Abstract
We present a method for using slopes and intercepts from a linear regression of a quantitative trait as outcomes in segregation and linkage analyses. We apply the method to the analysis of longitudinal systolic blood pressure (SBP) data from the Framingham Heart Study. A first-stage linear model was fit to each subject's SBP measurements to estimate both their slope over time and an intercept, the latter scaled to represent the mean SBP at the average observed age (53.7 years). The subject-specific intercepts and slopes were then analyzed using segregation and linkage analysis. We describe a method for using the standard errors of the first-stage intercepts and slopes as weights in the genetic analyses. For the intercepts, we found significant evidence of a Mendelian gene in segregation analysis and suggestive linkage results (with LOD scores ≥ 1.5) for specific markers on chromosomes 1, 3, 5, 9, 10, and 17. For the slopes, however, the data did not support a Mendelian model, and thus no formal linkage analyses were conducted.
Introduction
In the conventional epidemiology literature, much has been written about utilizing data in which measurement of quantitative traits are periodically taken from each subject over time [1-3]. However, relatively little has been written regarding the use of longitudinal data in genetic epidemiological studies. One method proposed by Levy et al. [4] utilized the within-subject mean SBP to summarize the longitudinal measurements for each subject. In Levy's analysis of data from the Framingham Heart Study, a sample-wide regression was used to model SBP over time as a linear function of age and body mass index after adjusting for sex, cohort group, and hypertension treatment in a first-stage analysis. Residuals for each subject from the regression analysis, averaged over time, were then used as continuous phenotype data in a linkage analysis in the second stage of the analysis.
In this paper, we also adopt a two-stage modeling approach. In our first stage, we fit a linear regression of SBP on age to obtain subject-specific intercepts and slopes. This first-stage model includes adjustment for any time-varying covariates of interest, such as calendar year, body mass index, and hypertension treatment. Also estimated in this first stage are the subject-specific standard errors of the corresponding intercepts and slopes. The second stage analysis consists of a segregation analysis of the subject-specific intercepts from the first stage model, and a separate segregation analysis of the slopes. We claim that the standard errors from the first stage should be used in weighting the contribution of each subject in the segregation analysis, and we describe how this can be accomplished. Based on the results of the segregation analyses, we conducted a genome screen using parametric linkage analysis applied to all pedigrees in the Framingham Heart Study. We demonstrate increased LOD scores using the weighed analysis, compared with the analogous approach that does not use weights.
Methods
General approach
Let Yij denote the SBP of the ith subject at the jth study visit, Tij be the corresponding age of the subject at the visit, and let Xij denote a matrix of time-dependent covariates. We propose using a first-stage model of the form
Yij = ai + bi (Tij - 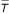 ) + γ'Xij + eij, (1)
) + γ'Xij + eij, (1)
where 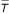 is the overall mean age in the sample and eij are residuals, assumed to be independent and normally distributed with mean 0 and variance ω2. The goal of this first-stage model is to estimate the subject-specific intercepts (ai) and slopes on age (bi), and their corresponding standard errors. We denote these standard errors by sai for intercepts and sbi for slopes. Note that the intercepts have the interpretation as the predicted mean Y at age
is the overall mean age in the sample and eij are residuals, assumed to be independent and normally distributed with mean 0 and variance ω2. The goal of this first-stage model is to estimate the subject-specific intercepts (ai) and slopes on age (bi), and their corresponding standard errors. We denote these standard errors by sai for intercepts and sbi for slopes. Note that the intercepts have the interpretation as the predicted mean Y at age 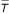 when all X values are zero. We center any continuous X variables on their corresponding sample means to increase interpretability of the intercepts.
when all X values are zero. We center any continuous X variables on their corresponding sample means to increase interpretability of the intercepts.
The second-stage model utilizes the first-stage intercepts ai and slopes bi as continuous phenotype data in a genetic analysis. We first perform segregation analysis to determine the evidence for a Mendelian gene and to estimate the associated model parameters. For analysis of the intercepts, the penetrance model used in the segregation analysis has the form
ai = α + βGi + η'Xi + ei, (2)
where Gi is a covariate based on an unobserved major gene gi, and Xi is a matrix of time-independent covariates. An analogous model was used for the slopes. The residual ei is assumed to be normally distributed with mean 0 and variance (σ2 + sai2), where sai2 is the square of the first-stage standard error of the intercept, and σ2 is the between-subject residual variance to be estimated. Note that this variance expression has the effect of weighting each subject's contribution to the genetic analysis based on the precision (standard error) of their intercept estimate. We thus denote the use of this variance for ei as a 'weighted' analysis. Generally speaking, these first-stage standard errors will be smallest for those with many measurements, and with measurements at ages that span the overall average age 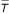 . One could also perform an 'unweighted' analysis by assuming that the variance of ei was simply σ2, which would treat the intercepts for all subjects as equally informative.
. One could also perform an 'unweighted' analysis by assuming that the variance of ei was simply σ2, which would treat the intercepts for all subjects as equally informative.
To estimate the parameters of the above model, we maximized the likelihood
![]()
where the F indexes family, gF is a vector of unobserved major genotypes, and YF and XF are the trait and covariate data for family F. The parameters Ω = {α, β, η, σ } are the parameters of the penetrance model, qA is the population frequency of the variant allele 'A', and τ = {τAA, τAa, τaa} are the probabilities that a parent with the subscripted genotype transmits an 'A' to their offspring. Computation of the above likelihood requires use of the peeling algorithm [5,6]. We considered six models in the segregation analysis: four Mendelian models (dominant, recessive, additive, and codominant), a no-major-gene model that included only measured covariates, and a general transmission model. In the general transmission model, τAA, τAa, and τaa were treated as free parameters to be estimated. This general model was compared to the Mendelian models, in which τAA, τAa, and τaa were constrained to their theoretical values of 1.0, 0.5, and 0.0, respectively. Likelihood ratio tests (LRTs) were used to compare the general model to the Mendelian models, and also to the no-major-gene model. We also computed Akaike's Information Criteria (AIC) for each model as -2(log-likelihood at the maximum likelihood estimator (MLE)) + 2(number of model parameters estimated). A lower AIC indicates a more parsimonious model.
Application to the Genetic Analysis Workshop 13 (GAW13) Framingham Heart Study data set
The GAW13 data set of the Framingham Heart Study included a total of 4692 subjects, of which 1213 subjects provided longitudinal observation data from the first cohort, and 1672 subjects from the offspring cohort. The outcome variable of interest in this paper was systolic blood pressure (SBP). A natural log transform was used to linearize the SBP relationship with age; thus Yij in equation (1) is ln(SBPij). Only observations with age in the range 30 to 80 were utilized, to further linearize the relationship between ln(SBP) and age. The average age was 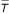 = 53.7. Time-dependent covariates defining Xij in equation (1) included body mass index (BMI), calendar year (CY), CY2, hypertension treatment (HRX), CY × HRX, CY × male, CY × cohort, CY × BMI, CY × age, male × age, and BMI × HRX. The continuous variables BMI and CY were centered on their respective sample means, while HRX and male were indicators of treatment status and male sex, respectively. The CY2 term was included to account for observed nonlinearity between SBP and CY. The intercepts from the first-stage model have interpretation as the subject-specific mean ln(SBP) adjusted to a female, untreated person of average age (53.7 years) and BMI (26.3 kg/m2) in calendar year 1969.5. PROC MIXED in SAS, Release 8.2 (SAS Institute, Cary NC), was used to fit the first-stage model and obtain person-specific intercepts and slopes, and their respective standard errors.
= 53.7. Time-dependent covariates defining Xij in equation (1) included body mass index (BMI), calendar year (CY), CY2, hypertension treatment (HRX), CY × HRX, CY × male, CY × cohort, CY × BMI, CY × age, male × age, and BMI × HRX. The continuous variables BMI and CY were centered on their respective sample means, while HRX and male were indicators of treatment status and male sex, respectively. The CY2 term was included to account for observed nonlinearity between SBP and CY. The intercepts from the first-stage model have interpretation as the subject-specific mean ln(SBP) adjusted to a female, untreated person of average age (53.7 years) and BMI (26.3 kg/m2) in calendar year 1969.5. PROC MIXED in SAS, Release 8.2 (SAS Institute, Cary NC), was used to fit the first-stage model and obtain person-specific intercepts and slopes, and their respective standard errors.
A total of 2883 person-specific intercepts (ai values) and 2787 person-specific slopes (bi values) were obtained from the first-stage analysis. These estimates were used as trait data in the second-stage segregation and linkage analyses. Covariates Xi in equation (2) included male sex and cohort, the latter an indicator of membership in Cohort 2. We fit the segregation and linkage models using a version of the Genetic Analysis Package (GAP, Epicenter Software, Pasadena, CA), modified by one of the authors (WJG) to utilize sai2 (and sbi2) in a weighted analysis. As will be demonstrated below, a Mendelian model was supported for the intercepts, but not for the slopes. We therefore focused our linkage analysis only on the intercepts. We fixed the segregation-model parameters to their MLEs from the weighted analysis, and performed two-point LOD-score linkage analysis to estimate the recombination fraction (θ) between g and each of 399 markers. Allele frequencies at each marker locus were fixed to the values provided with the data. For comparison, we also performed an unweighted linkage analysis, in which a segregation analysis was re-run without standard error weights, and these MLEs then used in linkage analysis.
Results
Segregation analysis
Segregation analysis of the intercepts supported a Mendelian codominant model, with strong evidence of a genetic effect (Table 1). Specifically, compared to the general model, the Mendelian codominant model did not fit significantly worse (χ2 = 1.7, p = 0.43, conservatively assuming two degrees of freedom). On the other hand, the remaining Mendelian models and the no-major-gene model could be rejected (p < 0.001 for each). The codominant model also provided the lowest AIC, again indicating that this model provided the best fit to the data. The estimated allele frequency from this model was qA = 0.31, translating into 48% of subjects with g = aa, 42% with g = Aa, and 10% with g = AA. Compared with subjects with g = aa, SBP was estimated to be 12% higher (exp(0.115)) for subjects with g = Aa, and 33% higher (exp (0.283)) for subjects with g = AA. Segregation analysis of the slopes, on the other hand, did not support evidence of Mendelian transmission (Table 2). Specifically, the general model fit significantly better than any of the Mendelian models (p < 0.001), and provided the lowest AIC. The estimate of the transmission parameter τAA was 0.0, far from its Mendelian expectation of 1.0.
Table 1.
Weighted segregation analysis of intercepts*
| Hypothesis | ||||||||||||
| Mendelian | ||||||||||||
| Segregation Parameter | General | Codominant | Dominant | Recessive | Additive | No Major Gene | ||||||
| Estimate | SE | Estimate | SE | Estimate | SE | Estimate | SE | Estimate | SE | Estimate | SE | |
| Intercept | 4.769 | 0.0078 | 4.771 | 0.0072 | 4.802 | 0.0052 | 4.801 | 0.0051 | 4.776 | 0.0065 | 4.846 | 0.0041 |
| βcohort | -0.088 | 0.0077 | -0.092 | 0.0043 | -0.092 | 0.0046 | -0.091 | 0.0047 | -0.092 | 0.0043 | -0.085 | 0.0049 |
| βSex | 0.011 | 0.0046 | 0.011 | 0.0046 | 0.012 | 0.0046 | 0.010 | 0.0047 | 0.012 | 0.0046 | 0.005 | 0.0049 |
| βAA | 0.288 | 0.0143 | 0.283 | 0.0142 | 0.165 | 0.0066 | 0.167 | 0.0072 | 0.269 | 0.0107 | — | — |
| βAa | 0.118 | 0.0076 | 0.115 | 0.0078 | 0.165A | — | 0.000B | — | 0.135C | — | — | — |
| q A | 0.323 | 0.0539 | 0.305 | 0.0373 | 0.139 | 0.0180 | 0.511 | 0.0304 | 0.257 | 0.0285 | — | — |
| σ2 | 0.004 | 0.0004 | 0.004 | 0.0004 | 0.006 | 0.0004 | 0.006 | 0.0004 | 0.004 | 0.0004 | 0.011 | 0.0004 |
| τaa | 0.000 | 0.0000 | 0.000D | — | 0.000D | — | 0.000D | — | 0.000D | — | — | — |
| τAa | 0.476 | 0.0610 | 0.500D | — | 0.500D | — | 0.500D | — | 0.500D | — | — | — |
| τAA | 0.935 | 0.0611 | 1.000D | — | 1.000D | — | 1.000D | — | 1.000D | — | — | — |
| -2(log-likelihood) | -3482.64 | -3480.94 | -3400.16 | -3376.52 | -3463.32 | -3155.59 | ||||||
| p-valueE | — | 0.43 | < 0.001 | < 0.001 | < 0.001 | < 0.001 | ||||||
| AICF | -3462.64 | -3466.94 | -3388.16 | -3364.52 | -3451.32 | -3147.59 | ||||||
*The outcome being modeled in equation (2) is ai from equation (1). AConstrained to equal βAA. BConstrained to equal 0. C Constrained to equal 1/2 βAA. D Parameter value is fixed. Ep-value based on a likelihood ratio test with the general model as the base model.FAIC = -2(log-likelihood) + 2(number of free parameters).
Table 2.
Weighted segregation analysis of slopes*
| Hypothesis | ||||||||||||
| Mendelian | ||||||||||||
| Segregation Parameter | General | Codominant | Dominant | Recessive | Additive | No Major Gene | ||||||
| Estimate | SE | Estimate | SE | Estimate | SE | Estimate | SE | Estimate | SE | Estimate | SE | |
| Intercept | 3.205 | 0.1814 | 3.500 | 0.1932 | 3.790 | 0.2246 | 4.139 | 0.1489 | 3.744 | 0.2081 | 4.265 | 0.1485 |
| βcohort | -3.541 | 0.2393 | -3.819 | 0.2094 | -3.785 | 0.2092 | -3.788 | 0.2143 | -3.793 | 0.2090 | -3.726 | 0.2113 |
| βSex | -1.621 | 0.1981 | -1.623 | 0.1965 | -1.584 | 0.2001 | -1.682 | 0.1907 | -1.580 | 0.1897 | -1.620 | 0.1936 |
| βAA | 16.614 | 1.7795 | 16.625 | 2.4421 | 6.742 | 1.2112 | 14.296 | 2.1622 | 12.821 | 2.1109 | — | — |
| βAa | 4.443 | 0.5003 | 3.525 | 0.8312 | 6.742A | — | 0.000B | — | 6.411C | — | — | — |
| q A | 0.199 | 0.0584 | 0.110 | 0.0265 | 0.042 | 0.0195 | 0.130 | 0.0269 | 0.047 | 0.0188 | — | — |
| σ2 | 0.485 | 0.1782 | 1.849 | 0.5964 | 2.384 | 0.7088 | 3.384 | 0.5886 | 2.206 | 0.5857 | 4.949 | 0.6492 |
| τaa | 0.000 | 0.0000 | 0.000D | — | 0.000D | — | 0.000D | — | 0.000D | — | — | — |
| τAa | 0.390 | 0.0694 | 0.500D | — | 0.500D | — | 0.500D | — | 0.500D | — | — | — |
| τAA | 0.000 | 0.0000 | 1.000D | — | 1.000D | — | 1.000D | — | 1.000D | — | — | — |
| -2(log-likelihood) | 17811.58 | 17824.66 | 17839.45 | 17828.49 | 17837.35 | 17867.83 | ||||||
| p-valueE | — | < 0.001 | < 0.001 | < 0.001 | < 0.001 | < 0.001 | ||||||
| AICF | 17831.58 | 17838.66 | 17851.45 | 17840.49 | 17849.35 | 17875.83 | ||||||
*The outcome being modeled in equation (2) is 1000 × bi, the subject-specific slope from equation (1). AConstrained to equal βAA. BConstrained to equal 0. CConstrained to equal 1/2 βAA. DParameter value is fixed. Ep-value based on a likelihood ratio test with the general model as the base model.FAIC = -2(log-likelihood) + 2(number of free parameters).
Linkage analysis
Given the findings in segregation analysis, linkage analysis was conducted only on the intercepts. The parameters of the segregation model were fixed to the values shown for the Mendelian codominant model in Table 1. Two-point linkage analysis was conducted, i.e., markers were considered one at a time in separate analyses. Each analysis utilized standard-error based weights as in the segregation analyses. For comparison, we also repeated the segregation and linkage analyses using unweighted analysis, i.e., not using first-stage standard errors as weights. Table 3 shows the markers that yielded a LOD score of at least 1.5 using either the weighted or unweighted approach. The strongest evidence of linkage was found at marker positions 202 (LOD = 2.3) and 212 (LOD = 2.9) on chromosome 1, position 32 (LOD = 2.3) on chromosome 9, and position 125 (LOD = 2.1) on chromosome 10 in the weighted analysis, and at position 153 (LOD = 2.0) on chromosome 3 in the unweighted analysis. Weaker evidence for linkage was observed on chromosomes 5 and 17 in the weighted analysis. The weighted analysis generally led to larger LOD scores than the unweighted analysis. To further compare the weighted and unweighted approaches, Figure 1 shows plots of the LOD scores at all markers on chromosomes 1, 5, 9, 10, and 17. While the LOD scores for the two approaches have similar trends across markers, the weighted analysis provides more striking peaks, particularly on chromosomes 9, 10, and 17.
Table 3.
Markers with LOD score ≥ 1.5 based on two-point linkage analysis* of subject-specific SBP intercepts
| Weighted | Unweighted | |||||
| Chromosome | Location (cM) | Marker | LOD | p-value | LOD | p-value |
| 1 | 202 | GATA7C01 | 2.31 | 0.0005 | 2.27 | 0.0006 |
| 1 | 212 | GATA48B01 | 2.93 | 0.0001 | 2.84 | 0.0002 |
| 3 | 153 | GATA4A10 | 0.83 | 0.026 | 2.00 | 0.0012 |
| 5 | 40 | GATA145D09 | 1.66 | 0.0028 | 0.17 | 0.19 |
| 9 | 32 | GATA27A11 | 2.30 | 0.0006 | 1.27 | 0.0078 |
| 10 | 125 | GATA64A09 | 2.10 | 0.0009 | 0.80 | 0.028 |
| 17 | 100 | GATA28D11 | 1.50 | 0.0042 | 0.64 | 0.0428 |
* Assuming the Mendelian codominant model (see Table 1)
Figure 1.
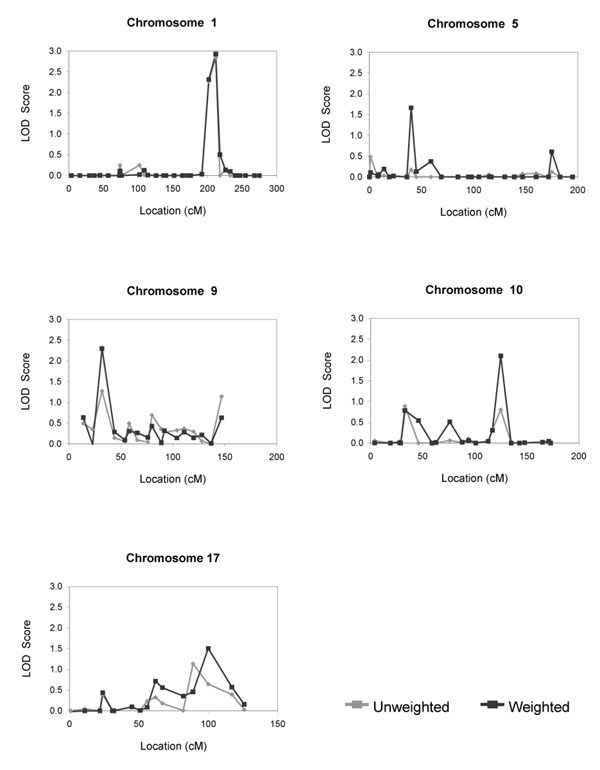
LOD scores on four chromosomes based on genetic analyses of intercepts that either do (weighted) or do not (unweighted) incorporate first-stage standard errors
Discussion
Our segregation analysis indicates that SBP, specifically average SBP at age 53, has a significant genetic basis. We estimated that approximately half of the population carries a genotype (g = Aa or AA) that leads to some elevation in average SBP relative to genetically normal (g = aa) individuals. Subsequent analysis revealed modest evidence of linkage on chromosomes 1 (202 and 212 cM), 5 (40 cM), 9 (32 cM), 10 (125 cM), and 17 (100 cM). Levy et al. [4] also found evidence of linkage at the same position on chromosome 10 (125 cM), but at different positions on chromosomes 5 (59 cM) and 10 (125 cM). Rao et al. [7] also reported linkage to position 125 cM on chromosome 10, while Briollais et al. [8] reported evidence of linkage at position 212 cM on chromosome 1.
The appropriate use of standard errors from the first stage model in the second stage model generally resulted in larger LOD scores than were obtained in an unweighted analysis. However, since our application was to a real data set for which we do not know the truth, we cannot conclude with certainty that the use of weights will generally lead to more significant linkage peaks. Our proposed two-stage approach should be evaluated further using simulated data.
We did not find support for an effect of a Mendelian gene on SBP slope in our segregation analysis. This may be a consequence of the model form we applied in our analysis, for example in our assumption that any genetic effect was mediated through a single major gene. If multiple genes affect SBP change over time, our model may have had low power to detect a genetic signal. As a 'fishing expedition', we ignored our lack of support for a Mendelian model and performed a genome screen for linkage to SBP slopes. The segregation model parameters were fixed to the values for the Mendelian codominant model shown in Table 2. This analysis revealed no LOD scores that exceeded 1.5 at any marker. This failure to find any linkage signals may again be a consequence of poor power, or it may reflect our segregation-analysis finding that slopes are not determined by a Mendelian gene.
Although we developed a two-stage modeling approach in this paper, we believe that it would be preferable to combine the first and second stage models into a single analysis. This would consist of performing a joint segregation and linkage analysis of the original, repeated SBP measurements on each subject. Some advantages of this approach are that parameter estimates in each model would be mutually adjusted for one another, and subjects with more observations would naturally contribute more information to parameter estimation and testing. To achieve this latter quality in a two-stage analysis required the incorporation of standard-error derived weights from the first-stage model into the second-stage model, as described in this paper. The primary deterrent to implementing a joint approach lies in the computational difficulty of simultaneously fitting a longitudinal model and summing over a large space of unobserved genotypes. One could consider using Markov chain Monte Carlo methods to solve this computational difficulty (see Palmer et al. [9] and Scurrah et al. [10]), and we would encourage formal comparison of these approaches to a two-stage approach to assess their relative merits.
There are some difficult issues in this particular data set that we have not addressed. First is the issue of how to best handle hypertension treatment (HRX). We chose to include HRX as a time-dependent covariate in our first-stage model. However, since the decision to treat is based on SBP, this approach may lead to invalid estimates of the HRX effect, and may ultimately affect our genetic inferences as well. Levy et al. [4] propose a different approach for dealing with HRX in the analysis of longitudinal SBP. Clearly, more work is required to better understand how to best adjust for covariates that are themselves determined by the outcome variable. Another important issue is the problem of missing data. In our analysis, we used only observations at each time point that had complete outcome and covariate data. The elimination of missing observations may introduce bias if the missingness is related to the condition of the subject at that time (see Kang et al. [11], for a longer discussion). Furthermore, if missingness patterns are correlated within families, results from segregation and linkage analyses may be further misrepresented.
We adopted a parametric modelling approach in our genetic analysis. An advantage of this approach is that it utilizes all available data in each pedigree. A disadvantage, however, is that the model form was likely misspecified, particularly if SBP is determined by several genes with differing allele frequencies and effects on the trait. As an alternative, one could replace our second-stage parametric model with a weighted nonparametric linkage approach, for example using a variance components (VC) [12] or Haseman-Elston (HE) [13,14] model. In a VC analysis of intercepts (or slopes), one could add a subject-specific component to the variance based on the first-stage standard-error. In the HE approach, one could regress some function of the first stage intercepts for a pair of relatives (e.g., the squared difference in intercepts between sib pairs) on the proportion of alleles shared identical by descent at a marker locus. The delta method can be utilized to calculate the variance of the squared sib-pair difference as a function of the first-stage, subject-specific standard errors. The inverse of the variances for each sib pair could then be used as weights in the HE regression. The performance of weighted VC and HE linkage analysis, relative to each other and to unweighted analysis, is a topic for future research.
In conclusion, we have proposed a two-stage modelling approach to the genetic analysis of longitudinal data for a quantitative trait. Additional work is necessary to evaluate the method, including simulation studies and comparisons to other two-stage and joint-analysis approaches.
Acknowledgments
Acknowledgments
This work was supported by NIH grants ES-10421 and CA-52862.
Contributor Information
Conway Gee, Email: cgee@usc.edu.
John L Morrison, Email: jmorr@usc.edu.
Duncan C Thomas, Email: dthomas@usc.edu.
W James Gauderman, Email: jimg@usc.edu.
References
- Laird NM, Donnelly C, Ware JH. Longitudinal studies with continuous responses. Stat Methods Med Res. 1992;1:225–247. doi: 10.1177/096228029200100302. [DOI] [PubMed] [Google Scholar]
- Diggle PJ, Liang KY, Zeger SL. Analysis of Longitudinal Data. Oxford Clarendon Press. 1995.
- Zeger SL, Liang K, Albert PS. Models for longitudinal data: a generalized estimating equation approach. Biometrics. 1998;44:1049–1060. doi: 10.2307/2531734. [DOI] [PubMed] [Google Scholar]
- Levy D, DeStefano AL, Larson MG, O'Donnel CJ, Lifton RP, Gavras H, Cupples LA, Myers R. Evidence for a gene influencing blood pressure on chromosome 17. Hypertension. 2000;36:477–483. doi: 10.1161/01.hyp.36.4.477. [DOI] [PubMed] [Google Scholar]
- Elston RC, Stewart J. A general model for the genetic analysis of pedigree data. Hum Hered. 1971;21:523–542. doi: 10.1159/000152448. [DOI] [PubMed] [Google Scholar]
- Lange K, Elston RC. Extensions to pedigree analysis. I. Likelihood calculations for simple and complex pedigrees. Hum Hered. 1975;25:95–105. doi: 10.1159/000152714. [DOI] [PubMed] [Google Scholar]
- Rao S, Li L, Li X, Moser KL, Guo Z, Shen G, Cannata R, Zirzow E, Topol EJ, Wang Q. Genetic linkage analysis of longitudinal hypertension phenotypes using three summary measures. BMC Genet. 2003;4(Suppl 1):S24. doi: 10.1186/1471-2156-4-S1-S24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briollais L, Tzontcheva A, Bull S. Multilevel modeling for the analysis of longitudinal blood pressure data in the Framingham Heart Study pedigrees. BMC Genet. 2003;4(Suppl 1):S19. doi: 10.1186/1471-2156-4-S1-S19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer LJ, Scurrah KJ, Tobin M, Patel SR, Celedon JC, Burton PR, Weiss ST. Genome wide linkage analysis of longitudinal phenotypes using σ2A random effects (SSARs) fitted by Gibbs sampling. BMC Genet. 2003;4(Suppl 1):S12. doi: 10.1186/1471-2156-4-S1-S12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scurrah K, Tobin M, Burton P. Longitudinal variance components models for systolic blood pressure, fitted using Gibbs sampling. BMC Genet. 2003;4(Suppl 1):S25. doi: 10.1186/1471-2156-4-S1-S25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang T, Kraft P, Gauderman WJ, Thomas D. Multiple imputation methods for longitudinal blood pressure measurements from the Framingham Heart Study. BMC Genet. 2003;4(Suppl 1):S43. doi: 10.1186/1471-2156-4-S1-S43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almasy L, Blangero J. Multipoint quantitative-trait linkage analysis in general pedigrees. Am J Hum Genet. 1998;62:1198–1211. doi: 10.1086/301844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haseman JK, Elston RC. The investigation of linkage between a quantitative trait and a marker locus. Behav Genet. 1972;2:3–19. doi: 10.1007/BF01066731. [DOI] [PubMed] [Google Scholar]
- Elston RC, Buxbaum S, Jacobs KB, Olson JM. Haseman and Elston revisited. Genet Epidemiol. 2000;19:1–17. doi: 10.1002/1098-2272(200007)19:1<1::AID-GEPI1>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]


