait (Table 3). No interactions among primary QTL (loci with significant and suggestive linkages listed in Table 2) were detected. In a few cases, one locus with no linkage modified the effect of another linked locus. This was observed for D17Mit51 modified by D2Mit61 (3% ethanol intake), D13Mit35 modified by D6Mit36 (10% ethanol intake and preference), and D3Mit89 modified by DXMit69 (10% ethanol preference). In most cases, none of the epistatically interacting loci alone showed significant linkage (correspondingly, they were not detected in the linkage analysis described above) and only their interaction effect was significant.
Table 3.
Epistatic Interactions Detected by the Epistat Program and Confirmed by ANOVA
| Marker 1 | Marker 2 | Epistat | ANOVA | ||||||
|---|---|---|---|---|---|---|---|---|---|
| interaction LLR | interaction P-value | marker 1 P-value | marker 2 P-value | R2 | d.f. | interaction F-value | interaction P-value | ||
| 3% Ethanol intake | |||||||||
| D2Mit61 | D17Mit51** | 8.2 | 0.0067 | 0.17 | 0.004** | 0.068 | 1, 25 | 7.9 | 0.0094 |
| D3Mit86* | D13Mit147 | 12.8 | 0.0002 | 0.02* | 0.65 | 0.13 | 1, 31 | 13.2 | 0.00099 |
| D6Mit55 | D13Mit35 | 9.3 | 0.0042 | 0.30 | 0.95 | 0.029 | 1, 35 | 9.7 | 0.0036 |
| D7Mit7* | D13Mit44 | 9.7 | 0.0031 | 0.009* | 0.71 | 0.13 | 1, 43 | 12.2 | 0.0011 |
| D10Mit194 | D14Mit52 | 21.6 | 0.0005 | 0.76 | 0.21 | 0.0034 | 1, 27 | 11.0 | 0.0026 |
| D13Mit97 | DXMit69 | 11.4 | 0.0005 | 0.90 | 0.40 | 0.00027 | 1, 46 | 11.9 | 0.0012 |
| D14Mit82 | D18Mit33 | 12.4 | 0.0009 | 0.53 | 0.66 | 0.014 | 1, 27 | 17.0 | 0.00032 |
| 3% Ethanol preference | |||||||||
| D1Mit19 | D16Mit6 | 8.1 | 0.0101 | 0.51 | 0.48 | 0.018 | 1, 22 | 9.8 | 0.0049 |
| D1Mit48 | D12Mit194 | 9.1 | 0.0026 | 0.39 | 0.89 | 0.022 | 1, 31 | 9.0 | 0.0052 |
| D1Mit37 | D15Mit96 | 9.4 | 0.0011 | 0.08 | 0.17 | 0.10 | 1, 27 | 14.1 | 0.00083 |
| D3Mit25 | D6Mit36 | 10.6 | 0.0008 | 0.69 | 0.41 | 0.0041 | 1, 35 | 10.4 | 0.0028 |
| D3Mit10 | D17Mit6 | 8.7 | 0.0045 | 0.37 | 0.91 | 0.027 | 1, 29 | 9.0 | 0.0054 |
| D4Mit4 | D8Mit271* | 11.2 | 0.0005 | 0.39 | 0.01* | 0.021 | 1, 33 | 8.4 | 0.0067 |
| D4Mit58* | D19Mit35 | 8.7 | 0.0020 | 0.03* | 0.39 | 0.11 | 1, 33 | 10.8 | 0.0024 |
| D6Mit36 | D13Mit35 | 8.6 | 0.0021 | 0.46 | 0.42 | 0.017 | 1, 30 | 15.1 | 0.00053 |
| D8Mit271 | D9Mit218 | 20.3 | 0.0003 | 0.22 | 0.63 | 0.043 | 1, 32 | 13.1 | 0.00099 |
| D9Mit306 | D19Mit10 | 12.1 | 0.0032 | 0.43 | 0.96 | 0.017 | 1, 35 | 10.5 | 0.0026 |
| D11Mit199 | D16Mit6 | 10.9 | 0.0044 | 0.50 | 0.53 | 0.014 | 1, 31 | 10.7 | 0.0026 |
| 10% Ethanol intake | |||||||||
| D2Mit61 | D18Mit144 | 8.6 | 0.0007 | 0.18 | 0.25 | 0.052 | 1, 32 | 15.9 | 0.00037 |
| D6Mit36 | D13Mit35** | 29.2 | <0.0001 | 0.42 | 0.03** | 0.020 | 1, 30 | 15.3 | 0.00049 |
| D7Mit15 | D13Mit44 | 9.1 | 0.0012 | 0.94 | 0.79 | 0.00019 | 1, 27 | 15.6 | 0.00051 |
| D8Mit56 | D16Mit71* | 9.1 | 0.0045 | 0.41 | 0.02* | 0.018 | 1, 35 | 7.8 | 0.0080 |
| D11Mit184 | D19Mit10 | 8.5 | 0.0025 | 0.25 | 0.92 | 0.032 | 1, 38 | 8.9 | 0.0050 |
| 10% Ethanol preference | |||||||||
| D1Mit48 | D12Mit20 | 16.8 | 0.0010 | 0.23 | 0.38 | 0.041 | 1, 34 | 12.9 | 0.0010 |
| D1Mit37* | D15Mit96 | 17.3 | 0.0005 | 0.03* | 0.79 | 0.15 | 1, 27 | 19.1 | 0.00016 |
| D2Mit61 | D18Mit144 | 9.1 | 0.0013 | 0.22 | 0.06 | 0.044 | 1, 32 | 16.4 | 0.00031 |
| D3Mit89** | DXMit69 | 9.2 | 0.0030 | 0.007** | 0.23 | 0.14 | 1, 43 | 12.8 | 0.00089 |
| D5Mit214 | D8Mit190 | 41.7 | 0.0005 | 0.89 | 0.33 | 0.00053 | 1, 36 | 8.1 | 0.0071 |
| D6Mit36 | D13Mit35** | 18.7 | 0.0082 | 0.50 | 0.01** | 0.014 | 1, 30 | 21.4 | 0.000068 |
| D6Mit201 | D11Mit199 | 16.3 | 0.0057 | 0.97 | 0.45 | 0.00005 | 1, 35 | 7.7 | 0.0088 |
| D10Mit162 | D13Mit34 | 16.8 | 0.0002 | 0.89 | 0.17 | 0.00050 | 1, 42 | 9.0 | 0.0045 |
| D12Mit46 | D15Mit193 | 10.0 | 0.0063 | 0.73 | 0.78 | 0.0032 | 1, 36 | 7.8 | 0.0085 |
LLR, log likelihood ratio. R2 represents portion of phenotypical variation controlled by the interacting loci.
Single marker showing significant effect on a phenotype in the Epistat analysis (P < 0.05), but with no linkage to the trait in the Mapmaker analysis (LOD < 0.8).
Single marker showing significant effect on a phenotype in the Epistat analysis (P < 0.05) and located in a region of significant or suggestive linkage. LOD scores are: 2.1 (D17Mit51 and 3% ethanol intake), 1.0 (D13Mit35 and 10% ethanol intake), 1.4 (D13Mit35 and 10% ethanol preference), and 1.7 (D3Mit89 and 10% ethanol preference).
Most of the interacting pairs of markers were trait-specific, but some of them affected several phenotypes. An interaction of loci on chromosomes 6 (D6Mit36 or D6Mit55, two linked markers) and 13 (D13Mit35) affected all four indexes of ethanol consumption. An interaction of loci on chromosomes 7 (D7Mit7 or D7Mit15, two linked markers) and 13 (D13Mit44) affected 3% and 10% ethanol intakes. Interactions of loci on chromosomes 1 (D1Mit48) and 12 (D12Mit194 or D12Mit20, two linked markers), and 1 (D1Mit37) and 15 (D15Mit96) affected 3% and 10% ethanol preferences. An interaction of loci on chromosomes 2 (D2Mit61) and 18 (D18Mit144) affected 10% ethanol intakes and preferences.
DISCUSSION
In this study, using F2 hybrids between the B6 and 129 mouse strains, we characterized the inheritance of indexes of ethanol consumption in two-bottle tests, mapped quantitative trait loci underlying the strain differences, and detected epistatically interacting loci. High ethanol preferences and intakes of the B6 mice were inherited as additive or dominant traits in the F2, which is consistent with the results of our previous study (Bachmanov et al. 1996a). A genome screen using these F2 mice identified three significant linkages. Two loci, on distal chromosome 4 (Ap3q) and proximal chromosome 7 (Ap7q), strongly affected 10% ethanol intake and weakly affected 3% ethanol intake. A male-specific locus on proximal chromosome 8 (Ap8q) affected 3% ethanol preference, but not indexes of 10% ethanol consumption. In addition, six suggestive linkages (on chromosomes 2, 9, 12, 13, 17, and 18) affecting indexes of 3% and/or 10% ethanol consumption were detected. The loci with significant and suggestive linkages accounted for 35–44% of the genetic variation of ethanol consumption phenotypes. None of the primary loci with significant and suggestive linkages interacted epistatically. However, a few modifiers for the primary linkages and a larger number of interactions among unlinked loci were found, which demonstrates a significant role of genetic background in the variation of ethanol consumption.
The 9.0-cM confidence interval (75.0–84.0 cM from the centromere) for the Ap3q locus on distal chromosome 4 includes 67 genes, cDNA segments, and phenotypical loci included in the Mouse Genome Database ([MGD], www.informatics.jax.org). It corresponds to 13 Mb of genomic sequence in the Celera database (www.celera.com), including 338 transcripts. This region contains the saccharin preference (Sac) locus (Phillips et al. 1994; Lush et al. 1995; Bachmanov et al. 1997; Blizard et al. 1999; Li et al. 2001), which recently has been cloned positionally (Bachmanov et al. 2001b) and corresponds to a sweet-taste receptor gene, Tas1r3 (Kitagawa et al. 2001; Max et al. 2001; Montmayeur et al. 2001; Nelson et al. 2001; Sainz et al. 2001). There is considerable evidence showing genetic association between consumption of ethanol and sweeteners (for review, see Kampov-Polevoy et al. 1999). This association was detected in the B6 × 129 F2 intercross (Bachmanov et al. 1996a). Therefore, the Sac and Ap3q loci probably are identical, suggesting that the Tas1r3 gene is a candidate for the Ap3q locus (Bachmanov et al. 2001a). This region has conserved synteny with a subtelomeric region of a short arm of human chromosome 1 (1p36). A locus influencing vulnerability to alcoholism has been mapped to human chromosome 1 (1p13–35) (Nurnberger et al. 2001), however the linkage peak for this locus is substantially more centromeric than the region of conserved synteny for the Ap3q locus.
The 14.1-cM confidence interval (11.4–25.5 cM from the centromere) for the Ap7q locus on proximal chromosome 7 includes 83 genes, cDNA segments and phenotypical loci (MGD). It corresponds to 18 Mb of genomic sequence, including 497 transcripts (Celera). The only QTL with neurological effects mapped to this region is Bis4 (beta-carboline induced seizures 4) (Gershenfeld et al. 1999). This region has conserved synteny with human chromosomes 11p and 19q. A locus influencing alcohol dependence has been mapped to human chromosome 11p15 (Long et al. 1998) and thus may be a human ortholog of the mouse Ap7q locus.
The 27.0-cM confidence interval (0.0–27.0 cM from the centromere) for the Ap8q locus on proximal chromosome 8 includes 90 genes, cDNA segments, and phenotypical loci (MGD). It corresponds to 39 Mb of genomic sequence, including 605 transcripts (Celera). This region has conserved synteny with human chromosomes 4q, 8p, 13q, and 19p. Linkages of alcohol-related phenotypes to human chromosomes 4 (Long et al. 1998; Reich et al. 1998) and 13 (Schuckit et al. 2001) may be orthologs of the mouse Ap8q locus. The Ap8q locus exerts its effect only in males, which adds another example to gender-specific ethanol consumption loci detected in previous studies (Melo et al. 1996; Peirce et al. 1998; Fernandez et al. 1999).
In previous studies, at least eight QTL with significant effects on voluntary 10% ethanol consumption have been described, including those on chromosomes 1 (proximal and middle), 2 (proximal and distal), 3, 4, 9, and 11 (Table 4; for review, see Crabbe et al. 1999; Belknap and Atkins 2001). Of the three significant linkages identified in this study, only one, on distal chromosome 4, also was found to be linked to ethanol intake in the previous studies (Tarantino et al. 1998). The suggestive linkage on chromosome 2 (3% and 10% ethanol preference) may be equivalent to Alcp1/Pref2 (Phillips et al. 1994, 1998; Melo et al. 1996; Belknap et al. 1997; Whatley et al. 1999; Belknap and Atkins 2001), and the suggestive linkage on chromosome 9 (3% ethanol intake) may be equivalent to Ap5q/Pref1 (Phillips et al. 1994, 1998; Belknap et al. 1997; Tarantino et al. 1998; Belknap and Atkins 2001). Several epistatically interacting loci (Table 3) also may correspond to some of these previously detected linkages.
Table 4.
Summary of Significant (Lander and Kruglyak 1995) Linkages for Indexes of Voluntary Ethanol Consumption
| QTL symbol | Chra | Position, cMb | Ethanol concentration | Mapping panel | Reference |
|---|---|---|---|---|---|
| Alcp5 | 1 | 0–16 | 10% | B6.D2 phenotype-based congenic strains | Whatley et al. 1999 |
| Ap1q | 1 | 45–95 | 10% | B6 × D2 F2 intercross | Tarantino et al. 1998 |
| Alcp1 | 2 | 16–54 | 10% | B6.D2 phenotype-based congenic strains | Whatley et al. 1999 |
| Alcp1 | 2 | 20–35 | 10% | (B6 × D2) × B6 backcrosss | Melo et al. 1996 |
| Pref2 | 2 | 28 | 10% | BXD RI strains, B6 × D2 F2 intercross, a selected line | Phillips et al. 1994, 1998; Belknap et al. 1997 |
| – | 2 | 107 | 10% | AXB and BXA RI strains | Gill et al. 1998 |
| Ap6q/Alcp3 | 3 | 48–83 | 10% | BXD RI strains, B6 × D2 F2 intercross, (B6 × D2) × B6 backcross, a selected line | Belknap and Atkins 2001 |
| Ap3q | 4 | 65–tel | 10% | B6 × D2 F2 intercross | Tarantino et al. 1998 |
| Ap3q | 4 | 75–tel | 10% | B6 × 129 F2 intercross | This study |
| Ap7q | 7 | 11–26 | 10% | B6 × 129 F2 intercross | This study |
| Ap8q | 8 | cen–27 | 3% | B6 × 129 F2 intercross | This study |
| Pref1 | 9 | 29 | 10% | BXD RI strains, B6 × D2 F2 intercross, a selected line | Phillips et al. 1994, 1998; Belknap et al. 1997 |
| Ap5q | 9 | 10–35 | 10% | B6 × D2 F2 intercross | Tarantino et al. 1998 |
| Alcp2 | 11 | 35–52 | 10% | (B6 × D2) × B6 backcross | Melo et al. 1996 |
Chr, chromosome
Confidence interval range or peak of a linkage statistic.
The moderate correspondence between the results of our study and previous studies is consistent with having one common parental strain (B6), and the other strain being different in our (129) and the other (DBA/2 or A) studies. Mechanisms underlying ethanol avoidance may be different in different low-preferring strains; correspondingly, loci detected in crosses of the B6 strain with different strains also may be different. Furthermore, loci determining ethanol preference of the B6 mice may have different effects on different genetic backgrounds (i.e., in crosses with different strains), and thus can be detected only in some crosses. In addition, mapping using backcross onto the B6 strain (Melo et al. 1996) must have precluded detection of B6-dominant loci, but provided higher statistical power to detect B6-recessive loci compared with F2 intercross; this also may account for differences in identified sets of loci. Age potentially can modify the effects of alcohol-related QTL (McClearn et al. 1998). In most ethanol preference QTL studies, ∼2–3-month-old mice were phenotyped (Phillips et al. 1994; Belknap et al. 1997; Gill et al. 1998; Tarantino et al. 1998; Whatley et al. 1999), although sometimes up to 9-month-old mice were used (Peirce et al. 1998). Here, we used ∼7-month-old mice, so this also may have contributed to the different loci detected compared with the other studies. This possibility, however, appears to be remote because ethanol intakes and preferences of the B6 and 129 mice remained stable over a period of 1–12 months (Tordoff and Bachmanov, www.monell.org/MMTPP). Overall, our choices of the parental strains and the phenotyping procedure were efficient for discovering new linkages for ethanol preference.
Compared with the 129 mice, the B6 mice have higher indexes of ethanol consumption for a range of ethanol concentrations (Bachmanov et al. 1996b). However, there are indications that the genetic determination of the strain differences may be concentration-dependent. First, correlations between indexes of consumption for 3% and 10% ethanol in F2 were only moderate (+0.36 – +0.57 for corresponding indexes, Table 1), which is consistent with previous data on inbred strains for these concentrations (r = +0.34, n = 15, P = 0.2, [Belknap et al. 1993]). Accordingly, only partially overlapping sets of loci were detected for these two ethanol concentrations, suggesting that consumption of different concentrations of ethanol involves different, albeit overlapping, genetic controls. Finally, all loci involved in 10% ethanol consumption had similar effects in both genders, whereas some of the loci involved in 3% ethanol consumption were male-specific. This is consistent with larger parental strain differences in 3% ethanol intakes and preferences in males compared with females, whereas gender had little effect on strain differences in indexes of 10% ethanol consumption.
Approaches to analyze epistatic interactions among QTL are still evolving (Cheverud and Routman 1995; Frankel and Schork 1996; Brockmann et al. 2000; Fernandez et al. 2000; Cheverud et al. 2001; Cordell et al. 2001; Crabbe 2001; Hood et al. 2001; Jannink and Jansen 2001; Shimomura et al. 2001). These recent studies demonstrate the importance of epistatic interactions, as well as methodological difficulties involved in their detection. One complicated aspect is that interactions between already detected primary QTL may not encompass all epistatic interactions present. For example, an individual locus may affect a trait only when it is combined with a particular genotype at another locus, or it may have opposite effects depending on the genotype of another locus, revealing genetic background effects (Cheverud and Routman 1995). Therefore, when all individuals are analyzed as a single group, the overall effect of this locus may go undetected, and it thus would not be identified as a primary QTL. However, such hidden loci can be unveiled in a genome-wide screen for epistasis, in which the effects of combinations between all pairs of all genotyped markers are assessed. The problem with genome-wide screens is that they involve multiple comparisons resulting in higher thresholds of statistical significance, but group sizes are reduced compared with single-locus analyses. We approached the genome-wide analysis of epistatic interactions among QTL using a combination of specialized software, Epistat, and ANOVA. Our results show that epistatic interactions represent a substantial component of the genetic variation in ethanol consumption. This is consistent with strong effects of genetic background, such as those observed when targeted mutations are backcrossed on different inbred partner strains (e.g., Crawley et al. 1997; Thiele et al. 2000; Hoffman et al. 2001). The B6 and 129 strains used in this study are also commonly used in “knockout” experiments, forming mixed genetic background for mutant mice, and thus loci identified in this cross may modify effects of the genes with targeted mutations.
In conclusion, our mapping study using a novel cross and phenotypes of voluntary consumption of two different ethanol concentrations detected the presence of several primary loci and a substantial contribution of genetic background as revealed in multiple epistatic interactions. This study completes an initial step toward positional cloning of ethanol consumption QTL.
METHODS
Animals
129P3/J (129) and C57BL/6ByJ (B6) inbred mice were obtained from The Jackson Laboratory. The mice were housed in a temperature-controlled vivarium at 23°C on a 12:12 h light:dark cycle and had free access to water and Teklad Rodent Diet 8604. The F1 and F2 hybrids between the B6 and 129 strains were bred at Monell. Pups were weaned at 21–30 d of age and reared in groups of the same gender (in most cases, four to six mice per cage, but never more than six in one cage). During two-bottle tests, the mice were housed in individual cages.
450 (224 female and 226 male) F2 mice were obtained from three types of reciprocal crosses: (129♀ × B6♂) F1♀ × (129♀ × B6♂) F1♂ (92 females and 101 males), (B6♀ × 129♂) F1♀ × (B6♀ × 129♂) F1♂ (103 females and 91 males), and (B6♀ × 129♂) F1♀ × (129♀ × B6♂) F1♂ (29 females and 34 males). Mice from the parental B6 (9 females and 10 males) and 129 (10 females and 10 males) strains were tested together with the F2 mice with the same stimuli. When two-bottle tests with ethanol began, the F2 mice were on average 7.26 ± 0.04 mo old (with ages ranging from 5.7 to 8.6 mo); the B6 and 129 mice were 5.79 ± 0.14 mo old (ranging from 4.8 to 6.7 mo).
Two-Bottle Tests
Construction of the drinking tubes and other experimental procedures have been described previously (Bachmanov et al. 1996b, in press) and are given in detail on our web site (Tordoff and Bachmanov, www.monell.org/MMTPP). Individually housed mice were presented with one tube containing ethanol solution in deionized water and the other tube containing deionized water. Daily measurements were made in the middle of the light period by reading fluid volume to the nearest 0.2 mL. The positions of the tubes were switched every 24 h to control for side preferences. Ethanol solutions were tested for 8 d: during the first 4 d, mice received 3% ethanol, and during the next 4 d they received 10% ethanol. As part of another experiment, the mice were phenotyped in 4-d, 2-bottle tests with 30 mM glycine; 30 mM D-phenylalanine; 20 and 1 mM saccharin; 120, 300, and 3 mM sucrose; and 1 and 300 mM monosodium glutamate before the tests with ethanol. The tests were designed to avoid any possible “carry-over” effects from taste solutions on ethanol intake.
Analyses of Phenotypical Data
Based on daily solution and water intakes, 4-d averages were calculated for each mouse for each solution concentration. In preliminary statistical and linkage analyses, averages for the first two days and last two days of each 4-d test were analyzed separately. Results for the 2-d averages were similar to each other and to those for the 4-d averages, with the 4-d averages typically showing more significant effects. In another study, we have shown that the 4-d test duration provides optimal statistical power for detecting genetic differences (Tordoff and Bachmanov, in press). Therefore, only 4-d averages are reported here. Preference scores were calculated as the ratio of the average daily solution intake to average daily total fluid (solution + water) intake, in percent. The phenotype data were analyzed using Pearson correlation coefficients, ANOVA, and planned comparisons. When appropriate, Bonferroni corrections for multiple comparisons were used. Significance of deviation of experimental distributions from the normal distribution was tested using Kolmogorov-Smirnov tests. The analyses were conducted using STATISTICA software (StatSoft, Inc.).
To offset any difference between males and females, ethanol intakes and preferences were standardized within each gender to a group mean (0) and standard deviation (1). This standardization was conducted for all 450 F2 mice (224 females and 226 males); the standardized scores for a subset of 169 F2 mice (88 females and 81 males) were used in linkage analyses.
Because fluid intakes can depend on body weight (Bachmanov et al., in press; National Center for Environmental Assessment, www.epa.gov/ncea/pdfs/allometr.pdf), and because B6, 129, and F2 mice differed in body weights, in preliminary analyses we assessed the relationships between ethanol intake and body weight. Mouse body weight was measured before and after the tests with ethanol, averaged, and used to calculate adjusted intakes (mL per 30 g of body weight, the approximate weight of an adult mouse). Average body weights of females were 28.4 ± 0.4 (B6), 23.3 ± 0.8 (129), and 29.9 ± 0.3 (F2), and those of males were 35.1 ± 0.4, 27.1 ± 0.6, and 39.9 ± 0.4, respectively [effects of genotype F(2, 483) = 29.8, P<0.001; gender F(1, 483) = 31.6, P<0.001; and genotype × gender interaction F(2, 483) = 3.0, n.s.; two-way ANOVA]. Adjustment of ethanol intakes for body weight had little effect on the differences among B6, 129, and F2 mice. There were no positive correlations between body weights and unadjusted intakes in the F2 mice (r calculated for males and females separately ranged from −0.16 to −0.01, P≥0.018, n.s. after Bonferroni correction). Intakes adjusted for body weight showed significant negative correlations with body weights (r ranged from −0.41 to −0.32, P<0.001, significant after Bonferroni correction), indicating that the ratio became dependent on the denominator. Because of this dependence, linkages of intakes corrected for body weight may be from loci primarily affecting body size rather than ethanol consumption, and so unadjusted intakes were used in linkage analyses along with preference scores, which are independent of body size. As an additional measure to assess the possibility that linkages for ethanol intakes may be affected by body size, we also conducted a genome screen for body weight. Body weight was affected by loci on chromosomes 2, 9 (significant linkages), and 6 (suggestive linkages); see more details in D.R. Reed et al. (in prep.). Although confidence intervals for body weight and ethanol intake linkages on chromosome 9 overlapped, their distinct modes of inheritance showed that different loci affected these two traits.
Dominance in the F2 generation was detected when the F2 value significantly differed from a midparental value. This test was achieved using planned comparisons in ANOVA by collapsing B6 and 129 phenotypical values, assigning coefficient 1 for each of them, and assigning a coefficient −2 to the F2 value.
Heritability in the broad sense (the degree of genetic determination [Falconer and Mackay 1996]) was estimated based on variances in the parental strains and F2. The environmental (nongenetic) variance was calculated as an average between the phenotypical (total) variances for the two parental strains, VARE = ½ (VARB6 + VAR129). The genetic variance was calculated as a difference between the phenotypic variance of the F2 generation and the environmental variance, VARG = VARF2 − VARE. The heritability estimate was calculated as a percent of the genetic variance from the phenotypical variance of F2, h2 = VARG/VARF2 × 100 (Wright 1968; Falconer and Mackay 1996).
Genotyping
Mouse genomic DNA was purified from tails by NaOH/Tris (Truett et al. 2000) or phenol/chloroform (Hogan et al. 1986) extraction. Microsatellite (simple sequence length polymorphism [SSLP]) markers were purchased from Research Genetics Inc. and tested using a standard protocol (Dietrich et al. 1992), with minor modifications (Bachmanov et al. 1997; Li et al. 2001). The D18346 marker was genotyped using single-strand conformation polymorphism (SSCP) protocol (Li et al. 2001). The polymorphic bands were visualized by autoradiography. The bands were scored by two independent readers and discrepancies were resolved by regenotyping.
In addition to the semidominant SSLP and SSCP markers, several dominant coat and eye color markers were used: agouti (A) on chromosome 2, and tyrosinase (Tyr, formerly albino) and pink-eyed dilution (P) on chromosome 7. The B6 mice have black eyes and fur determined by genotypes a/a, Tyr/Tyr, and P/P. The 129 mice have pink eyes and albino (genotype Aw/Aw, Tyrc/Tyrc, p/p) or cream (light chinchilla; genotype Aw/Aw, Tyrc-ch/Tyrc, p/p) fur (Festing et al., www.informatics.jax.org/mgihome/nomen/strain_129.shtml; Roderick and Guidi 1989; Witham 1990). The F2 mice had several eye and coat color phenotypes. The F2 mice with pink eyes were albino (Tyrc/Tyrc), cream (Tyrc-ch/Tyrc) or light buff (Tyrc-ch/Tyrc-ch); agouti alleles could not be determined in these mice and were scored as unknown. The other variants were: white bellied agouti coat and black eyes (Aw/-, Tyr/-, P/-), black coat and black eyes (a/a, Tyr/-, P/-), yellow coat and pink eyes (Aw/-, Tyr/-, p/p), blue-gray coat and pink eyes (a/a, Tyr/-, p/p), chinchilla coat and black eyes (Aw/-, Tyrc-ch/Tyrc-ch, P/- and Aw/-, Tyrc-ch/Tyrc, P/-), and chocolate coat and black eyes (a/a, Tyrc-ch/Tyrc-ch, P/-) (Silvers 1979). For each coat and eye color marker, two genotypes could be distinguished, a homozygote for a recessive allele (nonagouti for the B6 strain, and albino and pink-eyed dilution for the 129 strain), and a heterozygote/homozygote for a dominant allele.
For genotyping, we have randomly selected a cohort of 169 F2 mice (88 females and 81 males). The mice were selected randomly, rather than from extremes of the phenotypical distributions because several phenotypes were assessed in the same mice (see Two-Bottle Tests section above). Thus, random selection of the mice for genotyping allowed us to use their genotypes for all tested phenotypes; it also allowed us to avoid problems with non-normal phenotypical distributions of the extreme subsets. We have genotyped 137 markers on all autosomes and the X chromosome (the list of markers is available online at www.genome.org). On average, distances between adjacent markers were 11 cM (based on MIT/whitehead database positions, www.genome.wi.mit.edu).
Linkage Analyses
Interval mapping was conducted using MAPMAKER software (Lander et al. 1987) with phenotypical scores of the F2 mice standardized within each gender. Linkages were analyzed using all 169 F2 mice and separately for the subsets of 88 females and 81 males. Thresholds for significant and suggestive linkages were applied as described in Lander and Kruglyak (1995). The confidence interval was defined as a LOD score drops of 1.0 from the LOD peak. The mode of inheritance (the character of allelic interactions) was determined by comparing LOD scores obtained under unconstrained, additive, recessive, and dominant models. The percent of phenotypical variance explained by several loci was determined using MAPMAKER/QTL by fitting all linked loci simultaneously under the unconstrained model. Using this value and heritability estimates (described above), the percent of genetic variance explained by several loci was calculated as: “% of phenotypical variance explained by QTLs”/“heritability” × 100 (heritability was calculated for males and females separately; mean of both genders was used in this calculation).
All genotyped X chromosome markers were in the non-pseudoautosomal region. To conduct linkage analyses using MAPMAKER, male hemizygotes were coded as homozygotes. Like for the autosomes, the MAPMAKER analysis for the X chromosome markers was conducted in males and females separately, as well as in a pooled set of both genders. Because coding male hemizygotes as homozygotes is an approximation, we also conducted a single-marker analysis of the X chromosome linkages using one-way ANOVA with three female genotypes (B6/B6 and 129/129 homozygotes and B6/129 heterozygotes) and two male genotypes (B6 and 129 hemizygotes) as five gradations of the genotype factor.
Analysis of Epistatic Interactions
Epistatic interactions were analyzed using Epistat software (Chase et al. 1997; Lark, www.larklab.4biz.net;) followed by two-way ANOVA. Our primary interest was in detecting additive-by-additive interactions revealing themselves in animals homozygous for the interacting markers. These interactions are of practical importance in inbred, recombinant inbred, and congenic strains. The Epistat software originally designed for analyzing recombinant inbred strains is suitable for detecting such interactions. An automated search for coadaptive interactions among all possible pairs of the genotyped markers was conducted using a minimal subgroup number of 5 (there were four subgroups homozygous for each marker in the pair) and log-likelihood ratio (LLR) threshold of 8.0. This LLR corresponds to a P-value of 0.0000634 (Lark, www.larklab.4biz.net), which approximates a threshold of statistical significance for a genome-wide epistasis search based on a Bonferroni correction (two unlinked regions per chromosome, 40 unlinked regions for the whole genome, 780 independent comparisons, P<0.05/780 ≈ 0.000064). Significance levels for the detected interactions were estimated based on Monte Carlo simulations with 10,000 permutations (this program is included in the Epistat package). The pairs of markers with significant (P<0.05) effects on a phenotype were next analyzed using two-way ANOVA with genotypes of each marker in the pair as between-group factors (only mice homozygous for marker genotypes were included in the analysis). Pairs of markers with significant interaction effects of P<0.01 in ANOVA were reported here as interacting epistatically. This threshold value was chosen arbitrarily but has been used in several other studies of epistatic interactions (Hood et al. 2001; Shimomura et al. 2001).
Bioinformatic Analyses
Genomic regions corresponding to confidence intervals of significantly linked QTL and respective regions of conserved synteny on human chromosomes were defined by consulting the MGD (www.informatics.jax.org). Markers at the ends of the confidence intervals were used to estimate physical size and gene content of the regions using the Celera database (www.celera.com).
WEB SITE REFERENCES
http://www.celera.com; Celera database.
http://www.epa.gov/ncea/pdfs/allometr.pdf; National Center for Environmental Assessment of the Environmental Protection Agency. Allometric Equations.
http://www.informatics.jax.org; Mouse Genome Database (MGD). The Jackson Laboratory, Mouse Genome Informatics.
http://www.informatics.jax.org/mgihome/nomen/strain_129.shtml; Festing, M.F.W., Simpson, E.M., Davisson, M.T., and Mobraaten, L.E. The Jackson Laboratory. Revised nomenclature for strain 129 mice.
http://www.larklab.4biz.net/; Lark, K.G. The University of Utah. Epistat: A computer program for identifying and testing interactions between pairs of quantitative trait loci.
http://www.monell.org/MMTPP, Tordoff, M.G. and Bachmanov, A.A. Monell Chemical Senses Center. Monell Mouse Taste Phenotyping Project.
www.genome.wi.mit.edu/; Massachusetts Institute of Technology (MIT)/Whitehead Institute for Biomedical Research database.
Acknowledgments
We thank R. Arlen Price and Maja Bucan for advice on genotyping and genetic analyses, and Josephine Yee, Mala Palkhivala, Jessica Santo, and Ke Lu for technical assistance with genotyping. This work was supported by National Institutes of Health grants R01AA11028 and R01DK46791 (MGT), R01DC00882 (GKB), R03DC03509, R01DC04188 and R01DK55853 (DRR), and R03DC03854 (AAB).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL bachmanov@monell.org; FAX (215) 898-2084.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.129702.
REFERENCES
- Arola L, Roig R, Cascon E, Brunet MJ, Fornos N, Sabate M, Raga X, Batista J, Salvado MJ, Blade C. Model for voluntary wine and alcohol consumption in rats. Physiol Behav. 1997;62:353–357. doi: 10.1016/s0031-9384(97)00023-1. [DOI] [PubMed] [Google Scholar]
- Bachmanov AA, Reed DR, Tordoff MG, Price RA, Beauchamp GK. Intake of ethanol, sodium chloride, sucrose, citric acid, and quinine hydrochloride solutions by mice: A genetic analysis. Behav Genet. 1996a;26:563–573. doi: 10.1007/BF02361229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmanov AA, Tordoff MG, Beauchamp GK. Ethanol consumption and taste preferences in C57BL/6ByJ and 129/J mice. Alcohol Clin Exp Res. 1996b;20:201–206. doi: 10.1111/j.1530-0277.1996.tb01630.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmanov AA, Reed DR, Ninomiya Y, Inoue M, Tordoff MG, Price RA, Beauchamp GK. Sucrose consumption in mice: Major influence of two genetic loci affecting peripheral sensory responses. Mamm Genome. 1997;8:545–548. doi: 10.1007/s003359900500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmanov AA, Beauchamp GK, Tordoff MG. Voluntary consumption of ethanol in 28 inbred mouse strains (Abstract) Alcohol Clin Exp Res. 2000;24:58A. [Google Scholar]
- Bachmanov AA, Li X, Reed DR, Li S, Chen Z, de Jong PJ, Wu C, West DB, Chatterjee A, Ross DA, et al. Sac (saccharin preference) locus: Positional cloning and effects on ethanol consumption (Abstract) Alcohol Clin Exp Res. 2001a;25:118A. [Google Scholar]
- Bachmanov AA, Li X, Reed DR, Ohmen JD, Li S, Chen Z, Tordoff MG, de Jong PJ, Wu C, West DB, et al. Positional cloning of the mouse saccharin preference (Sac) locus. Chem Senses. 2001b;26:925–933. doi: 10.1093/chemse/26.7.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmanov, A.A., Reed, D.R., Beauchamp, G.K., and Tordoff, H.G., in press. Food intake, water intake, and drinking spout side preference of 28 mouse strains. Behav. Genet. (in press). [DOI] [PMC free article] [PubMed]
- Belknap JK, Atkins AL. The replicability of QTLs for murine alcohol preference drinking behavior across eight independent studies. Mamm Genome. 2001;12:893–899. doi: 10.1007/s00335-001-2074-2. [DOI] [PubMed] [Google Scholar]
- Belknap JK, Crabbe JC, Young ER. Voluntary consumption of alcohol in 15 inbred mouse strains. Psychopharmacology. 1993;112:503–510. doi: 10.1007/BF02244901. [DOI] [PubMed] [Google Scholar]
- Belknap JK, Richards SP, O'Toole LA, Helms ML, Phillips TJ. Short-term selective breeding as a tool for QTL mapping: Ethanol preference drinking in mice. Behav Genet. 1997;27:55–66. doi: 10.1023/a:1025615409383. [DOI] [PubMed] [Google Scholar]
- Blizard DA, Kotlus B, Frank ME. Quantitative trait loci associated with short-term intake of sucrose, saccharin and quinine solutions in laboratory mice. Chem Senses. 1999;24:373–385. doi: 10.1093/chemse/24.4.373. [DOI] [PubMed] [Google Scholar]
- Brockmann GA, Kratzsch J, Haley CS, Renne U, Schwerin M, Karle S. Single QTL effects, epistasis, and pleiotropy account for two-thirds of the phenotypic F(2) variance of growth and obesity in DU6i x DBA/2 mice. Genome Res. 2000;10:1941–1957. doi: 10.1101/gr.gr1499r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chase K, Adler FR, Lark KG. Epistat: A computer program for identifying and testing interactions between pairs of quantitative trait loci. Theor Appl Genet. 1997;94:724–730. [Google Scholar]
- Cheverud JM, Routman EJ. Epistasis and its contribution to genetic variance components. Genetics. 1995;139:1455–1461. doi: 10.1093/genetics/139.3.1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheverud JM, Vaughn TT, Pletscher LS, Peripato AC, Adams ES, Erikson CF, King-Ellison KJ. Genetic architecture of adiposity in the cross of LG/J and SM/J inbred mice. Mamm Genome. 2001;12:3–12. doi: 10.1007/s003350010218. [DOI] [PubMed] [Google Scholar]
- Cordell HJ, Todd JA, Hill NJ, Lord CJ, Lyons PA, Peterson LB, Wicker LS, Clayton DG. Statistical modeling of interlocus interactions in a complex disease: Rejection of the multiplicative model of epistasis in type 1 diabetes. Genetics. 2001;158:357–367. doi: 10.1093/genetics/158.1.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crabbe JC. Use of genetic analyses to refine phenotypes related to alcohol tolerance and dependence. Alcohol Clin Exp Res. 2001;25:288–292. [PubMed] [Google Scholar]
- Crabbe JC, Belknap JK, Buck KJ. Genetic animal models of alcohol and drug abuse. Science. 1994;264:1715–1723. doi: 10.1126/science.8209252. [DOI] [PubMed] [Google Scholar]
- Crabbe JC, Phillips TJ, Buck KJ, Cunningham CL, Belknap JK. Identifying genes for alcohol and drug sensitivity: Recent progress and future directions. Trends Neurosci. 1999;22:173–179. doi: 10.1016/s0166-2236(99)01393-4. [DOI] [PubMed] [Google Scholar]
- Crawley JN, Belknap JK, Collins A, Crabbe JC, Frankel W, Henderson N, Hitzemann RJ, Maxson SC, Miner LL, Silva AJ, et al. Behavioral phenotypes of inbred mouse strains: Implications and recommendations for molecular studies. Psychopharmacology (Berl) 1997;132:107–124. doi: 10.1007/s002130050327. [DOI] [PubMed] [Google Scholar]
- Dietrich W, Katz H, Lincoln SE, Shin H-S, Friedman J, Dracopoli N, Lander ES. A genetic map of the mouse suitable for typing intraspecific crosses. Genetics. 1992;131:423–447. doi: 10.1093/genetics/131.2.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dole VP. On the relevance of animal models to alcoholism in humans. Alcohol Clin Exp Res. 1986;10:361–363. doi: 10.1111/j.1530-0277.1986.tb05107.x. [DOI] [PubMed] [Google Scholar]
- Dole VP, Gentry RT. Toward an analogue of alcoholism in mice. Proc Natl Acad Sci. 1984;81:3543–3546. doi: 10.1073/pnas.81.11.3543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson CK. Voice of the Victims. Alcohol Clin Exp Res. 1996;20:403. doi: 10.1111/j.1530-0277.1996.tb01659.x. [DOI] [PubMed] [Google Scholar]
- Falconer DS, Mackay TFC. Introduction to quantitative genetics. Essex, England: Longman; 1996. [Google Scholar]
- Fernandez JR, Vogler GP, Tarantino LM, Vignetti S, Plomin R, McClearn GE. Sex-exclusive quantitative trait loci influences in alcohol-related phenotypes. Am J Med Genet. 1999;88:647–652. doi: 10.1002/(sici)1096-8628(19991215)88:6<647::aid-ajmg13>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- Fernandez JR, Tarantino LM, Hofer SM, Vogler GP, McClearn GE. Epistatic quantitative trait loci for alcohol preference in mice. Behav Genet. 2000;30:431–437. doi: 10.1023/a:1010232900342. [DOI] [PubMed] [Google Scholar]
- Foroud T, Li TK. Genetics of alcoholism: a review of recent studies in human and animal models. Am J Addict. 1999;8:261–278. doi: 10.1080/105504999305677. [DOI] [PubMed] [Google Scholar]
- Foroud T, Edenberg HJ, Goate A, Rice J, Flury L, Koller DL, Bierut LJ, Conneally PM, Nurnberger JI, Bucholz KK, et al. Alcoholism susceptibility loci: Confirmation studies in a replicate sample and further mapping. Alcohol Clin Exp Res. 2000;24:933–945. [PubMed] [Google Scholar]
- Frankel WN, Schork NJ. Who's afraid of epistasis? Nat Genet. 1996;14:371–373. doi: 10.1038/ng1296-371. [DOI] [PubMed] [Google Scholar]
- Gershenfeld HK, Neumann PE, Li X, St. Jean PL, Paul SM. Mapping quantitative trait loci for seizure response to a GABAA receptor inverse agonist in mice. J Neurosci. 1999;19:3731–3738. doi: 10.1523/JNEUROSCI.19-10-03731.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill K, Desaulniers N, Desjardins P, Lake K. Alcohol preference in AXB/BXA recombinant inbred mice: Gender differences and gender-specific quantitative trait loci. Mamm Genome. 1998;9:929–935. doi: 10.1007/s003359900902. [DOI] [PubMed] [Google Scholar]
- Hoffman PL, Yagi T, Tabakoff B, Phillips TJ, Kono H, Messing RO, Choi DS. Transgenic and gene “knockout” models in alcohol research. Alcohol Clin Exp Res. 2001;25:60S–66S. doi: 10.1097/00000374-200105051-00011. [DOI] [PubMed] [Google Scholar]
- Hogan B, Costantini F, Lacy E. Manipulating the mouse embryo: A laboratory manual. Cold Spring Harbor, NY: Cold Spring Harbor; 1986. [Google Scholar]
- Hood HM, Belknap JK, Crabbe JC, Buck KJ. Genomewide search for epistasis in a complex trait: Pentobarbital withdrawal convulsions in mice. Behav Genet. 2001;31:93–100. doi: 10.1023/a:1010214026692. [DOI] [PubMed] [Google Scholar]
- Jannink JL, Jansen R. Mapping epistatic quantitative trait loci with one-dimensional genome searches. Genetics. 2001;157:445–454. doi: 10.1093/genetics/157.1.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kampov-Polevoy AB, Garbutt JC, Janowsky DS. Association between preference for sweets and excessive alcohol intake: A review of animal and human studies. Alcohol Alcohol. 1999;34:386–395. doi: 10.1093/alcalc/34.3.386. [DOI] [PubMed] [Google Scholar]
- Kitagawa M, Kusakabe Y, Miura H, Ninomiya Y, Hino A. Molecular genetic identification of a candidate receptor gene for sweet taste. Biochem Biophys Res Commun. 2001;283:236–242. doi: 10.1006/bbrc.2001.4760. [DOI] [PubMed] [Google Scholar]
- Lander E, Green P, Abrahamson J, Barlow A, Daley M, Lincoln S, Newburg L. MAPMAKER: An interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics. 1987;1:174–181. doi: 10.1016/0888-7543(87)90010-3. [DOI] [PubMed] [Google Scholar]
- Lander ES, Kruglyak L. Genetic dissection of complex traits: Guidelines for interpreting and reporting linkage findings. Nat Genet. 1995;11:241–247. doi: 10.1038/ng1195-241. [DOI] [PubMed] [Google Scholar]
- Lester D, Freed EX. Criteria for an animal model of alcoholism. Pharmacol Biochem Behav. 1973;1:103–107. doi: 10.1016/0091-3057(73)90062-2. [DOI] [PubMed] [Google Scholar]
- Li TK, Lumeng L, McBride WJ, Waller MB. Progress toward a voluntary oral consumption model of alcoholism. Drug Alcohol Depend. 1979;4:45–60. doi: 10.1016/0376-8716(79)90040-1. [DOI] [PubMed] [Google Scholar]
- Li X, Inoue M, Reed DR, Huque T, Puchalski RB, Tordoff MG, Ninomiya Y, Beauchamp GK, Bachmanov AA. High-resolution genetic mapping of the saccharin preference locus (Sac) and the putative sweet taste receptor (T1R1) gene (Gpr70) to mouse distal chromosome 4. Mamm Genome. 2001;12:13–16. doi: 10.1007/s003350010236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logue SF, Swartz RJ, Wehner JM. Genetic correlation between performance on an appetitive-signaled nosepoke task and voluntary ethanol consumption. Alcohol Clin Exp Res. 1998;22:1912–1920. [PubMed] [Google Scholar]
- Long JC, Knowler WC, Hanson RL, Robin RW, Urbanek M, Moore E, Bennett PH, Goldman D. Evidence for genetic linkage to alcohol dependence on chromosomes 4 and 11 from an autosome-wide scan in an American Indian population. Am J Med Genet. 1998;81:216–221. doi: 10.1002/(sici)1096-8628(19980508)81:3<216::aid-ajmg2>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- Lush IE, Hornigold N, King P, Stoye JP. The genetics of tasting in mice. VII. Glycine revisited, and the chromosomal location of Sac and Soa. Genet Res. 1995;66:167–174. doi: 10.1017/s0016672300034510. [DOI] [PubMed] [Google Scholar]
- Max M, Shanker YG, Huang L, Rong M, Liu Z, Campagne F, Weinstein H, Damak S, Margolskee RF. Tas1r3, encoding a new candidate taste receptor, is allelic to the sweet responsiveness locus Sac. Nat Genet. 2001;28:58–63. doi: 10.1038/ng0501-58. [DOI] [PubMed] [Google Scholar]
- McClearn GE, Tarantino LM, Hofer SM, Jones B, Plomin R. Developmental loss of effect of a chromosome 15 QTL on alcohol acceptance. Mamm Genome. 1998;9:991–994. doi: 10.1007/s003359900912. [DOI] [PubMed] [Google Scholar]
- Melo JA, Shendure J, Pociask K, Silver LM. Identification of sex-specific quantitative trait loci controlling alcohol preference in C57BL/6 mice. Nat Genet. 1996;13:147–153. doi: 10.1038/ng0696-147. [DOI] [PubMed] [Google Scholar]
- Montmayeur JP, Liberles SD, Matsunami H, Buck LB. A candidate taste receptor gene near a sweet taste locus. Nat Neurosci. 2001;4:492–498. doi: 10.1038/87440. [DOI] [PubMed] [Google Scholar]
- Myers RD. Experimental alcoholism: World wide need to standardize animal models. Alcohol. 1996;13:109–110. doi: 10.1016/0741-8329(95)02092-6. [DOI] [PubMed] [Google Scholar]
- Nelson G, Hoon MA, Chandrashekar J, Zhang Y, Ryba NJ, Zuker CS. Mammalian sweet taste receptors. Cell. 2001;106:381–390. doi: 10.1016/s0092-8674(01)00451-2. [DOI] [PubMed] [Google Scholar]
- Nurnberger JI, Jr, Foroud T, Flury L, Su J, Meyer ET, Hu K, Crowe R, Edenberg H, Goate A, Bierut L, et al. Evidence for a locus on chromosome 1 that influences vulnerability to alcoholism and affective disorder. Am J Psychiatry. 2001;158:718–724. doi: 10.1176/appi.ajp.158.5.718. [DOI] [PubMed] [Google Scholar]
- Peirce JL, Derr R, Shendure J, Kolata T, Silver LM. A major influence of sex-specific loci on alcohol preference in C57Bl/6 and DBA/2 inbred mice. Mamm Genome. 1998;9:942–948. doi: 10.1007/s003359900904. [DOI] [PubMed] [Google Scholar]
- Phillips TJ, Crabbe JC, Metten P, Belknap JK. Localization of genes affecting alcohol drinking in mice. Alcohol Clin Exp Res. 1994;18:931–941. doi: 10.1111/j.1530-0277.1994.tb00062.x. [DOI] [PubMed] [Google Scholar]
- Phillips TJ, Belknap JK, Buck KJ, Cunningham CL. Genes on mouse chromosomes 2 and 9 determine variation in ethanol consumption. Mamm Genome. 1998;9:936–941. doi: 10.1007/s003359900903. [DOI] [PubMed] [Google Scholar]
- Reed, D.R., Li, X., Lu, K., Li, S., Tordoff, M.G., Price, R.A., Beauchamp, G.K., and Bachmanov, A.A., in prep. Loci on chromosomes 2, 4, 9, and 16 for body weight, body length, and adioposity identified in a genome scan of an F2 intercross between the 129P3/J and C57BL/6ByJ mouse strains. Mamm. Genome. [DOI] [PMC free article] [PubMed]
- Reich T, Edenberg HJ, Goate A, Williams JT, Rice JP, Van Eerdewegh P, Foroud T, Hesselbrock V, Schuckit MA, Bucholz K, et al. Genome-wide search for genes affecting the risk for alcohol dependence. Am J Med Genet. 1998;81:207–215. [PubMed] [Google Scholar]
- Roderick TH, Guidi JN. Strain distribution of polymorphic variants. In: Lyon MF, Searle AG, editors. Genetic variants and strains of the laboratory mouse, 2nd ed. New York: Oxford University Press; 1989. pp. 663–772. [Google Scholar]
- Sainz E, Korley JN, Battey JF, Sullivan SL. Identification of a novel member of the T1R family of putative taste receptors. J Neurochem. 2001;77:896–903. doi: 10.1046/j.1471-4159.2001.00292.x. [DOI] [PubMed] [Google Scholar]
- Schuckit MA, Edenberg HJ, Kalmijn J, Flury L, Smith TL, Reich T, Bierut L, Goate A, Foroud T. A genome-wide search for genes that relate to a low level of response to alcohol. Alcohol Clin Exp Res. 2001;25:323–329. [PubMed] [Google Scholar]
- Shimomura K, Low-Zeddies SS, King DP, Steeves TD, Whiteley A, Kushla J, Zemenides PD, Lin A, Vitaterna MH, Churchill GA, et al. Genome-wide epistatic interaction analysis reveals complex genetic determinants of circadian behavior in mice. Genome Res. 2001;11:959–980. doi: 10.1101/gr.171601. [DOI] [PubMed] [Google Scholar]
- Silvers WK. The coat colors of mice, a model for mammalian gene action and interaction. New York: Springer-Verlag; 1979. [Google Scholar]
- Tarantino LM, McClearn GE, Rodriguez LA, Plomin R. Confirmation of quantitative trait loci for alcohol preference in mice. Alcohol Clin Exp Res. 1998;22:1099–1105. [PubMed] [Google Scholar]
- Thiele TE, Miura GI, Marsh DJ, Bernstein IL, Palmiter RD. Neurobiological responses to ethanol in mutant mice lacking neuropeptide Y or the Y5 receptor. Pharmacol Biochem Behav. 2000;67:683–691. doi: 10.1016/s0091-3057(00)00413-5. [DOI] [PubMed] [Google Scholar]
- Tordoff, M.G. and Bachmanov, A.A. Influence of test duration on the sensitivity of two-bottle choice tests. Chem. Senses. (in press). [DOI] [PubMed]
- Truett GE, Heeger P, Mynatt RL, Truett AA, Walker JA, Warman ML. Preparation of PCR-quality mouse genomic DNA with hot sodium hydroxide and tris (HotSHOT) Biotechniques. 2000;29:52. doi: 10.2144/00291bm09. , 54. [DOI] [PubMed] [Google Scholar]
- Vadasz C, Saito M, Balla A, Kiraly I, Vadasz 2nd C, Gyetvai B, Mikics E, Pierson D, Brown D, Nelson JC. Mapping of quantitative trait loci for ethanol preference in quasi-congenic strains. Alcohol. 2000;20:161–171. doi: 10.1016/s0741-8329(99)00068-3. [DOI] [PubMed] [Google Scholar]
- Whatley VJ, Johnson TE, Erwin VG. Identification and confirmation of quantitative trait loci regulating alcohol consumption in congenic strains of mice. Alcohol Clin Exp Res. 1999;23:1262–1271. doi: 10.1111/j.1530-0277.1999.tb04287.x. [DOI] [PubMed] [Google Scholar]
- Witham BA. Coat colors of sublines of 129 mice. JAX Notes. 1990;No:441. [Google Scholar]
- Wright S. Genetics and biometrical foundations. Vol. 1. Chicago: University of Chicago Press; 1968. Evolution and the genetics of populations. [Google Scholar]