Abstract
During development, the Notch signaling pathway is essential for the appropriate differentiation of many cell types in organisms across the phylogenetic scale, including humans. Notch signaling is also implicated in human diseases, including a leukemia and two hereditary syndromes known as Alagille and CADASIL. To generate tools for pursuing the role of the Notch pathway in human disease and development, we have cloned and analyzed the expression of three human homologues of the Notch ligands Delta and Serrate, human Jagged1 (HJ1), human Jagged2 (HJ2), and human Delta1 (H-Delta-1), and determined their chromosomal localizations. We have also raised antibodies to HJ1, and used these antibodies in conjunction with in situ hybridization to examine the expression of these ligands in normal and cancerous cervical tissue. We find that, as reported previously for Notch, the ligands are up-regulated in certain neoplastic tissues. This observation is consistent with the notion that Notch signaling is an important element in these pathogenic conditions, raising the possibility that modulation of Notch activity could be used to influence the fate of the cells and offering a conceivable therapeutic avenue.
In recent years, considerable advances have been made in understanding the molecular interactions underlying cell fate decisions. Across the phylogenetic spectrum, the Notch signaling system appears to play a major role in regulating such decisions. 1-4 In humans, in vitro studies have shown the Notch pathway to be important in hematopoiesis 5-7 and, recently, two Notch pathway elements have been identified as the culprit genes in two human congenital syndromes known as Alagille (HJ1) and CADASIL (Notch 3). 8-12 In addition, Notch signaling has been shown to be important in certain cancers. Constitutively active aberrant forms of Notch are associated with a human leukemia and mouse epithelial tumors. 13,14 In a previous study from this laboratory, Notch pathway components were shown to be up-regulated in cervical cancers. 15
Because of the likely importance of the Notch pathway to a variety of human diseases, we have begun to clone the human homologues of genes belonging to the Notch signal transduction system and to evaluate their expression patterns in humans. Here we present the cloning of three homologues of Notch ligands, specifically, one that codes for a protein similar to Drosophila DELTA and two that are most similar to the Drosophila protein SERRATE. These genes have been named, respectively, Human Delta-1 (H-Delta-1) and Human Jagged1 and 2 (HJ1 and HJ2). We have analyzed their expression patterns on Northern blots, have raised antibodies to HJ1, and have determined their chromosomal localizations, thus aquiring the molecular tools and information needed to better pursue the role of the Notch signaling pathway in human disease. Using these tools, we have determined the expression patterns of these ligands in normal and cancerous cervical tissue. Like Notch 1 and 2, Notch ligands are up-regulated in cervical cancers. Thus, the Notch pathway may be activated in cervical cancer through receptor-ligand interactions. 15
Materials and Methods
Cloning
HJ1 and HJ2 clones were originally identified by probing a Stratagene Human Fetal Brain cDNA library with a 32P-dCTP-labeled polymerase chain reaction (PCR) fragment. The PCR probe was made by using degenerate primers (see reference 15 for primer sequences) with high homology to fly Delta and Serrate to amplify cDNA from a human placenta library. The fragments were sequenced and determined to contain sequences similar to the 5′ ends of the fly ligands. The entire HJ1 cDNA was retrieved as a single insert in LambdaZap. Two clones encoding partial sequence of HJ2 were isolated and rescued from LambdaZap as inserts in pBlueScript sk−. One of these (pBS15) contained 3 kb of sequence and included coding for the start methionine, the other clone was 1.5 kb long and contained sequence internal to pBS15. The 3.5-kb insert from pBS15 was used as a probe to screen a Clontech human fetal brain cDNA library. Two clones of about 3 kb were isolated, one of these (pBS3–2) started at about base 1200 of pBS15 and extended through the polyadenylation signal; the other was internal to it. Together pBS15 and pBS3–2 appeared to contain the entire coding sequence of HJ2. However, subsequent sequencing showed that there was a deletion and frame shift at base 240 of pBS15, where sequence coding for about 70 amino acids appeared to be missing, on the basis of comparison with HJ1. A genomic library (a generous gift from Dr. Richard Lifton) was screened using as probes short PCR products that spanned the deleted region. Three positive clones were identified and appeared to be identical. Sequencing of one of these clones revealed a large intron at the point of the deletion. Coding sequence for the missing region was identified by sequencing through this intron. This sequence codes for amino acids 70–140.
A probe for the H-Delta-1 clone was made using nested degenerate PCR primers derived from amino acid sequences in C-δ-1. 16 The primers were ACIATGAA(C/T)AA(C/T)CTIGCIAAC/TTG (sense), AC(A/G)TAIACI GA(C/T)TG(A/G)TA(C/T)TTIGT, and GC(A/G/T)ATIAC(A/G)CA(CT)TC(A/G)TC(C/T)TT(C/T)TC (both antisense) and corresponded to the peptides TMNNLANC, TKYQSVYV, and EKDECVIA, respectively. They were used to amplify DNA from a human genomic clone in a pAC vector. 17 (The pAC clone was identified using low-stringency hybridization with a mouse probe.) The amplified DNA was sequenced for verification, and the larger piece was used to probe a Clontech human fetal brain cDNA library. Three positive clones were identified, and two contained sequence coding for a polypeptide with high homology to mouse and chicken Delta-1. The two clones were slightly overlapping and represented the 3′ half of the protein. One of the clones (pBS24) was used to probe a Stratagene human fetal brain cDNA library, and two additional clones were identified. Both contained sequence 5′ to pBS24, one of these (pBS18) contained sequence that coded for the entire molecule 3′ of amino acid 120, based on the mouse and chicken sequences. It also contained two introns, the first intron was located within the first EGF repeat and the second was found within the fourth EGF repeat. A full-length clone was then isolated using a probe made from the 5′ end of pBS18, upstream of the first splice site, to rescreen the Stratagene library.
Northern Blots
Two Northern blots that had been purchased from Clontech were probed according to their instructions. (The same filter was used for analyzing HJ1 and HJ2 expression; the other filter was used for H-Delta-1.) For HJ1, the probe was made from the full length cDNA. For HJ2 the probe was made from the full-length insert of pBS15. For H-Delta-1, the probe was made by restriction digestion (XhoI to XbaI) of the HDL18, which gave rise to a 1533-bp nucleotide fragment corresponding to the 3′ end of the clone. A probe for human β-actin was made from DNA supplied by Clontech and hybridized to the stripped blots as a control for loading errors. No differences in the strengths of the β-actin signals were seen (data not shown).
Production of HJ1 and HJ2 Antibodies
A PCR product spanning the entire intracellular coding region was amplified from the HJ1 cDNA and cloned into pGEX to make a construct coding for a fusion protein with glutathioneS-transferase. Fusion protein was made using standard methods and injected into three rats and three rabbits. None of the rabbits produced usable antibodies, but a good rat polyclonal antibody was identified, TS1. Screening was carried out first against the fusion proteins and then against HaCaT and NIH 3T3 cells transfected with HJ1 cDNA. Co-transfections were done using Lipofectamine (Gibco) following the manufacturer’s instructions. Western blots were done under standard conditions using TS1 at a dilution of 1:500–1:1000. The second antibody was goat anti-rat labeled with horseradish peroxidase used at 1:5000 (Jackson). Western blots were visualized using ECL, according to the manufacturer’s instructions. In Western blots, TS1 recognized a band at 130 kd in transfected cells but not in control cells (see Figure 3A ▶ ). Some nonspecific labeling was also seen in both control and transfected cells.
Figure 3.
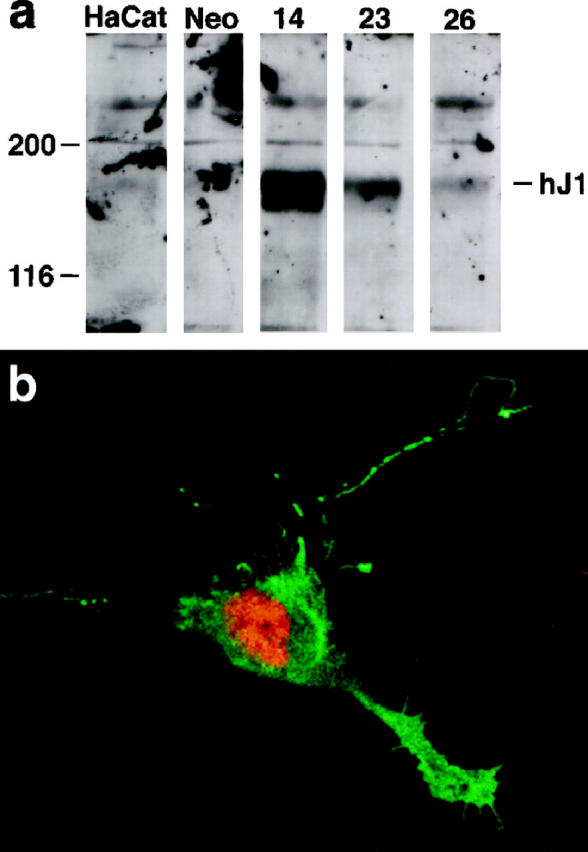
Anti-HJ1 antibody TS1. a: Western blot demonstrates specific labeling of band at 130 kd. The first two lanes represent untransfected HaCat cells and ones transfected with empty vector, respectively. Three independently transfected HaCat cell lines, 14, 23, and 26, were tested, and all expressed a specifically labeled 130-kd band. b: rat polyclonal labeling of HaCaT cells co-transfected with HJ1 and intracellular Notch cDNA. Notch is labeled red and HJ1 green. Only cells that were immunoreactive for the Notch antibody were also labeled with TS1, showing that there was no nonspecific cross-reactivity with untransfected cells in culture. Original magnification, ×650.
In immunocytochemical experiments, TS1 antibodies were used at a dilution of 1:1000 to label paraformaldehyde (3% in phosphate buffered saline (PBS)-fixed transfected and control NIH 3T3 cells. Lissamine rhodamine goat anti-rat secondary antibody (Jackson) was used at 1:250. Visualization under a Leitz Orthoplan microscope showed that TS1 bound to cells that had been transfected with HJ1, but no labeling was seen in control cells. To be certain that the fluorescent labeling detected in our transiently transfected plates was due to specific immunoreactivity of TS1 with HJ1, we co-transfected cells with a construct coding for a truncated Notch construct that is localized to the nucleus. Only cells whose nuclei were stained with an anti-Notch-1 antibody were also labeled with HJ1, demonstrating that there was no detectable nonspecific cross-reactivity in these cultures (see Figure 3B ▶ ).
Immunohistochemistry
Cervical tissues were fixed in formalin, embedded in paraffin, and sectioned on a paraffin microtome. Sections were mounted on slides, deparaffinized in xylene, and rehydrated in graded alcohols. The samples were incubated in 0.2% bovine serum albumin (BSA) in PBS for 10 minutes at room temperature and then 30 minutes in 0.2% BSA, 10% normal goat serum, and 0.1% Triton X-100. Primary antibody was added to 0.2% BSA in PBS, and the slides were incubated for 2 hours at room temperature. Antibody dilutions were as follows: anti-HJ1 (TS1) 1:200–1:400; anti-Notch1 (bTAN20) 1:10-1:20; anti-Notch2 (bhN6) 1:20-1:50. (Notch monoclonal antibodies were rat monoclonal supernatants.) Staining with primary antibodies was followed with three 10-minute washes in 0.2% BSA-PBS. Dab staining was done using a Vectastain kit following the kit directions. For immunofluorescent staining, CY3-labeled goat anti rat was used at 1:70 and DAFT-labeled goat anti-mouse was used at 1:30. Specimens were then washed in PBS and viewed under a Leitz Orthoplan microscope. Secondary-only control sections were prepared, and no specific staining pattern was seen, although there was light background staining. As an additional control, the staining patterns of TS1 in cervical tissue and in human fetal tissue were compared with those obtained by in situ hybridization; the patterns were identical (data not shown).
In Situ Probes and Protocols
The protocol for the in situ hybridizations has been described elsewhere. 18 The probe for HJ2 was transcribed from a 333-bp fragment, coding for the 10th EGF repeat, where there was little homology with HJ1. The results obtained using this probe were identical to results obtained using a different probe corresponding to 310 bp of the 3′ untranslated region (not shown). The probe for H-Delta-1 was transcribed from a 2-kb cDNA fragment (clone HDL24) encoding the 3′ half of H-Delta-1. For comparison with TS1 antibody staining, a probe was transcribed from a 3-kb fragment comprising the 5′ half of HJ1 (see above). For all in situ experiments, the sense strand was also labeled and used as a control for nonspecific staining. The control slides were either blank or showed some staining at the edges of the tissue, a common artifact.
Chromosomal Localization
The chromosomal localizations of H-Delta-1, HJ1, and HJ2 were determined by fluorescence in situ hybridization as described previously. 19 Briefly, DNA was labeled with biotin-dUTP (Boehringer Mannheim) by a standard nick translation reaction yielding probe fragments between 200 and 500 nucleotides. Metaphase spreads were prepared by standard techniques and denatured in 70% formamide/2× SSC at 75°C for 90 seconds. After dehydration through a cold ethanol series, the chromosomes were incubated with heat-denatured probe (500 ng of each), 3 μg of Cot-1 DNA (Gibco BRL), and 7 μg of salmon sperm DNA in 10 μl of 50% formamide/10% dextran sulfate/2× SSC for 16–18 hours at 37°C. Posthybridization washes and blocking were done as previously reported. 19 Biotin-labeled probe was detected with fluorescein avidin DCS (Vector). Chromosomes were stained with DAPI, producing a “G-like” banding pattern, allowing chromosomal identification. The DNA for the probes was as follows: HJ1, the full-length cDNA in pBlueScript sk−; HJ2, the genomic clone described above; H-Delta-1, the genomic pAC clone described above.
Results
Cloning
Human fetal brain cDNA libraries were screened for homologues of fly Notch ligands using probes described under Materials and Methods. Sequences were obtained for three different cDNAs. The deduced amino acid sequences of these genes show that all of them code for transmembrane proteins that bear EGF-like domains. They have a signal peptide sequence and a partial EGF-like repeat that contains conserved residues defining the “DSL domain,” a region that has been described in all Notch ligands and their homologues. 20,21
The deduced amino acid sequence of the first ligand is most similar to fly DELTA, and it shares 83% identity with C-Delta-1 and 88% with mouse Delta (Figure 1) ▶ . 19,22 It appears to code for the human orthologue of these molecules, and we have named it Human Delta-1 (H-Delta-1). The H-Delta-1 cDNA codes for a transmembrane protein bearing 8 tandem EGF-like repeats and has a short cytoplasmic C-terminal domain, like the other vertebrate DELTAs.
Figure 1.

The deduced amino acid sequence for H-Delta-1, shown in alignment with C-Delta-1, 16 mouse Dll-1, 21 and Xenopus Delta-1. 28 Boxed and shaded residues are identical. Shaded unboxed residues are conserved. Dotted lines indicate the signal peptide and transmembrane regions. Dashed line marks the DSL domain. Solid lines demarcate EGF repeats.
The two other deduced amino acid sequences were most similar to the Drosophila ligand SERRATE. The overall domain structure of these proteins was identical to that of the rat Notch ligands Jagged1 and 2, 23,24 and we therefore assigned them the names Human Jagged 1 and 2 (HJ1 and HJ2). HJ1 is 95% identical to Jagged 1 and HJ2 is 88% identical to Jagged 2, strongly suggesting that HJ1 and HJ2 are the human orthologues of these molecules. (The accession numbers for the sequences of HJ1, HJ2, and H-Delta-1 are HSU61276, AF003521, and AF003522, respectively.) Human homologues of Jagged1 and Jagged2 have also recently been cloned independently in other labs. 5,8,9,25,26
Expression
Northern Analysis
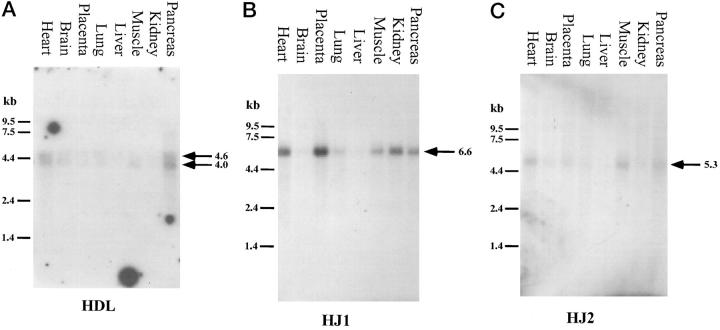
To analyze the expression of these ligands in human tissues, we probed Northern blots containing adult human mRNA from several tissues (Clontech). The H-Delta-1 probe hybridized to two bands, at 4.6 and 4.0 kb (Figure 2A) ▶ . H-Delta-1 was expressed most highly in heart and pancreas, with very little expression seen in brain and muscle, and virtually none in placenta, lung, liver, and kidney. The HJ1 probe bound an mRNA of approximately 6.6 kb and hybridized most strongly to heart, placenta, and kidney, with a slightly weaker signal detected in lung, muscle, and pancreas. Almost no expression was seen in brain and liver (Figure 2A) ▶ . The expression of HJ1 in kidney is in accordance with the finding of kidney disease in a subset of Alagille patients, who have missing or aberrant HJ1 sequences. 7,8 The HJ2 probe recognized a 5.3-kb mRNA and hybridized most strongly to heart, placenta, and skeletal muscle, with a weaker signal in placenta and pancreas, and almost no discernible expression in, brain, lung, liver, and kidney (Figure 2B) ▶ .
Figure 2.
Northern blots. A–C: H-Delta-1 and HJ1 and HJ2, respectively. The sizes of the labeled bands are given in the figure.
Expression in Normal and Cancerous Cervical Tissue
In a previous study from this group, 15 Notch receptor expression was shown to be up-regulated in cervical and colon cancers. We were interested in examining whether the ligand components of the Notch signaling system might also be expressed in transformed cells, specifically in cervical cancers.
To study the expression of Notch ligands in normal and cancerous cervical tissue, we raised polyclonal antibodies to the cytoplasmic portion of HJ1 as described under Materials and Methods. Western blot analysis of cells transfected with HJ1 DNA and untransfected cells revealed a strongly staining band in the transfected cells at about 130 kd, the expected molecular weight (Figure 3a) ▶ . Immunocytochemistry showed specific labeling of transfected cells and revealed no background staining in the untransfected cells (Figure 3b) ▶ . In addition, the pattern of expression determined using our anti-HJ1 antibodies was identical to that seen using in situ hybridization (not shown). To detect HJ2 and H-Delta-1 expression, we used in situ hybridization. Notch1 and 2 were detected using previously reported antibodies. 15
The cervical epithelium has two forms, a squamous epithelium that covers the ectocervix, and a colum-nar epithelium that lines the cervical canal. The squamous epithelium arises from basal undifferentiated cells and forms several layers of committed, proliferating cells that differentiate into keratinocytes. The columnar epithelium arises from undifferentiated subcolumnar reserve cells to form the fully differentiated, mucin-secreting cells that make up the columnar epithelium. The junction between the two types of epithelium is known as the squamocolumnar junction. During reproductive life a combination of ingrowth of the exocervical squamous epithelium and squamous metaplasia of the subcolumnar reserve cells transforms this area into squamous epithelium.
In exocervical squamous epithelium, Notch1 and Notch2 are expressed only in the stratum spinosum, which contains proliferating cells that are committed to the squamous fate, but not in the underlying layer of undifferentiated cells or in the upper layer of keratinized, fully differentiated cells. 15 This pattern was also seen for each of the ligands (not shown). Likewise, in the columnar epithelium, HJ1 and H-Delta-1, like Notch1 and 2, are expressed only in the reserve cells, but not in the differentiated cells, and HJ2 expression was not seen even in the reserve cells (not shown). None of the Notch components is expressed in normal differentiated cervical glands (see Figure 5 ▶ ).
Figure 5.
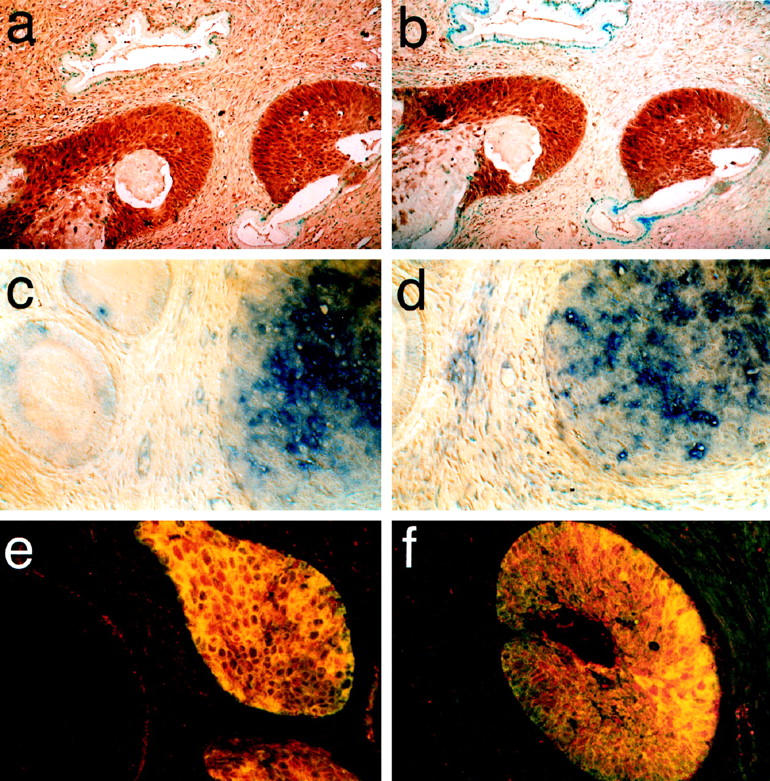
Expression of Notch and its ligands in squamous cell carcinoma of the uterine cervix is up-regulated. a: Notch2, immunohistochemistry; b: HJ1, immunohistochemistry; c: H-Delta-1,; in situ hybridization; d: HJ2, in situ hybridization. Note in a–d the presence of unlabeled normal cervical glands. e, f: Double-labeled fluorescent photomicrographs of squamous carcinomas. Anti-Notch1 antibodies are shown in red; anti-HJ1 labeling is shown in green. Overlapping regions are yellow and include virtually all of the labeled tissue. Original magnification, ×300.
In metaplastic squamous epithelia, the patterns of expression are similar to those seen for nonmetaplastic squamous epithelium. Notch 1 (not shown) and 2 were highly expressed as was HJ1 (Figure 4a and b) ▶ . H-Delta-1 was expressed patchily (Figure 4c) ▶ . HJ2, whose expression is not seen in normal columnar epithelium, was not expressed at all (Figure 4d) ▶ .
Figure 4.
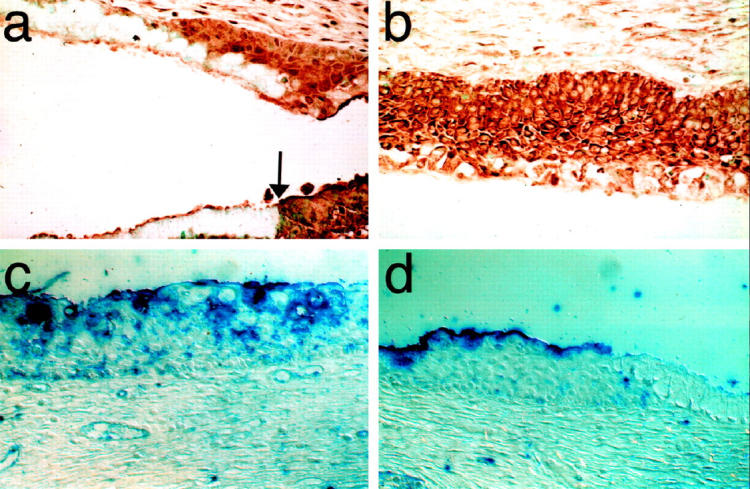
Expression of Notch and its ligands in squamous metaplasia of the uterine cervix. a: Notch2. Cervical sections reacted with anti-Notch antibody. Both the columnar epithelium, which is unstained except for the basal reserve cells, and the metaplastic tissue can be seen in this view. Note the sharp border between the strongly stained metaplastic squamous cells and the columnar cells epithelium (arrow). b; Immature squamous metaplasia immunolabeled for HJ1. HJ1 is strongly expressed only in the metaplastic tissue. c: In situ hybridization for H-Delta-1 shows patchy expression. d: In situ hybridization for HJ2 is negative in squamous metaplasia. Original magnification, ×300.
We investigated expression of our ligands in two types of cervical cancer, squamous carcinoma, which is the most prevalent, and adenocarcinoma, which is relatively rare. The numbers of cases analyzed for each Notch receptor and ligand is given in Table 1 ▶ . In squamous cell carcinoma (Figure 5) ▶ , Notch 1 (not shown) and 2, HJ1, H-Delta-1, and HJ2 were all up-regulated. Double labeling with antibodies to Notch1 and HJ1 showed that the expression of the receptor and its ligand completely overlapped (Figure 5, e and f) ▶ . This result was seen in both in situ and invasive squamous cell carcinoma.
Table 1.
Number of Cervical Cancer Cases Analyzed for Expression of Notch Ligands and Receptors
| Cancer | Ligands | Receptors | |||
|---|---|---|---|---|---|
| HJ1 | HJ2 | h-Delta-1 | TAN-1 | HN2 | |
| Adenocarcinoma | 3 | 3 | 2 | 4 | 5 |
| Squamous cell carcinoma | 4 | 2 | 1 | 3 | 2 |
TAN-1 and HN2 refer to human Notch 1 and 2, respectively. Immunohistochemistry was used to analyze the expression patterns of TAN-1, HJ2, and HJ1. In situ hybridization was used to examine the expression patterns of HJ2 and h-Delta-1.
In both in situ and invasive adenocarcinoma, Notch1 and 2, HJ1, and H-Delta-1 all showed a dramatic up-regulation (Figure 6) ▶ . This result is in sharp contrast to the complete lack of expression in normal cervical glands (See Figure 5 ▶ ).
Figure 6.

Expression of Notch, HJ1, and H-Delta-1 in cervical adenocarcinoma is up-regulated. a: Notch1, immunohistochemistry; b: HJ1, immunohistochemistry; c: H-Delta-1, in situ hybridization. d: HJ1, in situ hybridization. Original magnification, ×300.
These results show that in cervical cancer, the ligands and receptors for the Notch signaling system are expressed in the same tissues and at the same time. Thus, the potential exists for the Notch pathway to be activated in cervical cancer through receptor-ligand interactions.
Chromosomal Localization
The chromosomal localizations for all three ligands were determined using fluorescence in situ hybridization (see Figure 7 ▶ ). H-Delta-1 was localized to 6q27, HJ1 to 20p11–12, and HJ2 to 14q32. Subsequently, our finding for HJ1 was confirmed when two groups independently cloned HJ1 from 20p12, and identified it as the culprit gene for Alagille syndrome, a congenital disorder characterized by a paucity of intrahepatic bile ducts associated with other abnormalities, including cholestasis, cardiac disease, skeletal abnormalities including “butterfly” vertebrae, ocular abnormalities, and a characteristic facial appearance. 7,8 Interestingly, the locations of the other two ligands are both “hot spots” for breakpoints involved with tumor development (see under Discussion).
Figure 7.
Chromosomal localizations identified by FISH. a: H-Delta-1 is localized to 6q27. b: HJ1 is localized to 20p11–12. c: HJ2 is localized to 14q32.
Discussion
We have presented here the cloning of three human homologues of Drosophila Notch ligands, their expression patterns in human Northern blots and in normal and cancerous cervical tissue, and their chromosomal localizations. Like all of the known ligands of Notch homologues, these bear the conserved “DSL domain,” a cryptic EGF repeat near the amino terminus of the molecule (see Figure 1 ▶ ). 16,19,20,22,26,27,28 Like other members of the Jagged family, HJ1 and HJ2 bear 16 EGF repeats with an insertion in the 10th repeat. Likewise, H-Delta-1 has the eight tandem EGF repeats that characterize vertebrate homologues of Delta. 16,28
The functional roles of the EGF repeats and other conserved domains of the Notch ligands are still being elucidated. The DSL domains are believed to be the portion of the ligands that activate the receptor, and swapping experiments in Caenorhabditis elegans suggest that these domains may be functionally interchangeable. 30 The importance, if any, of the number of EGF repeats to the function of the molecules is currently unknown. It is known that Drosophila Notch requires specific EGF repeats to bind to Delta and Serrate, but the roles, if any, of the other EGF repeats have not been established. 31 Recent work from our lab indicates that proteolytic processing of the Drosophila ligands is required for normal Notch signaling (unpublished data). It seems likely that the ligands described here will prove to function much as the Drosophila ligands do, and they may well undergo the same types of processing.
Previous work from our lab showed that Notch 1 and 2 were expressed at high levels in cervical cancers, and that likely down-stream members of the pathway, the TLEs, were expressed in the same cells as Notch, except in the case of invasive squamous cell carcinomas, where the TLEs were absent. 15 It was impossible to know, of course, whether the Notch expressed in the cervical tissues was activated or not.
Since the Notch pathway plays a fundamental role in controlling the fates of undifferentiated, proliferative cell populations, its role in cancer may be similar. Specifically, the activated Notch pathway appears to prevent cells from taking on their normal, differentiated fate. Activation of the Notch pathway, then, might be important in preventing neoplastic cells from responding properly to the differentiation signals in their environment. The present study shows that all three known Notch ligands are expressed in cervical cancers, providing a possible means for activating the Notch receptors, which are also expressed at high levels. It is conceivable that the high expression levels of a variety of ligands make certain that the Notch receptors are constitutively activated, thus promoting the growth of the cancer by blocking differentiation.
Hints at other possible roles for Notch components in human disease states may be gleaned from knowing their chromosomal localizations. The HJ1 gene is now known to cause Alagille syndrome when deleted or mutated at any of several places. The phenotype of Alagille indicates an important role for HJ1 in the development of bile ducts, heart, skeleton, and eye, and neurological and renal defects are also seen, although less commonly. 8,10 HJ1 also appears to have a role in hematopoiesis. 5-7
The localizations of H-Delta-1 and HJ2 to 6q27 and 14q32, respectively, raise the possibility that these may turn out to play important roles in human diseases. There are two known diseases for which H-Delta-1 may be a good candidate culprit gene. Allelic deletions of chromosome 6 are frequently associated with ovarian carcinomas, and much effort has been made to closely define the region that bears a putative tumor suppressor gene. 32 It will be important to discover whether H-Delta-1 falls within the suspect interval, especially in light of the fact that the Notch signaling pathway appears to be active in certain cancers. 13-15 Similarly, linkage and association studies have placed a locus for type I diabetes at 6q27 near marker D6S281 33 As H-Delta-1 is strongly expressed in pancreas, it could be important to follow up on this clue as well.
HJ2 is located at 14q32 which is a well-studied “hot spot” for lymphomas. Many of these breakpoints occur in the IGH locus, and are probably not relevant for HJ2. However, tumor suppressor genes at this site remain to be identified (eg, see reference 34). Interestingly, one recent study places a putative suppressor gene for ovarian cancer at this locus, defined by the markers D14S65 and D14S67. 34,35 Other diseases that have been linked to 14q32 include Usher syndrome 1 and childhood neuroblastomas.36,37
In sum, the roles of the various Notch components in human disease are still being elucidated, and there is still much to be learned about how the Notch pathway may contribute to the development of cancer. However, from the extensive developmental analysis in Drosophila, we know that inappropriate activation or inactivation of the Notch pathway will derail cells from their normal course of differentiation. It is therefore conceivable that altering its activity in neoplastic tissue could divert the cells into a different fate, suggesting that the Notch pathway might offer a novel route for therapeutic intervention.
Acknowledgments
We thank Dr. Richard Lifton for giving us the human genomic library, Dr. Gerry Weinmaster for sharing her Jagged-2 sequence before publication, Dr. Jeremy Quirk for screening the PAC library, and Dr. Vincent Marchesi for critical reading.
Footnotes
Address reprint requests to Spyros Artavanis-Tsakonas, Harvard Medical School, MGH Cancer Center, Bldg. 149, 13th St., Charlestown, MA 02129.
Supported in part by the Howard Hughes Medical Institute (S.A.-T.), the National Institutes of Health (Grant 2R01NS26804–06A1 to S.A.-T.), the HHMI International Research Scholars Program (D.I.-H.), the Imperial Cancer Research Fund (D.I.-H.), and the American Cancer Society (Grant PF-4036 to G.E.G.).
The first two authors contributed equally to this paper.
E.M.’s current address is Institut de Biologie du Developpement de Marseille, Campus de Luminy case 907, F-13288 Marseille Cedex 9, France.
D.H.’s current address is Instituto Histologia e Embriologia, Faculdade de Medicina de Lisboa, Av. Prof. Egas Moniz, 1699 Lisboa Codex, Portugal.
References
- 1.Artavanis-Tsakonas S, Matsuno K, Fortini ME: Notch signaling. Science 1995, 268:225-232 [DOI] [PubMed] [Google Scholar]
- 2.Lardelli M, Williams R, Lendahl U: Notch-related genes in animal development. Int J Dev Biol 1995, 39(5):769-780 [PubMed] [Google Scholar]
- 3.Greenwald I: Structure/function studies of lin-12/Notch proteins. Curr Opin Genet Dev 1994, 4:556-562 [DOI] [PubMed] [Google Scholar]
- 4.Fleming RJ, Purcell K, Artavanis-Tsakonas S: The NOTCH receptor and its ligands. Trends Cell Biol 1997, 7:437-441 [DOI] [PubMed] [Google Scholar]
- 5.Zimrin AB, Pepper MS, McMahon GA, Nguyen F, Montesano R, Maciag T: An antisense oligonucleotide to the Notch ligand Jagged enhances fibroblast growth factor-induced angiogenesis in vitro. J Biol Chem 1996, 51:32499-32502 [DOI] [PubMed] [Google Scholar]
- 6.Varnum-Finney B, Purton LE, Yu M, Brashem-Stein C, Flowers D, Staats S, Moore KA, Le Roux I, Mann R, Gray G, Artavanis-Tsakonas S, Bernstein ID: The notch ligand, Jagged-1, influences the development of primitive hematopoietic precursor cells. Blood 1998, 11:4084-4091 [PubMed] [Google Scholar]
- 7.Li L, Krantz ID, Deng Y, Genin A, Banta AB, Collins CC, Qi M, Trask BJ, Kuo WL, Cochran J, Costa T, Pierpont ME, Rand EB, Piccoli DA, Hood L, Spinner NB: Alagille syndrome is caused by mutations in human Jagged1, which encodes a ligand for Notch 1. Nature Genet 1997, 16:243-251 [DOI] [PubMed] [Google Scholar]
- 8.Oda T, Elkahloun AG, Meltzer PS, Chandrasekharappa SC: Identification and cloning of the human homolog (Jag1) of the rat Jagged1 gene from the Alagille syndrome critical region at 20p12. Genomics 1997, 43:376-379 [DOI] [PubMed] [Google Scholar]
- 9.Oda T, Elkahloun AG, Pike BL, Okajima K, Krantz ID, Genin A, Piccoli DA, Meltzer PS, Spinner NB, Collins FS, Chandrasekharappa SC: Mutations in the human Jagged1 gene are responsible for Alagille syndrome. Nature Genet 1997, 16:235-242 [DOI] [PubMed] [Google Scholar]
- 10.Artavanis-Tsakonas S: Alagille syndrome—a notch up for the Notch receptor. Nature Genetics 1997, 16:212-213 [DOI] [PubMed] [Google Scholar]
- 11.Joutel A, Corpechot C, Ducros A, Vahedi K, Chabriat H, Mouton P, Alamowitch S, Domenga V, Cecillion M, Marechal E, J, Vayssiere C, Cruaud C, Cabanis E-A, Ruchoux MM, Weissenbach J, Bach JF, Bousser MG, Tournier-Lasserve E: Notch3 mutations in CADASIL, a hereditary adult-onset condition causing stroke and dementia. Nature 1997, 383:707–710 [DOI] [PubMed]
- 12.Ellisen LW, Bird J, West DC, Soreng AL, Reynolds TC, Smith SD, Sklar J: Tan-1, the human homolog of the drosophila Notch gene, is broken by chromosomal translocations in T lymphoblastic neoplasms. Cell 1991, 66:649-661 [DOI] [PubMed] [Google Scholar]
- 13.Jhappan C, Gallahan D, Stahle C, Chu E, Smith GH, Merlino G, Callahan R: Expression of an activated Notch-related int-3 transgene interferes with cell differentiation and induces neoplastic transformation in mammary and salivary glands. Genes Dev 1992, 6:345-355 [DOI] [PubMed] [Google Scholar]
- 14.Zagouras P, Stifani S, Blaumueller CM, Carcangiu ML, Artavanis-Tsakonas S: Alterations in Notch signaling in neoplastic lesions of the human cervix. Proc Natl Acad Sci USA 1995, 92:6414-6418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Henrique D, Adam J, Myat A, Chitnis A, Lewis J, Ish-Horowicz D: Expression of a Delta homologue in prospective neurons in the chick. Nature 1995, 375:787-790 [DOI] [PubMed] [Google Scholar]
- 16.Mitsiadis TA, Lardelli M, Lendahl U, Thesleff I: Expression of Notch 1, 2, and 3 is regulated by epithelial-mesenchymal interactions and retinoic acid in the developing mouse tooth and associated with determination of ameloblast cell fate. J Cell Biol 1995, 130:407-418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Iannou PA, Amemiya CT, Garnes J, Kroisel PM, Shizuga H, Chen C, Batzer MA, De Jong PA: A new bacteriophage P1-derived vector for the propagation of large human DNA fragments. Nat Genet 1994, 16:84-89 [DOI] [PubMed] [Google Scholar]
- 18.Lichter P, Tang CJ, Call K, Hermanson G, Evans GA, Housman D, Ward DC: High-resolution mapping of human chromosome 11 by in situ hybridization with cosmid clones. Science 1990, 247:64-69 [DOI] [PubMed] [Google Scholar]
- 19.Tax FE, Yeargers JJ, Thomas JH: Sequence of C. elegans lag-2 reveals a cell-signalling domain shared with Delta and Serrate of Drosophila. Nature 1994, 368:150-154 [DOI] [PubMed] [Google Scholar]
- 20.Mello CC, Draper BW, Priess JR: The maternal genes apx-1 and glp-1 and establishment of dorsal-ventral polarity in the early C. elegans embryo. Cell 1994, 77:95-106 [DOI] [PubMed] [Google Scholar]
- 21.Bettenhausen B, Hrabe de Angelis M, Simon D, Guenet JL, Gossler A: Transient and restricted expression during mouse embryogenesis of Dll1, a murine gene closely related to Drosophila Delta. Development 1995, 121:2407-2418 [DOI] [PubMed] [Google Scholar]
- 22.Lindsell CE, Shawber CJ, Boulter J, Weinmaster G: Jagged: A mammalian ligand that activates Notch1. Cell 1995, 80:909-917 [DOI] [PubMed] [Google Scholar]
- 23.Shawber C, Boulter J, Lindsell CE, Weinmaster G: Jagged2: a serrate-like gene expressed during rat embryogenesis. Dev Biol 1996, 180:370-376 [DOI] [PubMed] [Google Scholar]
- 24.Luo B, Aster JC, Hasserjian RP, Kuo F, Sklar J: Isolation and functional analysis of a cDNA for human. Jagged2, a gene encoding a ligand for the Notch1 receptor. Mol Cell Biol 1997, 17:6057-6067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Valsecchi V, Ghezzi C, Ballabio A, Rugarli EI: Jagged2—a putative Notch ligand expressed in the apical ectodermal ridge and I sites of epithelial-mesenchymal interactions. Mech Dev 1997, 69:203-207 [DOI] [PubMed] [Google Scholar]
- 26.Fleming RJ, Scottgale TN, Diederich RJ, Artavanis-Tsakonas S: The gene Serrate encodes a putative EGF-like transmembrane protein essential for proper ectodermal development in Drosophila melanogaster. Genes Dev 1990, 4:2188-2201 [DOI] [PubMed] [Google Scholar]
- 27.Kopczynski CC, Alton AK, Fechtel K, Kooh PJ, Muskavitch MAT: Delta, a Drosophila neurogenic gene, is transcriptionally complex and encodes a protein related to blood coagulation factors and epidermal growth factor of vertebrates. Genes Dev 1988, 2:1723-1735 [DOI] [PubMed] [Google Scholar]
- 28.Chitnis A, Henrique D, Lewis J, Ish-Horowicz D, Kintner C: Primary neurogenesis in Xenopus embryos regulated by a homologue of the Drosophila neurogenic gene Delta. Nature 1995, 375:761-766 [DOI] [PubMed] [Google Scholar]
- 29.Fitzgerald K, Greenwald I: Interchangeability of Caenorhabditis elegans DSL proteins and intrinsic signalling activity of their extracellular domains in vivo. Development 1995, 121:4275-4282 [DOI] [PubMed] [Google Scholar]
- 30.Rebay I, Fleming RJ, Fehon RG, Cherbas L, Cherbas P, Artavanis-Tsakonas S: Specific EGF repeats of Notch mediate interactions with Delta and Serrate: implications for Notch as a multifunctional receptor. Cell 1991, 67:687-699 [DOI] [PubMed] [Google Scholar]
- 31.Saito S, Sirahama S, Matsushima M, Suzuki M, Sagae S, Kudo R, Saito J, Noda K, Nakamura Y: Definition of a commonly deleted region in ovarian cancers to a 300-kb segment of chromosome 6q27. Cancer Res 1996, 56:5586-5589 [PubMed] [Google Scholar]
- 32.Todd JA: Genetics of type 1 diabetes. Pathol Biol 1997, 45:219-227 [PubMed] [Google Scholar]
- 33.Khokhar MT, Brito-Babapulle V, Matute E, Catovsky D: Cytogenetic abnormalities in the leukemic phase of non-Hodgkin lymphoma. Cancer Genet Cytogenet 1995, 83:18-24 [DOI] [PubMed] [Google Scholar]
- 34.Bandera CA, Takahashi H, Behbakht K, Liu PC, LiVolsi VA, Benjamin I, Morgan MA, King SA, Rubin SC, Boyd J: Deletion mapping of two potential chromosome 14 tumor suppressor gene loci in ovarian carcinoma. Cancer Res 1997, 57:513-515 [PubMed] [Google Scholar]
- 35.Gerber S, Larget-Piet D, Rozet JM, Bonneau D, Mathieu M, Der Kaloustian V, Munnich A, Kaplan J: Evidence for a forth locus in Usher syndrome type I. J Med Genet 1996, 33:77-79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brodeur GM: Genetics of embyonal tumours of childhood: retinoblastoma, Wilms’ tumour and neuroblastoma. Cancer Surv 1995, 25:67-99 [PubMed] [Google Scholar]