Abstract
Genomic imprinting refers to an epigenetic marking of genes that results in monoallelic expression. This parent-of-origin dependent phenomenon is a notable exception to the laws of Mendelian genetics. Imprinted genes are intricately involved in fetal and behavioral development. Consequently, abnormal expression of these genes results in numerous human genetic disorders including carcinogenesis. This paper reviews genomic imprinting and its role in human disease. Additional information about imprinted genes can be found on the Genomic Imprinting Website at http://www.geneimprint.com.
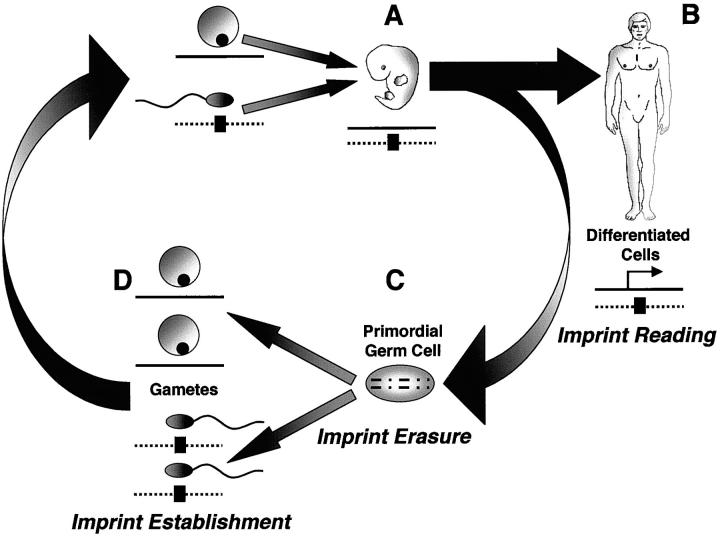
Genomic imprinting (also referred to as gametic or parental imprinting) is the epigenetic marking of a gene based on its parental origin that results in monoallelic expression. Genomic imprinting differs from classical genetics in the sense that the parental complement of imprinted genes are not equivalent with respect to their expression, despite both parents contributing equally to the genetic content of their progeny. The mechanism of imprinting is complex and not completely understood; however, evidence suggests that the “imprint mark” is a parental-specific methylation of CpG-rich domains that is established during gametogenesis. The imprint marks on a gene must be erasable in the germline when transmitted through individuals of the opposite sex, but maintained during somatic cell division (Figure 1) ▶ .
Figure 1.
Imprint establishment and propagation during gametogenesis and development. The paternal allele (dashed line) is imprinted and the maternal allele is expressed (solid line). The “imprint mark” (black box) represents a parental-specific methylation established during gametogenesis. A: The maternal and paternal genomes have different imprint patterns following fertilization. B: Both “imprint marks” and imprint reading are maintained during somatic cell division. C: The parental specific imprints are erased in the primordial germ cells. D: The appropriate “imprint marks” are reestablished for the next generation.
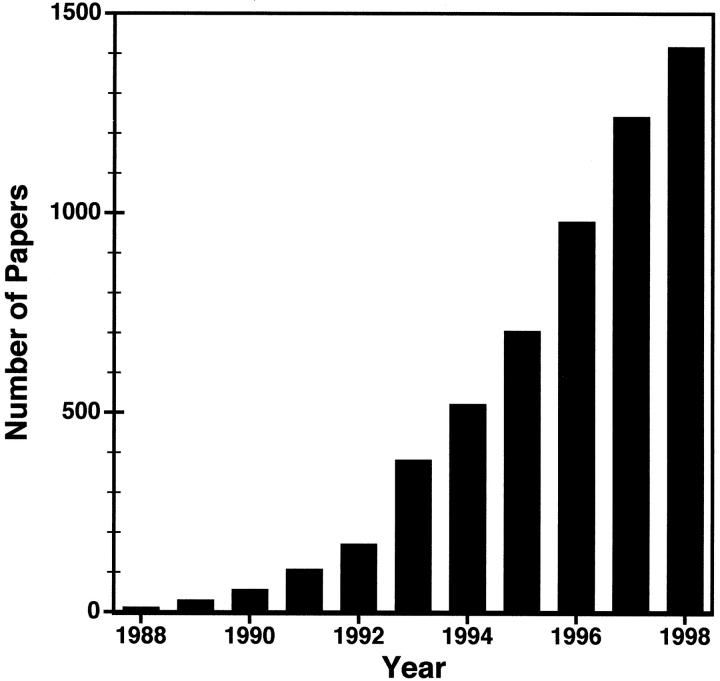
The total number of publications on genomic imprinting has increased markedly over the past 10 years and has now reached almost 1500 (Figure 2) ▶ . There are now more than 25 identified imprinted genes (Table 1) ▶ , and estimates based on mouse models indicate that as many as 100 to 200 may exist. 1 Imprinted genes are involved in many aspects of development including fetal and placental growth, cell proliferation, and adult behavior. Consequently, alteration of normal imprinting patterns gives rise to numerous human genetic diseases including cancer. This review examines the role of genomic imprinting in several human genetic diseases such as the Beckwith-Wiedemann, Prader-Willi, and Angelman syndromes, as well as the evidence implicating genomic imprinting in behavioral disorders and carcinogenesis. For excellent reviews on the mechanistic models of genomic imprinting, consult Reik and Walter, 2 Constancia et al3, and Barlow. 4
Figure 2.
Total number of papers published on genomic imprinting versus time.
Table 1.
Identified Imprinted Genes and Transcripts
| Human | Mouse | References | ||||
|---|---|---|---|---|---|---|
| Gene | Location | Expressed allele | Gene | Location | Expressed allele | |
| NOEY2 (ARHI) | 1p31 | Paternal | 129 | |||
| p73 | 1p36 | Maternal | 147, 148 | |||
| U2AFBPL | 5q22-q31 | Biallelic | U2afbp-rs | Proximal 11 | Paternal | 25, 149, 150 |
| MAS1 | 6q25.3-q26 | Biallelic/Monoallelic in breast | Mas | Proximal 17 | Paternal | 151–153 |
| M6P/IGF2R | 6q26-q27 | Biallelic/Maternal* | M6p/Igf2r | Proximal 17 | Maternal | 18, 136–139 |
| Igf2r-AS | Proximal 17 | Paternal | 4, 140 | |||
| GRB10 | 7p11.2-12 | NR | Meg1/Grb10 | Proximal 11 | Maternal | 31 |
| PEG1/MEST | 7q32 | Paternal | Peg1/Mest | Proximal 6 | Paternal | 21, 154, 155 |
| WT1 | 11p13 | Biallelic/Maternal* | Wt1 | 2 | NR | 120, 156 |
| ASCL2/HASH2 | 11p15.5 | Maternal | Mash2 | Distal 7 | Maternal | 157, 158 |
| H19 | 11p15.5 | Maternal | H19 | Distal 7 | Maternal | 30, 159 |
| IGF2 | 11p15.5 | Paternal | Igf2 | Distal 7 | Paternal | 17, 36, 160–162 |
| Igf2-AS | Distal 7 | Paternal | 36 | |||
| IMPT1/BWR1A/ORCTL2/TSSC5 | 11p15.5 | Maternal | Impt1 | Distal 7 | Maternal | 163–166 |
| INS | 11p15.5 | Biallelic | Ins2 | Distal 7 | Paternal | 167–169 |
| IPL/TSSC3/BWR1C | 11p15.5 | Maternal | Ipl | Distal 7 | Maternal | 164, 170, 171 |
| ITM | 11p15.5 | NR | Itm | Distal 7 | Maternal | 172 |
| KvLQT1 | 11p15.5 | Maternal | Kvlqt1 | Distal 7 | Maternal | 62, 173 |
| p57KIP2/CDKN1C | 11p15.5 | Maternal | p57KIP2 | Distal 7 | Maternal | 48, 122, 174 |
| TAPA1 | 11p15.5 | Biallelic† | Tapa1 | Distal 7 | Maternal? | 27, 67, 104 |
| HTR2A | 13q14 | Biallelic/Maternal* | Htr2 | 14,Band D3 | Maternal | 145, 175, 176 |
| FNZ127 | 15q11-q13 | Paternal | 177 | |||
| GABRA5 | 15q11-q13 | Paternal?† | Gabra5 | Central 7 | Biallelic | 26, 27, 178 |
| GABRB3 | 15q11-q13 | Paternal?† | Gabrb3 | Central 7 | Biallelic | 26, 27, 179 |
| GABRG3 | 15q11-q13 | Paternal?† | Gabrg3 | Central 7 | Biallelic | 26, 27, 178 |
| IPW | 15q11-q13 | Paternal | Ipw | Central 7 | Paternal | 35, 177, 180, 181 |
| NDN (necdin) | 15q11-q13 | Paternal | Ndn | Central 7 | Paternal | 82, 181, 182 |
| PAR1 | 15q11-q13 | Paternal | 177, 180 | |||
| PAR5 | 15q11-q13 | Paternal | 177, 180 | |||
| PAR-SN | 15q11-q13 | Paternal | 183 | |||
| SNRPN | 15q11-q13 | Paternal | Snrpn | Central 7 | Paternal | 84, 184–186 |
| UBE3A | 15q11-q13 | Maternal | Ube3a | Central 7 | Maternal | 77–79 |
| ZNF127 | 15q11-q13 | Paternal | Zfp127 | Central 7 | Paternal | 80, 181, 187 |
| PEG3 | 19q13.4 | Paternal | Peg3/Apoc2 | Proximal 7 | Paternal | 22, 188 |
| Neuronatin | 20q11.2-q12 | NR | Peg5/Nnat | Distal 2 | Paternal | 23, 189, 190 |
| GNAS1 | 20q13 | Paternal | Gnas1 | Distal 2 | Maternal/Paternal | 191–194 |
| XIST | Xq13.2 (XIC)‡ | Paternal? | Xist | Xic | Paternal | 195–200 |
| Grf1/Cdc25Mm | Distal 9 | Paternal | 24 | |||
| Impact | Proximal 18 | Paternal | 201 | |||
| Ins1 | Distal 19 | Paternal | 167, 202 |
NR, not reported.
* Polymorphic imprinting.
† Determined in vitro.
‡ X-inactivation center.
Background
Genomic imprinting plays a critical role in embryogenesis as evidenced by certain aberrations of human pregnancy. The complete hydatidiform mole arises from the fertilization of an anuclear egg either by a haploid sperm (followed by duplication of the paternal genome) or two haploid sperm (diandric diploidy). 5 This trophoblastic disease is characterized by a completely androgenetic (Ag) genome and results in reduced or absent fetal growth coupled with hyperplastic extraembryonic growth. 6,7 In contrast, ovarian dermoid cysts arise from the spontaneous activation of an ovarian oocyte resulting in the duplication of the maternal genome. 8 These abnormalities indicate that normal human development proceeds only when a complete complement of the paternal and maternal genomes is present.
Experimental evidence for the requirement of both the maternal and paternal chromosomal complements was demonstrated through the manipulation of mouse embryos. 9,10 Mouse embryos were altered in vitro to produce diploid Ag or diploid parthenogenetic (Pg) embryos, possessing only paternal or maternal chromosomes, respectively. Similarities to the human pregnancy aberrations were apparent since Ag mouse embryos had reduced fetal growth and proliferative extraembryonic growth while Pg embryos maintained relatively normal fetal growth but exhibited poor extraembryonic growth. Neither Ag nor Pg embryos were viable to term. 9,10 This demonstrates that genes expressed exclusively from one parental genome exist, and abnormal embryonic development results from the loss of function of these monoallelically expressed genes. A mark or imprint conferring parental memory must therefore differentiate between the parental genomes, be present on the parental chromosomes through cell division, and be inheritable. This was confirmed when nuclei from early haploid preimplantation embryos were transplanted into fertilized eggs following the removal of one pronucleus. The embryo was viable only if the sex of the donor nucleus was opposite that of the remaining pronucleus. 11
The chromosomal regions responsible for the genomic imprinting effects observed in mouse embryos were mapped to specific mouse chromosomes by artificially generating uniparental disomies (UPD) in mice. Certain regions of distinct chromosomes were responsible for markedly different phenotypes ranging from embryonic lethality to various growth and developmental defects apparent only after birth. These effects were dependent on whether the two copies were inherited entirely from one parent, resulting in either duplication or deficiency of genes in these chromosomal regions. 12-14 It was initially postulated that only mouse chromosomes 2, 6, 7, 11, 12, and 17 harbored imprinted chromosomal regions. 15 However, there are now reports of other chromosomes either containing more localized areas of genomic imprinting or harboring genes that show more subtle imprinted effects.
UPD also results in phenotypic abnormalities in humans. These include maternal UPD for chromosomes 2, 7, 14, 15, and 16, and paternal UPD for chromosomes 6, 11, 14, 15, and 20. 16 Classic examples of diseases associated with regional maternal and paternal UPD on chromosome 15 include the Prader-Willi syndrome and Angelman syndrome, respectively. Investigations of these genetic diseases are now helping to elucidate the mechanisms of genomic imprinting in humans.
Imprinting of Specific Genes
The first endogenous imprinted gene identified was mouse insulin-like growth factor 2 (Igf2), which encodes for a critical fetal-specific growth factor. A targeted mutation in Igf2 gave rise to a heterozygous dwarfing phenotype when the mutation was passed from the father while the offspring were normal when the mutation was inherited from the mother. 17 Furthermore, the dwarfing phenotype was observed in paternal heterozygotes and homozygotes suggesting that Igf2 gene expression is exclusively from the paternal allele. At about the same time, the mannose 6-phosphate/insulin-like growth factor type 2 receptor (M6p/Igf2r) gene was shown to be imprinted and maternally expressed in mice. 18 Interestingly, the products of these oppositely imprinted genes interact at the biochemical level since the degradation of Igf2 occurs via the M6p/Igf2r. 19 When a mutation was targeted to the M6p/Igf2r in mice, maternal heterozygotes or homozygotes showed a 30% increase in fetal growth, but they were not viable at birth. 20 Thus, the reciprocally imprinted Igf2 and M6p/Igf2r genes both play an important role in regulating embryonic development and fetal growth. 17,20
Numerous techniques have now been used to identify additional imprinted genes. Positional cloning coupled with candidate gene testing has identified novel human imprinted genes located in imprinted clusters at chromosome positions 11p15.5 and 15q11-q13. Techniques have also used parental differences in DNA methylation and expression to identify imprinted genes. Subtractive hybridization or differential display using cDNA from Pg, Ag, and fertilized embryos have yielded novel imprinted genes such as Peg1/Mest, a mesoderm restricted hydrolase at mouse chromosome 6; Peg3, a novel zinc-finger protein on proximal mouse chromosome 7; and Peg5/Nnat located on mouse chromosome 2. 21-23 The Grf1 and U2afl-rs1 imprinted genes were identified by a genome-wide screen termed restriction landmark genome screening (RLGS). 24,25 Finally, three GABAA receptor subunit genes (GABRB3, GABRA5, and GABRG3) were shown to be exclusively expressed from the paternal allele by microcell-mediated chromosome transfer. 26 More recently, results from a somatic-cell hybrid system indicated that these receptor subunit genes were not imprinted. 27
Characteristics of Imprinted Genes
Several theories have been proposed for the endogenous function of genomic imprinting. Moore and Haig 28 have suggested that genomic imprinting in mammals has evolved from a conflict of interest between the paternal and maternal genome in regulating fetal growth. Whereas benefits of a large placenta and fetus might ensure future propagation of a paternal line, the result may tax the resources of the mother, thereby compromising future pregnancies. Conversely, if fetal and placental growth is held in check, more offspring from the mother’s (and possibly different father’s) lineage may be produced. Accordingly, the mother would be predicted to imprint or silence genes that promote placental and fetal growth, whereas the father would imprint genes that inhibit growth.
In support of this theory, the gene encoding the fetal growth factor, Igf2, is maternally imprinted, whereas H19, which encodes for an untranslated RNA involved in silencing Igf2 expression, is paternally imprinted. 17,29,30 The result of this reciprocal imprinting is parent-of-origin, monoallelic paternal expression of the gene encoding for Igf2. Interestingly, the genes that encode for the M6p/Igf2r which degrades Igf2, and Meg1/Grb10 which inhibits Igf2 signaling are both paternally imprinted, adding further support for this theory. 18,19,31
An alternative proposal for imprinting suggests that the cytosine methylation involved in imprint regulation evolved as a defense mechanism for the inactivation of parasitic sequences such as transposable elements and proviral DNA. 32 This is supported by the finding that 5-aza-deoxycytidine, an inhibitor of cytosine DNA methyltransferase, activates silent retroviruses. 33 Irrespective of the reason for the evolution of genomic imprinting in mammals, the functional consequences of genomic imprinting include the inhibition of parthenogenesis and the loss of protection from deleterious recessive mutations.
As more imprinted genes are identified, the characteristics of imprinting are becoming apparent. For example, two chromosomal regions harbor more than one imprinted gene. These imprinting clusters reside at human chromosome 11p15.5 (syntenic to the distal region of mouse chromosome 7) and human chromosome 15q11-q13 (syntenic to the central region of mouse chromosome 7). Within these imprinted gene clusters, genes have been identified that encode for untranslated RNA 34,35 and antisense RNA 36,37 that may be involved in imprint control. Some imprinted genes, such as H19 and IGF2, that are located in imprinted clusters show coordinate regulation. Imprinted genes also often reside in chromosomal regions that undergo asynchronous replication, 38,39 and the meiotic recombination frequencies in these regions may differ between the male and female germ cells. 40 Another characteristic of imprinted genes is an associated allele-specific DNA methylation of cytosine residues in CpG dinucleotides that appears to distinguish the parental alleles. 41-43 Repetitive elements associated with the areas of differential methylation have also been identified in several imprinted genes (ie, H19, M6p/Igf2r, U2afbp-rs, and p57KIP2). 44-48
Imprinting in Genetic Diseases
Beckwith-Wiedemann Syndrome
There are a number of human genetic diseases associated with imprinting defects (reviewed in Refs. 49 and 50 ). Beckwith-Wiedemann syndrome (BWS) maps to 11p15 and is characterized by general overgrowth with symptoms including hemihypertrophy, macroglossia, and visceromegaly. Genomic imprinting in BWS was first suspected when preferential maternal transmission of mutations was observed in some BWS families. 51 Additionally, approximately 10–20% of BWS individuals are predisposed to embryonal tumors, the most frequent of which are Wilms’ tumors and adenocortical carcinoma. 52 The rate of Wilms’ tumor formation in the BWS population is 1000-fold higher than in the normal population, and these tumors often show preferential loss of maternal 11p15. 53 The majority of BWS cases arise sporadically; however, in both sporadic and familial forms, a small percentage exhibits UPD at chromosome 11p15. In these cases, the remainder of the chromosome is biparental in inheritance, indicative of somatic mosaicism through a postfertilization mitotic recombination event. 54,55
The most common molecular event occurring in BWS patients that do not have cytogenetic abnormalities is the biallelic expression of IGF2 due to loss of imprinting (LOI). 56,57 LOI at the IGF2 locus may be accompanied by the methylation and/or silencing of the active maternal allele of H19. 58,59 This H19-dependent event is consistent with an enhancer-competition model for the co-regulation of these genes. 60
Translocations in BWS patients may also lead to LOI at the IGF2 locus, but without loss of H19 imprinting. 61 These translocations affect imprinting by disrupting a gene involved in imprint control, or by altering the function of an imprinting center (IC). Therefore, disruption of IGF2 imprinting in BWS may also occur via an H19-independent event. 56,57 The imprinted KvLQT1 gene located centromeric to IGF2 spans a common breakpoint region in BWS, and has been proposed to maintain regional imprint control at 11p15.5. 60 KvLQT1 shows preferential expression from the maternal allele in most tissues examined except the heart where it is biallelically expressed. 62 This explains why KvLQT1, responsible for the autosomal dominant cardiac arrhythmia long QT syndrome, shows no parent-of-origin effect in this disorder. The maternally expressed p57KIP2, which encodes for a cyclin-dependent kinase inhibitor, also maps to 11p15.5. Abnormal imprinting and epigenetic silencing of p57KIP2 is found in some individuals with BWS, 63 and mutations are present in about 5% of BWS patients. 64-66
To date, ten imprinted genes have been mapped to 11p15.5 (Table 1) ▶ . Flanking these imprinted genes are the non-imprinted NAP2 (centromeric border) and L23MRP (telomeric border) genes. 67 The syntenic region in the mouse, distal chromosome 7, confirms the existence of an imprinting cluster at this chromosomal location. 68 A possible explanation for the involvement of multiple genes in BWS (even if IGF2 overexpression is directly responsible for BWS) is that one or more of the adjacent genes (eg, H19, p57KIP2, KvLQT1) are involved in the regulation of IGF2 expression. Experimental evidence supports this postulate since transgenic mice that overexpress Igf2 develop symptoms similar to BWS. 69
Prader-Willi and Angelman Syndromes
Two clinically distinct genetic diseases associated with genomic imprinting on chromosome 15q11-q13 are the Prader-Willi syndrome (PWS) and the Angelman syndrome (AS). Each syndrome is associated with deficiencies in sexual development and growth, and behavioral and mental problems including retardation. 70,71 Major diagnostic criteria for PWS patients include hypotonia, hyperphagia and obesity, hypogonadism and developmental delay. 72 AS patients often display ataxia, tremulousness, sleep disorders, seizures, and hyperactivity. Severe mental retardation accompanied with a lack of speech may also be present, but AS individuals often display a happy disposition with outbreaks of laughter. 73
PWS and AS are autosomal dominant disorders showing parent-of-origin effects since the inherited diseases are transmitted from only one of the parents. Approximately 70% of PWS and AS individuals have a de novo 3- to 4-megabase deletion in their paternal or maternal chromosome 15q11-q13, respectively. Maternal UPD occurs in most of the remaining PWS patients (25%); however, paternal UPD only occurs in about 4% of AS patients. 16,74 The preferential loss of parental alleles associated with different phenotypes, coupled with the instances of UPD indicate the involvement of imprinted genes (ie, paternally expressed gene(s) for PWS and maternally expressed gene(s) for AS). 70 Recently, approximately 20% of the AS patients without a chromosomal deletion were found to have truncating mutations in UBE3A, a gene encoding a ubiquitin protein ligase involved in protein turnover. 75,76 UBE3A, mapped to 15q11-q13, has now been reported to be maternally expressed in the human brain. 77,78 Thus, abnormalities in the maternal-specific expression of UBE3A during brain development has been proposed for AS. 79 This region also harbors four imprinted, paternally expressed candidate PWS genes: small nuclear riboprotein-associated polypeptide N (SNRPN), Imprinted in Prader-Willi (IPW), zinc finger 127 (ZNF127), and necdin (NDN). 35,80-82 The imprinted, paternally expressed transcripts of PAR1, PAR5, and PAR-SN may also be involved in PWS.
Imprinting defects resulting from microdeletions targeted to the SNRPN gene have been identified in a small percentage of PWS patients that maintain both parental complements of 15q11-q13. 80,83,84 These deletions alter SNRPN promoter methylation and prevent expression of its paternal allele. This results in the silencing of other paternally expressed genes in the cluster. 83,85 These microdeletions apparently disrupt an imprinting center 85 involved in resetting the correct imprinting pattern during gametogenesis. 84,85 The alternate use of SNRPN transcripts (BD exons) may be involved in the normal imprinting process. 86 Offspring inheriting microdeletions from their mother exhibit no apparent phenotype; however, a subsequent paternal transmission results in PWS. In comparison, a small percentage of AS patients have similar microdeletions in the SNRPN gene (albeit in a region farther upstream) that disrupt the resetting of the imprinting pattern. In this case, progeny inheriting paternal microdeletions do not develop AS, but maternal transmission to offspring results in AS. These PWS and AS microdeletion results support the IC hypothesis, but a bipartite structure must be present since the minimally deleted regions responsible for PWS and AS are distinct. 87 An alternate mechanism for imprinting maintenance in this region relies on an enhancer-competition model between cis-linked genes; 4,88 however, methylation analysis of the PWS/AS region reported by Schumacher et al 89 does not support this.
Imprinting in Brain and Behavior Development
The paternally expressed human MEST gene maps to 7q32, a region where maternal UPD is associated with intrauterine and postnatal growth retardation. 21,90 Recently, a targeted deletion was introduced into the coding sequence of the mouse Mest gene to determine its function. 91 When the deletion was paternally derived, Mest +/− mice were viable and fertile; however, they exhibited growth retardation and increased lethality. Mest−/+ animals (deletion maternally derived) showed none of these effects indicating that the phenotypic consequences of this mutation are detected only through paternal inheritance. Interestingly, Lefebvre et al 91 found decreased reproductive fitness in the females that inherited the targeted disruption from their father. This effect was not based on the genotype of the progeny, but rather was due to an abnormal nurturing behavior of the mutant parturient females. Aberrant behavior of the mothers included failure to ingest the extraembryonic tissues (a normal behavior in most mammals), reduced rate of nest building, and pup neglecting. When the pups were fostered to wild-type females, no phenotypic differences between wild-type pups and Mest−/+ pups were apparent.
The results of this study demonstrate that the paternally expressed Mest is a positive regulator of embryonic growth, and is involved in the regulation of mammalian behavior associated with the rearing of offspring. These findings are consistent with the hypothesis that the imprinting of genes arises from the conflict of interest of the parental genomes in mammals, 28 and supports the importance of imprinted genes in brain development. Previous results using Pg and Ag mouse embryos suggested that both maternally and paternally derived genes contribute to the growth and function of specific brain regions in a complementary fashion. 92 Keverne et al 93 found that Ag cells primarily contributed to hypothalamic composition, whereas Pg cells localized to the cortex, striatum, and hippocampus, but not to the hypothalamus. Brain growth was enhanced by Pg cells and retarded by Ag cells, further supporting the postulate that genomic imprinting is critically involved in mammalian brain development.
Evidence for imprinting effects in human diseases associated with mental abnormalities includes the aforementioned Prader-Willi and Angelman syndromes. There is now also evidence of cognitive imprinting effects in humans displaying normal intelligence. Skuse et al 94 recently reported that an imprinted X-linked locus is potentially responsible for differences in cognitive function of females with Turner’s syndrome. Although normal females (46,XX) inherit an X chromosome from both their mother and father, only one X chromosome is inactivated. Turner’s syndrome is a sporadic disorder resulting when all or part of one X chromosome is deleted in females. These females display normal intelligence, but overall have a higher incidence of social difficulties. 95,96 Turner syndrome women who inherit the X chromosome from their mother (45,Xm) generally exhibit more behavioral difficulties than those inheriting the X chromosome from their father (45,Xp). This finding provides the first evidence of genomic imprinting on the human X chromosome. 94 Based on cytogenetic analysis of these patients, partial deletions of the short arm of the paternally derived X chromosome were found. This suggests that the putative imprinted locus escapes X-inactivation and potentially lies in Xp11.23-Xqter. Interestingly, Miller and Willard 97 have recently identified a 5.5 megabase region on the human Xp11.21-p11.22 that contains eight expressed sequences which escape X inactivation. However, an imprinted gene(s) in this region is yet to be identified.
Parent-of-origin effects involved in other behavioral and brain disorders have also been reported. Included among these are bipolar affective disorder, 98-100 schizophrenia, 101,102 and autism. 103 However, the involvement of genomic imprinting in these examples remains to be elucidated. For an extensive summary of parent-of-origin effects in human disease, consult Morison and Reeve. 104
Imprinting in Human Cancer
There are numerous reports of tumors showing a bias in allelic loss. On a genome-wide scale, the complete hydatidiform mole and benign ovarian dermoid cyst arise from cells that are completely Ag or Pg in origin, respectively. 105,106 In addition, numerous tumors are associated with the preferential loss of a particular parental chromosome, indicating the involvement of imprinted genes. Examples include neuroblastoma (maternal chromosome 1p36 and paternal chromosome 2), 107 acute myeloblastic leukemia (paternal chromosome 7), 108 Wilms’ tumor (maternal chromosome 11p15.5), 109 rhabdomyosarcoma (maternal chromosome 11p15.5), 110 and sporadic osteosarcoma (maternal chromosome 13). 111 A role for genomic imprinting has also been implicated in the development of familial glomus tumors based on inheritance patterns since tumor susceptibility is inherited paternally. 112
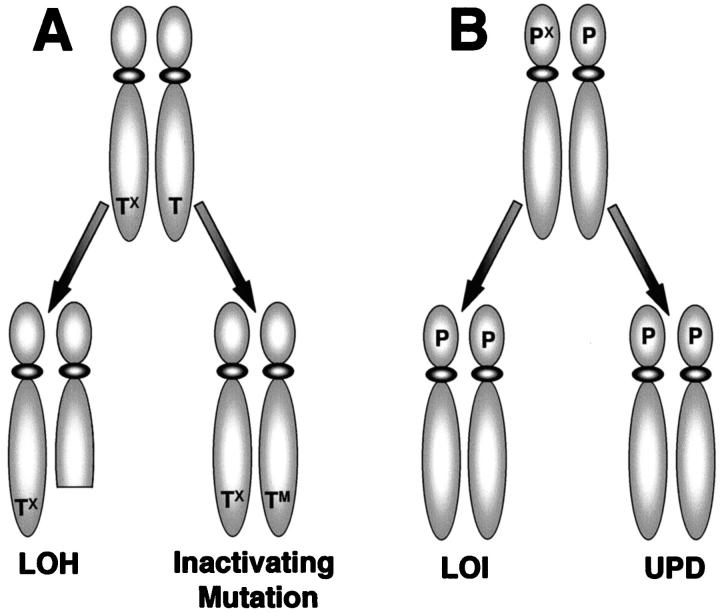
Imprinted genes can be involved in carcinogenesis in several ways (Figure 3) ▶ . Loss of heterozygosity or UPD at an imprinted region may result in the deletion of the only functional copy of a tumor suppressor gene. Alternatively, LOI or UPD of an imprinted gene that promoted cell growth may allow gene expression to be inappropriately increased. Finally, mutational inactivation of an IC might result in the aberrant expression of multiple imprinted oncogenes and/or tumor suppressor genes present in an imprinted chromosomal region.
Figure 3.
A: Only one allele of a tumor suppressor gene (T) is expressed because of genomic imprinting (TX). Loss of heterozygosity (LOH) of the expressed allele or an inactivating mutation in the expressed allele (TM) results in loss of tumor suppressor function. B: Only one allele of the proto-oncogene (P) is expressed because of genomic imprinting (PX). Loss of imprinting (LOI) or uniparental disomy (UPD) results in biallelic expression of the proto-oncogene.
Aberrant genomic imprinting and its role in cancer are best exemplified by studies on Wilms’ tumor, a childhood tumor that arises from metanephric blastemal cells. Direct genetic evidence linking tumorigenesis and aberrant imprinting was identified when 70% of Wilms’ tumors were found to have biallelic IGF2 expression. 113-115 Inactivation of H19 was also present in a number of these cases. 115 The H19 gene possesses a CpG island in its promoter that is normally methylated on the paternal allele and unmethylated on the maternal allele. 44,45,115 An enhancer competition model for the reciprocal control of expression of the imprinted IGF2 and H19 genes has recently been proposed. 116,117 Thus, LOI of the IGF2 gene in Wilms’ tumor could result from loss of H19 expression. 116,117 This scenario is supported by the finding that H19 null transgenic mice show biallelic expression of IGF2. 118 The coupling of biallelic IGF2 gene expression with H19 inactivation is even observed in phenotypically normal kidney tissue surrounding the Wilms’ tumor. This suggests that the inactivation of H19 and the biallelic expression of IGF2 are linked, and occur early in development. 119 Other human malignancies showing LOI at the IGF2 locus are presented in Table 2 ▶ . These results indicate deregulation of IGF2 imprinting is mechanistically involved in the development of a variety of tumors.
Table 2.
Aberrant Imprinting in Human Cancer
| Tumor type | Gene | Reference |
|---|---|---|
| Childhood Tumors | ||
| Wilms’ tumor | IGF2,H19,p57KIP2,M6P/IGF2R | 63, 113, 139, 162 |
| Rhabdomyosarcoma | IGF2 | 203 |
| Ewing’s sarcoma | IGF2 | 204 |
| Hepatoblastoma | IGF2 | 205, 206 |
| Adult Tumors | ||
| Bladder | IGF2,H19,IPW | 207, 208 |
| Breast | IGF2 | 209, 210 |
| Cervical | IGF2,H19 | 211 |
| Choriocarcinoma | IGF2,H19 | 212 |
| Colorectal | IGF2 | 213 |
| Esophageal | H19 | 214 |
| Gastric adenocarcinoma | IGF2 | 215 |
| Glioma | IGF2 | 216 |
| Hepatocellular | IGF2,H19 | 217, 218 |
| Leukemia-acute myeloid | IGF2 | 219 |
| Leukemia-chronic myelogenous | IGF2 | 220 |
| Lung | IGF2,H19,p73 | 221–223 |
| Medulloblastoma | IGF2,H19 | 224 |
| Mesothelioma | IGF2 | 225 |
| Ovarian | IGF2 | 226 |
| Prostate | IGF2 | 227 |
| Renal cell carcinoma | IGF2,p73 | 148, 228 |
| Testicular germ cell | IGF2,H19 | 229 |
| Uterine | IGF2 | 230 |
Because imprinted genes are functionally haploid, an imprinted tumor suppressor gene would be predicted to increase cancer susceptibility since the inactivation of only one allele would eliminate tumor suppressor function. WT1,120,121p57KIP2 122-124 and M6P/IGF2R 125-128 represent imprinted genes implicated in tumor suppression. p57KIP2, mapped to 11p15.5, encodes for a cyclin-dependent kinase inhibitor that is maternally expressed. Epigenetic silencing of the expressed allele has been reported in some tumors and BWS patients. 63 Additionally, approximately 5% of BWS patients have p57KIP2 mutations; 64 however, p57KIP2 mutations have not been identified in tumors. Thus, the putative tumor suppressor function of p57KIP2 remains to be clarified. Recently, NOEY2 (ARHI), a novel ras-related, maternally imprinted gene at 1p31, was identified as a putative tumor suppressor gene in breast and ovarian carcinomas. In the majority of cases, the functional allele was lost. 129
Recent reports demonstrate that the M6P/IGF2R at 6q26 is inactivated in a variety of tumors at the earliest stage of transformation. 126-128 The M6P/IGF2R plays an integral part in the intracellular sorting of lysosomal enzymes, the activation of the growth inhibitor transforming growth factor-β1 (TGF-β1), and the degradation of IGF2, but it is not directly involved in cell signaling. 19,130 The M6P/IGF2R is mutated in 60% of dysplastic liver lesions and hepatocellular carcinomas of patients with or without hepatitis virus infection. 125,126,128 The M6P/IGF2R is also mutated in 30% of breast tumors, 127 and the gene contains a polyG region that is a common mutational target in colon, gastric and endometrial tumors with mismatch repair deficiencies and microsatellite instability. 128,131,132 Moreover, it has recently been reported that the M6P/IGF2R is mutated in human glioma samples that do not contain mutations in the transforming growth factor-β type II receptor (TGFBRII) or Bax genes. 133 In both breast 127,134 and liver carcinogenesis, 128 the allelic inactivation of M6P/IGF2R occurs as an early event, during the initiation rather than the progression stage of transformation.
Although imprinting among individuals and mammalian species is generally conserved, the imprint status of M6P/IGF2R in humans and rodents is strikingly different. The M6p/Igf2r is imprinted in mice 18 and rats, 135 but imprinting at this locus appears to be a polymorphic trait in humans, with most individuals having biallelic expression. 136-138 The existence of individuals with an imprinted M6P/IGF2R tumor suppressor suggests that they may have increased susceptibility to tumor development because of aberrant imprint control. This postulate is supported by Xu et al 139 who recently reported partial imprinting of M6P/IGF2R in 50% of Wilms’ tumor patients.
The precise molecular mechanism for genomic imprinting of M6P/IGF2R is not completely defined. Methylation of a CpG rich region in intron 2 (Region 2) of the expressed maternal allele has been shown to carry the imprint signal for this gene in mice. 46,140 Birger et al 141 have identified a 113-bp sequence, in region 2 of the mouse M6p/Igf2r gene, that serves as a methylation imprinting box responsible for the establishment of differential methylation. Furthermore, this region appears to function as the promoter of an antisense transcript that originates only from the repressed paternal allele. This indicates that a form of expression competition regulates imprinting of the M6p/Igf2r gene in mice. 140 Region 2 of the human M6P/IGF2R also contains parent-of-origin methylation, but gene expression is biallelic. 142,143 Consequently, humans and mice appear to possess an altered ability to read the M6P/IGF2R imprint marks.
Functional polymorphic imprinting has also been observed for human genes encoding IGF2,144WT1, 120 and the human 5-HT2A receptor gene HTR2A. 145 Recently, the mouse Kvlqt1 gene has been shown to undergo developmental relaxation of imprinting in a strain-dependent fashion. 146 Whether polymorphic genomic imprinting occurs in other genes, and functions in determining individual and/or species differences in susceptibility to diseases remains to be determined.
Footnotes
Address reprint requests to Randy L. Jirtle, Box 3433, Duke University Medical Center, Durham, NC 27710. E-mail: jirtle@radonc.duke.edu.
Supported by National Institutes of Health grants CA25951 and ES08823, Department of Defense Grant DAMD17-98-1-8305 (to J.G.F. and R.L.J.), and Zeneca Pharmaceuticals (to D.J.P. and A.A.W).
References
- 1.Barlow DP: Gametic imprinting in mammals. Science 1995, 270:1610-1613 [DOI] [PubMed] [Google Scholar]
- 2.Reik W, Walter J: Imprinting mechanisms in mammals. Curr Opin Genet Dev 1998, 8:154-164 [DOI] [PubMed] [Google Scholar]
- 3.Constancia M, Pickard B, Kelsey G, Reik W: Imprinting mechanisms. Genome Res 1998, 8:881-900 [DOI] [PubMed] [Google Scholar]
- 4.Barlow DP: Competition—a common motif for the imprinting mechanism? EMBO J 1997, 16:6899-6905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lindor NM, Ney JA, Gaffey TA, Jenkins RB, Thibodeau SN, Dewald GW: A genetic review of complete and partial hydatidiform moles and nonmolar triploidy. Mayo Clin Proc 1992, 67:791-799 [DOI] [PubMed] [Google Scholar]
- 6.Kajii T, Ohama K: Androgenetic origin of hydatidiform mole. Nature 1977, 268:633-634 [DOI] [PubMed] [Google Scholar]
- 7.Ohama K, Kajii T, Okamoto E, Fukuda Y, Imaizumi K, Tsukahara M, Kobayashi K, Hagiwara K: Dispermic origin of XY hydatidiform moles. Nature 1981, 292:551-552 [DOI] [PubMed] [Google Scholar]
- 8.Ohama K, Nomura K, Okamoto E, Fukuda Y, Ihara T, Fujiwara A: Origin of immature teratoma of the ovary. Am J Obstet Gynecol 1985, 152:896-900 [DOI] [PubMed] [Google Scholar]
- 9.McGrath J, Solter D: Completion of mouse embryogenesis requires both the maternal and paternal genomes. Cell 1984, 37:179-183 [DOI] [PubMed] [Google Scholar]
- 10.Surani MA, Barton SC, Norris ML: Development of reconstituted mouse eggs suggests imprinting of the genome during gametogenesis. Nature 1984, 308:548-550 [DOI] [PubMed] [Google Scholar]
- 11.Surani MA, Reik W, Norris ML, Barton SC: Influence of germline modifications of homologous chromosomes on mouse development. J Embryol Exp Morphol 1986, 97(Suppl):123-136 [PubMed] [Google Scholar]
- 12.Cattanach BM, Kirk M: Differential activity of maternally and paternally derived chromosome regions in mice. Nature 1985, 315:496-498 [DOI] [PubMed] [Google Scholar]
- 13.Searle AG, Beechey CV: Complementation studies with mouse translocations. Cytogenet Cell Genet 1978, 20:282-303 [DOI] [PubMed] [Google Scholar]
- 14.Searle AG, Beechey CV: Genome imprinting phenomena on mouse chromosome 7. Genet Res 1990, 56:237-244 [DOI] [PubMed] [Google Scholar]
- 15.Cattanach BM, Jones J: Genetic imprinting in the mouse: implications for gene regulation. J Inherit Metab Dis 1994, 17:403-420 [DOI] [PubMed] [Google Scholar]
- 16.Ledbetter DH, Engel E: Uniparental disomy in humans: development of an imprinting map and its implications for prenatal diagnosis. Hum Mol Genet 1995, 4:1757-1764 [DOI] [PubMed] [Google Scholar]
- 17.DeChiara TM, Robertson EJ, Efstratiadis A: Parental imprinting of the mouse insulin-like growth factor II gene. Cell 1991, 64:849-859 [DOI] [PubMed] [Google Scholar]
- 18.Barlow DP, Stöger R, Herrmann BG, Saito K, Schweifer N: The mouse insulin-like growth factor type-2 receptor is imprinted and closely linked to the Tme locus. Nature 1991, 349:84-87 [DOI] [PubMed] [Google Scholar]
- 19.Kornfeld S: Structure and function of the mannose 6-phosphate/insulinlike growth factor II receptors. Annu Rev Biochem 1992, 61:307-330 [DOI] [PubMed] [Google Scholar]
- 20.Wang ZQ, Fung MR, Barlow DP, Wagner EF: Regulation of embryonic growth and lysosomal targeting by the imprinted Igf2/Mpr gene. Nature 1994, 372:464-467 [DOI] [PubMed] [Google Scholar]
- 21.Kaneko-Ishino T, Kuroiwa Y, Miyoshi N, Kohda T, Suzuki R, Yokoyama M, Viville S, Barton SC, Ishino F, Surani MA: Peg1/Mest imprinted gene on chromosome 6 identified by cDNA subtraction hybridization. Nat Genet 1995, 11:52-59 [DOI] [PubMed] [Google Scholar]
- 22.Kuroiwa Y, Kaneko-Ishino T, Kagitani F, Kohda T, Li LL, Tada M, Suzuki R, Yokoyama M, Shiroishi T, Wakana S, Barton SC, Ishino F, Surani MA: Peg3 imprinted gene on proximal chromosome 7 encodes for a zinc finger protein. Nat Genet 1996, 12:186-190 [DOI] [PubMed] [Google Scholar]
- 23.Kagitani F, Kuroiwa Y, Wakana S, Shiroishi T, Miyoshi N, Kobayashi S, Nishida M, Kohda T, Kaneko-Ishino T, Ishino F: Peg5/Neuronatin is an imprinted gene located on sub-distal chromosome 2 in the mouse. Nucleic Acids Res 1997, 25:3428-3432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Plass C, Shibata H, Kalcheva I, Mullins L, Kotelevtseva N, Mullins J, Kato R, Sasaki H, Hirotsune S, Okazaki Y, Held WA, Hayashizaki Y, Chapman VM: Identification of Grf1 on mouse chromosome 9 as an imprinted gene by RLGS-M. Nat Genet 1996, 14:106-109 [DOI] [PubMed] [Google Scholar]
- 25.Hatada I, Sugama T, Mukai T: A new imprinted gene cloned by a methylation-sensitive genome scanning method. Nucleic Acids Res 1993, 21:5577-5582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meguro M, Mitsuya K, Sui H, Shigenami K, Kugoh H, Nakao M, Oshimura M: Evidence for uniparental, paternal expression of the human GABAA receptor subunit genes, using microcell-mediated chromosome transfer. Hum Mol Genet 1997, 6:2127-2133 [DOI] [PubMed] [Google Scholar]
- 27.Gabriel JM, Higgins MJ, Gebuhr TC, Shows TB, Saitoh S, Nicholls RD: A model system to study genomic imprinting of human genes. Proc Natl Acad Sci USA 1998, 95:14857-14862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moore T, Haig D: Genomic imprinting in mammalian development: a parental tug-of-war. Trends Genet 1991, 7:45-49 [DOI] [PubMed] [Google Scholar]
- 29.DeChiara TM, Efstratiadis A, Robertson EJ: A growth-deficiency phenotype in heterozygous mice carrying an insulin-like growth factor II gene disrupted by targeting. Nature 1990, 345:78-80 [DOI] [PubMed] [Google Scholar]
- 30.Bartolomei MS, Zemel S, Tilghman SM: Parental imprinting of the mouse H19 gene. Nature 1991, 351:153-155 [DOI] [PubMed] [Google Scholar]
- 31.Miyoshi N, Kuroiwa Y, Kohda T, Shitara H, Yonekawa H, Kawabe T, Hasegawa H, Barton SC, Surani MA, Kaneko-Ishino T, Ishino F: Identification of the Meg1/Grb10 imprinted gene on mouse proximal chromosome 11, a candidate for the Silver-Russell syndrome gene. Proc Natl Acad Sci USA 1998, 95:1102-1107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bestor TH, Tycko B: Creation of genomic methylation patterns. Nat Genet 1996, 12:363-367 [DOI] [PubMed] [Google Scholar]
- 33.Jaenisch R, Schnieke A, Harbers K: Treatment of mice with 5-azacytidine efficiently activates silent retroviral genomes in different tissues. Proc Natl Acad Sci USA 1985, 82:1451-1455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang Y, Tycko B: Monoallelic expression of the human H19 gene. Nature Genet 1992, 1:40-44 [DOI] [PubMed] [Google Scholar]
- 35.Wevrick R, Kerns JA, Francke U: Identification of a novel paternally expressed gene in the Prader-Willi syndrome region. Hum Mol Genet 1994, 3:1877-1882 [DOI] [PubMed] [Google Scholar]
- 36.Moore T, Constancia M, Zubair M, Bailleul B, Feil R, Sasaki H, Reik W: Multiple imprinted sense and antisense transcripts, differential methylation and tandem repeats in a putative imprinting control region upstream of mouse Igf2. Proc Natl Acad Sci USA 1997, 94:12509-12514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wutz A, Smrzka OW, Barlow DP: Making sense of imprinting the mouse and human IGF2R loci. Novartis Found Symp 1998, 214:251-259 [DOI] [PubMed] [Google Scholar]
- 38.Knoll JH, Cheng SD, Lalande M: Allele specificity of DNA replication timing in the Angelman/Prader-Willi syndrome imprinted chromosomal region. Nat Genet 1994, 6:41-46 [DOI] [PubMed] [Google Scholar]
- 39.Kitsberg D, Selig S, Brandeis M, Simon I, Keshet I, Driscoll DJ, Nicholls RD, Cedar H: Allele-specific replication timing of imprinted gene regions. Nature 1993, 364:459-463 [DOI] [PubMed] [Google Scholar]
- 40.Robinson WP, Lalande M: Sex-specific meiotic recombination in the Prader–Willi/Angelman syndrome imprinted region. Hum Mol Genet 1995, 4:801-806 [DOI] [PubMed] [Google Scholar]
- 41.Laird PW, Jaenisch R: The role of DNA methylation in cancer genetics and epigenetics. Annu Rev Genet 1996, 30:441-464 [DOI] [PubMed] [Google Scholar]
- 42.Tycko B: DNA methylation in genomic imprinting. Mutat Res 1997, 386:103-105 [DOI] [PubMed] [Google Scholar]
- 43.Razin A, Cedar H: DNA methylation and genomic imprinting. Cell 1994, 77:473-476 [DOI] [PubMed] [Google Scholar]
- 44.Bartolomei MS, Webber AL, Brunkow ME, Tilghman SM: Epigenetic mechanisms underlying the imprinting of the mouse H19 gene. Genes Dev 1993, 7:1663-1673 [DOI] [PubMed] [Google Scholar]
- 45.Ferguson-Smith AC, Sasaki H, Cattanach BM, Surani MA: Parental-origin-specific epigenetic modification of the mouse H19 gene. Nature 1993, 362:751-755 [DOI] [PubMed] [Google Scholar]
- 46.Stöger R, Kubicka P, Liu CG, Kafri T, Razin A, Cedar H, Barlow DP: Maternal-specific methylation of the imprinted mouse Igf2r locus identifies the expressed locus as carrying the imprinting signal. Cell 1993, 73:61-71 [DOI] [PubMed] [Google Scholar]
- 47.Hatada I, Kitagawa K, Yamaoka T, Wang X, Arai Y, Hashido K, Ohishi S, Masuda J, Ogata J, Mukai T: Allele-specific methylation and expression of an imprinted U2af1-rs1 (SP2) gene. Nucleic Acids Res 1995, 23:36-41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hatada I, Mukai T: Genomic imprinting of p57KIP2, cyclin-dependent kinase inhibitor, in mouse. Nat Genet 1995, 11:204-206 [DOI] [PubMed] [Google Scholar]
- 49.Lalande M: Parental imprinting and human disease. Annu Rev Genet 1996, 30:173-195 [DOI] [PubMed] [Google Scholar]
- 50.Hall JG: Genomic imprinting: nature and clinical relevance. Annu Rev Med 1997, 48:35-44 [DOI] [PubMed] [Google Scholar]
- 51.Viljoen D, Ramesar R: Evidence for paternal imprinting in familial Beckwith-Wiedemann syndrome. J Med Genet 1992, 29:221-225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ward A: Beck-Wiedemann syndrome, and Wilms’ tumour. Mol Hum Reprod 1997, 3:157-168 [DOI] [PubMed] [Google Scholar]
- 53.Feinberg AP: Genomic Imprinting: The Genetic Basis of Human Cancer. Edited by Vogelstein B, Kinzler KW. New York, McGraw-Hill Health Professions Division, 1998, pp 95–107
- 54.Bischoff FZ, Feldman GL, McCaskill C, Subramanian S, Hughes MR, Shaffer LG: Single cell analysis demonstrating somatic mosaicism involving 11p in a patient with paternal isodisomy and Beckwith-Wiedemann syndrome. Hum Mol Genet 1995, 4:395-399 [DOI] [PubMed] [Google Scholar]
- 55.Henry I, Puech A, Riesewijk A, Ahnine L, Mannens M, Beldjord C, Bitoun P, Tournade MF, Landrieu P, Junien C: Somatic mosaicism for partial paternal isodisomy in Wiedemann-Beckwith syndrome: a post-fertilization event. Eur J Hum Genet 1993, 1:19-29 [DOI] [PubMed] [Google Scholar]
- 56.Weksberg R, Shen DR, Fei YL, Song QL, Squire J: Disruption of insulin-like growth factor 2 imprinting in Beckwith-Wiedemann syndrome. Nat Genet 1993, 5:143-150 [DOI] [PubMed] [Google Scholar]
- 57.Joyce JA, Lam WK, Catchpoole DJ, Jenks P, Reik W, Maher ER, Schofield PN: Imprinting of IGF2 and H19: lack of reciprocity in sporadic Beckwith-Wiedemann syndrome. Hum Mol Genet 1997, 6:1543-1548 [DOI] [PubMed] [Google Scholar]
- 58.Reik W, Brown KW, Schneid H, Le Bouc Y, Bickmore W, Maher ER: Imprinting mutations in the Beckwith-Wiedemann syndrome suggested by altered imprinting pattern in the IGF2–H19 domain. Hum Mol Genet 1995, 4:2379-2385 [DOI] [PubMed] [Google Scholar]
- 59.Catchpoole D, Lam WW, Valler D, Temple IK, Joyce JA, Reik W, Schofield PN, Maher ER: Epigenetic modification and uniparental inheritance of H19 in Beckwith-Wiedemann syndrome. J Med Genet 1997, 34:353-359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Reik W, Maher ER: Imprinting in clusters: lessons from Beckwith-Wiedemann syndrome. Trends Genet 1997, 13:330-334 [DOI] [PubMed] [Google Scholar]
- 61.Brown KW, Villar AJ, Bickmore W, Clayton-Smith J, Catchpoole D, Maher ER, Reik W: Imprinting mutation in the Beckwith-Wiedemann syndrome leads to biallelic IGF2 expression through an H19-independent pathway. Hum Mol Genet 1996, 5:2027-2032 [DOI] [PubMed] [Google Scholar]
- 62.Lee MP, Hu RJ, Johnson LA, Feinberg AP: Human KVLQT1 gene shows tissue-specific imprinting, and encompasses Beckwith-Wiedemann syndrome chromosomal rearrangements. Nat Genet 1997, 15:181-185 [DOI] [PubMed] [Google Scholar]
- 63.Thompson JS, Reese KJ, DeBaun MR, Perlman EJ, Feinberg AP: Reduced expression of the cyclin-dependent kinase inhibitor gene p57KIP2 in Wilms’ tumor. Cancer Res 1996, 56:5723-5727 [PubMed] [Google Scholar]
- 64.Hatada I, Ohashi H, Fukushima Y, Kaneko Y, Inoue M, Komoto Y, Okada A, Ohishi S, Nabetani A, Morisaki H, Nakayama M, Niikawa N, Mukai T: An imprinted gene p57KIP2 is mutated in Beckwith-Wiedemann syndrome. Nat Genet 1996, 14:171-173 [DOI] [PubMed] [Google Scholar]
- 65.O’Keefe D, Dao D, Zhao L, Sanderson R, Warburton D, Weiss L, Anyane-Yeboa K, Tycko B: Coding mutations in p57KIP2 are present in some cases of Beckwith-Wiedemann syndrome but are rare or absent in Wilms’ tumors. Am J Hum Genet 1997, 61:295-303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lee MP, DeBaun M, Randhawa G, Reichard BA, Elledge SJ, Feinberg AP: Low frequency of p57KIP2 mutation in Beckwith-Wiedemann syndrome. Am J Hum Genet 1997, 61:304-309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Reid LH, Davies C, Cooper PR, Crider-Miller SJ, Sait SN, Nowak NJ, Evans G, Stanbridge EJ, deJong P, Shows TB, Weissman BE, Higgins MJ: A 1-Mb physical map, and PAC contig of the imprinted domain in 11p15.5 that contains TAPA1, and the BWSCR1/WT2 region. Genomics 1997, 43:366-375 [DOI] [PubMed] [Google Scholar]
- 68.Paulsen M, Davies KR, Bowden LM, Villar AJ, Franck O, Fuermann M, Dean WL, Moore TF, Rodrigues N, Davies KE, Hu RJ, Feinberg AP, Maher ER, Reik W, Walter J: Syntenic organization of the mouse distal chromosome 7 imprinting cluster and the Beckwith-Wiedemann syndrome region in chromosome 11p15.5. Hum Mol Genet 1998, 7:1149-1159 [DOI] [PubMed] [Google Scholar]
- 69.Sun FL, Dean WL, Kelsey G, Allen ND, Reik W: Transactivation of Igf2 in a mouse model of Beckwith-Wiedemann syndrome. Nature 1997, 389:809-815 [DOI] [PubMed] [Google Scholar]
- 70.Bartolomei MS, Tilghman SM: Genomic imprinting in mammals. Annu Rev Genet 1997, 31:493-525 [DOI] [PubMed] [Google Scholar]
- 71.Nicholls RD, Saitoh S, Horsthemke B: Imprinting in Prader-Willi and Angelman syndromes. Trends Genet 1998, 14:194-200 [DOI] [PubMed] [Google Scholar]
- 72.Holm VA, Cassidy SB, Butler MG, Hanchett JM, Greenswag LR, Whitman BY, Greenberg F: Prader-Willi syndrome: consensus diagnostic criteria. Pediatrics 1993, 91:398-402 [PMC free article] [PubMed] [Google Scholar]
- 73.Williams CA, Angelman H, Clayton-Smith J, Driscoll DJ, Hendrickson JE, Knoll JH, Magenis RE, Schinzel A, Wagstaff J, Whidden EM, et al: Angelman syndrome: consensus for diagnostic criteria. Angelman Syndrome Foundation. Am J Med Genet 1995, 56:237-238 [DOI] [PubMed] [Google Scholar]
- 74.Nicholls RD: Genomic imprinting and uniparental disomy in Angelman and Prader-Willi syndromes: a review. Am J Med Genet 1993, 46:16-25 [DOI] [PubMed] [Google Scholar]
- 75.Matsuura T, Sutcliffe JS, Fang P, Galjaard RJ, Jiang YH, Benton CS, Rommens JM, Beaudet AL: De novo truncating mutations in E6-AP ubiquitin-protein ligase gene (UBE3A) in Angelman syndrome. Nat Genet 1997, 15:74-77 [DOI] [PubMed] [Google Scholar]
- 76.Kishino T, Lalande M, Wagstaff J: UBE3A/E6-AP mutations cause Angelman syndrome. Nat Genet 1997, 15:70-73 [DOI] [PubMed] [Google Scholar]
- 77.Rougeulle C, Glatt H, Lalande M: The Angelman syndrome candidate gene, UBE3A/E6-AP, is imprinted in brain. Nat Genet 1997, 17:14-15 [DOI] [PubMed] [Google Scholar]
- 78.Vu TH, Hoffman AR: Imprinting of the Angelman syndrome gene, UBE3A, is restricted to brain. Nat Genet 1997, 17:12-13 [DOI] [PubMed] [Google Scholar]
- 79.Albrecht U, Sutcliffe JS, Cattanach BM, Beechey CV, Armstrong D, Eichele G, Beaudet AL: Imprinted expression of the murine Angelman syndrome gene, Ube3a, in hippocampal and Purkinje neurons. Nat Genet 1997, 17:75-78 [DOI] [PubMed] [Google Scholar]
- 80.Glenn CC, Nicholls RD, Robinson WP, Saitoh S, Niikawa N, Schinzel A, Horsthemke B, Driscoll DJ: Modification of 15q11–q13 DNA methylation imprints in unique Angelman and Prader-Willi patients. Hum Mol Genet 1993, 2:1377-1382 [DOI] [PubMed] [Google Scholar]
- 81.Reed ML, Leff SE: Maternal imprinting of human SNRPN, a gene deleted in Prader-Willi syndrome. Nat Genet 1994, 6:163-167 [DOI] [PubMed] [Google Scholar]
- 82.Sutcliffe JS, Han M, Christian SL, Ledbetter DH: Neuronally-expressed necdin gene: an imprinted candidate gene in Prader-Willi syndrome. Lancet 1997, 350:1520-1521 [DOI] [PubMed] [Google Scholar]
- 83.Reis A, Dittrich B, Greger V, Buiting K, Lalande M, Gillessen-Kaesbach G, Anvret M, Horsthemke B: Imprinting mutations suggested by abnormal DNA methylation patterns in familial Angelman and Prader-Willi syndromes. Am J Hum Genet 1994, 54:741-747 [PMC free article] [PubMed] [Google Scholar]
- 84.Sutcliffe JS, Nakao M, Christian S, Orstavik KH, Tommerup N, Ledbetter DH, Beaudet AL: Deletions of a differentially methylated CpG island at the SNRPN gene define a putative imprinting control region. Nat Genet 1994, 8:52-58 [DOI] [PubMed] [Google Scholar]
- 85.Buiting K, Saitoh S, Gross S, Dittrich B, Schwartz S, Nicholls RD, Horsthemke B: Inherited microdeletions in the Angelman and Prader-Willi syndromes define an imprinting centre on human chromosome 15. Nat Genet 1995, 9:395-400 [DOI] [PubMed] [Google Scholar]
- 86.Dittrich B, Buiting K, Korn B, Rickard S, Buxton J, Saitoh S, Nicholls RD, Poustka A, Winterpacht A, Zabel B, Horsthemke B: Imprint switching on human chromosome 15 may involve alternative transcripts of the SNRPN gene. Nat Genet 1996, 14:163-170 [DOI] [PubMed] [Google Scholar]
- 87.Horsthemke B: Structure and function of the human chromosome 15 imprinting center. J Cell Physiol 1997, 173:237-241 [DOI] [PubMed] [Google Scholar]
- 88.Tilghman SM, Caspary T, Ingram RS: Competitive edge at the imprinted Prader-Willi/Angelman region? Nat Genet 1998, 18:206-208 [DOI] [PubMed] [Google Scholar]
- 89.Schumacher A, Buiting K, Zeschnigk M, Doerfler W, Horsthemke B: Methylation analysis of the PWS/AS region does not support an enhancer- competition model. Nat Genet 1998, 19:324-325 [DOI] [PubMed] [Google Scholar]
- 90.Spotila LD, Sereda L, Prockop DJ: Partial isodisomy for maternal chromosome 7 and short stature in an individual with a mutation at the COL1A2 locus. Am J Hum Genet 1992, 51:1396-1405 [PMC free article] [PubMed] [Google Scholar]
- 91.Lefebvre L, Viville S, Barton SC, Ishino F, Keverne EB, Surani MA: Abnormal maternal behaviour and growth retardation associated with loss of the imprinted gene Mest. Nat Genet 1998, 20:163-169 [DOI] [PubMed] [Google Scholar]
- 92.Fundele RH, Surani MA: Experimental embryological analysis of genetic imprinting in mouse development. Dev Genet 1994, 15:515-522 [DOI] [PubMed] [Google Scholar]
- 93.Keverne EB, Fundele R, Narasimha M, Barton SC, Surani MA: Genomic imprinting and the differential roles of parental genomes in brain development. Brain Res Dev Brain Res 1996, 92:91-100 [DOI] [PubMed] [Google Scholar]
- 94.Skuse DH, James RS, Bishop DVM, Coppin B, Dalton P, Aamodt-Leeper G, Bacarese-Hamilton M, Creswell C, McGurk R, Jacobs PA: Evidence from Turner’s syndrome of an imprinted X-linked locus affecting cognitive function. Nature 1997, 387:705-708 [DOI] [PubMed] [Google Scholar]
- 95.McCauley E, Ito J, Kay T: Psychosocial functioning in girls with Turner’s syndrome and short stature: social skills, behavior problems, and self-concept. J Am Acad Child Psychiatry 1986, 25:105-112 [DOI] [PubMed] [Google Scholar]
- 96.McCauley E, Kay T, Ito J, Treder R: The Turner syndrome: cognitive deficits, affective discrimination, and behavior problems. Child Dev 1987, 58:464-473 [PubMed] [Google Scholar]
- 97.Miller AP, Willard HF: Chromosomal basis of X chromosome inactivation: identification of a multigene domain in Xp11.21-p11.22 that escapes X inactivation. Proc Natl Acad Sci USA 1998, 95:8709-8714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.McMahon FJ, Stine OC, Meyers DA, Simpson SG, DePaulo JR: Patterns of maternal transmission in bipolar affective disorder. Am J Hum Genet 1995, 56:1277-1286 [PMC free article] [PubMed] [Google Scholar]
- 99.Kato T, Winokur G, Coryell W, Keller MB, Endicott J, Rice J: Parent-of-origin effect in transmission of bipolar disorder. Am J Med Genet 1996, 67:546-550 [DOI] [PubMed] [Google Scholar]
- 100.Grigoroiu-Serbanescu M, Wickramaratne PJ, Hodge SE, Milea S, Mihailescu R: Genetic anticipation and imprinting in bipolar I illness. Br J Psychiatry 1997, 170:162-166 [DOI] [PubMed] [Google Scholar]
- 101.Asherson P, Walsh C, Williams J, Sargeant M, Taylor C, Clements A, Gill M, Owen M, McGuffin P: Imprinting and anticipation. Are they relevant to genetic studies of schizophrenia? Br J Psychiatry 1994, 164:619-624 [DOI] [PubMed] [Google Scholar]
- 102.Ohara K, Xu HD, Mori N, Suzuki Y, Xu DS, Wang ZC: Anticipation and imprinting in schizophrenia. Biol Psychiatry 1997, 42:760-766 [DOI] [PubMed] [Google Scholar]
- 103.Cook EH, Jr, Lindgren V, Leventhal BL, Courchesne R, Lincoln A, Shulman C, Lord C, Courchesne E: Autism or atypical autism in maternally but not paternally derived proximal 15q duplication. Am J Hum Genet 1997, 60:928-934 [PMC free article] [PubMed] [Google Scholar]
- 104.Morison IM, Reeve AE: A catalogue of imprinted genes and parent-of-origin effects in humans and animals. Hum Mol Genet 1998, 7:1599-1609 [DOI] [PubMed] [Google Scholar]
- 105.Wake N, Fujino T, Hoshi S, Shinkai N, Sakai K, Kato H, Hashimoto M, Yasuda T, Yamada H, Ichinoe K: The propensity to malignancy of dispermic heterozygous moles. Placenta 1987, 8:319-326 [DOI] [PubMed] [Google Scholar]
- 106.Linder D, McCaw BK, Hecht F: Parthenogenetic origin of benign ovarian teratomers. N Engl J Med 1975, 292:63-66 [DOI] [PubMed] [Google Scholar]
- 107.Caron H, van Sluis P, van Hoeve M, de Kraker J, Bras J, Slater R, Mannens M, Voute PS, Westerveld A, Versteeg R: Allelic loss of chromosome 1p36 in neuroblastoma is preferential maternal origin and correlates with N-myc amplification. Nat Genet 1993, 4:187-190 [DOI] [PubMed] [Google Scholar]
- 108.Katz F, Webb D, Gibbons B, Reeves B, McMahon C, Cheselles J, Mitchell C: Possible evidence for genomic imprinting in childhood acute myeloblastic leukaemia associated with monosomy for chromosome 7. Br J Haematol 1992, 80:332-336 [DOI] [PubMed] [Google Scholar]
- 109.Tycko B: Genomic imprinting in Wilms’ tumor. 1996. Med Ped Oncol [DOI] [PubMed]
- 110.Scrable H, Cavenne W, Ghavimi F, Lovell M, Morgan K, Sapienza C: A model for embryonal rhabdomyosarcoma tumorigenesis that involves genome imprinting. Proc Natl Acad Sci USA 1989, 86:7480-7484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Toguchida J, Ishizaki K, Sasaki MS, Nakamura Y, Ikenaga M, Kato M, Sugimot M, Kotoura Y, Yamamuro T: Preferential mutation of paternally derived RB gene as the initial event in sporadic osteosarcoma. Nature 1989, 338:156-158 [DOI] [PubMed] [Google Scholar]
- 112.Milunsky J, DeStefano AL, Huang XL, Baldwin CT, Michels VV, Jako G, Milunsky A: Familial paragangliomas: linkage to chromosome 11q23 and clinical implications. Am J Med Genet 1997, 72:66-70 [DOI] [PubMed] [Google Scholar]
- 113.Rainier S, Johnson LA, Dobry CJ, Ping AJ, Grundy PE, Feinberg AP: Relaxation of imprinted genes in human cancer. Nature 1993, 362:747. [DOI] [PubMed] [Google Scholar]
- 114.Ogawa O, Eccles MR, Szeto J, McNoe LA, Yun K, Maw MA, Smith PJ, Reeve AE: Relaxation of insulin-like growth factor II gene imprinting implicated in Wilms’ tumour. Nature 1993, 362:749-751 [DOI] [PubMed] [Google Scholar]
- 115.Steenman MJC, Rainier S, Dobry CJ, Grundy P, Horon IL, Feinberg AP: Loss of imprinting of IGF2 is linked to reduced expression and abnormal methylation of H19 in Wilms’ tumour. Nat Genet 1994, 7:433-439 [DOI] [PubMed] [Google Scholar]
- 116.Hao Y, Crenshaw T, Moulton T, Newcomb E, Tycko B: Tumor-suppressor activity of H19. Nature 1993, 365:764-767 [DOI] [PubMed] [Google Scholar]
- 117.Webber AL, Ingram RS, Levorse JM, Tilghman SM: Location of enhancers is essential for the imprinting of H19 and Igf2 genes. Nature 1998, 391:711-715 [DOI] [PubMed] [Google Scholar]
- 118.Leighton PA, Ingram RS, Eggenschwiler J, Efstratiatis A, Tilghman SM: Disruption of imprinting caused by deletion of the H19 gene region in mice. Nature 1995, 375:34-39 [DOI] [PubMed] [Google Scholar]
- 119.Okamoto K, Morison IM, Taniguchi T, Reeve AE: Epigenetic changes at the insulin-like growth factor II/H19 locus in developing kidney is an early event in Wilms’ tumorigenesis. Proc Natl Acad Sci USA 1997, 94:5367-5371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Jinno Y, Yun K, Nishiwaki K, Kubota T, Ogawa O, Reeve AE, Nikawa N: Mosaic and polymorphic imprinting of the WT1 gene in humans. Nat Genet 1994, 6:305-309 [DOI] [PubMed] [Google Scholar]
- 121.Rauscher FJd: The Wt1 Wilms’ tumor gene product: a developmentally regulated transcription factor in the kidney that functions as a tumor suppressor. FASEB J 1993, 7:896-903 [PubMed] [Google Scholar]
- 122.Chung WY, Yuan L, Feng L, Hensle T, Tycko B: Chromosome 11p15.5 regional imprinting: comparative analysis of KIP2 and H19 in human tissues and Wilms’ tumors. Hum Mol Genet 1996, 5:1101-1108 [DOI] [PubMed] [Google Scholar]
- 123.Hatada I, Inazawa J, Abe T, Nakayama M, Kaneko Y, Jinno Y, Niikawa N, Ohashi H, Fukushima Y, Iida K, Yutani C, Takahashi S, Chiba Y, Ohishi S, Mukai T: Genomic imprinting of human p57KIP2 and its reduced expression in Wilms’ tumors. Hum Mol Genet 1996, 5:783-788 [DOI] [PubMed] [Google Scholar]
- 124.Kondo M, Matsuoka S, Uchida K, Osada H, Nagatake M, Takagi K, Harper JW, Takahashi T, Elledge SJ, Takahashi T: Selective maternal-allele loss in human lung cancers of the maternally expressed p57KIP2 gene at 11p15.5. Oncogene 1996, 12:1365-1368 [PubMed] [Google Scholar]
- 125.De Souza AT, Hankins GR, Washington MK, Fine RL, Orton TC, Jirtle RL: Frequent loss of heterozygosity on 6q at the mannose 6- phosphate/insulin-like growth factor II receptor locus in human hepatocellular tumors. Oncogene 1995, 10:1725-1729 [PubMed] [Google Scholar]
- 126.De Souza AT, Hankins GR, Washington MK, Orton TC, Jirtle RL: M6P/IGF2R gene is mutated in human hepatocellular carcinomas with loss of heterozygosity. Nat Genet 1995, 11:447-449 [DOI] [PubMed] [Google Scholar]
- 127.Hankins GR, De Souza AT, Bentley RC, Patel MR, Marks JR, Iglehart JD, Jirtle RL: M6P/IGF2 receptor: a candidate breast tumor suppressor gene. Oncogene 1996, 12:2003-2009 [PubMed] [Google Scholar]
- 128.Yamada T, De Souza AT, Finkelstein S, Jirtle RL: Loss of the gene encoding mannose 6-phosphate/insulin-like growth factor II receptor is an early event in liver carcinogenesis. Proc Natl Acad Sci USA 1997, 94:10351-10355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Yu Y, Xu F, Peng H, Fang X, Zhao S, Li Y, Cuevas B, Kuo WL, Gray JW, Siciliano M, Mills GB, Bast RC, Jr: NOEY2 (ARHI), an imprinted putative tumor suppressor gene in ovarian, and breast carcinomas. Proc Natl Acad Sci USA 1999, 96:214-219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Korner C, Nurnberg B, Uhde M, Braulke T: Mannose 6-phosphate/insulin-like growth factor II receptor fails to interact with G-proteins: analysis of mutant cytoplasmic receptor domains. J Biol Chem 1995, 270:287-295 [DOI] [PubMed] [Google Scholar]
- 131.Souza RF, Appel R, Yin J, Wang S, Smolinski KN, Abraham JM, Zou T-T, Shi Y-Q, Lei J, Cottrell J, Cymes K, Biden K, Simms L, Leggett B, Lynch PM, Frazier M, Powell SM, Harpaz N, Sugimura H, Young J, Meltzer SJ: The insulin-like growth factor II receptor gene is a target of microsatellite instability in human gastrointestinal tumours. Nat Genet 1996, 14:255-257 [DOI] [PubMed] [Google Scholar]
- 132.Ouyang H, Shiwaku HO, Hagiwara H, Miura K, Abe T, Kato Y, Ohtani H, Shiiba K, Souza RF, Meltzer SJ, Horii A: The insulin-like growth factor II receptor gene is mutated in genetically unstable cancers of the endometrium, stomach, and colorectum. Cancer Res 1997, 57:1851-1854 [PubMed] [Google Scholar]
- 133.Leung SY, Chan TL, Chung LP, Chan AS, Fan YW, Hung KN, Kwong WK, Ho JW, Yuen ST: Microsatellite instability and mutation of DNA mismatch repair genes in gliomas. Am J Pathol 1998, 153:1181-1188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Chappell SA, Walsh T, Walker RA, Shaw JA: Loss of heterozygosity at the mannose 6-phosphate insulin-like growth factor 2 receptor gene correlates with poor differentiation in early breast carcinomas. Br J Cancer 1997, 76:1558-1561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Mills JJ, Falls JG, De Souza AT, Jirtle RL: Imprinted M6p/Igf2 receptor is mutated in rat liver tumors. Oncogene 1998, 16:2797-2802 [DOI] [PubMed] [Google Scholar]
- 136.Xu Y, Goodyer CG, Deal C, Polychronakos C: Functional polymorphism in the parental imprinting of the human IGF2R gene. Biochem Biophys Res Commun 1993, 197:747-754 [DOI] [PubMed] [Google Scholar]
- 137.Ogawa O, McNoe LA, Eccles MR, Morison IM, Reeve AE: Human insulin-like growth factor type I and type II receptors are not imprinted. Hum Mol Genet 1993, 2:2163-2165 [DOI] [PubMed] [Google Scholar]
- 138.Kalscheuer VM, Mariman EC, Schepens MT, Rehder H, Ropers H-H: The insulin-like growth factor type-2 receptor gene is imprinted in the mouse but not in humans. Nat Genet 1993, 5:74-78 [DOI] [PubMed] [Google Scholar]
- 139.Xu YQ, Grundy P, Polychronakos C: Aberrant imprinting of the insulin-like growth factor II receptor gene in Wilms’ tumor. Oncogene 1997, 14:1041-1046 [DOI] [PubMed] [Google Scholar]
- 140.Wutz A, Smrzka OW, Schweifer N, Schellander K, Wagner EF, Barlow DP: Imprinted expression of the Igf2r gene depends on an intronic CpG island. Nature 1997, 389:745-749 [DOI] [PubMed] [Google Scholar]
- 141.Birger Y, Shemer R, Perk J, Razin A: The imprinting box of the mouse Igf2r gene. Nature 1999, 397:84-88 [DOI] [PubMed] [Google Scholar]
- 142.Smrzka OW, Fae I, Stöger R, Kruzbauer R, Fischer GF, Henn T, Weith A, Barlow DP: Conservation of a maternal-specific methylation signal at the human IGF2R locus. Hum Mol Genet 1995, 4:1945-1952 [DOI] [PubMed] [Google Scholar]
- 143.Riesewijk AM, Schepens MT, Welch TR: Van den Berg-Loonen EM, Mariman EM, Ropers HH, Kalscheuer VM: Maternal-specific methylation of the human IGF2R gene is not accompanied by allele-specific transcription. Genomics 1996, 31:158-166 [DOI] [PubMed] [Google Scholar]
- 144.Giannoukakis N, Deal C, Paquette J, Kukuvitis A, Polychronakos C: Polymorphic functional imprinting of the human IGF2 gene among individuals, in blood cells, is associated with H19 expression. Biochem Biophys Res Commun 1996, 220:1014-1019 [DOI] [PubMed] [Google Scholar]
- 145.Bunzel R, Blumcke I, Cichon S, Normann S, Schramm J, Propping P, Nothen MM: Polymorphic imprinting of the serotonin-2A (5-HT2A) receptor gene in human adult brain. Brain Res Mol Brain Res 1998, 59:90-92 [DOI] [PubMed] [Google Scholar]
- 146.Jiang S, Hemann MA, Lee MP, Feinberg AP: Strain-dependent developmental relaxation of imprinting of an endogenous mouse gene, kvlqt1. Genomics 1998, 53:395-399 [DOI] [PubMed] [Google Scholar]
- 147.Kaghad M, Bonnet H, Yang A, Creancier L, Biscan J-C, Valent A, Minty A, Chalon P, Lelias J-M, Dumont X, Ferrara P, McKeon F, Caput D: Monoallelically expressed gene related to p53 at 1p36, a region frequently deleted in neuroblastoma and other human cancers. Cell 1997, 90:809-819 [DOI] [PubMed] [Google Scholar]
- 148.Mai M, Qian C, Yokomizo A, Tindall DJ, Bostwick D, Polychronakos C, Smith DI, Liu W: Loss of imprinting and allele switching of p73 in renal cell carcinoma. Oncogene 1998, 17:1739-1741 [DOI] [PubMed] [Google Scholar]
- 149.Pearsall RS, Shibata H, Brozowska A, Yoshino K, Okuda K, deJong PJ, Plass C, Chapman VM, Hayashizaki Y, Held WA: Absence of imprinting in U2AFBPL, a human homologue of the imprinted mouse gene U2afbp-rs. Biochem Biophys Res Commun 1996, 222:171-177 [DOI] [PubMed] [Google Scholar]
- 150.Hayashizaki Y, Shibata H, Hirotsune S, Sugino H, Okazaki Y, Sasaki N, Hirose K, Imoto H, Okuizumi H, Muramatsu M, et al: Identification of an imprinted U2af binding protein related sequence on mouse chromosome 11 using the RLGS method. Nat Genet 1994, 6:33-40 [DOI] [PubMed] [Google Scholar]
- 151.Riesewijk AM, Schepens MT, Mariman EM, Ropers HH, Kalscheuer VM: The MAS proto-oncogene is not imprinted in humans. Genomics 1996, 35:380-382 [DOI] [PubMed] [Google Scholar]
- 152.Miller N, McCann AH, O’Connell D, Pedersen IS, Spiers V, Gorey T, Dervan PA: The MAS proto-oncogene is imprinted in human breast tissue. Genomics 1997, 46:509-512 [DOI] [PubMed] [Google Scholar]
- 153.Villar AJ, Pedersen RA: Parental imprinting of the Mas protooncogene in mouse. Nat Genet 1994, 8:373-379 [DOI] [PubMed] [Google Scholar]
- 154.Kobayashi S, Kohda T, Miyoshi N, Kuroiwa Y, Aisaka K, Tsutsumi O, Kaneko-Ishino T, Ishino F: Human PEG1/MEST, an imprinted gene on chromosome 7. Hum Mol Genet 1997, 6:781-786 [DOI] [PubMed] [Google Scholar]
- 155.Riesewijk AM, Hu L, Schulz U, Tariverdian G, Hoglund P, Kere J, Ropers HH, Kalscheuer VM: Monoallelic expression of human PEG1/MEST is paralleled by parent-specific methylation in fetuses. Genomics 1997, 42:236-244 [DOI] [PubMed] [Google Scholar]
- 156.Mitsuya K, Sui H, Meguro M, Kugoh H, Jinno Y, Niikawa N, Oshimura M: Paternal expression of WT1 in human fibroblasts and lymphocytes. Hum Mol Genet 1997, 6:2243-2246 [DOI] [PubMed] [Google Scholar]
- 157.Alders M, Hodges M, Hadjantonakis AK, Postmus J, van Wijk I, Bliek J, de Meulemeester M, Westerveld A, Guillemot F, Oudejans C, Little P, Mannens M: The human Achaete-Scute homologue 2 (ASCL2,HASH2) maps to chromosome 11p15.5, close to IGF2 and is expressed in extravillus trophoblasts. Hum Mol Genet 1997, 6:859-867 [DOI] [PubMed] [Google Scholar]
- 158.Guillemot F, Caspary T, Tilghman SM, Copeland NG, Gilbert DJ, Jenkins NA, Anderson DJ, Joyner AL, Rossant J, Nagy A: Genomic imprinting of Mash2, a mouse gene required for trophoblast development. Nat Genet 1995, 9:235-242 [DOI] [PubMed] [Google Scholar]
- 159.Rachmilewitz J, Goshen R, Ariel I, Schneider T, De Groot N, Hochberg A: Parental imprinting of the human H19 gene. FEBS Lett 1992, 309:25-28 [DOI] [PubMed] [Google Scholar]
- 160.Ohlsson R, Nystrom A, Pfeifer-Ohlsson S, Tobonen V, Hedbourg F, Schofield P, Flam F, Ekstrom TJ: IGF2 is parentally imprinted during human embryogenesis, and in the Beckwith-Wiedemann syndrome. Nat Genet 1993, 4:94-97 [DOI] [PubMed] [Google Scholar]
- 161.Giannoukakis N, Deal C, Paquette J, Goodyer CG, Polychronakos C: Parental genomic imprinting of the human IFG2 gene. Nat Genet 1993, 4:98-101 [DOI] [PubMed] [Google Scholar]
- 162.Ogawa O, Becroft DM, Morison IM, Eccles MR, Skeen JE, Mauger DC, Reeve AE: Constitutional relaxation of insulin-like growth factor II gene imprinting associated with Wilms’ tumour and gigantism. Nat Genet 1993, 5:408-412 [DOI] [PubMed] [Google Scholar]
- 163.Dao D, Frank D, Qian N, O’Keefe D, Vosatka RJ, Walsh CP, Tycko B: IMPT1, an imprinted gene similar to polyspecific transporter, and multi-drug resistance genes. Hum Mol Genet 1998, 7:597-608 [DOI] [PubMed] [Google Scholar]
- 164.Schwienbacher C, Sabbioni S, Campi M, Veronese A, Bernardi G, Menegatti A, Hatada I, Mukai T, Ohashi H, Barbanti-Brodano G, Croce CM, Negrini M: Transcriptional map of 170-kb region at chromosome 11p15.5: identification and mutational analysis of the BWR1A gene reveals the presence of mutations in tumor samples. Proc Natl Acad Sci USA 1998, 95:3873-3878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Cooper PR, Smilinich NJ, Day CD, Nowak NJ, Reid LH, Pearsall RS, Reece M, Prawitt D, Landers J, Housman DE, Winterpacht A, Zabel BU, Pelletier J, Weissman BE, Shows TB, Higgins MJ: Divergently transcribed overlapping genes expressed in liver and kidney and located in the 11p15.5 imprinted domain. Genomics 1998, 49:38-51 [DOI] [PubMed] [Google Scholar]
- 166.Lee MP, Reeves C, Schmitt A, Su K, Connors TD, Hu RJ, Brandenburg S, Lee MJ, Miller G, Feinberg AP: Somatic mutation of TSSC5, a novel imprinted gene from human chromosome 11p15.5. Cancer Res 1998, 58:4155-4159 [PubMed] [Google Scholar]
- 167.Giddings SJ, King CD, Harman KW, Flood JF, Carnaghi LR: Allele specific inactivation of insulin 1 and 2, in the mouse yolk sac, indicates imprinting. Nat Genet 1994, 6:310-313 [DOI] [PubMed] [Google Scholar]
- 168.Bennett ST, Wilson AJ, Esposito L, Bouzekri N, Undlien DE, Cucca F, Nistico L, Buzzetti R, Bosi E, Pociot F, Nerup J, Cambon-Thomsen A, Pugliese A, Shield JP, McKinney PA, Bain SC, Polychronakos C, Todd JA: Insulin VNTR allele-specific effect in type 1 diabetes depends on identity of untransmitted paternal allele. The IMDIAB Group. Nat Genet 1997, 17:350-352 [DOI] [PubMed] [Google Scholar]
- 169.Bennett ST, Lucassen AM, Gough SC, Powell EE, Undlien DE, Pritchard LE, Merriman ME, Kawaguchi Y, Dronsfield MJ, Pociot F, et al: Susceptibility to human type 1 diabetes at IDDM2 is determined by tandem repeat variation at the insulin gene minisatellite locus. Nat Genet 1995, 9:284-292 [DOI] [PubMed] [Google Scholar]
- 170.Qian N, Frank D, O’Keefe D, Dao D, Zhao L, Yuan L, Wang Q, Keating M, Walsh C, Tycko B: The IPL gene on chromosome 11p15.5 is imprinted in humans, and mice, and is similar to TDAG51, implicated in Fas expression, and apoptosis. Hum Mol Genet 1997, 6:2021-2029 [DOI] [PubMed] [Google Scholar]
- 171.Lee MP, Feinberg AP: Genomic imprinting of a human apoptosis gene homologue, TSSC3. Cancer Res 1998, 58:1052-1056 [PubMed] [Google Scholar]
- 172.Morisaki H, Hatada I, Morisaki T, Mukai T: A novel gene, ITM, located between p57KIP2 and IPL, is imprinted in mice. DNA Res 1998, 5:235-240 [DOI] [PubMed] [Google Scholar]
- 173.Gould TD, Pfeifer K: Imprinting of mouse Kvlqt1 is developmentally regulated. Hum Mol Genet 1998, 7:483-487 [DOI] [PubMed] [Google Scholar]
- 174.Matsuoka S, Thompson JS, Edwards MC, Barletta JM, Grundy P, Kalikin LM, Harper JW, Elledge SJ, Feinberg AP: Imprinting of the gene encoding a human cyclin-dependent kinase inhibitor, p57KIP2, on chromosome 11p15. Proc Natl Acad Sci USA 1996, 93:3026-3030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Kato MV, Shimizu T, Nagayoshi M, Kaneko A, Sasaki MS, Ikawa Y: Genomic imprinting of the human serotonin-receptor (HTR2) gene involved in development of retinoblastoma. Am J Hum Genet 1996, 59:1084-1090 [PMC free article] [PubMed] [Google Scholar]
- 176.Kato MV, Ikawa Y, Hayashizaki Y, Shibata H: Paternal imprinting of mouse serotonin receptor 2A gene Htr2 in embryonic eye: a conserved imprinting regulation on the RB/Rb locus. Genomics 1998, 47:146-148 [DOI] [PubMed] [Google Scholar]
- 177.Glenn CC, Driscoll DJ, Yang TP, Nicholls RD: Genomic imprinting: potential function and mechanisms revealed by the Prader-Willi and Angelman syndromes. Mol Hum Reprod 1997, 3:321-332 [DOI] [PubMed] [Google Scholar]
- 178.Culiat CT, Stubbs LJ, Montgomery CS, Russell LB, Rinchik EM: Phenotypic consequences of deletion of the γ 3, α 5, or β 3 subunit of the type A γ-aminobutyric acid receptor in mice. Proc Natl Acad Sci USA 1994, 91:2815-2818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Nicholls RD, Gottlieb W, Russell LB, Davda M, Horsthemke B, Rinchik EM: Evaluation of potential models for imprinted and nonimprinted components of human chromosome 15q11–q13 syndromes by fine-structure homology mapping in the mouse. Proc Natl Acad Sci USA 1993, 90:2050-2054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Saitoh S, Buiting K, Rogan PK, Buxton JL, Driscoll DJ, Arnemann J, Konig R, Malcolm S, Horsthemke B, Nicholls RD: Minimal definition of the imprinting center and fixation of chromosome 15q11–q13 epigenotype by imprinting mutations. Proc Natl Acad Sci USA 1996, 93:7811-7815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Yang T, Adamson TE, Resnick JL, Leff S, Wevrick R, Francke U, Jenkins NA, Copeland NG, Brannan CI: A mouse model for Prader-Willi syndrome imprinting-centre mutations. Nat Genet 1998, 19:25-31 [DOI] [PubMed] [Google Scholar]
- 182.MacDonald HR, Wevrick R: The necdin gene is deleted in Prader-Willi syndrome and is imprinted in human and mouse. Hum Mol Genet 1997, 6:1873-1878 [DOI] [PubMed] [Google Scholar]
- 183.Ning Y, Roschke A, Christian SL, Lesser J, Sutcliffe JS, Ledbetter DH: Identification of a novel paternally expressed transcript adjacent to snRPN in the Prader-Willi syndrome critical region. Genome Res 1996, 6:742-746 [DOI] [PubMed] [Google Scholar]
- 184.Glenn CC, Porter KA, Jong MT, Nicholls RD, Driscoll DJ: Functional imprinting and epigenetic modification of the human SNRPN gene. Hum Mol Genet 1993, 2:2001-2005 [DOI] [PubMed] [Google Scholar]
- 185.Leff SE, Brannan CI, Reed ML, Ozcelik T, Francke U, Copeland NG, Jenkins NA: Maternal imprinting of mouse Snrpn gene and conserved linkage homology with the human Prader-Willi syndrome region. Nat Genet 1992, 4:259-264 [DOI] [PubMed] [Google Scholar]
- 186.Cattanach BM, Barr JA, Evans EP, Burtenshaw M, Beechey CV, Leff SE, Brannan CI, Copeland NG, Jenkins NA, Jones J: A candidate mouse model for Prader-Willi syndrome which shows an absence of Snrpn expression. Nat Genet 1992, 2:270-274 [DOI] [PubMed] [Google Scholar]
- 187.Driscoll DJ, Waters MF, Williams CA, Zori RT, Glenn CC, Avidano KM, Nicholls RD: A DNA methylation imprint, determined by the sex of the parent, distinguishes the Angelman and Prader-Willi syndromes. Genomics 1992, 13:917-924 [DOI] [PubMed] [Google Scholar]
- 188.Kim J, Ashworth L, Branscomb E, Stubbs L: The human homolog of a mouse-imprinted gene, Peg3, maps to a zinc finger gene-rich region of human chromosome 19q13.4. Genome Res 1997, 7:532-540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Joseph R, Dou D, Tsang W: Molecular cloning of a novel mRNA (neuronatin) that is highly expressed in neonatal mammalian brain. Biochem Biophys Res Commun 1994, 201:1227-1234 [DOI] [PubMed] [Google Scholar]
- 190.Dou D, Joseph R: Cloning of human neuronatin gene and its localization to chromosome-20q 11.2–12: the deduced protein is a novel “proteolipid’. Brain Res 1996, 723:8-22 [DOI] [PubMed] [Google Scholar]
- 191.Hayward BE, Kamiya M, Strain L, Moran V, Campbell R, Hayashizaki Y, Bonthron DT: The human GNAS1 gene is imprinted and encodes distinct paternally and biallelically expressed G proteins. Proc Natl Acad Sci USA 1998, 95:10038-10043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Campbell R, Gosden CM, Bonthron DT: Parental origin of transcription from the human GNAS1 gene. J Med Genet 1994, 31:607-614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Williamson CM, Schofield J, Dutton ER, Seymour A, Beechey CV, Edwards YH, Peters J: Glomerular-specific imprinting of the mouse gsalpha gene: how does this relate to hormone resistance in albright hereditary osteodystrophy? Genomics 1996, 36:280-287 [DOI] [PubMed] [Google Scholar]
- 194.Yu S, Yu D, Lee E, Eckhaus M, Lee R, Corria Z, Accili D, Westphal H, Weinstein LS: Variable and tissue-specific hormone resistance in heterotrimeric Gs protein α-subunit (Gsalpha) knockout mice is due to tissue-specific imprinting of the gsalpha gene. Proc Natl Acad Sci USA 1998, 95:8715-8720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Daniels R, Zuccotti M, Kinis T, Serhal P, Monk M: XIST expression in human oocytes and preimplantation embryos. Am J Hum Genet 1997, 61:33-39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Goto T, Wright E, Monk M: Paternal X-chromosome inactivation in human trophoblastic cells. Mol Hum Reprod 1997, 3:77-80 [DOI] [PubMed] [Google Scholar]
- 197.Ray PF, Winston RM, Handyside AH: XIST expression from the maternal X chromosome in human male preimplantation embryos at the blastocyst stage. Hum Mol Genet 1997, 6:1323-1327 [DOI] [PubMed] [Google Scholar]
- 198.Kay GF, Barton SC, Surani MA, Rastan S: Imprinting and X chromosome counting mechanisms determine Xist expression in early mouse development. Cell 1994, 77:639-650 [DOI] [PubMed] [Google Scholar]
- 199.Norris DP, Patel D, Kay GF, Penny GD, Brockdorff N, Sheardown SA, Rastan S: Evidence that random and imprinted Xist expression is controlled by preemptive methylation. Cell 1994, 77:41-51 [DOI] [PubMed] [Google Scholar]
- 200.Zuccotti M, Monk M: Methylation of the mouse Xist gene in sperm and eggs correlates with imprinted Xist expression and paternal X-inactivation. Nat Genet 1995, 9:316-320 [DOI] [PubMed] [Google Scholar]
- 201.Hagiwara Y, Hirai M, Nishiyama K, Kanazawa I, Ueda T, Sakaki Y, Ito T: Screening for imprinted genes by allelic message display: identification of a paternally expressed gene impact on mouse chromosome 18. Proc Natl Acad Sci USA 1997, 94:9249-9254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Deltour L, Montagutelli X, Guenet JL, Jami J, Paldi A: Tissue-and developmental stage-specific imprinting of the mouse proinsulin gene, Ins2. Dev Biol 1995, 168:686-688 [DOI] [PubMed] [Google Scholar]
- 203.Zhan S, Shapiro DN, Helman LJ: Activation of an imprinted allele of the insulin-like growth factor II gene implicated in rhabdomyosarcoma. J Clin Invest 1994, 94:445-448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Zhan S, Shapiro DN, Helman LJ: Loss of imprinting of IGF2 in Ewing’s sarcoma. Oncogene 1995, 11:2503-2507 [PubMed] [Google Scholar]
- 205.Rainier S, Dobry CJ, Feinberg AP: Loss of imprinting in hepatoblastoma. Cancer Res 1995, 55:1836-1838 [PubMed] [Google Scholar]
- 206.Li X, Adam G, Cui H, Sandstedt B, Ohlsson R, Ekstrom TJ: Expression, promoter usage, and parental imprinting status of insulin-like growth factor II (IGF2) in human hepatoblastoma: uncoupling of IGF2 and H19 imprinting. Oncogene 1995, 11:221-229 [PubMed] [Google Scholar]
- 207.Elkin M, Shevelev A, Schulze E, Tykocinsky M, Cooper M, Ariel I, Pode D, Kopf E, de Groot N, Hochberg A: The expression of the imprinted H19 and IGF-2 genes in human bladder carcinoma. FEBS Lett 1995, 374:57-61 [DOI] [PubMed] [Google Scholar]
- 208.Rachmilewitz J, Elkin M, Looijenga LH, Verkerk AJ, Gonik B, Lustig O, Werner D, de Groot N, Hochberg A: Characterization of the imprinted IPW gene: allelic expression in normal and tumorigenic human tissues. Oncogene 1996, 13:1687-1692 [PubMed] [Google Scholar]
- 209.McCann AH, Miller N, O’Meara A, Pedersen I, Keogh K, Gorey T, Dervan PA: Biallelic expression of the IGF2 gene in human breast disease. Hum Mol Genet 1996, 5:1123-1127 [DOI] [PubMed] [Google Scholar]
- 210.Wu HK, Squire JA, Catzavelos CG, Weksberg R: Relaxation of imprinting of human insulin-like growth factor II gene, IGF2, in sporadic breast carcinomas. Biochem Biophys Res Commun 1997, 235:123-129 [DOI] [PubMed] [Google Scholar]
- 211.Douc-Rasy S, Barrois M, Fogel S, Ahomadegbe JC, Stéhelin D, Coll J, Riou G: High incidence of loss of heterozygosity and abnormal imprinting of H19 and IGF2 genes in invasive cervical carcinomas. Uncoupling of H19 and IGF2 expression and biallelic hypomethylation of H19. Oncogene 1996, 12:423-430 [PubMed] [Google Scholar]
- 212.Arima T, Matsuda T, Takagi N, Wake N: Association of IGF2 and H19 imprinting with choriocarcinoma development. Cancer Genet Cytogenet 1997, 93:39-47 [DOI] [PubMed] [Google Scholar]
- 213.Kinouchi Y, Hiwatashi N, Higashioka S, Nagashima F, Chida M, Toyota T: Relaxation of imprinting of the insulin-like growth factor II gene in colorectal cancer. Cancer Lett 1996, 107:105-108 [DOI] [PubMed] [Google Scholar]
- 214.Hibi K, Nakamura H, Hirai A, Fujikake Y, Kasai Y, Akiyama S, Ito K, Takagi H: Loss of H19 imprinting in esophageal cancer. Cancer Res 1996, 56:480-482 [PubMed] [Google Scholar]
- 215.Wu MS, Wang HP, Lin CC, Sheu JC, Shun CT, Lee WJ, Lin JT: Loss of imprinting and overexpression of IGF2 gene in gastric adenocarcinoma. Cancer Lett 1997, 120:9-14 [DOI] [PubMed] [Google Scholar]
- 216.Uyeno S, Aoki Y, Nata M, Sagisaka K, Kayama T, Yoshimoto T, Ono T: IGF2 but not H19 shows loss of imprinting in human glioma. Cancer Res 1996, 56:5356-5359 [PubMed] [Google Scholar]
- 217.Takeda S, Kondo M, Kumada T, Koshikawa T, Ueda R, Nishio M, Osada H, Suzuki H, Nagatake M, Washimi O, Takagi K, Takahashi T, Nakao A: Allelic-expression imbalance of the insulin-like growth factor 2 gene in hepatocellular carcinoma and underlying disease. Oncogene 1996, 12:1589-1592 [PubMed] [Google Scholar]
- 218.Kim KS, Lee YI: Biallelic expression of the H19 and IGF2 genes in hepatocellular carcinoma. Cancer Lett 1997, 119:143-148 [DOI] [PubMed] [Google Scholar]
- 219.Wu HK, Weksberg R, Minden MD, Squire JA: Loss of imprinting of human insulin-like growth factor II gene, IGF2, in acute myeloid leukemia. Biochem Biophys Res Commun 1997, 231:466-472 [DOI] [PubMed] [Google Scholar]
- 220.Randhawa GS, Cui H, Barletta JA, Strichman-Almashanu LZ, Talpaz M, Kantarjian H, Deisseroth AB, Champlin RC, Feinberg AP: Loss of imprinting in disease progression in chronic myelogenous leukemia. Blood 1998, 91:3144-3147 [PubMed] [Google Scholar]
- 221.Suzuki H, Veda R, Takahashi T: Altered imprinting in lung cancer. Nat Genet 1994, 6:332-333 [DOI] [PubMed] [Google Scholar]
- 222.Kondo M, Suzuki H, Ueda R, Osada H, Takagi K, Takahashi T: Frequent loss of imprinting of the H19 gene is often associated with its overexpression in human lung cancers. Oncogene 1995, 10:1193-1198 [PubMed] [Google Scholar]
- 223.Mai M, Yokomizo A, Qian C, Yang P, Tindall DJ, Smith DI, Liu W: Activation of p73 silent allele in lung cancer. Cancer Res 1998, 58:2347-2349 [PubMed] [Google Scholar]
- 224.Albrecht S, Waha A, Koch A, Kraus JA, Goodyer CG, Pietsch T: Variable imprinting of H19 and IGF2 in fetal cerebellum and medulloblastoma. J Neuropathol Exp Neurol 1996, 55:1270-1276 [DOI] [PubMed] [Google Scholar]
- 225.Hodzic D, Delacroix L, Willemsen P, Bensbaho K, Collette J, Broux R, Lefebvre P, Legros JJ, Grooteclaes M, Winkler R: Characterization of the IGF system and analysis of the possible molecular mech-anisms leading to IGF-II overexpression in a mesothelioma. Horm Metab Res 1997, 29:549-555 [DOI] [PubMed] [Google Scholar]
- 226.Yaginuma Y, Nishiwaki K, Kitamura S, Hayashi H, Sengoku K, Ishikawa M: Relaxation of insulin-like growth factor-II gene imprinting in human gynecologic tumors. Oncology 1997, 54:502-507 [DOI] [PubMed] [Google Scholar]
- 227.Jarrard DF, Bussemakers MJG, Bova GS, Isaacs WB: Regional loss of imprinting of the insulin-like growth factor II gene occurs in human prostate tissues. Clin Cancer Res 1995, 1:1471-1478 [PubMed] [Google Scholar]
- 228.Nonomura N, Nishimura K, Miki T, Kanno N, Kojima Y, Yokoyama M, Okuyama A: Loss of imprinting of the insulin-like growth factor II gene in renal cell carcinoma. Cancer Res 1997, 57:2575-2577 [PubMed] [Google Scholar]
- 229.van Gurp RJ, Oosterhuis JW, Kalscheuer V, Mariman EC, Looijenga LH: Biallelic expression of the H19 and IGF2 genes in human testicular germ cell tumors. J Natl Cancer Inst 1994, 86:1070-1075 [DOI] [PubMed] [Google Scholar]
- 230.Vu TH, Yballe C, Boonyanit S, Hoffman AR: Insulin-like growth factor II in uterine smooth-muscle tumors: maintenance of genomic imprinting in leiomyomata and loss of imprinting in leiomyosarcomata. J Clin Endocrinol Metab 1995, 80:1670-1676 [DOI] [PubMed] [Google Scholar]