Abstract
Olfaction is of considerable importance to many insects in behaviors critical for survival and reproduction, including location of food sources, selection of mates, recognition of colony con-specifics, and determination of oviposition sites. An ubiquitous, but poorly understood, component of the insect's olfactory system is a group of odorant-binding proteins (OBPs) that are present at high concentrations in the aqueous lymph surrounding the dendrites of olfactory receptor neurons. OBPs are believed to shuttle odorants from the environment to the underlying odorant receptors, for which they could potentially serve as odorant presenters. Here we show that the Drosophila genome carries 51 potential OBP genes, a number comparable to that of its odorant-receptor genes. We find that the majority (73%) of these OBP-like genes occur in clusters of as many as nine genes, in contrast to what has been observed for the Drosophila odorant-receptor genes. Two of the presumptive OBP gene clusters each carries an odorant-receptor gene. We also report an intriguing subfamily of 12 putative OBPs that share a unique C-terminal structure with three conserved cysteines and a conserved proline. Members of this subfamily have not previously been described for any insect. We have performed phylogenetic analyses of the OBP-related proteins in Drosophila as well as other insects, and we discuss the duplication and divergence of the genes for this large family.
[The sequence data from this study have been submitted to FlyBase. Annotations for these sequences are available as supplementary material at http://www.genome.org.]
Olfactory signal transduction has been well-studied and is generally similar in vertebrates, insects, crustaceans, and nematodes (Ache 1994; Hildebrand and Shepherd 1997; Prasad and Reed 1999). In all of these systems, odorant molecules are detected through interactions with specific G-protein-linked receptors present on the dendrites of olfactory receptor neurons. G-protein activation then produces a second-messenger cascade leading to ion channel activation and receptor neuron depolarization.
How is the olfactory system capable of perceiving and discriminating among a myriad of different airborne odorants? One possibility is that these odorants are recognized by a correspondingly large number of receptors. In fact, large numbers of different odorant-receptor genes are found in both mammals (∼1000 genes in mice and rats; Mombaerts 1999) and the roundworm Caenorhabditis elegans (∼800 genes; Bargmann 1998; Robertson 2000). In contrast, recent analyses of the Drosophila melanogaster genome revealed far fewer potential odorant-receptor genes: 60 genes of which only 43 are expressed in the antenna or maxillary palp (Clyne et al. 1999; Gao and Chess 1999; Vosshall et al. 1999, 2000; Vosshall 2001). A related family of 56 receptors is expressed primarily in gustatory neurons (Scott et al. 2001).
Why is the variety of odorant-receptor diversity in Drosophila more than an order of magnitude lower than it is in either mammals or C. elegans? Perhaps odorant receptors are not the only molecules involved in odorant recognition by insects. One attractive possibility is that another class of molecules, the odorant-binding proteins (OBPs), contributes substantially to the recognition of odorants in insects. OBPs are small, soluble proteins present at high levels in the fluid surrounding olfactory-receptor neurons (Pelosi 1994). They are generally thought to solubilize hydrophobic odorants and shuttle them to the underlying receptors (Vogt et al. 1991; Pelosi 1994; Prestwich et al. 1995). However, they could potentially function in odorant recognition, perhaps by presentation of the odorant molecule to the underlying receptor (Pelosi 1994; Prestwich et al. 1995).
In fact, there is increasing evidence that OBPs do play an active role in odorant recognition rather than merely serving as passive odorant shuttles. One line of evidence is the large number of OBPs present within a variety of insect species. For example, five OBPs have been described in the moth Antheraea pernyi (Breer et al. 1990; Raming et al. 1990; Krieger et al. 1991, 1997). Several studies have shown that the different OBPs found within a single insect species display distinct odorant-binding specificities (Du and Prestwich 1995; Prestwich et al. 1995; Maïbeche-Coisne et al. 1997; Plettner et al. 2000). Furthermore, Drosophila that lack the “LUSH” OBP show specific deficits in response to the odorants ethanol or benzaldehyde (Kim et al. 1998; Wang et al. 2001). Also, different OBPs show differential expression patterns in distinct subsets of the olfactory sensory hairs (sensilla) on an insect's antenna (Steinbrecht et al. 1995; Steinbrecht 1996; Park et al. 2000). Each sensillum carries a limited number of olfactory receptor neurons that are exposed only to OBPs present within that particular sensillum. If OBPs and odorant receptors are expressed within different, but overlapping subsets of sensilla, the result would be a mosaic of sensilla with different odorant thresholds. Thus, a moderate number of OBPs could act in a combinatorial manner with a moderate number of odorant receptors to greatly increase the discriminating power of an insect's olfactory system.
This combinatorial strategy does not appear to be the case for mammals. Odorant discrimination appears to be largely due to the diversity of olfactory receptors (∼1000; Mombaerts 1999) because only one or a few OBPs are present in the mammalian olfactory mucosa (Tegoni et al. 2000), and they show fairly broad odorant specificities (Löbel et al. 2002). C. elegans also resembles the mammalian system with a large olfactory receptor population (∼800; Bargmann 1998; Robertson 2000). In the case of C. elegans, no OBP has been described (Rubin et al. 2000). Hence, we have two seemingly contrasting situations: Some organisms (mammals and nematodes) have large numbers of olfactory receptors and few or no OBPs, whereas insects have a moderate number of receptors coupled with a moderate number of OBPs.
Exactly how many OBPs are there in insects, and how are their genes organized? In this study, we provide a comprehensive examination of OBP-like genes in Drosophila. We find that the Drosophila genome carries 51 potential OBP genes, a number comparable to that of its odorant-receptor genes (Clyne et al. 1999; Gao and Chess 1999; Vosshall et al. 1999, 2000; Vosshall 2001). We find that the majority (73%) of OBP-like genes occur in clusters of four to nine genes; two of these presumptive OBP gene clusters also include an odorant-receptor gene. Our analysis also reveals an apparently monophyletic subfamily of OBP-like proteins whose 12 members have a conserved C terminus.
RESULTS
The Drosophila Genome Carries 51 OBP-Gene-Family Members
The Drosophila genome search used here identified 51 members of the odorant-binding protein (OBP) gene family (Table 1). This included the seven previously identified Drosophila OBP genes: the PBPRP (pheromone-binding protein related protein) genes, the OS (olfactory-specific) genes, and LUSH (McKenna et al. 1994; Pikielny et al. 1994; Kim et al. 1998). Also identified in our search were 44 additional OBP-like sequences, of which 28 had been noted previously (FlyBase 1999; Robertson et al. 1999; Rubin et al. 2000; Galindo and Smith 2001) and 16 were not previously recognized as potential OBPs. Of the 35 CG (computational gene) sequences (FlyBase 1999), 13 were correctly annotated to give an OBP-like product, and the remaining 22 required a different pattern of splicing to produce the OBP-like product. Three sequences were previously unrecognized.
Table 1.
Drosophila Odorant-Binding Protein Genes
| New name | Previous name(s) | Position | Expression |
|---|---|---|---|
| Obp8a | CG12665.m | 8D34 | |
| Obp18a | CG15883.m, OBP18a.m | 18B1 | |
| Obp19a | CG11748.m, OBP19a | 19D2 | A4 |
| Obp19b | CG1670.m, clot #2470, OBP19b.m | 19D2 | O4 |
| Obp19c | CG15457, OBP19c | 19D2 | P, L4 |
| Obp19d | PBPRP-2, OBP19d | 19D2 | A, M, P14 |
| Obp28a | PBPRP-5, OBP28a | 28A1 | A, L14 |
| Obp44a | CG2297, clot #1214 | 44A8 | |
| Obp46a | CG12905.m | 46F11 | |
| Obp47a | CG12944.m, OBP47a.m | 47C6 | |
| Obp47b | CG13208 | 47E5 | |
| Obp49a | CG8769.m | 49B9 | |
| Obp50a | 50F6 | ||
| Obp50b | CG13940.m | 50F6 | |
| Obp50c | 50F6 | ||
| Obp50d | 50F6 | N5 | |
| Obp50e | CG13939.m | 50F6 | |
| Obp51a | OBP51a | 51D10-11 | |
| Obp56a | CG11797, clot #14015, OBP56a | 56E4 | L4 |
| Obp56b | CG15129.m2, OBP56b | 56E4 | P, L4 |
| Obp56c | CG15129.m1, OBP56c.m | 56E4 | A, O, L4 |
| Obp56d | CG11218.m, clot #1175, OBP56d.m | 56E4 | A, M, O, L4, P5 |
| Obp56e | CG8462, clot #16275, OBP56e | 56E5 | A, P4 |
| Obp56f | OBP56f | 56E5 | |
| Obp56g | CG13873.m, OBP56g | 56F1 | P, L4 |
| Obp56h | CG13874.m, OBP56h | 56F2 | A, P, L4 |
| Obp56i | OBP56i | 56F2 | |
| Obp57a | CG13421, GH01026, OBP57c | 57A6 | A, M, P, L4 |
| Obp57b | OBP57b | 57A6 | A, M, O4 |
| Obp57c | OBP57a | 57A6-7 | A, M4 |
| Obp57d | CG13429.m, OBP57e | 57A8 | O4 |
| Obp57e | OBP57d | 57A8 | O4 |
| Obp58a | CG13517 | 58F6 | |
| Obp58b | CG13518.m | 58F6 | N5 |
| Obp58c | CG13524 | 58F6 | N5 |
| Obp58d | CG13519 | 58F6-7 | |
| Obp69a | PBPRP-1, OBP69a | 69B2 | A1 |
| Obp76a | LUSH, OBP76c | 76B9-C1 | A, L4 |
| Obp83a | OS-F, PBPRP-3, OBP83a | 83C8-D1 | A12 |
| Obp83b | OS-E, OBP83b | 83D1 | A2 |
| Obp83c | CG15582.m1, OBP83c | 83D4 | P4 |
| Obp83d | CG15582.m2, OBP83d.m | 83D4 | |
| Obp83e | CG15583.m2, OBP83f.m | 83D4 | |
| Obp83f | CG15583.m1, OBP83g.m | 83D4 | |
| Obp84a | PBPRP-4, OBP84a | 84C7 | A1 |
| Obp85a | CG11732.m | 85A1 | |
| Obp93a | CG17284 | 93C1 | |
| Obp99a | CG18111.m, clot#14334, OBP99a | 99B9 | L4 |
| Obp99b | CG7584m clot #1575 | 99B10 | |
| Obp99c | CG15505 | 99B11 | |
| Obp99d | CG7592, clot #11054, OBP99b | 99C1 | A, M4 |
This table lists all 51 odorant-binding protein genes described in this paper along with their cytological positions and previous names where applicable. Previous names are: PBPRP (pheromone-binding protein related protein gene, Pikielny et al. 1994), OS (olfactory-specific gene, McKenna et al. 1994), LUSH (Kim et al. 1998), CG (computational gene, Flybase 1999), and OBP (odorant-binding protein gene, Galindo and Smith 2001). We have modified annotations to some of the CG and OBP genes as indicated by the suffix “.m.” Clot # is the Berkeley Drosophila Genome Project designation for a set of homologous Drosophila expressed sequence tags (dESTs) likely to come from the same gene. Many of the Drosophila Obp genes are expressed in one or more chemosensory organs: antenna (A), maxillary palp (M), proboscis (P), other adult chemosensory (O), or larval chemosensory organ (L). N indicates that no expression in adult heads (i.e., A, M, or P) was seen. Expression data are from: 1Pikielny et al. 1994, 2McKenna et al. 1994, 3Kim et al. 1998, 4Galindo and Smith 2001, and/or 5this study.
Because the number of OBP genes is large and because family members have previously been known by several conflicting names, we are proposing a single new nomenclature. The nomenclature is analogous to the one used for the large family of Drosophila odorant-receptor genes (Drosophila Odorant Receptor Nomenclature Committee 2000). Here, we use the preface “Obp” to reflect the fact that a gene is a member of the family of OBP-like genes. This is followed by a number conveying the gene's cytogenetic location. In cases where there is only one OBP gene within a given numbered region, it is appended with the letter “a.” In cases where there are multiple OBP genes within a single numbered region, each gene is appended with a letter that conveys its relative position on the cytogenetic map. Thus, the new name Obp56d refers to the fourth OBP-like sequence in cytological region 56. A similar nomenclature has been proposed by Galindo and Smith (2001).
Expression data for approximately half of the Drosophila Obp genes is available (Table 1) and indicate that these genes are expressed in olfactory and/or gustatory tissues (McKenna et al. 1994; Pikielny et al. 1994; Kim et al. 1998; Galindo and Smith 2001). Additional data on the gene products will be required to determine which of these 51 Obp genes encode bona fide OBPs (Steinbrecht et al. 1992, 1995; Du and Prestwich 1995; Ozaki et al. 1995; Hekmat-Scafe et al. 1997; Kim et al. 1998; Park et al. 2000; Plettner et al. 2000).
Most Obp Genes Are Clustered Within the Drosophila Genome
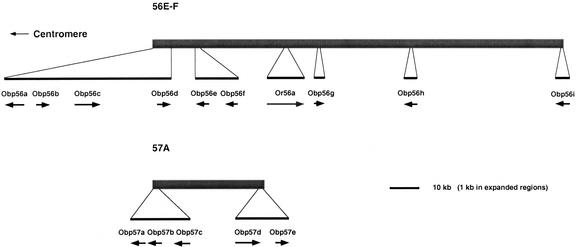
The Obp genes are dispersed throughout the genome, although a disproportionate number (29/51) are located on Chromosome 2R, which has several clusters of Obp genes (Table 1). Of the 51 Obp genes, 37 are organized into seven clusters of four or more Obp genes. A striking example is the nine Obp genes organized into a cluster located in chromosomal region 56E-F (Fig. 1). Five related Obp genes are present in a nearby cluster at 57A (Fig. 1). The genes are not present in tandem arrays; rather they occur in both orientations, indicating they were formed by a complex series of duplication and rearrangement events.
Figure 1.
Organization of two Drosophila odorant-binding protein gene clusters in chromosomal regions 56E-F and 57A. The upper panel depicts a cluster of nine Obp genes and one odorant receptor (Or) gene located in chromosomal region 56E-F. The length of each arrow corresponds to the length of the coding sequence, with arrowheads indicating direction of transcription. Note that Or56a is located near the center of the Obp56 gene cluster. There are also two other nonolfactory genes in this region that have been omitted for clarity. (A predicted chaperone protein gene lies between Obp56f and Or56a, and a predicted cell-adhesion protein gene lies between Obp56h and Obp56i.) The lower panel depicts a cluster of five Obp genes located in chromosomal region 57A. For clarity, six other nonolfactory genes in this region (located between Obp57c and Obp57d) have been omitted.
Two clusters of Obp genes each contain an odorant-receptor gene. The odorant-receptor gene Or56a (Vosshall et al. 2000) is located within the Obp56 cluster between Obp56f and Obp56g (Fig. 1). Or83a (Vosshall et al. 2000) is located within the Obp83 cluster between Obp83b and Obp83c (data not shown). The significance of this clustering is at present unclear but may indicate a functional linkage between the clustered Or and Obp genes.
Obp genes present in the same genomic cluster generally show different patterns of expression in chemosensory organs (Table 1). Perhaps not unexpectedly, the 5′-flanking regions of these clustered Obp genes share few repeated motifs that might serve as binding sites for common regulatory elements. One possible exception would be Obp83a and Obp83b, which are coexpressed in olfactory sensilla (Hekmat-Scafe et al. 1997) and which share a few repeated motifs in their 5′-flanking regions. The sequence GTGTC/TTCTA is present twice in the 1000 kb of DNA upstream of Obp83b and once upstream of Obp83a; and the sequence GAAGCGCA/CAATTGG is present once upstream of both genes. More tenuous possibilities include the sequences AGTTCCAGCT/GGG (present once upstream of both Obp19b and Obp19d) and GAACTTTA/TAAC (present once upstream of both Obp56d and Obp56e). None of these repeated motifs constitutes a known transcription factor binding site in Drosophila.
The Drosophila Obp Genes Encode a Diverse Family of Proteins
An alignment of the deduced Drosophila Obps shows a diverse family of proteins (13.4–28 kD) that display several notable conserved features (Fig. 2). Each of the Obps has a hydrophobic N terminus that could serve as a signal sequence (von Heijne 1986). The overall pairwise sequence identity is modest (median identity=20.4%); the N termini are particularly divergent. The Drosophila Obp genes carry 0–3 introns, the majority of which are located in one of nine conserved positions (Fig. 2). In the preponderance of cases (86%), introns occur precisely between codons.
Figure 2.
Amino acid alignment of the various Drosophila OBP family members. The alignment was produced using CLUSTAL X; the order is identical to that depicted in the phylogenetic tree (Fig. 3). The program BOXSHADE (K. Hofmann and M.D. Boron, unpubl.; available at http://ludwig-sunl.unil.ch:8080/software/Box_form. html) was used with a threshold of 0.5 to identify those residues either identical or similar to a consensus (shown as white on black background or black on gray background, respectively). Six cysteine residues (dark arrowheads) are highly conserved; C3, C4, and C6 (asterisks) are absolutely conserved. Dark boxes indicate conserved cysteine residues; open boxes indicate other notable cysteine conservations. Open arrowheads indicate highly conserved positions where introns are inserted (cf. Fig. 3): an arrowhead between two amino acids signifies that the intron occurs between the two corresponding codons, whereas an arrowhead over a particular amino acid signifies that the intron occurs within its codon. Bombyx mori pheromone-binding protein (B. mor PBP, GenBank accession no. X94987) was shown by X-ray crystallography to carry seven α-helices (Sandler et al. 2000), indicated here by horizontal boxes (α1a–α6). Obp83e, which is composed of two tandem, in-frame OBP-like sequences, was omitted from the alignment. To simplify the presentation of the figure, four noninformative stretches present in single OBPs were omitted, as indicated by horizontal lines: At the N terminus, Obp56c contains the sequence [MYFRASLMALLC]-, and at the C terminus, Obp50b contains the sequence -[NGCGHIDSEGSNLLMACFLTLMIAKFISDH]. Between C1 and C2, Obp56c contains the sequence QSIGD-[VNNLGDLDFNGNSQMPYL]-DLKH, and Obp58a contains the sequence EDRG-[RGGQGRQGNGY]-EYGY, where the omitted sequence is contained within the brackets.
The most striking conservation is six cysteine residues that are present in characteristic positions in all known insect OBPs (Pelosi and Maida 1995) and that are conserved in the Drosophila sequences described here (Fig. 2). In the pheromone-binding protein (PBP) of the moth Bombyx mori, the conserved cysteine residues each contribute to an α-helical structure (Sandler et al. 2000). In addition to the conserved cysteines, the Drosophila Obps show modest sequence similarity, principally in the regions corresponding to five of the α-helices, α2–α6 (Fig. 2). The most extensive sequence similarity occurs in the region extending from α3 to α4 and includes a number of residues that correspond to ones in the hydrophobic odorant-binding pocket of B. mori PBP (Sandler et al. 2000). The spacing of conserved cysteines in the Drosophila Obps is: (X22–68–C1–X25–68–C2–X3–C3–X31–46–C4–X8–29–C5–X8–9–C6–X5–71), in which Xp stands for any p amino acids as described in Pikielny et al. (1994). All six cysteine residues are present at conserved positions in 44 of the Drosophila Obps. Three of the Drosophila Obps (Obp44a, Obp99b, and Obp8a) are missing C2 and C5, which are thought to form a disulfide (Sandler et al. 2000). Two of the Obps (Obp19c and Obp84a) are missing C1, but each has a conserved cysteine 15–16 residues away that might act as an alternative. One Obp (Obp99c) lacks C2, C5, and C1, but contains the alternate C1.
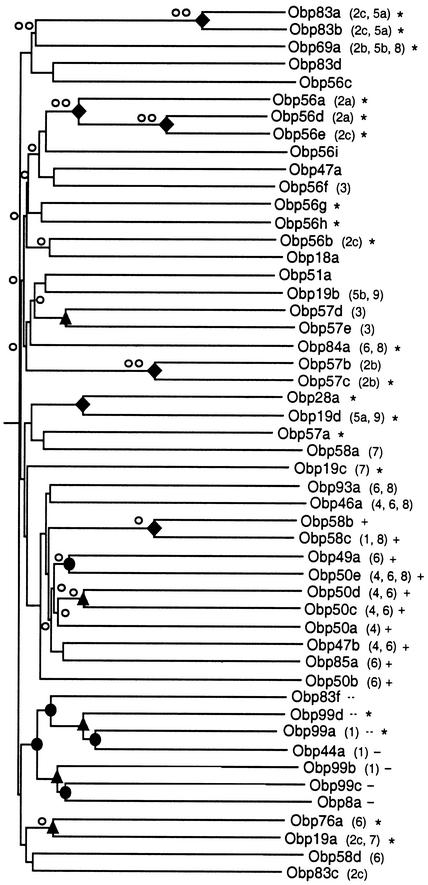
A phylogenetic tree based on neighbor-joining is shown for the Drosophila Obps in Figure 3. There is good bootstrap support for many terminal relationships and for one subfamily that we call the Minus-C subfamily. There is little bootstrap support for the other subfamilies, relationships between subfamilies, and the overall tree architecture. However, subfamily groupings are generally supported by genomic clustering and/or common intron insertion sites of the corresponding Obp genes. The conserved cysteines contribute little to the overall tree architecture, which is essentially the same when these residues are eliminated from the alignment (data not shown). In general, neither did masking of gaps within the alignment alter the predicted subfamily groupings; we have noted exceptions to this rule below. The average pairwise ratio of nonsynonymous to synonymous substitutions (dn/ds) for sequences under each of the nodes was ≤1. The observation that dn/ds values for the Obp sequences under the various nodes of the Drosophila phylogenetic tree are ≤1 (Fig. 3) is consistent with the notion of purifying selection.
Figure 3.
Phylogenetic comparison of Drosophila OBP family members. Depicted is an unrooted distance (neighbor-joining) tree that represents the relatedness of the various Drosophila OBP family members. The tree was constructed using an alignment of Drosophila OBPs that was identical to the one shown in Figure 2 with the divergent signal sequence-bearing N terminus removed from each OBP sequence. Each truncated protein starts at the same relative position at the beginning of helix α1a (i.e., ENY for Obp83a; cf. Fig. 2). Conserved intron insertion sites in the corresponding genes (cf. Fig. 2) are given in parentheses. Branch lengths are proportional and reflect the mean distance (percentage difference) between the various sequences. Symbols indicate nodes whose bootstrap values are at least 90% (♦), 70% (▴), or 50% (●). Nodes whose mean dn/ds ratios are <0.25 are indicated by two open circles (○○) and those between 0.5 and 0.25 by a single open circle (○). Plus-C OBPs (+) and Minus-C OBPs lacking C2 and C5 (−) or carrying all six conserved cysteines (…) are identified. Each OBP for which olfactory system expression has been shown (cf. Table 1) is indicated by an asterisk.
We have named one Obp subfamily Minus-C because some of its members do not contain all six conserved cysteine residues. The Minus-C subfamily has seven members: Obp8a, Obp44a, Obp83f, Obp99a, Obp99b, Obp99c, and Obp99d (Fig. 3). The average pairwise sequence identity for these seven Obps is 20.2%. The products of all four Obp genes located in cytogenetic region 99B-C are contained in the Minus-C subfamily, as are three additional Obps encoded by Obp genes in scattered locations. The Obp83f gene is located in the large 83C-D cluster, but Obp83f is more closely related to the Obps whose genes are found in cytogenetic region 99B-C. Three members of the Minus-C subfamily (Obp83f, Obp99a, and Obp99d) carry all six conserved cysteines, whereas four members of the subfamily (Obp8a, Obp44a, Obp99b, and Obp99c) are missing C2 and C5 (Fig. 2). The two most closely related subfamily members are Obp99a and Obp44a. One of the two is encoded by a gene located in the 99B-C cluster, whereas the other is not; one of the two (Obp99a) has all six conserved cysteines, whereas the other (Obp44a) lacks C2 and C5 (Figs. 2 and 3). Both Obp99a and Obp44a have an intron inserted at site 1, as does Obp99b (Fig. 3).
We have named one subfamily Plus-C because its members carry more than six conserved cysteine residues. This subfamily contains a total of 12 members, which share on average 17.4% identity. The Plus-C Obps are encoded by the products of all five Obp genes in the cluster at cytogenetic region 50F (Obp50a, Obp50b, Obp50c, Obp50d, and Obp50e), two Obp genes in the 58F cluster (Obp58b and Obp58c), and five Obp genes in scattered locations (Obp46a, Obp47b, Obp49a, Obp85a, and Obp93a). All 12 Plus-C subfamily members show the six conserved cysteine residues, C1–C6, as described above. All 12 also carry an additional three conserved cysteines and a conserved proline located downstream of C6 (C6a, C6b, and C6c) with the spacing: C6–X8–C6a–P–X10–11–C6b–X9–C6c–X3–50 (Fig. 2). Ten of the 12 have an additional three conserved cysteine residues that cluster around C1 (C1a, C1b, and C1c) with the spacing: X21–27–C1a–X11–13–C1–C1b–X11–13–C1c (Fig. 2). In all of the Plus-C subfamily members, C5 and C6 are separated by nine residues; they are separated by eight residues in all of the non-Plus-C Drosophila Obps with the sole exception of Obp58d. A phylogenetic tree of Drosophila Obps constructed after masking all gaps in the CLUSTAL X alignment actually groups Obp58d with the Plus-C Obps, albeit with extremely limited bootstrap support (data not shown). Obp93a is the most divergent member of the Plus-C subfamily, and it is not grouped with the other 11 Plus-C subfamily members in a phylogenetic tree constructed from an alignment with gaps masked (data not shown). However, the Obp93a gene does have two intron insertion sites found in genes encoding many of the other subfamily members (Fig. 3). Almost all of the Plus-C Obp genes have introns at sites 4, 6, and/or 8; none of the other Drosophila Obp genes has an intron at site 4 (Fig. 3). Our analysis reveals a preponderance of synonymous codon substitutions throughout much of the Plus-C Obps, consistent with the notion of negative (purifying) selection. We analyzed the pairwise dn/ds ratios for four exon segments: (1) N terminus–intron site 4, (2) site 4–site 6, (3) site 6–site 8, and (4) site 8–C terminus (cf. Fig. 2), and found the corresponding mean dn/ds ratios to be 0.71, 0.55, 0.47, and 0.57. This indicates that exon 3, and to lesser extents exons 2 and 4, are under strong purifying selection. The region of B. mori PBP that corresponds to exon 3 forms much of its hydrophobic odorant-binding core (Sandler et al. 2000).
Three Obps lack C1 (Obp19c, Obp84a/PBPRP-2, and Obp99c). All three contain a cysteine at the position of C1c, which may serve as an alternate C1 (Fig. 2). These three Obps do not constitute clades within a cluster. Rather, each of these three Obps shares more sequence similarity with other Drosophila Obps than with any of those carrying the C1c alternate (Fig. 3). Two of the alternative C1 Obps (Obp84a/PBPRP-4 and Obp19c) are known to be expressed in the olfactory system (Pikielny et al. 1994; Galindo and Smith 2001).
Drosophila Obps Within the Insect OBP Family
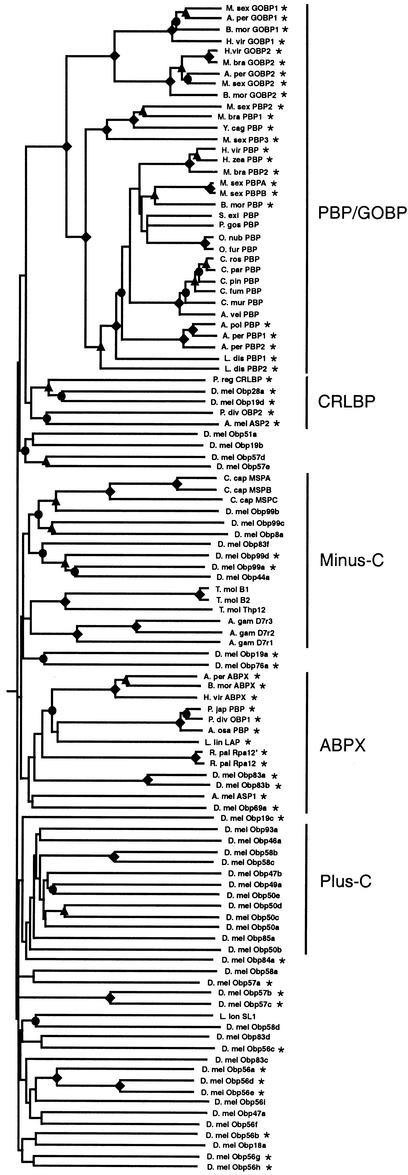
The Drosophila Obps were also examined in the broader context of the insect OBP family. A distance (neighbor-joining) tree represents a hypothesis of relationship among the various insect OBPs and OBP-like proteins (Fig. 4). Generally, there is bootstrap support for terminal relationships and several subfamilies, whereas there is little support for the greater tree architecture. We have noted several other possible subfamilies to facilitate their description.
Figure 4.
Phylogenetic tree of the family of insect OBPs and related proteins. Sequences were aligned stepwise using the CLUSTAL X package. The proteins were then truncated by removing the signal sequence-bearing N termini as in Figure 3. This alignment was then used to construct the unrooted distance (neighbor-joining) tree shown. Branch lengths are proportional and reflect the mean distance (percentage difference) between the various sequences. The symbols for bootstrap values are as shown in Figure 3. Proteins with known olfactory system expression are indicated by asterisks. GenBank accession nos. of the non-Drosophila insect protein sequences are as follows: T. mol B1 (Tenebrio molitor B1, M97916); T. mol B2, M97917; T. molThp12, 6573610; G. mel SER, (Galleria mellonella sericotropin, L41640); P. reg CRLBP, Phormia regina CRLBP, S78710; A. per PBP1, (Antheraea pernyi PBP1, X96773); A. pol PBP (Antheraea polyphemus PBP, X17559; A. per PBP2, X96860); M. sex PBPA (Manduca sexta PBPA, M21797; M. sex PBPB, M21798); B. mor PBP (Bombyx mori PBP, X94987); H. vir PBP (Heliothis virescens PBP, X96861); L. dis PBP2 (Lymantria dispar PBP2, 2444187); A. per GOBP2, X96772); M. sex GOBP2, M73798; H. vir GOBP2, 96863; B. mor GOBP2, X94989; B. mor GOBP1, X94988; M. sex GOBP1, M73797; H. vir GOBP1, X96862; B. mor ABPX, X94990, H. zea PBP (Helicoverpa zea PBP, 3639083); L. lin LAP (Lygus lineolaris, 3644030); L. dis PBP1, 2444185; H. vir ABPX, AJ002518); P. japPBP (Popillia japonica PBP, 3721994); A. osa PBP (Anomala osakana PBP, 3721996); A. per ABPX, AJ002519; M. bra GOBP2 (Mamestra brassicae GOBP2, 2961244); R. pal Rpa12′ (Rhynchophorus palmarum Rpa12‘, AF141865); R.palRpa12, AF139912; M. bra PBP2, 2961240; C. cap MSP (Ceratitis capitata MSP, Y08954); M. bra PBP1, 2961242; A. gam D7r1 (Anopheles gambiae D7r1, AJ133852); A. gam D7r3, AJ000035; A. gam D7r2, AJ000036; L. lon SL1 (Lutzomyia longipalpias SL1, AF132517); A. mel ASP1 Apis mellifera ASP1, AF166496; A. mel ASP2, AF166497; Y. cag PBP (Yponomeuta cagnagellus PBP, AF177661); P. gos PBP (Pectinophora gossypiella PBP, AF177656); C. ros PBP (Choristoneura rosaceana PBP, AF177654); A. vel PBP (Argyrotaenia velutinana PBP, AF177639); S. exi PBP (Synanthedon exitiosa PBP, AF177660); O. nub PBP (Ostrinia nubilalis PBP, AF133631); C. pin PBP (Choristoneura pinus PBP, AF177650); C. mur PBP (Choristoneura murinana PBP, AF177645; C. par PBP (Choristoneura parallela PBP, AF177648); P. div OBP1 (Phyllopertha diversa PBP, PC7028; P. div OBP2, PC7029.
Two insect OBP subfamilies appear monophyletic. A large subfamily of general odorant-binding proteins (GOBPs) and pheromone-binding proteins (PBPs) appears specific to Lepidoptera (Fig. 4). The Plus-C Drosophila subfamily also appears to be monophyletic. The 12 members of this subfamily are the only insect OBPs described thus far with additional conserved cysteines (C1a, C1b, C1c, C6a, C6b, and C6c).
We have named one subfamily the ABPX subfamily (Fig. 4) because it includes a group of moth antennal binding proteins referred to as ABPXs (Krieger et al. 1996, 1997). The 13 members of this subfamily share an average of 30.8% amino acid identity. The ABPX subfamily includes three Drosophila Obps (Obp83a, Obp83b, and Obp69a), each of which is encoded by a gene that has introns inserted at both sites 2 and 5 (Fig. 3). The ABPX subfamily also includes three beetle OBPs, the queen pheromone-binding protein from the honey bee Apis mellifera, and an OBP from the “true bug” Lygus lineolaris (Wojtasek et al. 1998, 1999; Danty et al. 1999; Vogt et al. 1999). We have named another OBP subfamily the CRLBP family (Fig. 4) because it includes an OBP called CRLBP (chemical-sense-related lipophilic-ligand-binding protein) from the fly Phormia regina (P. reg CRLBP; Ozaki et al. 1995). The five CRLBP OBPs share an average of 22% identity. The CRLBP subfamily is polyphyletic and includes two Drosophila Obps (Obp19d and Obp28a) along with OBPs from the beetle Phyllopertha diversa (P. div OBP2) and the honey bee A. mellifera (A. mel ASP2; Danty et al. 1999; Wojtasek et al. 1999). The sand fly Lutzomyia longipalpis salivary protein SL1 (Charlab et al. 1999) clusters with a group of Drosophila OBPs, including most of those encoded by the 14 Obp genes in cytological region 56E-57A.
The Minus-C subfamily of Drosophila OBPs is part of a larger subfamily of insect OBPs that have been identified in the Mediterranean fruit fly (Ceratitis capitata), beetle (Tenebrio molitor), and mosquito (Anopheles gambiae). Three of the seven Drosophila members of this subfamily (Obp99a, Obp99d, and Obp83f) carry all six conserved cysteines, and two of these (Obp99a and Obp99d) are expressed in the olfactory system (Fig. 4). All of the non-Drosophila subfamily members described thus far lack conserved cysteines C2 and C5 and are nonolfactory OBP-like proteins (Kodrik et al. 1995; Paesen and Happ 1995; Thymianou et al. 1998; Arca et al. 1999; Graham et al. 2001). Some of the Drosophila Minus-C OBPs may also have adapted to a nonolfactory function. It is conceivable that Obp99b, Obp99c, and/or Obp44a represent unprocessed pseudogenes. However, we consider this unlikely as true pseudogenes are quite rare in Drosophila, with only ∼100 present in the entire genome (Harrison et al. 2002), and there are no obvious disabling mutations in any of the open-reading frames.
Many of the insect OBP family members are expressed in the olfactory system, as would be expected for bona fide OBPs (Fig. 4). These include representatives of four of the five insect OBP subfamilies (PBP/GOBP, CRLBP, Minus-C, and ABPX), as well as members of the 56E-F and 57A clusters. The two OBPs most related to the Plus-C subfamily (Obp19c and Obp84a/PBPRP-4) also show olfactory system expression (Fig. 4). On the other hand, our in situ analysis of three different Plus-C family members (Obp50d, Obp58b, and Obp58c) consistently revealed no expression in Drosophila heads (i.e., antennae, maxillary palps, and proboscis; data not shown). It is possible that these OBPs are expressed in the larval olfactory organ and/or in an adult chemosensory organ (i.e., chemosensory bristles on the leg or wing) that is less amenable to visualization by in situ hybridization. It is also possible that the Plus-C OBPs have adapted to serve a nonolfactory function. Nonetheless, we believe that the 12 Plus-C genes do, indeed, encode members of the same family as the other Drosophila Obps based on their shared sequence similarity, intron insertion sites, and presence within Obp gene clusters. A number of the Plus-C genes carry an intron at site 6 and/or 8, as do several other Obp genes with known olfactory system expression: Obp69a/PBPRP-1, Obp84a/PBPRP-4, and Obp76a/LUSH (Fig. 3). Furthermore, two of the Plus-C Obp genes (Obp58b and Obp58c) are members of a cluster of four OBP-like genes at cytological region 58F.
DISCUSSION
An analysis of the paralogous members of large gene families can provide considerable insight into a genome's evolutionary dynamics. In this paper we describe the results of our genome-wide analysis of the Obp gene family in Drosophila melanogaster. We show that the Drosophila genome carries 51 potential Obp genes. The majority of the Obp genes occur in clusters, two of which also include an odorant-receptor gene. Phylogenetic analysis of the family provides evidence for a series of ancient and complex gene duplication events. Finally, we describe an apparently monophyletic Drosophila OBP subfamily, whose 12 members have conserved C termini.
The Drosophila Obp Gene Family Is Composed of 51 Members, Most of Which Occur in Gene Clusters
We have identified 51 Drosophila genes likely to encode this insect's entire repertoire of OBPs and related proteins. This number is significantly greater than the 14 OBP-like genes found in the original annotation of the Drosophila genome (Rubin et al. 2000) and includes 18 genes not discerned in a recent TBLASTN search for OBP-like proteins in the Drosophila genome (Galindo and Smith 2001). The larger number of Obp genes identified by our search is primarily a reflection of our combination of a PSI-BLAST search for protein family members, TBLASTN search to identify additional unannotated family members, and a careful examination of the conceptually translated protein sequences for OBP features (an N-terminal signal sequence, small size, and landmark cysteine residues) to identify more plausible alternative splicing patterns. It has been estimated that approximately half of the computer-annotated “computational genes” in the Drosophila genome have been incorrectly spliced (Reese et al. 2000; Karlin et al. 2001). Twelve of the additional genes in our set of 51 encode the Plus-C subfamily members. Four others (Obp8a, Obp44a, Obp99b, and Obp99d) encode atypical OBP family members that lack the second and fifth of six conserved cysteines. Although counterparts of such OBP-like proteins in other insect species have been implicated in nonolfactory functions (Kodrik et al. 1995; Paesen and Happ 1995; Thymianou et al. 1998; Arca et al. 1999; Graham et al. 2001), we believe that these four Drosophila genes should be annotated as Obp genes based on sequence similarity (Obp44a) and conservation of an intron insertion site or (Obp99b and Obp99c) presence within the same genomic cluster as canonical Obp genes that do show olfactory system expression (Figs. 3 and 5).
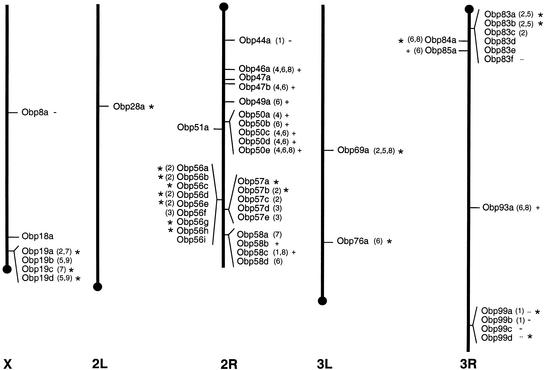
Figure 5.
Genomic locations of the 51 potential Drosophila odorant-binding protein genes. The location of each of the 51 Obp genes is indicated by a horizontal line. Chromosomal arms are scaled as in Rubin et al. (2000). Conserved intron insertion sites (cf. Fig. 2) are given in parentheses; for clarity of presentation, “2” is used for 2a-c and “5” for 5a-b. Plus-C and Minus-C subfamily members are identified as in Figure 3. Olfactory system expression (cf. Table 1) is indicated by an asterisk.
The majority of Drosophila Obp genes occur in clusters: 37 of the 51 Obp genes (73%) are located near at least three other Obp genes (Table 1). The Obp genes are dispersed throughout the genome, although a disproportionate number (30/51) are located on the second chromosome, which contains several of the larger Obp gene clusters (Fig. 5). The clustered Obp genes occur in both orientations, indicating a complex series of duplication and divergence events. A cluster of nine Obp genes located in chromosomal region 56E-F (Fig. 1) is one of the largest gene clusters found in Drosophila; the only larger clusters are one of 18 tetraspanin genes, another of 17 genes of unknown function, and two that each contain 10 glutathione S-transferase genes (Rubin et al. 2000; Todres et al. 2000).
Two of the Obp gene clusters also contain an odorant receptor (Or) gene. The cluster of nine Obp genes at chromosomal region 56E-F contains Or56a (Fig. 1), and the cluster of six Obp genes at 83C-D contains Or83c. Interestingly, similar expression patterns have been observed for Obp83a, Obp83b, and Or83c: All are found in sensory hairs on the ventro-lateral aspect of the antenna (McKenna et al. 1994; Pikielny et al. 1994; Hekmat-Scafe et al. 1997; Vosshall et al. 2000). On the other hand, the expression pattern of Or56a differs from those of the Obp56 genes (Vosshall et al. 2000; Galindo and Smith 2001).
A Monophyletic Drosophila Subfamily of OBPs With Conserved N and C Termini
The 12 Drosophila OBPs in the Plus-C subfamily share a conserved C-terminal structure with three conserved cysteines downstream of C6 (C6a, C6b, and C6c); 10 of the 12 also have a conserved N-terminal structure with three conserved cysteines that cluster around C1 (C1a, C1b, and C1c). These conserved N and C termini likely have functional significance as the corresponding regions of B. mori PBP (Fig. 2) are precisely the ones that differ between the liganded and unliganded PBP structures (Sandler et al. 2000; Horst et al. 2001), and which consequently may mediate odorant release. The B. mori pheromone bombykal binds to the PBP's hydrophobic pocket formed primarily by four of the six α-helices: α3, α4, α5, and α6 (Sandler et al. 2000). Whereas in the pheromone-liganded form of B. mori PBP the N-terminal helix α1a is part of a lid covering the pheromone-binding cavity and the C terminus is in an extended conformation on the PBP surface, in the unliganded PBP the N-terminal helix α1a is flexibly disordered (open lid) and the C terminus forms an additional α-helix that serves as a plug, occupying the PBP's pheromone-binding pocket (Horst et al. 2001). The significance of the six additional conserved cysteines in the specialized termini of the Plus-C subfamily members is unclear, but one possibility is that they serve to stabilize the unliganded form of the OBP through disulfide bonding. However, detailed structural information is not yet available for any Plus-C OBP, and consequently it is not known whether the conserved N and C termini mediate odorant binding and/or release. It is also possible that the Plus-C OBPs have adapted to transport hydrophobic ligands other than odorants.
Phylogenetic Analysis of the Insect OBP Family
Our analysis revealed that the insect OBP family is, indeed, specific to insects, consistent with previous observations (Rubin et al. 2000). Vertebrate OBPs, which are members of the lipocalin family of carrier proteins, resemble insect OBPs in the sense that they are also small, secreted proteins with a series of conserved, disulfide-bonded cysteines. However, they show no homology to the insect OBPs in terms of either their primary or secondary structures and are presumed to have arisen by convergent evolution (Pelosi 1994; Tegoni et al. 2000). OBP-family representatives occur in one Paraneoptera, of the order Hemiptera (“true bug”), as well as in a variety of Endopterygotan orders: the Lepidoptera (moths), the Diptera (flies and mosquitoes), and the Coleoptera (beetles; Vogt et al. 1999). It is most likely that insect OBP progenitors were present in ancient Neoptera (one subgroup of the winged, terrestrial insects, Pterygota).
Our phylogenetic analysis of the large insect OBP family (Fig. 4) reveals a number of subfamilies, all but one of which include Drosophila members. Two subgroups appear monophyletic: A large group of pheromone-binding proteins (PBPs) and related general odorant-binding proteins (GOBPs) comprise a large Lepidoptera-specific subfamily, and the Plus-C subfamily is specific to Drosophila.
Most of the Drosophila OBPs share orthologs in other insects. The ABPX subfamily includes three Drosophila OBPs (Obp83a, Obp83b, and Obp69a) as well as antennal binding proteins termed ABPXs from a variety of Lepidoptera species (Krieger et al. 1996, 1997), and related antennal proteins from multiple species of beetle (Wojtasek et al. 1998, 1999), the honey bee A. mellifera (Danty et al. 1999), and the Hemiptera (“true bug”) L. lineolaris (Vogt et al. 1999). Insect OBP progenitors were likely present in ancient Neoptera, and the ABPX subfamily evidently diverged from other insect OBPs before the Endopterygota–Paraneoptera split. The Minus-C subfamily of OBP-like proteins includes four Drosophila proteins that lack the conserved cysteines C2 and C5 (Obp44a, Obp99b, Obp99c, and Obp8a) as well as related nonolfactory proteins from the medfly (Ceratitis capitata), the beetle (Tenebrio molitor), and the mosquito (Anopheles gambiae; Paesen and Happ 1995; Thymianou et al. 1998; Arca et al. 1999). This family also includes three Drosophila proteins that carry all six conserved cysteines (Obp99a, Obp99d, and Obp83f), indicating that the loss of cysteines C2 and C5 happened after the family diverged from the rest of the insect OBPs. The progenitor Drosophila Minus-C Obp gene was most likely located in cytological region 99B-C. As shown in Figure 5, four of the seven Drosophila Minus-C Obp genes are located in this region, including two of three that encode OBPs with all six conserved cysteines (Obp99a and Obp99d) and two of the three that have conserved intron insertion site 1 (Obp99a and Obp99b).
Duplication and Divergence of the Drosophila Obp Gene Family
The genomic clustering, sequence conservation, and common intron insertion sites of the 51 Drosophila Obp genes summarized in Figure 5 reflect a complicated history of gene duplication and divergence for this large gene family. Expansion of the Drosophila Obp gene family may not be ongoing as the two most closely related Obp genes (Obp83a and Obp83b) likely diverged from each other more than 60 million years ago (Hekmat-Scafe et al. 2000). The Plus-C subfamily is monophyletic and thus appears to be the most recently derived Drosophila OBP subfamily. Eleven of the 12 Plus-C Obp genes are located on Chromosome 2R, including five genes clustered in cytological region 50F and two in 58F (Fig. 5). The 14 Obp genes in cytological region 56E–57A also appear to have arisen more recently because there is no known ortholog for any of these genes in another insect species. Furthermore, most genes in the 56/57 cluster have an intron inserted at site 2 or 3, with site 3 unique to genes in the 56/57 cluster (Figs. 3 and 5). Nonetheless, both the Plus-C and the 56/57 Obp genes display a great deal of sequence divergence (Fig. 2), indicating considerable elapsed time.
The Drosophila Obp gene family appears to have evolved by a series of gene duplication and divergence events starting with a progenitor gene at 58F. The distal portion of Drosophila Chromosome 2R carries three large Obp clusters containing a total of 18 genes (Fig. 5). These are the Obp56 cluster (9 genes), the Obp57 cluster (5 genes), and the Obp58 cluster (4 genes). The genes in the Obp58 cluster are notable with respect to their diversity in both coding sequence and intron insertion sites (Figs. 3 and 5). Obp58b and Obp58c are members of the Plus-C subfamily. The Obp58c gene has introns inserted at site 8 (found in some of the other Plus-C genes as well as ABPX subfamily members Obp69a/PBPRP-1 and Obp84a/PBPRP-4) and at site 1 (found in the Minus-C genes Obp44a and Obp99a). Obp58d is most similar to L. longipalpis SL1 (Charlab et al. 1999) and D. melanogaster Obp57a (Fig. 4). The Obp58d gene has an intron inserted at site 7, which is also found in two genes in the cluster at cytological region 19D. The Obp58d gene also shares an intron insertion site with PBP1 from Antherea pernyi (Krieger et al. 1991) and with GOBP2 from Manduca sexta (GenBank accession no. AF323972), although it shows little coding sequence similarity with either of these moth OBPs. Taken together, it seems plausible that one of the progenitor Obp genes occurred at 58F, and then gave rise to much of the subsequent diversity observed for the Drosophila Obp genes.
The OBPs and related proteins described here are a family of proteins that are ancient, numerous, and completely insect-specific. There are 51 putative OBPs present in Drosophila, and we expect that a careful examination of other insect species will reveal an astonishing degree of monophyletic and polyphyletic diversity. The richness of this diversity indicates that OBPs must play a fundamentally important role in odorant detection, although it is unclear whether this is for odorant recognition, discrimination, and/or sensitivity. The few OBPs of mammals bear no sequence relationship to the insect OBPs and therefore must have arisen by convergent evolution (Pelosi 1994; Tegoni et al. 2000). Thus, although olfactory transduction is similar in both cases, there may be fundamental mechanistic differences in odorant detection between mammals and insects. Ultimately, an understanding of OBP complexity, odorant receptor complexity, and how these two systems interplay will be required for a complete appreciation of olfactory sensation in insects.
METHODS
Identification of Drosophila OBP Family Members
Thirty-one potential OBP family members were identified through three iterations of a PSI-BLAST search (Altschul et al. 1997) of Drosophila genomic sequences at the National Center for Biotechnology Information (NCBI) beginning with the OS-E protein sequence. The E-values for proteins identified by this convergent PSI-BLAST search ranged from 1e-45 to 3e-10. The corresponding amino acid sequences were extracted from GenBank and examined for three features characteristic of insect OBPs: (1) a predicted size of ∼14–20 kD, (2) an N-terminal signal sequence (determined by a Kyte–Doolittle hydrophobicity plot with a window of 20 amino acids), and (3) four–six stereotypically placed cysteine residues (Pelosi and Maida 1995). In three cases (CG15129, CG15582, and CG15583) it was apparent that the gene predicted by Celera Genomics in collaboration with the Berkeley Drosophila Genome Project (BDGP; FlyBase 1999) actually consisted of multiple adjacent genes, each of which could encode an OBP-like protein. In 11 other cases (CG11218, CG13873, CG13874, CG13429, CG18111, CG13518, CG11748, CG1670, CG12944, CG15883, and CG12665) the predicted protein lacked one or more OBP hallmarks (generally an N-terminal signal sequence). In these cases, the corresponding DNA sequence, along with ∼400 bp of flanking sequence on either end, was extracted from GenBank. In each case, an alternative splice form that would generate an OBP-like protein was found using FGENESH (http://genomic.sanger.ac.uk/gf/gf.html) or Splice Site Prediction by Neural Network (http://www.fruitfly.org/seq_tools/splice.html). The 31 OBP-like protein sequences were used to scan Drosophila genomic sequences at NCBI using the TBLASTN program with an E-value threshold of 10. This search revealed 13 additional sequences encoding OBP-like products with stereotypically placed cysteines. Subsequent phylogenetic analysis (described below) indicated that one of these genes, located at cytological region 22B, which has been described previously (Robertson et al. 1999; Galindo and Smith 2001) and which shows no discernible expression (Galindo and Smith 2001), encodes a protein that lacks two α-helices found in other insect OBPs and consequently is unlikely to be a bona fide OBP. The remaining 12 new OBP-like protein sequences were then used in a TBLASTN scan of the Drosophila genome. This search produced four additional OBP family members. A TBLASTN search of Drosophila genomic sequences with the four final OBP protein sequences revealed no additional OBP family members. Hence, our search revealed a total of 51 potential Drosophila Obp genes. For clarity, we refer to the genes encoding all 51 of these OBP family members as “Obp” genes, although for many this inclusion is presumptive and is based only on sequence similarity; odorant-binding activity has not yet been shown. We have communicated annotations for all 51 Drosophila Obp genes to those responsible for updating FlyBase.
Genomic locations of the 51 predicted Obp genes were assembled from the Genome Annotation Database of Drosophila (GadFly) database (http://www.fruitfly.org/annot/bands.html; also available as supplementary material at http://www.genome.org.). The locations of nearby genes were used to align the nine Obp genes that had not previously been assigned to a cytogenetic location. We expect that the cytological positions of the various Obp genes may change slightly as the alignment of the D. melanogaster genomic sequence to its cytogenetic map is further refined. We also searched the Drosophila Expressed Sequence Tag (dEST) database at the BDGP (http://www.fruitfly.org/blast/) with protein sequences of selected Drosophila Obp using the TBLASTN program. These searches revealed dESTs encoded by some of the Obp genes that the BDGP had organized into clots (sets of homologous Drosophila dESTs likely to come from the same gene); these are listed in Table 1.
Phylogenetic Analysis of Drosophila OBP Family Members
The CLUSTAL X program (version 8.0; Thompson et al. 1997) was used to produce an initial alignment of the products of the various Drosophila Obp genes with the exception of Obp83e, which represents a double OBP protein. Subgroups of sequences that appeared most similar were aligned to each other in multiple alignment mode, and these various alignments were successively added using the profile alignment function. In this way, we were able to drive the alignment order of these relatively divergent sequences such that the most closely related sequences were aligned first. This method produced an initial alignment of 34 of the Drosophila Obp sequences. We produced an alignment of 10 OBP sequences that have a conserved N terminus as well as a conserved C terminus (Obp46a, Obp47b, Obp49a, Obp50a, Obp50b, Obp50c, Obp50d, Obp50e, Obp58b, and Obp58c). Two sequences (Obp85a and Obp93a) have the same conserved C terminus, but different N termini; these were aligned to each other. These two subgroups were then aligned to each other and subsequently added to the main alignment using the profile alignment function. Four particularly divergent sequences (Obp19c, Obp83c, Obp84a, and Obp99c) were then added successively to produce an overall alignment. At each step, protein alignments were inspected to ensure the alignment of landmark cysteine residues, and misaligned sequences or subregions were realigned using either the realign selected sequence or realign selected residue range functions, respectively. The recently solved B. mori pheromone-binding protein (B. mori PBP) secondary structure (Sandler et al. 2000) was used to create a gap penalty mask, which was added in profile mode. Finally, the overall alignment was further refined through minor manual adjustment. An unrooted, distance (neighbor-joining) tree (Saitou and Nei 1987) was constructed using our final Drosophila Obp alignment, except that the signal sequence-bearing N terminus was removed from each OBP sequence, as described in the legend to Figure 2. We used the PHYLIP program (Felsenstein 1993) to produce a majority rule consensus distance tree derived from equivalent-length trees using tree-bisection–reconnection. Bootstrap analysis of the reliability of branching used 1000 neighbor-joining replicates.
Identification and Phylogenetic Analysis of Insect OBP Family Members
OBP family members present in insects other than Drosophila were obtained by performing two iterations of a PSI-BLAST search of the Non-Redundant GenBank CDS starting with the OS-E protein sequence. The corresponding amino acid sequences were then extracted from GenBank, and redundant sequences were eliminated from the data set. This analysis revealed 62 non-Drosophila insect OBP-like protein sequences. We aligned 52 of these sequences using CLUSTAL X (version 8.0; Thompson et al. 1997). We added one of the remaining sequences (L. lon SL1) to the Drosophila alignment described above with the profile alignment function. We independently aligned three mosquito sequences (A. gam D7r1, D7r2, and D7r3) and the remaining six sequences (C. cap MSPA, MSPB, and MSPC; T. mol B1, B2, and Thp12) with the multiple alignment function. Using the profile alignment function, we aligned the two subgroups to each other, then to the main set of insect OBP sequences, and finally to the Drosophila alignment. We used our insect OBP alignment to create an unrooted distance (neighbor-joining) tree (Saitou and Nei 1987). As before, the signal sequence-bearing N terminus was removed from each OBP sequence as described in the legend to Figure 2. We used the PHYLIP program (Felsenstein 1993) to derive a majority rule consensus distance tree. Bootstrap analysis used 1000 neighbor-joining replicates.
Additional Computational Methods
Our protein alignment for the Drosophila OBPs was used to guide the alignment of the corresponding coding nucleotide sequences by the Perl program protal2dna (K. Schuerer and C. Letondal, unpubl.; available at ftp://ftp.pasteur.fr/pub/GenSoft/unix/alignment/protal2dna). All pairwise dn/ds values were then calculated using the program SNAP.pl (http://hiv-web.lanl.gov/seq-db.html; Nei and Gojobori 1986). Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions, and the average pairwise dn/ds values were calculated for the sequences under each node in the phenogram tree.
Then 1 kb of genomic sequence upstream of each of the 16 Obp genes that have corresponding cDNA clones was extracted from GadFly (http://hedgehog.lbl.gov:8002/cgi-bin/annot/query), and each sequence was subjected to self × self dot plot analysis with the program Dotter (http://www.cgr.ki.se/cgr/groups/sonnhammer/Dotter.html; Sonnhammer and Durbin 1995). This identified regions that are repeated multiple times within each putative regulatory region. Then, the upstream regions of four pairs of clustered Obp genes showing similar expression patterns (19b/19d, 56d/56e, 83a/83b, and 99b/99d) were subjected to a pairwise dot plot analysis to determine whether closely situated genes might share regulatory elements leading to similar patterns of gene expression.
Molecular Biology
The polymerase chain reaction (PCR) was used to amplify a fragment of Obp50d from genomic clone BACR16C17 and fragments of Obp58b and Obp58c from genomic clone BACR11M08. The PCR primers used were: 50d, 5′- GGAATTCC AGCTTTGAGTGCATCTTTCG)-3′, 5′- GCTCTAGAGCATGT CATCGCAGCGAATGC-3′; 58b, 5′-GGAATTCCGTGGCTG TCCGAGTTCATTGC-3′, 5′-GCTCTAGAGCATTCAGCA TTTCAGTCG-3′; 58c, 5′-GGAATTCCACATCCACTATTGC TGC-3′, 5′-GCTCTAGAGCGTTGATCATTTCCTTGG-3′ (5′ EcoRI and XbaI sites are underlined). The PCR conditions were: 95°C for 4 min, followed by 35 cycles of 95°C for 30 sec, 55°C for 45 sec, 72°C for 1 min, and then one cycle of 72°C for 5 min. AmpliTaq DNA polymerase (Perkin-Elmer Biosystems) was used for all PCR reactions. The purified Obp50d, Obp58b, and Obp58c PCR products were digested with EcoRI and XbaI and subcloned into pBluescript II (Stratagene) using standard methods (Sambrook et al. 1989) to create plasmids pDH145, pDH146, and pDH147, respectively.
Anti-sense and sense DIG-RNA probes (Roche Bioscience) were prepared for Obp50d, Obp58b, Obp58c, Obp56d, Obp99a, Obp99b, and (as a positive control) Obp83a from linearized pDH145, pDH146, pDH147, GH09027, GH16332, GH15449, and pDH50 (McKenna et al. 1994) according to the manufacturer's directions. Obp transcripts were examined in 8 μM Drosophila head sections as described (McKenna et al. 1994) with minor modifications. Drosophila heads were prefixed prior to sectioning as described (Clyne et al. 1999), hybridization was at 55°C, and levamisole (0.24 mg/mL) was added to the staining solution to inhibit endogenous alkaline phosphatases as recommended by the manufacturer.
WEB SITE REFERENCES
ftp://ftp.pasteur.fr/pub/GenSoft/unix/alignment/protal2dna; Perl program protal2dna.
http://evolution.genetics.washington.edu/phylip.html; Free package of programs for inferring phylogenics.
http://flybase.bio.indiana.edu/; FlyBase.
http://genomic.sanger.ac.uk/gf/gf.html; FGENESH program.
http://hedgehog.lbl.gov:8002/cgi-bin/annot/query; GadFly.
http://hiv-web.lanl.gov/seq-db.html; SNAP.pl program used to calculate all pairwise dn/ds values.
http://ludwig-sunl.unil.ch:8080/software/Box_form.html; BOXSHADE program.
http://www.cgr.ki.se/cgr/groups/sonnhammer/Dotter.html; Dotter program.
http://www.fruitfly.org/annot/bands.html; Genome Annotation Database of Drosophila (GadFly).
http://www.fruitfly.org/blast/; Fly BLAST search at BDGP.
http://www.fruitfly.org/seq_tools/splice.html; Splice Site Prediction by Neural Network.
Acknowledgments
This work was supported by an NIH grant (5R01NS31231) to M.T. Earlier unpublished portions of this work were supported by an AAUW Summer Faculty Fellowship and a Barrett Faculty Research Fellowship from Mills College to D.H. and a Barrett Undergraduate Research Fellowship to A.M. We are indebted to the Berkeley and European Drosophila Genome Projects and to Celera Genomics, whose publicly available Drosophila genomic sequence made this work possible.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL daria@nature.berkeley.edu; FAX (510) 643-6791.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.239402.
REFERENCES
- Ache BW. Towards a common strategy for transducing olfactory information. Sem Cell Biol. 1994;5:55–63. doi: 10.1006/scel.1994.1008. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arca B, Lombardo F, De Lara Capurro M, Della Torre A, Dimopoulos G, James AA, Colui M. Trapping cDNAs encoding secreted proteins from the salivary glands of the malarial vector Anopheles gambiae. Proc Natl Acad Sci. 1999;96:1516–1521. doi: 10.1073/pnas.96.4.1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bargmann CI. Neurobiology of the Caenorhabditis elegans genome. Science. 1998;282:2028–2033. doi: 10.1126/science.282.5396.2028. [DOI] [PubMed] [Google Scholar]
- Breer H, Krieger J, Raming K. A novel class of binding proteins in the antennae of the silk moth Antherea pernyi. Insect Biochem. 1990;20:735–740. [Google Scholar]
- Charlab R, Valenzuela JR, Rowton ED, Ribeiro JM. Towards an understanding of the biochemical and pharmacological complexity of the saliva of a hematophagous fly, Lutzomyia longipalpis. Proc Natl Acad Sci. 1999;96:15155–15160. doi: 10.1073/pnas.96.26.15155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clyne PJ, Warr CG, Freeman MR, Lessing D, Kim J, Carlson JR. A novel family of divergent seven-transmembrane proteins: Candidate odorant receptors in Drosophila. Neuron. 1999;22:327–338. doi: 10.1016/s0896-6273(00)81093-4. [DOI] [PubMed] [Google Scholar]
- Danty E, Briand L, Michard-Vanhée D, Perez V, Arnold G, Gaudemer O, Huet D, Huet J-C, Ouali C, Masson C, et al. Cloning and expression of a queen pheromone-binding protein in the honeybee: An olfactory-specific, developmentally regulated protein. J Neurosci. 1999;19:7468–7475. doi: 10.1523/JNEUROSCI.19-17-07468.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drosophila Odorant Receptor Nomenclature Committee. A unified nomenclature system for the Drosophila odorant receptors. Cell. 2000;102:145–146. doi: 10.1016/s0092-8674(00)00020-9. [DOI] [PubMed] [Google Scholar]
- Du G, Prestwich GD. Protein structure encodes the ligand binding specificity in pheromone binding proteins. Biochemistry. 1995;34:8726–8732. doi: 10.1021/bi00027a023. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. PHYLIP: Phylogeny interference package, Version 3.5. Seattle: University of Washington; 1993. [Google Scholar]
- FlyBase. The FlyBase database of the Drosophila genome projects and community literature. Available from http://flybase.bio.indiana.edu/ Nucleic Acids Res. 1999;27:85–88. doi: 10.1093/nar/27.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galindo K, Smith DP. A large family of divergent Drosophila odorant-binding proteins expressed in gustatory and olfactory sensilla. Genetics. 2001;159:1059–1072. doi: 10.1093/genetics/159.3.1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Q, Chess A. Identification of candidate Drosophila olfactory receptors from genomic DNA sequence. Genomics. 1999;60:31–39. doi: 10.1006/geno.1999.5894. [DOI] [PubMed] [Google Scholar]
- Graham LA, Tang W, Baust JG, Liou Y-C, Reid TS, Davies PL. Characterization and cloning of a Tenebrio molitor hemolymph protein with sequence similarity to insect odorant-binding proteins. Insect Biochem Mol Biol. 2001;31:691–702. doi: 10.1016/s0965-1748(00)00177-6. [DOI] [PubMed] [Google Scholar]
- Harrison PM, Kumar A, Lang N, Snyder M, Gerstein M. A question of size: The eucaryotic proteome and the problems in defining it. Nucleic Acids Res. 2002;30:1083–1090. doi: 10.1093/nar/30.5.1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hekmat-Scafe DS, Steinbrecht RA, Carlson JR. Coexpression of two odorant-binding protein homologs in Drosophila: Implications for olfactory coding. J Neurosci. 1997;17:1616–1624. doi: 10.1523/JNEUROSCI.17-05-01616.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hekmat-Scafe DS, Dorit RL, Carlson JR. Molecular evolution of odorant-binding protein genes OS-E and OS-F in Drosophila. Genetics. 2000;155:117–127. doi: 10.1093/genetics/155.1.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildebrand JG, Shepherd GM. Mechanisms of olfactory discrimination: Converging evidence for common principles across phyla. Ann Rev Neurosci. 1997;20:595–631. doi: 10.1146/annurev.neuro.20.1.595. [DOI] [PubMed] [Google Scholar]
- Horst R, Damberger F, Luginbühl P, Güntert P, Peng G, Nikonova L, Leal WS, Wüthrich K. NMR structure reveals intramolecular regulation mechanism for pheromone binding and release. Proc Natl Acad Sci. 2001;98:14374–14379. doi: 10.1073/pnas.251532998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlin S, Bergman A, Gentles AJ. Annotation of the Drosophila genome. Nature. 2001;411:259–260. doi: 10.1038/35077152. [DOI] [PubMed] [Google Scholar]
- Kim MS, Repp A, Smith DP. LUSH odorant-binding protein mediates chemosensory responses to alcohols in Drosophila melanogaster. Genetics. 1998;150:711–721. doi: 10.1093/genetics/150.2.711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodrik D, Filippov VA, Filippova MA, Sehnal F. Sericotropin: An insect neurohormone affecting RNA transcription. Netherlands J Zool. 1995;45:68–70. [Google Scholar]
- Krieger J, Raming K, Breer H. Cloning of genomic and complementary DNA encoding insect pheromone binding proteins: Evidence for microdiversity. Biochim Biophys Acta. 1991;1088:277–284. doi: 10.1016/0167-4781(91)90064-s. [DOI] [PubMed] [Google Scholar]
- Krieger J, von Nickisch-Rosenegk E, Mameli E, Pelosi P, Breer H. Binding proteins from the antennae of Bombyx mori. Insect Biochem Mol Biol. 1996;26:297–307. doi: 10.1016/0965-1748(95)00096-8. [DOI] [PubMed] [Google Scholar]
- Krieger J, Mameli M, Breer H. Elements of the olfactory signaling pathways in insect antennae. Invert Neurosci. 1997;3:137–144. doi: 10.1007/BF02480368. [DOI] [PubMed] [Google Scholar]
- Löbel D, Jacob M, Völkner M, Breer H. Odorants of different chemical classes interact with distinct odorant binding protein subtypes. Chem Senses. 2002;27:39–44. doi: 10.1093/chemse/27.1.39. [DOI] [PubMed] [Google Scholar]
- Maïbeche-Coisne M, Sobrio F, Delaunay T, Lettere M, Dubroca J, Jacquin-Joly E, Nagnan-Le Meillour P. Pheromone binding proteins of the moth Mamestra brassicae: Specificity of ligand binding. Insect Biochem Mol Biol. 1997;27:213–221. [Google Scholar]
- McKenna MP, Hekmat-Scafe DS, Gaines P, Carlson JR. Putative Drosophila pheromone-binding proteins expressed in a subregion of the olfactory system. J Biol Chem. 1994;269:16340–16347. [PubMed] [Google Scholar]
- Mombaerts P. Seven-transmembrane proteins as odorant and chemosensory receptors. Science. 1999;286:707–711. doi: 10.1126/science.286.5440.707. [DOI] [PubMed] [Google Scholar]
- Nei M, Gojobori T. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol Biol Evol. 1986;3:418–426. doi: 10.1093/oxfordjournals.molbev.a040410. [DOI] [PubMed] [Google Scholar]
- Ozaki M, Morisaki K, Idei W, Ozaki K, Tokunaga F. A putative lipophilic stimulant carrier protein commonly found in the taste and olfactory systems: A unique member of the pheromone-binding protein superfamily. Eur J Biochem. 1995;230:298–308. doi: 10.1111/j.1432-1033.1995.0298i.x. [DOI] [PubMed] [Google Scholar]
- Paesen GC, Happ GM. The B proteins secreted by the tubular accessory sex gland of the male mealworm beetle, Tenebrio molitor, have sequence similarity to moth pheromone-binding proteins. Insect Biochem Mol Biol. 1995;25:401–408. doi: 10.1016/0965-1748(94)00085-v. [DOI] [PubMed] [Google Scholar]
- Park S-K, Shanghag SR, Wang Q, Hasan G, Steinbrecht RA, Pikielny CW. Expression patterns of two putative odorant-binding proteins in the olfactory organs of Drosophila melanogaster have different implications for their functions. Cell Tissue Res. 2000;300:181–192. doi: 10.1007/s004410000187. [DOI] [PubMed] [Google Scholar]
- Pelosi P. Odorant-binding proteins. Crit Rev Biochem Mol Biol. 1994;29:199–228. doi: 10.3109/10409239409086801. [DOI] [PubMed] [Google Scholar]
- Pelosi P, Maida R. Odorant-binding proteins in insects. Comp Biochem Physiol B. 1995;111:503–514. doi: 10.1016/0305-0491(95)00019-5. [DOI] [PubMed] [Google Scholar]
- Pikielny CW, Hasan G, Rouyer F, Rosbash M. Members of a family of Drosophila putative odorant-binding proteins are expressed in different subsets of olfactory hairs. Neuron. 1994;12:35–49. doi: 10.1016/0896-6273(94)90150-3. [DOI] [PubMed] [Google Scholar]
- Plettner E, Lazar J, Prestwich EG, Prestwich GD. Discrimination of pheromone-enantiomers by two pheromone-binding proteins from the gypsy moth Lymantria dispar. Biochemistry. 2000;2000:8953–8962. doi: 10.1021/bi000461x. [DOI] [PubMed] [Google Scholar]
- Prasad BC, Reed RR. Chemosensation: Molecular mechanisms in worms and mammals. Trends Genet. 1999;15:150–153. doi: 10.1016/s0168-9525(99)01695-9. [DOI] [PubMed] [Google Scholar]
- Prestwich GD, Du G, LaForest S. How is pheromone specificity encoded in proteins? Chem Senses. 1995;20:461–469. doi: 10.1093/chemse/20.4.461. [DOI] [PubMed] [Google Scholar]
- Raming K, Krieger J, Breer H. Primary structure of a pheromone-binding protein from Antheraea pernyi: Homologies with other ligand-carrying proteins. J Comp Physiol B. 1990;160:503–509. doi: 10.1007/BF00258977. [DOI] [PubMed] [Google Scholar]
- Reese MG, Hartzell G, Harris NL, Ohler U, Abril JF, Lewis SE. Genome annotation assessment in Drosophila melanogaster. Genome Res. 2000;10:483–501. doi: 10.1101/gr.10.4.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson HM. The large srh family of chemoreceptor genes in Caenorhabditis nematodes reveals processes of genome evolution involving large duplications and deletions and intron gains and losses. Genome Res. 2000;10:192–203. doi: 10.1101/gr.10.2.192. [DOI] [PubMed] [Google Scholar]
- Robertson HM, Martos R, Sears CR, Todres EZ, Walden KKO, Nardi JB. Diversity of odourant binding proteins revealed by an expressed sequence tag project on male Manduca sexta moth antennae. Insect Mol Biol. 1999;8:501–518. doi: 10.1046/j.1365-2583.1999.00146.x. [DOI] [PubMed] [Google Scholar]
- Rubin GM, Yandell MD, Wortman JR, Miklos GLG, Nelson CR, Hariharan IK, Fortini ME, Li PW, Apweiler R, Fleischmann W, et al. Comparative genomics of the eucaryotes. Science. 2000;287:2204–2214. doi: 10.1126/science.287.5461.2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou N, Nei M. The neighbor joining method: A new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: A laboratory manual. 2nd ed. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Sandler BH, Nikonova L, Leal WS, Clardy J. Sexual attraction in the silkworm moth: Structure of the pheromone-binding-protein–bombykol complex. Biol Chem. 2000;7:143–151. doi: 10.1016/s1074-5521(00)00078-8. [DOI] [PubMed] [Google Scholar]
- Scott K, Brady R, Cravchik A, Morozov P, Rzhetsky A, Zuker C, Axel R. A chemosensory gene family encoding candidate gustatory and olfactory receptors in Drosophila. Cell. 2001;104:661–673. doi: 10.1016/s0092-8674(01)00263-x. [DOI] [PubMed] [Google Scholar]
- Sonnhammer ELL, Durbin R. A dot-matrix program with dynamic threshold control suited for genomic DNA and protein sequence analysis. Gene. 1995;167:GC1–GC10. doi: 10.1016/0378-1119(95)00714-8. [DOI] [PubMed] [Google Scholar]
- Steinbrecht RA. Structure and function of insect olfactory sensilla. In: Bock G, Cardew G, Hildebrand J, editors. Olfaction in mosquito–host interactions. Chichester, UK: Wiley; 1996. pp. 158–177. [DOI] [PubMed] [Google Scholar]
- Steinbrecht RA, Ozaki M, Ziegelberger G. Immunocytochemical localization of pheromone-binding protein in moth antennae. Cell Tissue Res. 1992;270:287–302. [Google Scholar]
- Steinbrecht RA, Laue M, Ziegelberger G. Immunolocalization of pheromone-binding and general odorant-binding protein in olfactory sensilla of the silkmoths Antheraea and Bombyx. Cell Tissue Res. 1995;282:203–217. [Google Scholar]
- Tegoni M, Pelosi P, Vincent F, Spinelli S, Campanacci V, Grolli S, Ramoni R, Cambillau C. Mammalian odorant binding proteins. Biochim Biophys Acta. 2000;1482:229–240. doi: 10.1016/s0167-4838(00)00167-9. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thymianou S, Mavroidis M, Kokolakis G, Komitopoulou K, Zacharopoulou A, Mintzas AC. Cloning and characterization of a cDNA encoding a male-specific serum protein of the Mediterranean fruit fly, Ceratitis capitata, with sequence similarity to odourant-binding proteins. Insect Mol Biol. 1998;7:345–353. doi: 10.1046/j.1365-2583.1998.740345.x. [DOI] [PubMed] [Google Scholar]
- Todres E, Nardi JB, Robertson HM. The tetraspanin superfamily in insects. Insect Mol Biol. 2000;9:581–590. doi: 10.1046/j.1365-2583.2000.00222.x. [DOI] [PubMed] [Google Scholar]
- Vogt RG, Prestwich GD, Lerner MR. Odorant-binding-protein subfamilies associate with distinct classes of olfactory receptor neurons in insects. J Neurobiol. 1991;22:74–84. doi: 10.1002/neu.480220108. [DOI] [PubMed] [Google Scholar]
- Vogt RG, Callahan FE, Rogers ME, Dickens JC. Odorant binding protein diversity and distribution among the insect orders, as indicated by LAP, an OBP-related protein of the true bug Lygus lineolaris (Hemiptera, Heteroptera) Chem Senses. 1999;24:481–495. doi: 10.1093/chemse/24.5.481. [DOI] [PubMed] [Google Scholar]
- von Heijne G. A method for predicting signal sequence cleavage sites. Nucleic Acids Res. 1986;14:4683–4690. doi: 10.1093/nar/14.11.4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vosshall LB. The molecular logic of olfaction in Drosophila. Chem Senses. 2001;26:207–213. doi: 10.1093/chemse/26.2.207. [DOI] [PubMed] [Google Scholar]
- Vosshall LB, Amrein H, Morozov PS, Rzhetsky A, Axel R. A spatial map of olfactory receptor expression in the Drosophila antenna. Cell. 1999;96:725–736. doi: 10.1016/s0092-8674(00)80582-6. [DOI] [PubMed] [Google Scholar]
- Vosshall LB, Wong AM, Axel R. An olfactory sensory map in the fly brain. Cell. 2000;102:147–159. doi: 10.1016/s0092-8674(00)00021-0. [DOI] [PubMed] [Google Scholar]
- Wang Y, Wright NJD, Guo H-F, Xie Z, Svoboda K, Malinow R, Smith DP, Zhong Y. Genetic manipulation of the odor-evoked distributed neural activity in the Drosophila mushroom body. Neuron. 2001;29:267–276. doi: 10.1016/s0896-6273(01)00196-9. [DOI] [PubMed] [Google Scholar]
- Wojtasek H, Hansson BS, Leal WS. Attracted or repelled? A matter of two neurons, one pheromone binding protein, and a chiral center. Biochem Biophys Res Commun. 1998;250:217–222. doi: 10.1006/bbrc.1998.9278. [DOI] [PubMed] [Google Scholar]
- Wojtasek H, Picimbon J-F, Leal WS. Identification and cloning of odorant binding proteins from the scarab beetle Phyllopertha diversa. Biochem Biophys Res Commun. 1999;263:832–837. doi: 10.1006/bbrc.1999.1448. [DOI] [PubMed] [Google Scholar]