Abstract
Distinction of malignant uterine leiomyosarcomas from benign leiomyomas by morphological criteria is not always possible. Leiomyosarcomas typically have complex cytogenetic abnormalities; in contrast, leiomyomas have simple or no cytogenetic abnormalities. To understand better the biological distinction(s) between these tumors, we analyzed two other potential markers of genomic instability, loss of heterozygosity (LOH) and microsatellite instability. We examined archival materials from 16 leiomyosarcomas and 13 benign leiomyomas by polymerase chain reaction for 26 microsatellite polymorphisms. Markers were selected based on previous reports of cytogenetic or molecular genetic abnormalities in leiomyosarcomas or leiomyomas and surveyed chromosomes 7, 9, 10, 11, 12, 14, 15, 16, 18, 21, and X. LOH for markers on chromosomes 15, 18, 21, and X was infrequent in leiomyosarcomas (1 of 6 tumors for each chromosome) and not observed for markers on chromosomes 7, 9, 11, 12, 14, or 16. Interestingly, 8 of 14 (57.2%) informative leiomyosarcomas had LOH for at least one marker on chromosome 10 and involved both chromosomal arms in 45.5% (5 of 11). In contrast to leiomyosarcomas, LOH for chromosome 10 was not found in 13 benign leiomyomas. Microsatellite instability was found infrequently in leiomyosarcomas and not detected in leiomyoma. Clinicopathological features (eg, atypia, necrosis, and clinical outcome) did not appear to correlate with LOH for chromosome 10. In contrast to other chromosomes studied, LOH on chromosome 10 was frequent in leiomyosarcomas and absent in benign leiomyomas.
Uterine smooth muscle tumors include benign leiomyomas, malignant leiomyosarcomas, and unusual “quasi-malignant” proliferations such as disseminated peritoneal and intravenous leiomyomatosis. The prevalence of uterine leiomyomas has been estimated to be as high as 77%, and each uterus may contain an average of 6.5 tumors. 1 Although most leiomyomas are asymptomatic, the remaining tumors are the most frequent indication for hysterectomy, accounting for nearly 1 in 3 cases or 175,000 procedures per year in the United States. 2 In contrast to benign leiomyomas, malignant leiomyosarcomas represent only 1 in 800 uterine smooth muscle tumors. 3
Pathological diagnosis of uterine smooth muscle tumors requires evaluation of mitotic activity, nuclear atypia, tumor necrosis, and perhaps to a lesser extent, cellularity and circumscription. 4,5 Uterine smooth muscle tumors may have any one of these features and still be considered benign variants of leiomyoma. When several of these features are present, histological distinction of uterine leiomyosarcomas from leiomyomas is not always possible. The diagnostic difficulty posed by this apparent overlap in morphological phenotype is reflected in diagnostic terms such as “smooth muscle tumor of uncertain malignant potential” and “atypical leiomyoma with recurring potential.” Whether smooth muscle tumors of uncertain malignant potential or atypical leiomyomas represent true biological intermediates, such as the relationship between colonic adenomas and carcinomas, is unknown.
One approach taken to understand the differences between benign and malignant uterine smooth tumors has been investigation of their respective cytogenetics. Leiomyosarcomas typically have complex karyotypic abnormalities. 6-12 Their karyotypes show both numerical and structural aberrations, which often preclude identification of some derivative chromosomes in given metaphases. These aberrations are often unstable, resulting in significant variation from metaphase to metaphase within a tumor. In contrast to leiomyosarcomas, benign leiomyomas have normal karyotypes or simple cytogenetic abnormalities in approximately 40% of tumors. 13-15 The most common aberrations in leiomyomas include a translocation between chromosomes 12 and 14 and deletions of the long arm of chromosome 7. 16,17 The t(12;14) results in aberrant expression of a member of the high mobility group (HMG) family of architectural factors, HMGIC, from chromosome 12. 18,19 This translocation breakpoint maps near the estrogen receptor β (ESR2) on chromosome 14, but does not significantly alter this gene’s expression. 20 Other chromosomal changes found in uterine leiomyomas include trisomy 12, and rearrangements involving chromosomes 3, 6, 10, or 13. 21-25 Of note, rearrangements involving chromosome 6 band p21 in uterine leiomyomas involve another high mobility group family member, HMGIY. 26 The cytogenetics of benign variants and smooth muscle tumors of uncertain malignant potential have yet to be studied in detail. 27-31
To understand better the molecular pathogenetic distinction(s) between these tumors, we analyzed uterine leiomyosarcomas and leiomyomas for two other potential markers of genomic instability, loss of heterozygosity (LOH) and microsatellite instability (MI).
Materials and Methods
Case Selection
Our study materials consisted of 29 archival hysterectomy and myomectomy specimens from the Division of Women’s and Perinatal Pathology, Brigham and Women’s Hospital, Boston, MA and from a previously described collection of uterine smooth muscle tumors (D.R.H. and W.A.P.). 32,33 Three pathologists (B.J.Q., A.P.P., and C.P.C.) reviewed hematoxylin and eosin stained histological sections to either confirm or reclassify the diagnosis of leiomyoma or benign variant, and leiomyosarcoma based on current criteria. Benign vari-ants of leiomyomas included mitotically active, cellular, and symplastic (atypical) types, as well as a lipoleiomyoma and an epithelioid leiomyoma. Leiomyosarcomas in our panel included spindle cell tumors with varying pleomorphism and tumors with epithelioid differentiation (Table 1) ▶ .
Table 1.
Clinicopathological Features of Leiomyosarcomas
| Case no. | Age (yr) | Parity* | Nuclear atypia (other histological features) | Tumor necrosis† | Mitotic count (MF/10HPF)‡ | Flow cytometry (S phase fraction) | Clinical outcome§ |
|---|---|---|---|---|---|---|---|
| 1 | 47 | — | Moderate | + | 50 | NA | |
| 2 | 51 | — | Moderate | — | 16 | NA | |
| 3 | 49 | — | Moderate | + | 20 | Recurred at 16 mo | |
| 4 | 38 | — | Mild to moderate | — | 25 | Recurred at 19 mo | |
| 5 | 68 | — | Moderate (focally epithelioid) | + | >50 | NA | |
| 6 | 56 | — | Moderate (epitheliod and myxoid) | + | 14 | NA | |
| 7 | 56 | — | Moderate | + | 14 | NA | |
| 8¶ | 69 | — | Focally moderate | + | 15 | NED at 84 mo | |
| 9¶ | 51 | P2 | Mild | — | 54 | Aneuploid (21%) | Recurred at 3 mo DOD at 5 mo |
| 10¶ | 44 | P4 | Mild | — | 8 | Aneuploid (8–14.4%) | NED at 22 mo |
| 11¶ | 67 | P1 | Severe | + | 12 | Aneuploid (16%) | Recurred at 9 mo DOD at 22 mo |
| 12¶ | 88 | P1 | Moderate to severe (epithelioid) | + | 24 | Aneuploid (26%) | Recurred at 10 mo DOD at 12 mo |
| 13¶ | 47 | — | Moderate (vascular invasion) | — | 5 | NED at 52 mo | |
| 14¶ | 51 | P3 | Severe | + | 43 | NED at 22 mo | |
| 15¶ | 54 | P4 | Moderate | + | 30 | Recurred at 22 mo NED at 46 mo | |
| 16¶ | 50 | P2 | Moderate to severe | — | 10–12 | Diploid (4.8%) | Recurred at 55 mo DOD at 79 mo |
*—, unknown parity.
†Presence (+) or absence (—) of tumor necrosis was histologically confirmed.
‡Mitotic counts are expressed in mitotic figures (MF) per 10 high power (×400) fields (HPF).
§NED, no evidence of disease; DOD, deceased of disease (leiomyosarcoma); NA, not available.
¶Previously described in Refs. 32 and 33.
Microsatellite Analysis by the Polymerase Chain Reaction
Five-micrometer sections were prepared from archival paraffin blocks. One section was stained with hematoxylin and eosin and used as a reference for dissection. Tumor or normal tissue was then collected by excising the paraffin embedded tissue from two additional sections with sterile surgical blades. DNA was extracted from paraffin fragments by incubation at 62°C in 300 μl of extraction buffer (50 mmol/L Tris-HCl, pH 8.5; 1 mmol/L EDTA; 0.5% Tween-20; and 0.2 μg/μl proteinase K) for 36 hours. The paraffin emulsion was microfuged briefly and 2 μl of clarified aqueous phase was removed for use as template for polymerase chain reaction (PCR). Primer pairs were selected primarily from the Cooperative Human Linkage Center’s Human Screening Set (Weber version 8, Research Genetics, Huntsville, AL) and analyzed the following microsatellite markers: D7S1824, D10S1435, D10S2325, D10S1432, D10S1418, D10S218, D10S188, D10S2327, D10S541, D10S1765, D10S1239, D10S1213, D11S2000, D12S375, D14S606, D14S1426, D15S643, D16S521, D16S291, D18S843, D18S464, D18S542, D21S1446, D21S2052, D21S2055, DXS6810. PCR amplification was carried out in 25 μl reaction volumes containing 2 μl of DNA solution, 50 mmol/L KCl, 10 mmol/L Tris-HCl, pH 8.3, 1.5 mmol/L MgCl2, 200 μmol of each dNTP (supplemented with 50 μmol of 32P-α-dCTP), 50 nmol of each forward and reverse microsatellite primer, and 1 U of AmpliTaq DNA polymerase (Perkin Elmer, Norwalk, CT). Twenty-nine cycles of amplification were performed in a thermocycler (DNA Thermocycler 480, Perkin-Elmer) with the following profile: denaturation at 95°C for 30 seconds, annealing at 55°C for 45 seconds, extension at 72°C for 1.5 minutes. The first three cycles were preceded by a 4-minute denaturation step at 95°C. The last cycle was followed by an extended incubation for 7 minutes at 72°C. PCR products were separated on 7% polyacrylamide gels and visualized by autoradiography.
Loss of heterozygosity at a microsatellite locus is manifested by partial or total absence of either one of the two PCR products in tumor compared to some normal tissue. Samples were scored positive for LOH if the ratio of autoradiogram intensity for the alleles changed by a factor of 2 or more between tumor and normal tissues. 34 LOH was obvious by visual inspection (without scanning densitometry) in most cases. MI was scored positive if novel bands were present in tumor samples compared to normal tissues. 35
Results
Analysis of Loss of Heterozygosity in Leiomyosarcomas
We examined archival material from 16 leiomyosarcomas and 13 leiomyomas by polymerase chain reaction and analyzed 26 microsatellites on 11 chromosomes. Markers surveying the 11 chromosomes (including 7, 9, 10, 11, 12, 14, 15, 16, 18, 21, and X) were selected for evaluation based on previous reports of cytogenetic or molecular genetic abnormalities in either leiomyosarcomas or leiomyomas. 10,11,16,17,36-38
We found LOH involving microsatellite markers on chromosomes 10, 15, 18, and 21 in uterine leiomyosarcomas. Interestingly, 57.2% (8 of 14) informative leiomyosarcomas had LOH for at least one marker on chromosome 10. In contrast, LOH at markers on chromosomes 15, 18, 21, and X was infrequent, involving 16.5% (1 of 6) of informative tumors tested for each of the microsatellite markers. The genomic region subject to LOH on chromosome 10 was large, involving markers on both p and q arms (viz., in cases 2, 3, 6, 8, and 12 in Figure 1 ▶ ) in 45.5% (5 of 11) of informative tumors. LOH was observed on the short arm of chromosome 10 in 54.5% (6 of 11) of informative tumors. The long arm also had LOH in 50% (7 of 14) of informative cases. In cases 9 and 10, the contiguous region of LOH included at least the 10q21–23 region. Multiple interstitial deletions were present in case 3.
Figure 1.
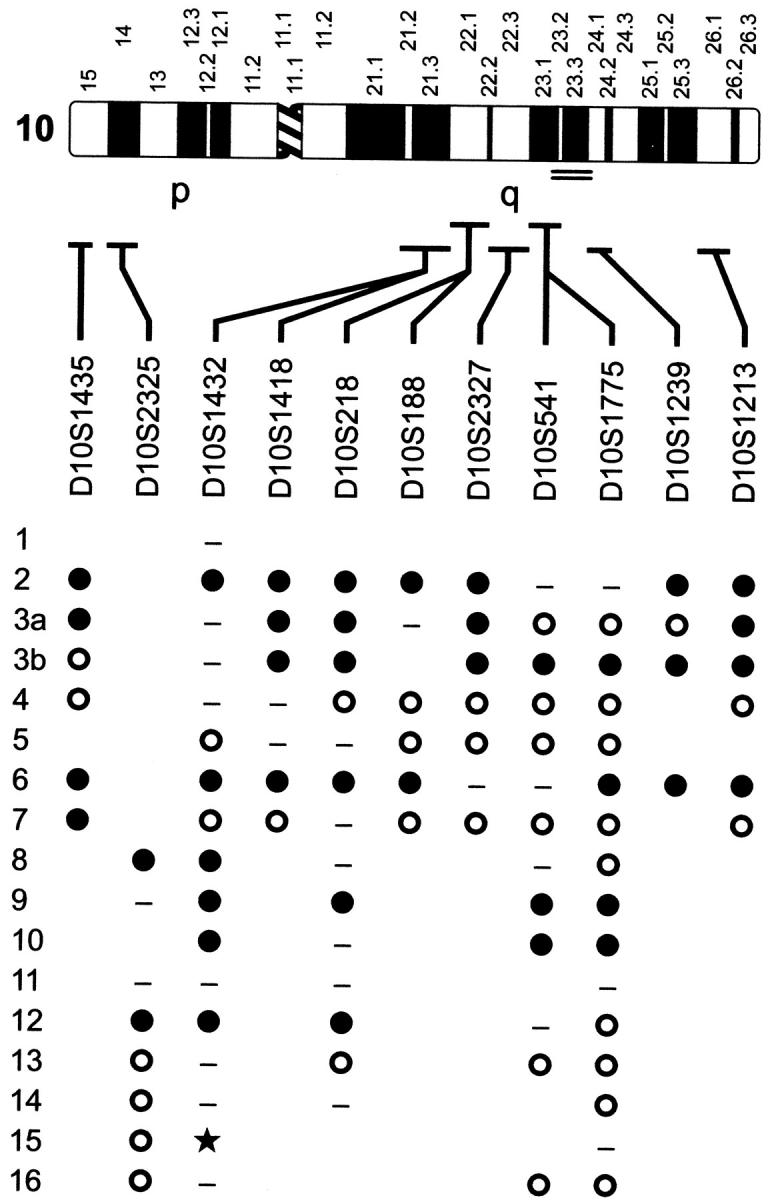
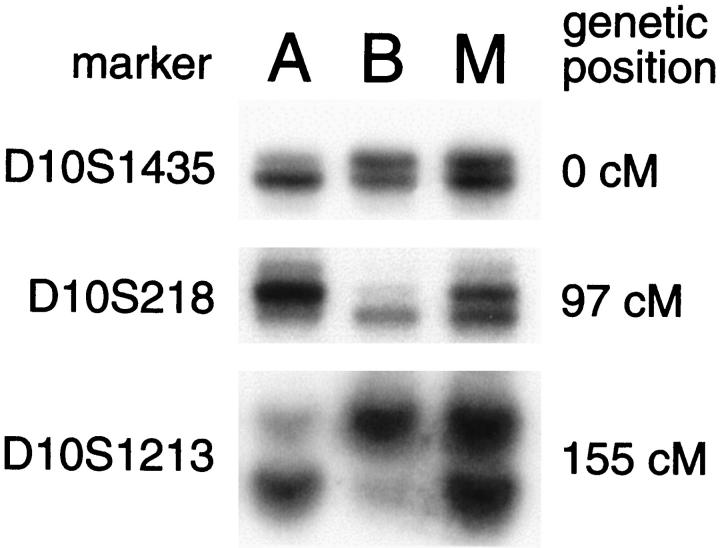
Distribution of loss of heterozygosity for chromosome 10 in uterine leiomyosarcomas. Case numbers are indicated in the left column. •, loss of heterozygosity; ○, retention of heterozygosity; —, uninformative (homozygosity); ★, microsatellite instability; blank, not determined. Numbers above the 550 band resolution idiogram of chromosome 10 label cytogenetic bands and bars below show the approximate cytogenetic localization of microsatellite markers. The diagonal hash marks on the ideogram correspond to the centromere between the short (p) and long (q) chromosomal arms. The double lines beneath the idiogram mark the cytogenetic localization of the PTEN gene in band q23. Rows 3a and 3b correspond to different portions of the same tumor and analysis of selected microsatellites is illustrated in Figure 2 ▶ (lanes A and B, respectively).
In one tumor with LOH for markers on chromosome 10 (case 3), the losses of alternative alleles at loci across chromosome 10 were unexpectedly heterogeneous (Figure 2) ▶ . One of two alleles for two different markers (eg, D10S218 and D10S1213) was absent in one portion of the tumor whereas the other allele was lost in a sample from another portion (lanes A and B). Both of these markers are on the long arm of chromosome 10. Loss of heterozygosity for D10S1435, a marker near the 10p telomere, was present in only one of two tumor samples (lane A). These results are consistent with two tumor subclones in this case.
Figure 2.
Heterogeneity of allelic losses for chromosome 10 markers in one leiomyosarcoma. Microsatellite polymorphisms from the telomeric region of 10p (D10S1435) and the central and subtelomeric regions of 10q (D10S218 and D10S1213) were analyzed from different portions (tumor, lanes A and B; normal myometrium, lane M) of case 3 (rows 3a and 3b, respectively, in Figure 1 ▶ ). LOH for D10S1435 was limited to sample A. In contrast, both tumor samples showed LOH for markers from 10q, but the tumor samples differed with respect to the particular copy of chromosome 10 that was lost. This complex pattern of allelic loss is not consistent with an initial event but rather suggests clonal evolution with several acquired losses of chromosome 10 material. Normal myometrium is analyzed in lane M.
Analysis of Loss of Heterozygosity in Leiomyomas
As a comparison to leiomyosarcomas, we tested 13 benign leiomyomas for LOH for markers on both 10p and 10q. These benign tumors included eight usual type leiomyomas, two cellular leiomyomas, one leiomyoma with epithelioid differentiation, one lipoleiomyoma, and one symplastic leiomyoma with significant nuclear atypia. No LOH was detected. By Fischer’s Exact test, the difference in frequency of LOH between leiomyomas and leiomyosarcomas was significant at a value of P < 0.001.
Analysis of Microsatellite Instability
MI was found only in three cases, and each occurred in different leiomyosarcomas and at different loci on different chromosomes (data not shown). No MI was detected in leiomyomas.
Correlation of Loss of Heterozygosity with Clinicopathological Features
Pathological and clinical parameters for the leiomyosarcomas are shown in Table 1 ▶ . Some of the cases (cases 8–16) have been analyzed previously with respect to flow cytometric parameters. 32,33 Consequently, the duration of clinical follow-up of these cases has increased. Correlation between LOH and these clinicopathological features was not apparent. In particular, LOH did not appear to be a strong prognostic factor. Slightly more than one-half (3 of 5) of leiomyosarcomas with long term follow-up and LOH for any marker on chromosome 10 had unfavorable clinical outcomes. A similar fraction (4 of 6) of leiomyosarcomas with long term follow-up but without LOH for any marker on chromosome 10 also had unfavorable clinical outcomes.
Discussion
To test the hypothesis that genomic instability apart from chromosomal aberrations might be manifested preferentially in leiomyosarcomas, we analyzed archival material from benign leiomyomas and malignant leiomyosarcomas. We identified frequent and selective LOH for markers on chromosome 10 in leiomyosarcomas compared to other chromosomes tested. Nearly 75% of leiomyosarcomas had LOH for at least one informative marker on chromosome 10. In contrast to LOH on chromosome 10, markers on other chromosomes, including two near the tuberous sclerosis gene TSC2 (D16S521 and D16S291) on chromosome 16, were not frequently deleted in spontaneous human uterine leiomyosarcomas. 39,40 Germline mutations of the TSC2 homolog predispose Eker rats to several tumors including uterine leiomyomas and leiomyosarcomas. 37,38,41 Although point mutations or submicroscopic rearrangements at the TSC2 locus cannot be excluded, absence of LOH at this locus suggests that this gene does not play a significant role in human uterine leiomyosarcomas. In addition, leiomyosarcomas with LOH of the chromosomal region most frequently deleted in benign leiomyomas (7q) 42 were not observed. In contrast to chromosomal instability, MI was infrequent and apparently random, suggesting that errors in DNA replication are not a prominent component of the genetic instability found in leiomyosarcomas.
Recent studies using a different approach (comparative genomic hybridization) to identify changes in genomic DNA copy number have also suggested that leiomyosarcomas are characterized by frequent losses and gains in chromosomal copy number. 43,44 In a study of uterine leiomyosarcomas, the most commonly observed genetic aberrations were gains on either arm of chromosome 1 (5 of 8 tumors). 43 Chromosomal gains (amplifications) and the corresponding allelic imbalance(s) potentially might be interpreted as LOH when analyzed by non-quantitative microsatellite PCR analysis. In contrast to our results, consistent losses were not detected by comparative genomic hybridization in that tumor collection and loss of material from chromosome 10 was found only in 1 of 8 leiomyosarcomas in that study. The chromosomal loss in this particular tumor did, however, involve the entire chromosome 10. In a study of extrauterine leiomyosarcomas, the most frequent loss was detected in 10q (20 of 29 tumors), with a minimal common overlapping region corresponding to 10q11-q24. 44 Frequent loss of 10q in leiomyosarcomas from a number of anatomical sites including the uterus suggests a common pathogenetic mechanism.
Significantly, LOH for markers on chromosome 10 was found in none of the benign leiomyomas in our study. Rearrangements of chromosome 10 band q22 has been described as a distinct subgroup of those non-random rearrangements found in benign leiomyomas. 24,25,45 Interestingly, rearrangements of 10q22 by translocation to 17p13 as the sole cytogenetic abnormality in one leiomyosarcoma and by translocation to 11p15 among other complex aberrations in another leiomyosarcoma have also been reported. 46,47 These translocations in leiomyomas and leiomyosarcomas are ostensibly balanced and would not be detectable by LOH analysis except for possibly at or near translocation breakpoints. Such an alternative mechanism potentially might account for tumors lacking LOH for chromosome 10 loci. The fact that LOH for this or any region on chromosome 10 is not frequent in leiomyomas, however, suggests that leiomyomas and leiomyosarcomas are the products of two different mechanisms.
It is possible that chromosome 10 deletion(s) occur later in a hypothetical progression from normal myometrium to leiomyoma, and ultimately in a small number of cases, to leiomyosarcomas. The heterogeneous loss of chromosome 10 observed in one of our malignant tumors (case 3) raises the possibility that deletion of chromosome 10 material is acquired after malignant transformation. LOH involving much or all of chromosome 10 has also been observed in glioblastoma and a gene, PTEN, on chromosome 10 in band q23 has been implicated. 48,49 PTEN has also been implicated in another tumor of the female genital tract, endometrial adenocarcinoma. 50 Frequent LOH on chromosome 10 in leiomyosarcomas may likewise point to this or another tumor suppressor gene and this observation merits further study.
Progressive acquisition of chromosomal aberrations potentially might explain the variable phenotypes found in leiomyosarcomas. For example, tumors with losses on chromosome 10 might have had greater histological pleomorphism, more frequent necrosis, or more aggressive clinical behavior. When we reviewed these features, however, we were unable to discern any correlation between these morphological or prognostic parameters and LOH involving chromosome 10. It therefore seems unlikely that any potential leiomyosarcoma-associated gene on chromosome 10 is sufficient to determine histopathological or clinical phenotypes. Nevertheless, detection of LOH for loci on chromosome 10 may complement histological criteria for distinguishing between biologically benign and malignant uterine smooth muscle neoplasms. Such a diagnostic adjunct could be helpful both as a marker influencing clinical management and classifying smooth muscle tumors of uncertain malignant potential for further study of these neoplasms.
Footnotes
Address reprint requests to Bradley J. Quade, M.D., Ph.D., Department of Pathology, Brigham and Women’s Hospital, 75 Francis Street, Boston, MA 02115. E-mail: bquade@rics.bwh.harvard.edu.
Supported in part by National Cancer Institute Grant CA72594–02 (to B.J.Q.). A.P.P. is a recipient of a Ph.D. fellowship from Conselho National de Desenvolvimento em Pesquisa, Brazil.
References
- 1.Cramer SF, Patel A: The frequency of uterine leiomyomas. Am J Clin Pathol 1990, 94:435-438 [DOI] [PubMed] [Google Scholar]
- 2.Pokras P, Hunfnagel VG: Hysterectomies in the United States, 1965–84. Series 13, No. 92: Vital and Health Statistics. Washington, DC, Government Printing Office, 1987 [PubMed]
- 3.Zaloudek C, Norris HJ: Mesenchymal tumors of the uterus. Kurman RJ eds. Pathology of the Female Genital Tract. 1994, :pp 487-528 Springer-Verlag, New York [Google Scholar]
- 4.Bell SW, Kempson RL, Hendrickson MR: Problematic uterine smooth muscle neoplasms: a clinicopathologic study of 213 cases. Am J Surg Pathol 1994, 18:535-558 [PubMed] [Google Scholar]
- 5.Quade BJ: Pathology, cytogenetics, and molecular biology of uterine leiomyomas and other smooth muscle lesions. Curr Opin Obstet Gynecol 1995, 7:35-42 [PubMed] [Google Scholar]
- 6.Boghosian L, Dal Cin P, Turc-Carel C, Rao U, Karakousis C, Sait SJ, Sandberg AA: Three possible cytogenetic subgroups of leiomyosarcoma. Cancer Genet Cytogenet 1989, 43:39-49 [DOI] [PubMed] [Google Scholar]
- 7.Fletcher JA, Morton CC, Paelka K, Lage JM: Chromosome aberration in uterine smooth muscle tumors: potential diagnostic relevance of cytogenetic instability. Cancer Res 1990, 50:4092-4097 [PubMed] [Google Scholar]
- 8.Laxman R, Currie JL, Kurman RJ, Dudzinski M, Griffin A: Cytogenetic profile of uterine sarcomas. Cancer 1993, 71:1283-1288 [DOI] [PubMed] [Google Scholar]
- 9.Nilbert M, Mandahl N, Heim S, Rydholm A, Willen H, Akerman M, Mitelman F: Chromosome abnormalities in leiomyosarcomas. Cancer Genet Cytogenet 1988, 34:209-218 [DOI] [PubMed] [Google Scholar]
- 10.Nilbert M, Mandahl N, Heim S, Rydholm A, Helm G, Willén H, Baldetorp B, Mitelman F: Complex karyotypic changes, including rearrangements of 12q13 and 14q24, in two leiomyosarcoma. Cancer Genet Cytogenet 1990, 48:217-223 [DOI] [PubMed] [Google Scholar]
- 11.Sait SNJ, Dal Cin P, Sandberg AA: Consistent chromosome changes in leiomyosarcoma. Cancer Genet Cytogenet 1988, 35:47-50 [DOI] [PubMed] [Google Scholar]
- 12.Sreekantaiah C, Davis JR, Sandberg AA: Chromosomal abnormalities in leiomyosarcomas. Am J Pathol 1993, 142:293-305 [PMC free article] [PubMed] [Google Scholar]
- 13.Meloni AM, Surti U, Contento AM, Davare J, Sandberg AA: Uterine leiomyomas: cytogenetic and histologic profile. Obstet Gynecol 1992, 80:209-217 [PubMed] [Google Scholar]
- 14.Pandis N, Heim S, Bardi G, Flodérus U-M, Willén H, Mandahl N, Mitelman F: Chromosome analysis of 96 uterine leiomyomas. Cancer Genet Cytogenet 1991, 55:11-18 [DOI] [PubMed] [Google Scholar]
- 15.Rein MS, Friedman AJ, Barbieri RL, Pavelka K, Fletcher JA, Morton CC: Cytogenetic abnormalities in uterine leiomyomata. Obstet Gynecol 1991, 77:923-926 [PubMed] [Google Scholar]
- 16.Heim S, Nilbert M, Vanni R, Flodérus U-M, Mandahl N, Liedgren S, Lecca U, Mitelman F: A specific translocation, t(12;14)(q14–15;q23–24), characterizes a subgroup of uterine leiomyomas. Cancer Genet Cytogenet 1988, 32:13-17 [DOI] [PubMed] [Google Scholar]
- 17.Ozisik YY, Meloni AM, Surti U, Sandberg AA: Deletion 7q22 in uterine leiomyoma: a cytogenetic review. Cancer Genet Cytogenet 1993, 71:1-6 [DOI] [PubMed] [Google Scholar]
- 18.Schoenberg Fejzo M, Ashar HR, Krauter KS, Powell WL, Rein MS, Weremowicz S, Yoon SJ, Kucherlapati RS, Chada K, Morton CC: Translocation breakpoints upstream of the HMGIC gene in uterine leiomyomata suggest dysregulation of this gene by a mechanism different from that in lipomas. Genes Chromosomes Cancer 1996, 17:1-6 [DOI] [PubMed] [Google Scholar]
- 19.Schoenmakers EF, Wanschura S, Mols R, Bullerdiek J, Van den Berghe H, Van de Ven WJ: Recurrent rearrangements in the high mobility group protein gene, HMGI-C, in benign mesenchymal tumours. Nat Genet 1995, 10:436-443 [DOI] [PubMed] [Google Scholar]
- 20.Pedeutour F, Quade BJ, Weremowicz S, Dal Cin P, Ali S, Morton CC: Localization and expression of the human estrogen receptor β gene in uterine leiomyomata. Genes Chromosomes Cancer 1998, 23:361-366 [DOI] [PubMed] [Google Scholar]
- 21.Van de Ven WJ, Schoenmakers EF, Wanschura S, Kazmierczak B, Kools PF, Geurts JM, Bartnitzke S, Van den Berghe H, Bullerdiek J: Molecular characterization of MAR, a multiple aberration region on human chromosome segment 12q13–q15 implicated in various solid tumors. Genes Chromosomes Cancer 1995, 12:296-303 [DOI] [PubMed] [Google Scholar]
- 22.Nilbert M, Heim S, Mandahl N, Foldrus UM, Willén H, Mitelman F: Characteristic chromosome abnormalities, including rearrangements of 6p, del(7q), +12, and t(12;14), in 44 uterine leiomyomas. Hum Genet 1990, 85:605-611 [DOI] [PubMed] [Google Scholar]
- 23.Nilbert M, Heim S, Mandahl N, Flodérus U-M, Willén H, Mitelman F: Trisomy 12 in uterine leiomyomas: a new cytogenetic subgroup. Cancer Genet Cytogenet 1990, 45:63-66 [DOI] [PubMed] [Google Scholar]
- 24.Ozisik YY, Meloni AM, Surti U, Sandberg AA: Involvement of 10q22 in leiomyoma. Cancer Genet Cytogenet 1993, 69:132-135 [DOI] [PubMed] [Google Scholar]
- 25.Ozisik YY, Meloni AM, Altungoz O, Surti U, Sandberg AA: Translocation (6;10)(p21;q22) in uterine leiomyomas. Cancer Genet Cytogenet 1995, 79:136-138 [DOI] [PubMed] [Google Scholar]
- 26.Williams AJ, Powell WL, Collins T, Morton CC: HMGI(Y) expression in human uterine leiomyomata: involvement of another high-mobility group architectural factor in a benign neoplasm. Am J Pathol 1997, 150:911-918 [PMC free article] [PubMed] [Google Scholar]
- 27.Karaiskos C, Pandis N, Bardi G, Sfikas K, Tserkezoglou A, Fotiou S, Heim S: Cytogenetic findings in uterine epithelioid leiomyomas. Cancer Genet Cytogenet 1995, 80:103-106 [DOI] [PubMed] [Google Scholar]
- 28.Havel G, Wedell B, Dahlenfors R, Mark J: Cytogenetic relationship between uterine lipoleiomyomas and typical leiomyomas. Virchows Arch B Cell Pathol 1989, 57:77-79 [DOI] [PubMed] [Google Scholar]
- 29.Hu J, Surti U, Tobon H: Cytogenetic analysis of a uterine lipoleiomyoma. Cancer Genet Cytogenet 1992, 62:200-202 [DOI] [PubMed] [Google Scholar]
- 30.Ozisik YY, Meloni AM, Sandberg AA, Surti U, Tobon H, Devare J: Cytogenetic findings in a symplastic leiomyoma. Cancer Genet Cytogenet 1993, 67:79-80 [DOI] [PubMed] [Google Scholar]
- 31.Nilbert M, Heim S, Mandahl N, Flodérus U-M, Willén H, Baldetorp B, Mitelman F: Complex karyotypic anomalies in a bizarre leiomyoma of the uterus. Genes Chromosomes Cancer 1989, 1:131-134 [DOI] [PubMed] [Google Scholar]
- 32.Peters WA, III, Howard DR, Andersen WA, Figge DC: Uterine smooth-muscle tumors of uncertain malignant potential. Obstet Gynecol 1994, 83:1015-1020 [DOI] [PubMed] [Google Scholar]
- 33.Peters WA, III, Howard DR, Andersen WA, Figge DC: DNA analysis by flow cytometry of uterine leiomyosarcomas and smooth muscle tumors of uncertain malignant potential. Am J Obstet Gynecol 1992, 166:1646-1654 [DOI] [PubMed] [Google Scholar]
- 34.Lin MC, Mutter GL, Trivijisilp P, Boynton KA, Sun D, Crum CP: Patterns of allelic loss (LOH) in vulvar squamous carcinomas and adjacent noninvasive epithelia. Am J Pathol 1998, 152:1313-1318 [PMC free article] [PubMed] [Google Scholar]
- 35.Mutter GL, Boynton KA, Faquin WC, Ruiz RE, Jovanovic AS: Allelotype mapping of unstable microsatellites establishes direct lineage continuity between endometrial precancers and cancer. Cancer Res 1996, 56:4483-4486 [PubMed] [Google Scholar]
- 36.Ozisik YY, Meloni AM, Surti U, Sandberg AA: Deletion 7q22 in uterine leiomyoma: a cytogenetic review. Cancer Genet Cytogenet 1993, 71:1-6 [DOI] [PubMed] [Google Scholar]
- 37.Everitt JI, Wolf DC, Howe SR, Goldsworthy TL, Walker C: Rodent model of reproductive tract leiomyomata: clinical and pathological features. Am J Pathol 1995, 146:1556-1567 [PMC free article] [PubMed] [Google Scholar]
- 38.Kubo Y, Kikuchi Y, Mitani H, Kobayashi E, Kobayashi T, Hino O: Allelic loss at the tuberous sclerosis (Tsc2) gene locus in spontaneous uterine leiomyosarcomas and pituitary adenomas in the Eker rat model. Jpn J Cancer Res 1995, 86:828-832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Henske EP, Neumann HP, Scheithauer BW, Herbst EW, Short MPKDJ: Loss of heterozygosity in the tuberous sclerosis (TSC2) region of chromosome band 16p13 occurs in sporadic as well as TSC-associated renal angiomyolipomas. Genes Chromosomes Cancer 1995, 13:295-298 [DOI] [PubMed] [Google Scholar]
- 40.Povey S, Burley MW, Attwood J, Benham F, Hunt D, Jeremiah SJFD, Gillett G, Malas S, Robson EB, Tippett P, Edwards JH, Kwiatkowski DJ, Super M, Mueller R, Fryer A, Clarke A, Webb D, Osborne J: Two loci for tuberous sclerosis: one on 9q34 and one on 16p13. Ann Hum Genet 1994, 58:107-127 [DOI] [PubMed] [Google Scholar]
- 41.Yeung RS, Xiao GH, Jin F, Lee WC, Testa JR, Knudson AG: Predisposition to renal carcinoma in the Eker rat is determined by germ-line mutation of the tuberous sclerosis 2 (TSC2) gene. Proc Natl Acad Sci USA 1994, 91:11413-11416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ishwad CS, Ferrell RE, Davare J, Meloni AM, Sandberg AA, Surti U: Molecular and cytogenetic analysis of chromosome 7 in uterine leiomyomas. Genes Chromosomes Cancer 1995, 14:51-55 [DOI] [PubMed] [Google Scholar]
- 43.Packenham JP, du MS, Schrock E, Risinger JI, Dixon D, Denz DN, Evans JA, Berchuck A, Barrett JC, Devereux TR, Ried T: Analysis of genetic alterations in uterine leiomyomas and leiomyosarcomas by comparative genomic hybridization. Mol Carcinog 1997, 19:273-279 [DOI] [PubMed] [Google Scholar]
- 44.El-Rifai W, Sarlomo-Rikala M, Knuutila S, Miettinen M: DNA copy number changes in development and progression in leiomyosarcomas of soft tissues. Am J Pathol 1998, 153:985-990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kiechle-Schwarz M, Berger CS, Surti U, Sandberg AA: Rearrangement of band 10q22 in leiomyoma and leiomyosarcoma of the uterus. Cancer Genet Cytogenet 1990, 47:95-100 [DOI] [PubMed] [Google Scholar]
- 46.Dal Cin P, Boghosian L, Crickard K, Sandberg AA: t(10;17) as the sole chromosome change in a uterine leiomyosarcoma. Cancer Genet Cytogenet 1988, 32:263-266 [DOI] [PubMed] [Google Scholar]
- 47.Nilbert M, Jin YS, Heim S, Mandahl N, Floderus UM, Willen H, Mitelman F: Chromosome rearrangements in two uterine sarcomas. Cancer Genet Cytogenet 1990, 44:27-35 [DOI] [PubMed] [Google Scholar]
- 48.Bostrom J, Cobbers JM, Wolter M, Tabatabai G, Weber RG, Lichter P, Collins VP, Reifenberger G: Mutation of the PTEN (MMAC1) tumor suppressor gene in a subset of glioblastomas but not in meningiomas with loss of chromosome arm 10q. Cancer Res 1998, 58:29-33 [PubMed] [Google Scholar]
- 49.Steck PA, Pershouse MA, Jasser SA, Yung WK, Lin H, Ligon AH, Langford LA, Baumgard ML, Hattier T, Davis T, Frye C, Hu R, Swedlund B, Teng DH, Tavtigian SV: Identification of a candidate tumour suppressor gene, MMAC1, at chromosome 10q23.3 that is mutated in multiple advanced cancers. Nat Genet 1997, 15:356-362 [DOI] [PubMed] [Google Scholar]
- 50.Risinger JI, Hayes AK, Berchuck A, Barrett JC: PTEN/MMAC1 mutations in endometrial cancers. Cancer Res 1997, 57:4736-4738 [PubMed] [Google Scholar]