Vascular endothelium forms a continuous cellular interface between the circulating blood elements and the surrounding tissues. Endothelium forms a nonthrombogenic surface and selective permeability barrier capable of modulating vascular reactivity and blood flow. The integrity of the endothelium is fundamental for the maintenance of normal homeostasis. Injury to this lining results in dramatic changes in the functional characteristics of the endothelium, rendering it adhesive and prothrombotic. These changes may result from molecules inducibly expressed or simply secreted by injured endothelium. These initial events are correlated with subsequent inflammatatory and proliferative cellular changes associated with the development of vascular pathology.
In this issue of the American Journal of Pathology, Santiago et al have added another component in the complex chain of molecular events linking endothelial cell injury and growth factor induction. 1 These observations revolve around an inducible transcriptional factor called the “early growth response factor-1” (Egr-1). 2 Evidence is accumulating to suggest that Egr-1 is involved in the regulation of multiple genes within diverse organ systems through protein-protein interactions with protein kinases and other transcription factors. Indeed, Egr-1 and its role in transcription regulation has potential relevance to the pathogenesis of a variety of vascular diseases. Egr-1-mediated gene transcription and its possible role in vascular disease will be reviewed here.
Egr-1, 3 also known as nerve growth factor induced-A (NGFI-A), 4 krox-24, ZIF268, and TIS8, is an 80- to 82-kd protein consisting of 533 amino acids, discovered independently by a number of laboratories searching for factors regulating cell growth and proliferation. 2,5 It is the prototype of a family of zinc-finger transcription factors that includes Egr-2, Egr-3, Egr-4, and NGFI-B. Egr-1 is an example of an “immediate-early response protein” because it is rapidly and transiently induced by a large number of growth factors, cytokines, and injurious stimuli. 2 Egr-1 contains a DNA binding domain consisting of three zinc fingers which are located between amino acids 332 to 416 toward the carboxy-terminal region of the protein. Through these zinc fingers Egr-1 binds specifically to the major groove of DNA at commonly encountered G+C-rich DNA sequences containing the consensus binding code GCG(T/G)GGGCG. Once bound to DNA, Egr-1 alters gene transcription through mechanisms dependent on both coactivators and corepressors. Mutational studies have identified a strong transcription activation zone within the amino-terminal region of the protein between amino acids 1 and 281. 2 Transcriptional coactivators, such as CREB-binding protein (CBP) and p300, can interact directly with the activation region of Egr-1 and increase Egr-1 trans-activation. 6 However, these interactions are relatively weak compared with some sequence-specific transcription factors, such as the p65 (Rel A) component of NF-κB, and their role in authentic Egr-1-mediated gene expression remains unclear. 6
Corepressors such as NGFI-A-binding proteins 1 and 2 (NAB1 and NAB2) negatively regulate Egr-1 activity. NAB1 was identified using a yeast two-hybrid system by its ability to bind a 34-aa inhibitory domain of Egr-1, called R1, located 5′ of the zinc finger binding domain. 7,8 Deletion of R1 results in a marked increase in Egr-1 transcriptional activity and overexpression of NAB1 markedly decreases Egr-1 transcriptional activity. The related protein NAB2 was subsequently discovered because of its strong homology to NAB1. 9 NAB2 functions similarly to NAB1; however, there are important differences between these related proteins. For example, NAB1 is constitutively expressed in most cell types, whereas NAB2 is rapidly and transiently induced by many of the same stimuli that induce Egr-1. Furthermore, the pattern of tissue expression for NAB2 seems to be more tissue-selective than NAB1. Because of these differences NAB2 may play a negative feedback role by down-regulating the burst of Egr-1 activity that accompanies mitogenic, differentiative, or noxious stimuli. Although the mechanism of action of these inhibitors is unknown, it is tempting to speculate that they interfere with Egr-1’s ability to bind coactivators such as CBP or p300.
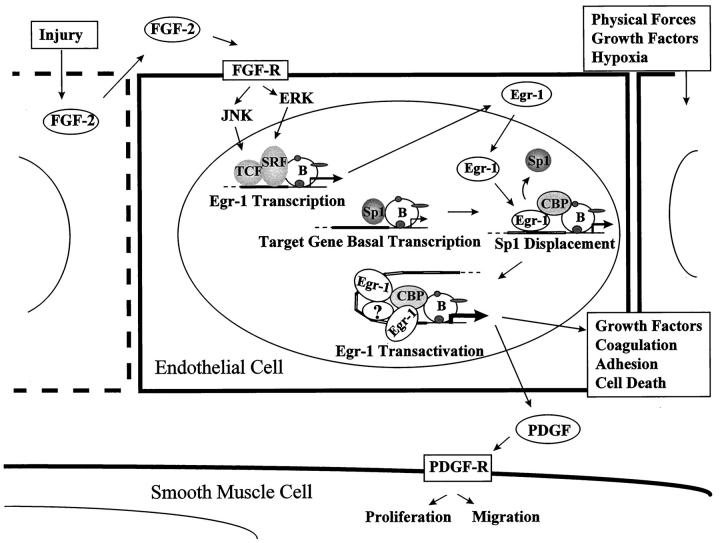
It has been hypothesized that Egr-1 may play a key regulatory role by linking injurious stimuli to the induction of genes directing the expression of effector molecules that ultimately result in vascular pathology. 10 Egr-1 is inducibly expressed in many different cell types; among the vascular cells known to express Egr-1 are endothelial cells, smooth muscle cells, fibroblasts, and leukocytes. Many stimuli associated with the development of vascular diseases, including shear stress, mechanical injury, platelet-derived growth factors (PDGF), hypoxia, reactive oxygen species, angiotensin II, and acidic fibroblast growth factor (FGF-1), are capable of inducing Egr-1 in tissue culture and, in some cases, in authentic blood vessels. 10,11 Santiago et al have provided compelling evidence to suggest that basic fibroblast growth factor (FGF-2) be included in this list of important mediators regulating Egr-1 expression in endothelial cells following injury. 1,11 They demonstrate that Egr-1 induction by injury involves liberation and paracrine activity of FGF-2 1 (Figure 1) ▶ . This growth factor rapidly activates signal transduction pathways involving the mitogen-activated protein kinases (MAPKs) that converge at the Egr-1 promoter. The authors also show that Egr-1 plays a necessary role in the reparative response of endothelial injury. These signaling events may underlie the pathogenesis of intravascular lesions following conventional coronary intervention in humans. 12
Figure 1.
Model of injury-induced PDGF A-chain gene transcription. Endothelial injury releases FGF-2, which binds to its tyrosine kinase receptor and activates ERK and JNK kinases in a paracrine fashion. These kinases phosphorylate ternary complex factors (TCF), which cooperate with serum response factor (SRF) to induce Egr-1 transcription. Egr-1 can displace Sp1 and other transcription factors from the G+C-rich region of target genes and increase transcription above basal levels through multiple protein-protein interactions with CBP/p300. The basal transcription apparatus (B) is indicated. The number of interactions is related to the number and position of Egr-1 consensus binding sites within the promoter. Other transcription factors (?) may be involved. Activated genes may alter cell growth, migration, adhesion, and survival. For example, the released PDGF A-chain binds to receptors on local cells, stimulating growth and migration of adjacent vascular smooth muscle cells.
Inducible Egr-1 gene expression is mediated through different subgroups of MAPKs, including the extracellular signal-regulated kinase (ERK), c-Jun NH2-terminal kinase (JNK) (also known as stress-activated protein kinases or SAPKs) and p38 pathways. Once activated, these pathways may lead to an interaction between ternary complex factors (TCF) and serum response factor (SRF) that activates Egr-1 gene transcription by binding to serum response elements (SRE) within its promoter 13,14 (Figure 1) ▶ . Santiago et al have shown that at least some of these events are activated following mechanical injury to cultured endothelium in an endogenous FGF-2-dependent manner. 1 It is possible that protein kinases involved in the induction of Egr-1 transcription may also affect Egr-1’s phosphorylation state and alter its ability to bind other proteins, as well as DNA.
FGF-2’s ability to induce Egr-1 is also noteworthy because it appears to be a factor responsible for a significant portion of Egr-1 production by endothelial cells after mechanical injury. If these findings hold true in authentic blood vessels, FGF-2 may represent a narrow focal point for a therapeutic intervention that breaks the cycle of injury-induced vascular proliferation that leads to lesion formation. However, in vivo, it is possible that other important factors and parallel pathways are involved, with perhaps several stimuli acting in an additive, synergistic, or inhibitory fashion to alter Egr-1 levels beyond those achievable by any single factor alone. In this regard, the role of the Egr-1 corepressors NAB1 and NAB2 in this context remains unknown. NAB2 is present in vascular endothelial cells and smooth muscle cells, and increases in response to injury closely following Egr-1 up-regulation [E.S. Silverman and T. Collins, unpublished data]. The time course suggests that NAB2 plays a role in modulating or counterbalancing the effect of increasing Egr-1 levels.
Following vascular cell activation or injury, Egr-1 is expressed primarily in the nucleus of cells and is capable of altering the transcription of several genes implicated in the pathogenesis of vascular disease, including PDGF-A, 15,16 PDGF-B, 11 FGF-2, 17 apolipoprotein A1, 18 macrophage colony-stimulating factor (M-CSF), 19 TNF-α, 20 tissue factor, 21 urokinase-type plasminogen activator (u-PA), 22 interleukin-2 (IL-2), 23 intracellular adhesion molecule-1 (ICAM-1), 24 copper-zinc superoxide dismutase gene (SOD1), 25 p53, 26 thrombospondin, 27 CD44, 28 and 5-lipoxygenase (5-LO). 29 All of these genes contain one or more Egr-1 consensus binding site within their promoter regions. Because many of these gene products also stimulate the expression of Egr-1, autocrine or paracrine loops within blood vessels are possible. These positive feedback loops serve to amplify and sustain gene transcription through Egr-1-mediated mechanisms. For example, PDGF A-chain and FGF can stimulate expression of Egr-1 and the increased Egr-1 can activate PDGF A-chain or FGF transcription, completing the amplification cycle. 16,30,31
The role of Egr-1 in the regulation of authentic genes in vivo remains to be determined. Most of the studies demonstrating Egr-1 inducibility have involved transient transfection analysis of cells using promoter-reporter constructs of genes suspected of being Egr-1 targets. However, promoter-reporter constructs can behave differently from authentic promoters in the context of other regulatory elements and chromatin. To our knowledge, the only genes definitively linked to Egr-1 expression in vivo have been luteinizing hormone-β (LH-β), tissue factor, and apoprotein A-I genes, based on studies using the Egr-1 knockout mouse developed by J. Milbrandt and colleagues. 32 For example, the human LH-β promoter contains two functional Egr-1 binding sites in the proximal promoter region. In vivo support of these observations is the finding that female homozygous Egr-1 null mice are infertile as a result of luteinizing hormone deficiency. 33 Another example with special relevance from a vascular perspective involves the tissue factor gene. The tissue factor promoter contains a serum response region that binds Egr-1 and activates transcription in vitro in response to hypoxia. 21 In contrast to wild-type mice, mice deficient in Egr-1 fail to produce tissue factor, nor do they deposit fibrin in the pulmonary vasculature under hypoxic conditions. These findings strongly suggest a role for Egr-1 in the transcriptional regulation of tissue factor in authentic blood vessels. 34 The final example of Egr-1’s transcriptional effects in vivo involves the apoprotein A-1 gene. In a mouse model of nephrotic syndrome, levels of apoprotein A-1 in Egr-1 null mice were half those of the wild-type mice. 35 Future studies involving these mice are likely to be particularly useful for identifying the roles of Egr-1 in an authentic biological system. However, attempts to demonstrate Egr-1-mediated transcription may be confounded by redundancy within the Egr family of transcription factors such that, in the absence of Egr-1, changes in gene expression may not be overtly manifested because related transcription factors may substitute and fulfill similar transcriptional roles. Therefore, in the absence of observable changes in transcription, one may not conclude that Egr-1 has no role in the transcriptional regulation of a given gene.
Egr-1’s potential involvement in the pathogenesis of vascular disease was first recognized by Khachigian et al through promoter analysis of the PDGF A-chain and B-chain genes in endothelial cells, and represents the first link between a transcription factor and a target gene in the context of vascular injury. 36 PDGFs are among the most potent mitogens and chemotaxins secreted by endothelial cells and vascular smooth muscle cells. Elevated levels of PDGFs are found in atheroscerotic plaques and are an important link between endothelial injury and the resulting fibroproliferative response that leads to atherosclerosis. 37,38 In an effort to understand the regulation of PDGFs as they relate to the pathogenesis of vascular disease, the promoter sequences were cloned and studied in endothelial cells. The human PDGF A-chain and B-chain gene promoters are highly G+C-rich, contain a TATA box, and have a single transcriptional start site. 39,40 The A-chain promoter is hypersensitive to cleavage by S1 nuclease and contains two overlapping Egr-1 binding sites between −71 and −50 bp from the transcription start site. 41 The B-chain promoter has a cryptic Egr-1 site located −30 to −19 bp from the transcription initiation site. Transient transfection analysis using promoter-reporter constructs containing deletions of the A- and B-chain promoters have determined that the G+C-rich regions are essential for promoter activity and inducibility in endothelial cells. 11,15 Phorbol ester 12-myristate 13-acetate (PMA) was the initial model agonist used to stimulate Egr-1 expression; however, these observations have subsequently been extended to several more pathophysiologically relevant stimuli such as mechanical injury, shear stress, and growth factors. 31,42 These G+C-rich promoter regions are capable of binding recombinant Egr-1 and purified Egr-1 from nuclear extracts of stimulated endothelial cells and vascular smooth muscle cells. Moreover, Egr-1 up-regulation in endothelial cells can significantly activate promoter-reporter constructs above basal levels. Inhibition of Egr-1 or modification of the consensus binding site mitigates promoter activation. These findings have been extended to vascular smooth muscle cells, which are known to be another important source of some PDGFs in blood vessels. 16,43 The role of the Egr-1 transcriptional pathway in authentic blood vessels is less clear. Although mechanical denudation of the endothelial lining of rat aortae leads to increased endothelial and smooth muscle Egr-1 expression before increased PDGF A-chain and B-chain expression, the causal association between Egr-1 and PDGF expression in vivo remains to be definitively proven. 11,16 The list of Egr-1 target genes includes not only growth and coagulation factors, but proteins that could influence growth retardation and cell survival. By stimulating the expression of the gene for TGF-β, Egr-1 may suppress growth of damaged endothelial cells. Additionally, induction of TGF-β may hinder leukocyte recruitment, modulate vascular tone, and increase expression of growth factors such as PDGF. 44 Injury-induced Egr-1 may also activate p53 via the Egr-1 site in its promoter. Induction of p53 genes associated with cell cycle arrest may provide modestly injured endothelial cells with the opportunity to respond to the injury. Lethally injured endothelial cells could be eliminated by induction of p53-dependent apoptotic genes, or by down-regulation of expression of bcl-2. 45 Determining the balance between Egr-1-dependent growth factors and those proteins involved in survival may play a key role in the cellular events associated with vascular injury. Again, future studies using Egr-1 knockout mice or transgenic mice overexpressing Egr-1 within vascular cells may help elucidate the significance of these possibilities in vivo, as they have for LH-β, tissue factor, and apoprotein A1.
The effect of Egr-1 on the enhancement or repression of reporter-gene transcription depends not only on the arrangement of DNA binding motifs within the promoter, but the cell type and nuclear milieu being studied. 5 Possible explanations include: 1) Egr-1’s ability to act in concert with multiple sequence-specific transcription factors; 2) variations in Egr-1’s ability to interact with transcriptional coactivators such as CBP/p300; 3) variations in Egr-1’s ability to interact with transcriptional corepressors such as NAB2; and 4) altered states of phosphorylation that affect Egr-1’s ability to interact with proteins or DNA. It is the sum of all of the transcriptional components and their phosphorylation status that ultimately determines the effect of Egr-1 on a specific promoter.
Egr-1’s ability to interact with other DNA binding proteins has particular relevance to vascular biology. These interactions may occur at one binding site in a gene; they may occur at mutually exclusive binding sites in the gene; or they may occur directly via protein-protein interactions in the absence of DNA. Of all these mechanisms, displacement by Egr-1 at overlapping Egr-1/Sp1 consensus binding sites, also termed Egr-1 displacement of Sp1, has been most thoroughly described in vascular systems 10,11,15 (Figure 1) ▶ . In this regard, Egr-1 shares similar consensus binding sites with transcription factors Sp1 (-GGGCGG-) and Wilm’s tumor suppressor, WT1 (-GNGNGGGNG-). In contrast to Egr-1 and WT1, Sp1 is a ubiquitous zinc finger protein expressed in nearly all cell types and is required for the expression of many essential genes. In general, highest levels of Sp1 expression are found in cells undergoing differentiation, and these high levels of Sp1 may be required for the subsequent induction of tissue-specific genes. 46 WT1 has the properties of a tumor suppressor gene and is expressed predominantly in the kidney and genital organs where it plays an important role in development. 47 Many promoters contain overlapping Egr-1, Sp1, and WT1 consensus binding sites. 5 In vitro studies using recombinant proteins suggest that these transcription factors can displace one another from many promoters, and binding site occupancy is dependent on an equilibrium determined by their concentration within the nucleus and affinity for the binding site. Sp1 is a relatively weak activator of PDGF A-chain and B-chain transcription and is responsible for basal levels of gene transcription in quiescent cultured endothelial cells. 48 When Egr-1 is up-regulated by injury or growth factors it may displace Sp1 from the A-chain promoter and increase transcription. 15 Egr-1’s ability to augment transcription above levels mediated by Sp1 may relate to its ability to interact with the transcriptional coactivator CBP/p300. Unlike Egr-1, Sp1 does not interact with CBP/p300 but directly with components of the basal transcription apparatus. 6 Sp1 may also have a structural role in transcription by maintaining chromatin in an accessible conformation. 46 Regardless of its mechanism of action, Sp1 appears to initiate transcription in a less efficient manner than Egr-1 in endothelial cells. It is conceivable, however, that Egr-1 may not always be an activator of transcription by this mechanism. If CBP/p300 is unavailable to interact with DNA bound Egr-1, due to sequestration or occupancy of binding sites by other transcription factors, the result of Egr-1 displacement of Sp1 could be transcription repression. In contrast to Egr-1 and Sp1, WT1 is a strong inhibitor of PDGF A-chain promoter-reporter gene expression and may function by occupying the G+C-rich promoter region and prevent Egr-1 or Sp1 from binding. 49 However, WT has not been demonstrated to regulate the corresponding gene, and it is unlikely to have a general role in vascular disease given its limited tissue distribution. This raises the possibility that other WT-like transcription factors may exist that play a role in the negative regulation of these genes.
The number and relative positions of the Egr-1 consensus binding sites are also essential determinants of Egr-1’s ability to activate gene transcription. This is true among different genes and within the same gene among different individuals. Studies involving the 5-lipoxygenase gene promoter illustrate this point. The wild-type human 5-LO gene promoter contains 5 tandem Egr-1 consensus binding sites that bind Egr-1 in vitro. 50 In transfection studies, the promoter-reporter constructs respond to overexpression of Egr-1 and these 5 tandem Egr-1 sites are essential for inducibility. 29 An interesting development in Egr-1-mediated transcription was the recent discovery of a family of naturally occurring promoter mutations within the human 5-LO gene by In et al. 51 These mutations are characterized by a variable number of tandem Egr-1 binding sites, from 3 to 6 sites for each allele. The frequency of mutant alleles was quite high in a sample of normal subjects, ranging from 2 to 18% depending on the particular mutation; the wild-type allele was found in 76% of subjects. These promoter mutations are capable of altering Egr-1 binding and reporter gene transcription such that the intensity of binding and degree of trans-activation are proportional to the number of consensus binding sites. 29 It has been hypothesized that this is due to Egr-1’s ability to interact with CBP/p300 through multiple protein-protein contact points 6 (Figure 1) ▶ . The greater the number of binding sites, the more stable is the platform for the recruitment of CBP/p300 and the greater is the effect on transcription activation. The effect of these promoter mutations on the expression of 5-LO in vivo is unknown at this time but is the subject of intense investigation. A similar phenomenon of genetic variability may relate to other promoters with multiple and variably spaced Egr-1 consensus binding sites.
There is ample evidence to suggest that Egr-1 functions in concert with other sequence-specific transcription factors binding to different sites that are appropriately spaced within the promoter; however, their role in vascular biology is less clear. For example, Egr-1 and the steroidogenic factor-1 can bind to different regions of the LH-β subunit gene promoter and synergistically increase transcription. 52 Egr-1 may also interact synergistically with the p65 protein to regulate transcription of the NF-κB1 (p50) gene. 53 Finally, Egr-1 may act synergistically with nuclear factor of activated T cells (NFAT) to augment IL-2 transcription. 54 Because some of these transcription factors are known to interact directly with CBP/p300, it is possible that together with Egr-1 they facilitate transcription by further stabilizing CBP/p300 at the promoter. Direct protein-protein interactions between sequence-specific transcription factors in the absence of DNA are also possible but less well documented. A study by Jain et al suggests that Egr-1 and Sp1 may bind directly to one another depending on Egr-1’s phosphorylation status as regulated by the casein kinase II-dependent phosphorylation pathway. 55 Bound together, they may effectively sequester each other and mitigate binding to gene promoters. 56 Phosphorylated Egr-1 binds less avidly to Sp1, resulting in higher levels of free transcription factor and the ability to interact with promoters.
Following vascular injury, a series of cellular changes takes place in the vessel wall that can result in the development of pathology. These events are preceded by the inducible expression of a series of genes in endothelial cells. Although we have only begun to dissect the signaling pathways activated by vascular injury, it appears that Egr-1 may play a key role in these initial changes. Egr-1 can be activated by multiple mechanisms, including injury-induced release and paracrine activation by FGF-2, as outlined in the paper by Santiago and coworkers. 1 Additionally, Egr-1 can activate several key groups of pathophysiologically relevant target genes, including growth and coagulation factors, cell surface adhesion molecules, and proteins that can alter cell survival. However, induction of Egr-1 and subsequent Egr-1-mediated transcription are complex processes and not a simple “on-off” switch, as sometimes perceived.
Acknowledgments
The authors thank Drs. Jeffrey M. Drazen, Jeffrey D. Parvin, and Levon M. Khachigian for their invaluable suggestions.
Footnotes
Address reprint requests to Eric S. Silverman, M.D., Pulmonary and Critical Care Division, Brigham and Women’s Hospital, 75 Francis Street, Boston, MA 02115. E-mail: esilverm@bustoff.bwh.harvard.edu.
Supported by National Institutes of Health grants K08 HL03827 to E.S.S. and R01 HL35716 to T.C.
References
- 1.Santiago FS, Lowe H, Day F, Chesterman CN, Khachigian LM: Egr-1 induction by injury is triggered by release and paracrine activation by fibroblastic growth factor-2. Am J Pathol 1999, 154:937-944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gashler A, Sukhatme VP: Early growth response protein 1 (Egr-1): prototype of a zinc-finger family of transcription factors. Prog Nucleic Acid Res Mol Biol 1995, 50:191-224 [DOI] [PubMed] [Google Scholar]
- 3.Sukhatme VP, Cao X, Chang LC, Tsai-Morris C-H, Stamenkovich D, Ferreira PCP, Cohen DR, Edwards SA, Shows TB, Curran T: A zinc finger-encoding gene coregulated with c-fos during growth and differentiation, and after cellular depolarization. Cell 1988, 53:37-43 [DOI] [PubMed] [Google Scholar]
- 4.Milbrandt J: A nerve growth factor-induced gene encodes a possible transcriptional regulatory factor. Science 1987, 238:797-799 [DOI] [PubMed] [Google Scholar]
- 5.Liu C, Ragnekar VM, Adamson E, Mercola D: Suppression of growth and transformation and induction of apoptosis by Egr-1. Cancer Gene Ther 1998, 5:3-28 [PubMed] [Google Scholar]
- 6.Silverman E, Du J, Williams A, Wadgaonkar R, Drazen J, Collins T: cAMP-response-element-binding-protein-binding protein (CBP), and p300 are transcriptional co-activators of Egr-1. Biochem J 1998, 336:183-189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Russo M, Sevetson B, Milbrandt J: Identification of NAB1, a repressor of NGFI-A and Krox20 mediated transcription. Proc Natl Acad Sci USA 1995, 92:6873-6877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Swirnoff AH, Apel ED, Svaren J, Sevetson BR, Zimonjic DB, Popescu NC, Milbrandt J: NAB1, a corepressor of NGFI-A (Egr-1), contains an active transcriptional repression domain. Mol Cell Biol 1998, 18:512-524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Svaren J, Sevetson B, Apel E, Zimonjic D, Popescu N, Milbrandt J: NAB2, a corepressor of NGFI-A (Egr-1), and Krox20, is induced by proliferative, and differentiative stimuli. Mol Cell Biol 1996, 16:3545-3553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Khachigian L, Collins T: Inducible expression of Egr-1-dependent genes: a paradigm of transcriptional activation in vascular endothelium. Circ Res 1997, 81:457-461 [DOI] [PubMed] [Google Scholar]
- 11.Khachigian LM, Linder V, Williams AJ, Collins T: Egr-1-Induced endothelial gene expression: a common theme in vascular injury. Science 1996, 271:1427-1431 [DOI] [PubMed] [Google Scholar]
- 12.Lowe HC, Chesterman CN, Khachigian LM: Left main coronary artery stenosis after percutaneous transluminal coronary angioplasty: importance of remaining “minimally invasive.” Cathet Cardiovasc Diagn 1999, in press [DOI] [PubMed]
- 13.Treisman R: Journey to the surface of the cell: Fos regulation and the SRE. EMBO J 1995, 14:4905-4913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lim C-P, Jain N, Cao X: Stress-induced immediate-early gene, egr-1, involves activation of p38/JNK1. Oncogene 1998, 16:2915-2926 [DOI] [PubMed] [Google Scholar]
- 15.Khachigian LM, Williams AJ, Collins T: Interplay of Sp1 and Egr-1 in the proximal platelet-derived growth factor A-chain promoter in cultured vascular endothelial cells. J Biol Chem 1995, 270:27679-27686 [DOI] [PubMed] [Google Scholar]
- 16.Silverman ES, Khachigian LM, Lindner V, Williams AJ, Collins T: Inducible PDGF A-chain transcription in smooth muscle cells is mediated by Egr-1 displacement of Sp1, and Sp3. Am J Physiol 1997, 273:H1415-H1426 [DOI] [PubMed] [Google Scholar]
- 17.Biesiada E, Razandi M, Levin E: Egr-1 activates basic fibroblastic growth factor transcription: mechanistic implications for astrocyte proliferation. J Biol Chem 1989, 271:18576-18581 [DOI] [PubMed] [Google Scholar]
- 18.Kilbourne E, Widom R, Harnish D, Malik S, Karathanasis S: Involvement of the early growth response factor Egr-1 in apolipoprotein AI gene transcription. J Biol Chem 1995, 270:7004-7010 [DOI] [PubMed] [Google Scholar]
- 19.Harrington M, Edenberg H, Saxman S, Pedigo L, Daub R, Broxmeyer H: Cloning and characterization of the murine promoter for the colony-stimulating factor-1-encoding gene. Gene 1991, 102:165-170 [DOI] [PubMed] [Google Scholar]
- 20.Yao J, Mackman N, Edgington TS, Fan ST: Lipopolysaccharide induction of the tumor necrosis factor-α promoter in human monocyte cells: regulation by egr-1, c-jun, NF-kappa B transcription factors. J Biol Chem 1997, 272:17795-17801 [DOI] [PubMed] [Google Scholar]
- 21.Cui M-Z, Parry G, Oeth P, Larson H, Smith M, Huang RP, Adamson ED, Mackman N: Transcriptional regulation of the tissue factor gene in human epithelial cells is mediated by Sp1 and Egr-1. J Biol Chem 1996, 271:2731-2739 [DOI] [PubMed] [Google Scholar]
- 22.Verde P, Boast S, Franze A, Robbiati F, Blasi F: An upstream enhancer and a negative element in the 5′ flanking region of the human urokinase plasminogen activator gene. Nucleic Acids Res 1988, 16:10699-10716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Skerka C, Decker E, Zipfel P: coordinate expression and distict DNA-binding characteristics of the four egr-1-zinc finger proteins in Jurkat T lymphocytes. J Biol Chem 1995, 270:22500-225067673240 [Google Scholar]
- 24.Maltzman J, Carmen J, Monroe J: Transcriptional regulation of the ICAM-1 gene in antigen receptor- and phorbol ester-stimulated B lymophocytes: role for transcription factor Egr-1. J Exp Med 1996, 183:1747-1759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Minc E, de Coppet P, Masson P, Thiery L, Dutertre S, Amor-Gueret M, Jaulin C: The human copper-zinc superoxide dismutase gene (SOD1) proximal promoter is regulated by Sp1 Egr-1, and WT1 via non-canonical binding sites. J Biol Chem 1999, 274:503-509 [DOI] [PubMed] [Google Scholar]
- 26.Nair P, Muthukkumar S, Sells SF, Han SS, Sukhatme VP, Rangnekar VM: Early growth response-1-dependent apoptosis is mediated by p53. J Biol Chem 1997, 272:20131-20138 [DOI] [PubMed] [Google Scholar]
- 27.Shingu T, Bornstein P: Overlapping egr-1 and Sp1 sites function in the regulation of transcription of the thrombospondin gene. J Biol Chem 1994, 269:32551-32557 [PubMed] [Google Scholar]
- 28.Maltzman J, Carman J, Monroe J: Role for Egr-1 in regulation of stimulus-dependent CD44 transcription in B lymphocytes. Mol Cell Biol 1996, 16:2283-2294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Silverman ES, Du J, De Sanctis GT, Radmark O, Samuelsson B, Drazen JM, Collins T: Egr-1 and Sp1 interact functionally with the 5-lipoxygenase promoter and its naturally occurring mutants. Am J Cell Mol Biol 1998, 19:316-323 [DOI] [PubMed] [Google Scholar]
- 30.Baldwin AJ, Mayo W: Basic fibroblastic growth factor transcriptional autoregulation requires Egr-1. Oncogene 1997, 14:2291-2299 [DOI] [PubMed] [Google Scholar]
- 31.Delbridge G, Khachigian LM: FGF-1-Induced platelet-derived growth factor-A chain gene expression in endothelial cells involves transcriptional activation by early growth response factor-1. Circ Res 1997, 81:282-288 [DOI] [PubMed] [Google Scholar]
- 32.Lee SL, Tourtellotte LC, Wesselschmidt RL, Milbrandt J: Growth and differentiation proceeds normally in cells deficient in the immediate early gene NGFI-A. J Biol Chem 1995, 270:9971-9977 [DOI] [PubMed] [Google Scholar]
- 33.Lee SL, Sadovsky A, Swirnoff AH, Polish JA, Goda P, Gavrilina G, Milbrandt J: Luteinizing hormone deficiency and female infertility in mice lacking the transcription factor NGFI-A. Science 1996, 273:1219-1221 [DOI] [PubMed] [Google Scholar]
- 34.Yan S, Zou Y, Gao Y, Zhai C, Mackman N, Lee SL, Milbrandt J, Pinsky D, Kisiel W, Stern D: Tissue factor transcription driven by Egr-1 is a critical mechanism of murine pulmonary fibrin deposition in hypoxia. Proc Natl Acad Sci USA 1998, 95:8298-8303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zaiou M, Azrolan N, Hayek T, Wang H, Wu L, Haghpassand M, Cizman B, Madaio MP, Milbrandt J, Marsh JB, Breslow JL, Fisher EA: The full induction of human apoprotein A-I gene expression by experimental nephrotic syndrome in transgenic mice depends on cis-acting elements in the proximal 256 base-pair promoter region and the trans-acting factor early growth response factor 1. J Clin Invest 1998, 101:1699-1707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Khachigian LM, Collins T: Early growth response factor 1: a pleiotropic mediator of inducible gene expression. J Mol Med 1998, 76:613-616 [DOI] [PubMed] [Google Scholar]
- 37.Ross R: The pathogenesis of atherosclerosis: a perspective for the 1990s. Nature 1993, 362:801-809 [DOI] [PubMed] [Google Scholar]
- 38.Schwartz S, deBlois D, O’Brien E: The intima: soil for atherosclerosis and restenosis. Circ Res 1995, 77:445-465 [DOI] [PubMed] [Google Scholar]
- 39.Bonthron D, Morton C, Orkin S, Collins T: Platelet-derived growth factor A chain: gene structure, chromosomal location, and basis for alternative mRNA splicing. Proc Natl Acad Sci USA 1988, 85:1492-1496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ratner L, Thielan B, Collins T: Sequences of the 5′ portion of the human c-sis gene: characterization of the transcriptional promoter and regulation of expression of the protein product by 5′ untranslated mRNA sequences. Nucleic Acids Res 1987, 15:6017-6036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang Z, Lin X-H, Qiu Q-Q, Deuel T: Modulation of transcription of the platelet-derived growth factor A-chain gene by a promoter region sensitive to S1 nuclease. J Biol Chem 1992, 267:17022-17031 [PubMed] [Google Scholar]
- 42.Khachigian LM, Anderson KR, Halnon NJ, Gimbrone MA, Resnick N, Collins T: Egr-1 is activated in endothelial cells exposed to fluid shear stress and interacts with a novel shear-stress-response element in the PDGF A-chain promoter. Arterioscler Thromb Vasc Biol 1997, 17:2280-2286 [DOI] [PubMed] [Google Scholar]
- 43.Rafty L, Khachigian L: Zinc finger transcription factors mediate high constitutive platelet-derived growth factor-B expression in smooth muscle cells derived from aortae of newborn rats. J Biol Chem 1998, 6:5778-5764 [DOI] [PubMed] [Google Scholar]
- 44.Pintavorn P, Ballermann BJ: TGF-β, and the endothelium during immune injury. Kidney Int 1997, 51:1401-1412 [DOI] [PubMed] [Google Scholar]
- 45.Huang RP, Fan Y, Peng A, Zeng ZL, Reed JC, Adamson ED, Boynton AL: Suppression of human fibrosarcoma cell growth by transcription factor, Egr-1, involves down-regulation of Bcl-2. Int J Cancer 1998, 77:8806. [DOI] [PubMed] [Google Scholar]
- 46.Courey AJ, Tjian R: Mechanisms of transcriptional control as revealed by studies of human transcription factor Sp1. In Transcriptional Regulation, Vol 28. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, 1992, pp 743–769
- 47.Rauscher FI: The WT1 Wilms tumor gene product: a developmentally regulated transcription factor in the kidney that functions as a tumor suppressor. FASEB J 1993, 7:896-903 [PubMed] [Google Scholar]
- 48.Khachigian LM, Fries JW, Benz MW, Bonthron DT, Collins T: Novel cis-acting elements in the human platelet-derived growth factor B-chain core promoter that mediate gene expression in cultured vascular endothelial cells. J Biol Chem 1994, 269:22647-22656 [PubMed] [Google Scholar]
- 49.Gashler A, Bonthron DT, Madden SL, Rauscher FJ, Collins T, Sukhatme VP: Human platelet-derived growth factor A-chain is transcriptionally repressed by the Wilm’s tumor suppressor WT1. Proc Natl Acad Sci USA 1992, 89:10984-10988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hoshiko S, Radmark O, Samuelsson B: Characterization of the human 5-lipoxygenase gene promoter. Proc Natl Acad Sci 1990, 87:9073-9077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.In KH, Asano K, Beier D, Grobholz J, Finn PW, Silverman EK, Silverman ES, Collins T, Fischer AR, Keith TP, Serino K, Kim SW, De Sanctis GT, Yandava C, Pillari A, Rubin P, Kemp J, Israel E, Busse W, Ledford D, Murray JJ, Segal A, Tinkleman D, Drazen JM: Naturally occurring mutations in the human 5-lipoxygenase gene promoter that modify transcription factor binding and reporter gene transcription. J Clin Invest 1997, 99:1130-1137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Halvorson L, Ito M, Jameson J, Chin W: Steroidogenic factor-1 and early growth response protein 1 act through two composite DNA binding sites to regulate luteinizing hormone β-subunit gene expression. J Biol Chem 1998, 273:14712-14720 [DOI] [PubMed] [Google Scholar]
- 53.Cogwell PC, Mayo MW, Baldwin AS, Jr: Involvement of Egr-1/Rel A synergy in distinguishing T cell activation from tumor necrosis factor-α-induced NF-kappa B1 transcription. J Exp Med 1997, 185:491-497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Decker E, Skerka C, Zipfel P: The early growth response protein (Egr-1) regulates interleukin-2 transcription by synergistic interactions with the nuclear factor of activated T cells. J Biol Chem 1998, 273:26923-26930 [DOI] [PubMed] [Google Scholar]
- 55.Jain N, Mahendran R, Philip R, Guy G, Tan Y, Cao X: Casein kinase II associates with Egr-1 and acts as a negative modulator of its DNA binding and transcription activities in NIH 3T3 cells. J Biol Chem 1996, 271:13530-13536 [DOI] [PubMed] [Google Scholar]
- 56.Srivastava S, Weitzmann MN, Kimble RB, Rizzo M, Zahner M, Milbrandt J, Ross FP, Pacifici R: Estrogen blocks M-CSF gene expression and osteoclast formation by regulating phosphorylation of Egr-1 and its interaction with Sp1. J Clin Invest 1998, 102:1850-1859 [DOI] [PMC free article] [PubMed] [Google Scholar]