Abstract
Several studies of substitution rate variation have indicated that the local mutation rate varies over the mammalian genome. In the present study, we show significant variation in substitution rates within the noncoding part of the human genome using 4.7 Mb of human-chimpanzee pairwise comparisons. Moreover, we find a significant positive covariation of lineage-specific chimpanzee and human local substitution rates, and very similar mean substitution rates down the two lineages. The substitution rate variation is probably not caused by selection or biased gene conversion, and so we conclude that mutation rates vary deterministically across the noncoding nonrepetitive regions of the human genome. We also show that noncoding substitution rates are significantly affected by G+C base composition, partly because the base composition is not at equilibrium.
Understanding human DNA sequence variation and molecular evolution requires detailed insight into the mutation processes that operate on the genome. Under the neutral theory of molecular evolution, the pattern and rate of substitutions, those mutations that have spread through the population and reached fixation, are determined by the pattern and rate of mutations (Kimura 1983). Thus, mutation processes in the human genome relate to a number of important topics in molecular evolution—compositional structure of the genome (Casane et al. 1997), variation in rates of protein evolution (Williams and Hurst 2000), the male mutation bias (Lercher et al. 2001), and mammalian regulatory sequences (Pennacchio and Rubin 2001)—besides their obvious importance in understanding human genetic variation and its contribution to phenotypic traits.
It is now recognized that genic point mutation rates vary across mammalian genomes (Wolfe et al. 1989; Casane et al. 1997; Matassi et al. 1999; Nachman and Crowell 2000; Williams and Hurst 2000; Chen et al. 2001; Lercher et al. 2001). Comparisons of protein coding genes have revealed extensive variation in synonymous substitution rates, suggestive of mutation rate variation across mammalian genomes (Chen et al. 2001; Lercher et al. 2001). However, genes only represent a small fraction of the human genome, and mutation rate variation across the noncoding regions of the human genome has not yet been examined in detail, although both Chen et al. (2001) and Fujiyama et al. (2002) have noted that in human-chimpanzee comparisons of genomic sequences, there is considerable variation in substitution rates.
We have adopted the approach of using many long genomic human-chimpanzee alignments from a single chromosome, namely, human chromosome 7, to study substitution rate variation. Long genomic alignments allow for reliable substitution rate estimation because of large amounts of data, as well as the possibility of considering substitution rate variation within alignments. Using publicly available chimpanzee bacterial artificial chromosome clone sequence data, we generated 77 human-chimpanzee genomic alignments (mean ungapped length, 61 kb; range, 5 to 153 kb), with a total length of 4.7 Mb, which was used to examine variation in noncoding substitution rates. In addition, orthologous baboon sequence data permitted the inference of lineage-specific human and chimpanzee substitution rates for 43 human-chimpanzee-baboon genomic alignments (mean ungapped length, 40 kb; range, 12 to 107 kb), with a total length of 1.7 Mb, thereby allowing tests for possible differences in the mutation processes of human and chimpanzee.
RESULTS
Validation of Alignments
Given that noncoding sequences in general (Smith and Hurst 1998b), and long genomic sequences in particular (Chen et al. 2001), can be hard to align correctly, we first consider whether our alignments are reliable. Our complete human-chimpanzee set of alignments, including all substitutions and all sequence data, yielded a mean uncorrected pairwise distance of 1.18%. This mean distance is similar to the value of 1.23% obtained by Fujiyama et al. (2002) in a genome-wide study involving short alignments. Furthermore, Chen et al. (2001) obtained a mean human-chimpanzee Jukes-Cantor (Jukes and Cantor 1969) distance of 1.19% for ∼2 Mb of human chromosome 7, the same chromosome that we consider here; their result is equivalent to an uncorrected distance of 1.18%, identical to our mean distance. Given that Chen et al. (2001) developed an alignment protocol to account for problems caused by repetitive elements and unalignable regions, this result indicates that our mean distance estimates are not inflated by such potential biases. Furthermore, it seems unlikely that our findings of substitution rate variation are caused by such alignment biases, because Chen et al. (2001) also found substitution rate variation.
As an additional check of alignment validity, we realigned our sequences using different alignment parameters with the same program, ClustalW (Thompson et al. 1994), and also using the default parameters of a different alignment program, Dialign 2, which is expected to outperform ClustalW when sequences are only locally related (Morgenstern 1999). We tested for ClustalW alignment parameter sensitivity by realigning 15 of the human-chimpanzee alignments under two alternative settings: (1) gap parameters (gap opening penalty and gap extension penalty) double the default values, and (2) gap parameters half the default values. In both cases, the mean human-chimpanzee distance was unchanged from the value of 1.23% (slightly different from the mean distance of 1.18% for all 77 human-chimpanzee alignments). We tested for alignment program sensitivity by realigning six of the human-chimpanzee-baboon alignments using Dialign 2 (Morgenstern 1999). With ClustalW, the mean human and chimpanzee lineage-specific distance (a measure dependent on the alignment of all three species) is 0.49%, and with Dialign 2, the mean distance is 0.54%. Such alignment parameter and alignment program insensitivity indicates that our alignments are reliable.
Substitution Rate Variation
Figure 1 shows the variation in substitution rates using nonoverlapping 5-kb blocks from the human-chimpanzee alignments. For these and all results reported below, we exclude potential CpG mutations from the estimation of substitution rates, thereby removing any variation caused by differences in levels of methylation. There is considerable substitution rate variation when all sequence data is considered. This variation is not solely owing to slow evolving protein coding sequences and fast evolving repetitive elements because there is similar variation in substitution rates for nonrepetitive noncoding intronic and intergenic DNA (for all results below, protein coding genes and repetitive elements have been excluded). The mean human-chimpanzee intronic and intergenic distances are significantly different (0.77% and 0.92%, respectively; P<0.001 Mann-Whitney U test). This difference in substitution rates means that intronic and intergenic data must be classed separately when variation in noncoding nonrepetitive genomic DNA is considered.
Figure 1.
Histograms of human-chimpanzee distances with frequencies given by numbers of nonoverlapping 5-kb blocks. Distance distributions are plotted for all sequence data and for noncoding nonrepetitive sequences divided into intronic and intergenic classes.
There is significantly more variation between the substitution rates of entire alignments than expected on the basis of the variation in substitution rates between nonoverlapping 1-kb blocks within alignments (one-way ANOVA: intronic, F=4.3; intergenic, F=1.8; P<0.001 for both; the intergenic data comprised 61 alignments containing 1272 blocks, and the intronic data comprised 48 alignments containing 1403 blocks). Because the alignments are distributed across human chromosome 7, the significant substitution rate differences between alignments show regional variation in substitution rates. Local similarity at the level of blocks within alignments was tested by a randomization study, which revealed that adjacent blocks have similar substitution rates (intronic, P<0.001; intergenic, P<0.001). Substitution rates of human noncoding DNA are thus characterized by regional variation and local similarity.
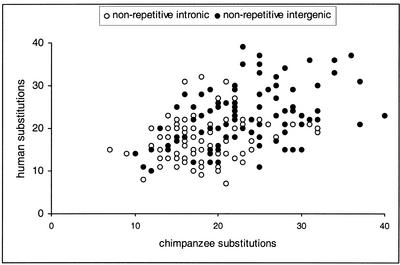
The repeatability of substitution rate variation (Smith and Hurst 1998a) was tested using human and chimpanzee lineage-specific substitution rates obtained from human-chimpanzee-baboon alignments. Figure 2 shows that the numbers of human and chimpanzee substitutions in nonoverlapping 5-kb blocks positively covary for both intronic and intergenic sequences. A Spearman's rank correlation test on the combined intronic and intergenic data (unbiased by the difference between the mean intronic and intergenic substitution rates because the two compared blocks are either both intronic or both intergenic) revealed a highly significant positive correlation (r=0.448 and P<0.001). The lineage-specific substitution data revealed no evidence of a difference in substitution rates between the human and chimpanzee lineages (as in Chen and Li 2001). Within the intronic data, there are 1976 chimpanzee substitutions and 2014 human substitutions, and within the intergenic data, there are 2418 chimpanzee substitutions and 2406 human substitutions.
Figure 2.
Repeatability of primate nonrepetitive noncoding substitution rates shown by a positive correlation between human and chimpanzee lineage-specific distances. Distances are given as substitutions per nonoverlapping 5-kb block. Intergenic and intronic data are treated as separate data points.
Controlling for Selection
A nonmutational explanation for our observations of substitution rate variation—including regional differences, local similarity, and substitution rate repeatability—is that certain regions within what we have identified as nonrepetitive noncoding regions might be subject to selection. Selectively constrained regions, those under strong negative selection, evolve slowly, and so substitution rate variation can be generated in the absence of mutation rate variation. Such regions may, for instance, include regulatory sequences (Pennacchio and Rubin 2001), unidentified protein-coding genes, and unidentified RNA genes (Eddy 2001). We address this issue by a theoretical argument that is then supported by an empirical analysis.
The theoretical argument rests on the relative effects of mutation and negative selection on substitution rate variation as the mean genetic distance changes. If we consider a high mean genetic distance, such as in the human-mouse comparison, then mutation rate differences will become lost as all unconstrained regions approach saturation. Thus, almost all substitution rate variation will be caused by strong negative selection—only selectively constrained regions will be conserved. But when the mean distance is very low, as in the human-chimpanzee comparison, then strong negative selection, which can only reduce substitution rates, cannot cause much variation in substitution rates. But mutation rate variation, which can result in local increases and decreases in substitution rates, can be a powerful force when substitution rates are low. This argument makes two assumptions concerning the distribution of selection coefficients in the noncoding part of the primate genome: (1) strong positive selection is rare, and (2) weak selection, when the multiple of the selection coefficient and the effective population size is small, is also rare. We discuss the issue of weak selection below in the context of our finding of compositional nonequilibrium.
Although the theoretical argument indicates that strong negative selection is unlikely to have generated our substitution rate observations, we tested for the effect of strong negative selection empirically by taking advantage of the fact that a large amount of pig sequence orthologous to our primate alignments is available. From the human-pig comparison, conserved and nonconserved regions were identified within our alignments. As expected, the conserved and nonconserved regions gave different human-chimpanzee distances (intronic, 0.58% versus 0.88%; intergenic, 0.63% versus 0.99%, conserved versus nonconserved, respectively). However, there is still significant variation within substitution rates in the nonconserved regions of human-chimpanzee alignments, for both intronic and intergenic data (nonoverlapping 1-kb blocks: intronic, F=2.7 and P<0.001; intergenic, F=1.6 and P<0.046). Thus, our finding of significant regional variation in intronic and intergenic regions is robust to the removal of putative conserved regions, despite the reductions in the amounts of data. We also used the nonconserved regions of the human-chimpanzee-baboon alignments to perform substitution rate repeatability tests: The combined intronic and intergenic data again revealed a highly significant positive correlation (Spearman's rank correlation, r=0.494 and P<0.001).
Base Composition and Substitution Rates
Base composition has been viewed as a potential correlate of mutation rates for a long time (Eyre-Walker and Hurst 2001), and the relationship between substitution rates and base composition has important implications for the understanding of the evolution of isochores (Piganeau et al. 2002). There is a significant positive correlation between summed human and chimpanzee lineage-specific noncoding nonrepetitive distances and G+C content using nonoverlapping 5-kb blocks with intronic and intergenic data combined (Spearman's rank correlation, r=0.337 and P<0.001; see Fig. 3). Note, however, that G+C content only explains ∼10% of the variation in substitution rates, so that most of the variation remains unexplained. The correlation between G+C content and divergence cannot be the result of CpG mutations, because potential CpG hypermutations were ignored in distance estimation (CpG mutations have been suggested as the cause of the relationship between G+C content and levels of polymorphism in the human genome [Sachidanandam et al. 2001]). It is also unlikely that this effect is caused by alignment biases, because a similar positive correlation is found between G+C content and substitution rates at fourfold degenerate sites in comparisons of human and mouse protein coding genes for which alignment is highly reliable (Hurst and Williams 2000). Therefore, we considered an alternative correlate of G+C content: The effect of genomic base composition is not at equilibrium.
Figure 3.
Positive correlation between G+C content and substitution rates. G+C content is the human-chimpanzee average. Substitution rates are summed over human and chimpanzee lineages, with data points representing nonoverlapping 5-kb blocks from the human-chimp-baboon alignments. The line gives the linear regression of substitution rates against G+C content.
It has recently been shown that the base composition of synonymous sites in mammalian protein coding genes is changing: Genes with high G+C content (GC) are decreasing in GC at the third codon position (L. Duret, M. Semon, G. Piganeau, D. Mouchiroud, and N. Galtier, unpubl.). This observation can be explained under a mutation bias model in terms of a change in the ratio of the GC→AT mutation rate to the AT→GC mutation rate: An increase in this ratio in regions of high GC leads to a decrease in GC. Alternatively, weak selection or biased gene conversion may have favored high GC in the past, higher than under mutation bias alone, but such forces are no longer effective. The importance of compositional nonequilibrium for our argument is that it increases the observed mutation rate: In high GC regions, the more mutable GC sites are at a higher level than the equilibrium level predicted on the basis of current mutation patterns. If such compositional nonequilibrium also affects noncoding nonrepetitive regions, then the effect of nonequilibrium would be consistent with our finding of higher substitution rates in high GC regions (see Piganeau et al. 2002).
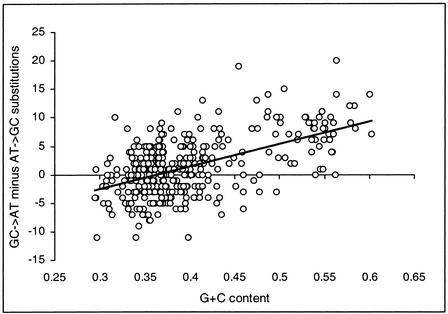
To test for compositional nonequilibrium, we used the human-chimpanzee-baboon data to infer GC→AT and AT→GC substitutions by parsimony. The number of GC→AT substitutions, NGC→AT, is expected to equal the number of AT→GC substitutions, NAT→GC, when the composition is at equilibrium, irrespective of G+C content (Eyre-Walker 1997), but we find that the number of GC→AT substitutions minus the number of AT→GC substitutions is positively correlated with G+C content (Spearman's rank correlation r=0.433 and P<0.001; see Fig. 4). This demonstration of compositional nonequilibrium is in accordance with reports of a GC→AT substitution bias in repetitive elements (see Fig. 27 in Lander et al. 2001). Because high G+C regions have a large excess of GC→AT substitutions, so that G+C content is decreasing, this result is consistent with compositional nonequilibrium being a major cause of the relationship between G+C content and substitution rates.
Figure 4.
Compositional nonequilibrium in the nonrepetitive noncoding regions of the primate genome. The difference between the number of GC→AT substitutions and the number of AT→GC substitutions (expected to be zero at equilibrium) is plotted against G+C content. Data points represent nonoverlapping 5-kb blocks of intronic and intergenic sequence, with human and chimpanzee lineage-specific substitutions summed. The line gives the linear regression, indicating the bias at high GC.
Compositional nonequilibrium has important implications for the interpretation of previous studies addressing whether weak selection affects the noncoding regions of the primate genome. The issue of weak selection is important both for the evolution of isochores and, more pertinently for this study, for the causes of substitution rate variation. Previous rejection of mutation bias hypotheses to explain isochores (Eyre-Walker 1999; Smith and Eyre-Walker 2001) indicates that weak selection or biased gene conversion (both of which generate a fixation bias in favor of AT→GC mutations) might be responsible for isochores. However, these studies relied on the assumption of compositional equilibrium. Given that this assumption does not hold, the observed patterns of compositional evolution can be explained in two ways: either (1) there was a fixation bias favoring high GC in certain parts of the genome in the ancient past, but such forces are no longer effective (perhaps owing to a reduction in effective population size in mammals); or (2) compositional evolution is simply caused by changes in mutation bias. In both explanations, there is no longer an effective fixation bias, and so we conclude that there is no evidence for weak selection or biased gene conversion currently having a major effect on the composition of the primate genome. Of course, our results do not rule out the possibility that there may be some low level of weak selection or biased gene conversion, but we see no reason to invoke such processes just to explain variation in substitution rates. Furthermore, we can rule out weak selection or biased gene conversion as the dominant force affecting substitution rates, because we would then predict a negative correlation between divergence and GC rather than the positive correlation observed (Eyre-Walker and Hurst 2001; Piganeau et al. 2002).
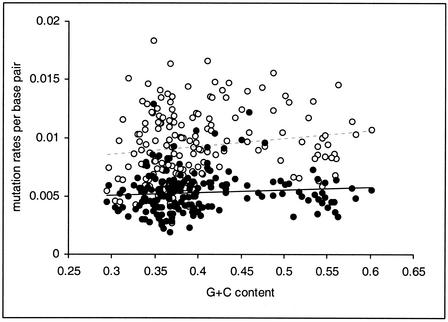
Returning to the relationship between G+C content and substitution rates, we can see if the positive correlation exists independently of the effect of compositional nonequilibrium by estimating the GC→AT mutation rate and the AT→GC mutation rate (assuming that substitution rates are unaffected by selection). As illustrated in Figure 5, both the GC→AT mutation rate and the AT→GC mutation rate show significant positive correlations with G+C content (Spearman's rank correlation: GC→AT mutation rate, r=0.177 and P=0.017; AT→GC mutation rate, r=0.162 and P=0.029), showing that compositional nonequilibrium is not a sufficient sole explanation of the positive correlation between divergence and G+C content.
Figure 5.
Mutation rates per base pair against G+C content for GC→AT (open circles) and AT→GC (closed circles) mutations. Data points represent nonoverlapping 5-kb blocks of intronic and intergenic sequence, with human and chimpanzee lineage-specific substitutions summed. The lines give linear regressions (dashed for GC→AT, solid for AT→GC).
DISCUSSION
We have provided three types of evidence of significant substitution rate variation in the noncoding nonrepetitive regions of the human genome: (1) regional variation, significant differences between alignments of tens of kilobases on the same chromosome; (2) local similarity, with adjacent 1-kb blocks within alignments tending to have similar substitution rates; and (3) repeatable variation, with the substitution rate variation down the human and chimpanzee lineages being positively correlated. From this substitution rate variation, we infer mutation rate variation, because there is no evidence to indicate that the observed substitution rate variation is owing to selection. We provide a theoretical argument against strong negative selection generating substitution rate variation in the human-chimpanzee comparison; we support this argument by showing that significant substitution rate variation remains in those regions that are not conserved in the human-pig comparison. We also argue that weak selection and biased gene conversion do not appear to explain the observed substitution rate variation.
Our conclusion of mutation rate variation in the human genome raises the question of its causes. G+C content is significantly positively correlated with substitution rates, and this is partly owing to compositional nonequilibrium; in the high GC regions of the noncoding nonrepetitive genome, the G+C content is decreasing. However, compositional nonequilibrium is not the only factor generating the positive correlation between G+C content and substitution rates; the mutation rates per base pair for both GC→AT and AT→GC mutations are higher in high G+C regions, indicating that some process of mutagenesis or repair covaries with G+C content.
Finally, we compare our work to that of Kumar and Subramanian (2002), who recently analyzed mutation rate variation in mammalian genomes and arrived at rather different conclusions to those presented here. Using the substitution rate at fourfold degenerate sites in protein coding genes as a measure of mutation rate, they report little mutation rate variation between genes spread throughout the genome. An important difference between their study and ours is that they excluded genes for which they had evidence of changing substitution matrices. Although changing substitution matrices does make rate estimation problematic, we believe that it worth considering all sequence data, as we have performed here, for several reasons. Most importantly, it seems unreasonable to make too strong a distinction between mutation patterns and rates when they are so interdependent. If changes in mutation pattern, as described by the instantaneous substitution matrix, are excluded, then the only sort of mutation rate changes considered will be those that affect all possible mutations in the substitution matrix equally. Such changes are probably rare, especially in the context of evidence of considerable mutation bias change during mammalian evolution (L. Duret, M. Semon, G. Piganeau, D. Mouchiroud, and N. Galtier, unpubl.). Additionally, we wish to understand substitution rate variation in all parts of the genome, including those regions in which base composition is not at equilibrium, and not just in the subset of the genome in which substitution matrices have been constant (Kumar and Subramanian excluded nearly half the genes in some of their pairwise species comparisons). Furthermore, it is possible to analyze substitution rates without assuming substitution matrix homogeneity (Galtier and Gouy 1998), although sequence data from multiple species is required.
In conclusion, our study shows that there is mutation rate variation across the primate genome, and that this variation is at least partially owing to compositional nonequilibrium that is caused by changes in selection or mutation biases. Our work, therefore, indicates the future importance of understanding mutation in mammalian genomes in the context of nonequilibrium processes.
METHODS
Primate genomic alignments were built in a number of stages. Chimpanzee and baboon sequence data orthologous to regions of human chromosome 7, generated by the National Institutes of Health Intramural Sequencing Center (NISC) Comparative Sequencing Initiative (http://www.nisc.nih.gov/), were retrieved using National Center for Biotechnology Information (NCBI) Entrez (http://www.ncbi.nlm.nih.gov/). Those sequences reported as “working draft sequence” were broken up into their constituent unordered pieces. Regions of the human genome orthologous to the chimpanzee sequences were identified by BLAST searches (Altschul et al. 1997) against the human genome. These BLAST searches provided positional information for the removal of overlapping chimpanzee sequence. Human-chimpanzee alignments were generated using the default values of ClustalW (Thompson et al. 1994). After the removal by eye of poorly aligned regions, only long contiguous alignments were retained. We obtained 77 human-chimpanzee genomic alignments, with a total ungapped length of 4.7 Mb. Standalone BLAST searches were used to identify those baboon sequences orthologous to regions of the human-chimpanzee sequences, and human-chimpanzee-baboon alignments were generated using the default values of ClustalW. We obtained 43 human-chimpanzee-baboon alignments, with a total ungapped length of 1.7 Mb. Details of the sequences in our alignments are available from the corresponding author.
We performed a number of analyses to categorize different sequence types within our genomic alignments. The positions of alignments in human contigs were determined by BLAST searches against the human genome, and comparison to the contig annotation files available at NCBI allowed the identification of coding regions within alignments. Repetitive sequence elements were identified by RepeatMasker (A.F.A. Smit and P. Green, unpubl.). Conserved and nonconserved regions in the primate alignments were classified on the basis of human-pig comparisons. We performed standalone BLAST searches between the human sequences from our alignments and pig sequences generated by the NISC Comparative Sequencing Initiative, checking for orthologous regions using a purpose-built graphical tool written in Pike. Conserved blocks were then identified using the VISTA alignment server (http://www-gsd.lbl.gov/vista/), setting the minimum block requirement as 75 bp at 85% similarity. These parameters lead to >10% of the regions studied being classified as conserved, far higher than other estimates of the level of conserved noncoding DNA (such as Meisler 2001), which makes the parameters conservative for our purposes. Alternative parameter settings yielded similar results. Because orthologous pig sequence was not available for all primate sequences, only those regions surrounded by conserved blocks were classified as nonconserved.
Lineage-specific substitutions were classified using parsimony (if the human-chimp-baboon sequences are A-C-C, then a C-to-A change is inferred down the human lineage). In the estimation of both pairwise and lineage-specific substitution rates, we ignored those substitutions that may be caused by the hypermutability of methylated CpG dinculeotides (CpG to TpG and CpG to CpA). Distances were not corrected for multiple hits (for such low distances, the use of a multiple hits correction model such as that of Jukes-Cantor [Jukes and Cantor 1969] as used by Chen and Li [2001] and Chen at al. [2001] has very little effect).
As a test of local similarity within alignments, a randomization test was performed to compare the observed sum of differences between adjacent blocks within alignments against the corresponding values generated by 1000 random shuffles of blocks within alignments.
WEB SITE REFERENCES
http://www.ncbi.nlm.nih.gov/; National Center for Biotechnology Information.
http://www.nisc.nih.gov/; National Institutes of Health Intramural Sequencing Center Comparative Sequencing Initiative.
http://www-gsd.lbl.gov/vista/; VISTA alignment server.
Acknowledgments
H.E. is a Royal Swedish Academy of Sciences Research Fellow supported by a grant from the Knut and Alice Wallenberg foundation. This study was supported by the Swedish Research Council. Thanks to Mikael Brandstrom for writing the graphical interpreter of BLAST output and to Laurent Duret for discussions on nonequilibrium composition.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL nick.smith@ebc.uu.se; FAX 46-18-4716310.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.220502.
REFERENCES
- Altschul SF, Madden TL, Schaffer AA, Zhang JH, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acid Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casane D, Boissinot S, Chang BHJ, Shimmin LC, Li WH. Mutation pattern variation among regions of the primate genome. J Mol Evol. 1997;45:216–226. doi: 10.1007/pl00006223. [DOI] [PubMed] [Google Scholar]
- Chen FC, Li WH. Genomic divergences between humans and other hominoids and the effective population size of the common ancestor of humans and chimpanzees. Am J Hum Genet. 2001;68:444–456. doi: 10.1086/318206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen FC, Vallender EJ, Wang H, Tzeng CS, Li WH. Genomic divergence between human and chimpanzee estimated from large-scale alignments of genomic sequences. J Hered. 2001;92:481–489. doi: 10.1093/jhered/92.6.481. [DOI] [PubMed] [Google Scholar]
- Eddy SR. Non-coding RNA genes and the modern RNA world. Nat Rev Genet. 2001;2:919–929. doi: 10.1038/35103511. [DOI] [PubMed] [Google Scholar]
- Eyre-Walker A. Differentiating between selection and mutation bias. Genetics. 1997;147:1983–1987. doi: 10.1093/genetics/147.4.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— Evidence of selection on silent site base composition in mammals: Potential implications for the evolution of isochores and junk DNA. Genetics. 1999;152:675–683. doi: 10.1093/genetics/152.2.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyre-Walker A, Hurst LD. The evolution of isochores. Nat Rev Genet. 2001;2:549–555. doi: 10.1038/35080577. [DOI] [PubMed] [Google Scholar]
- Fujiyama A, Watanabe H, Toyoda A, Taylor TD, Itoh T, Tsai SF, Park HS, Yaspo ML, Lehrach H, Chen Z, et al. Construction and analysis of a human-chimpanzee comparative clone map. Science. 2002;295:131–134. doi: 10.1126/science.1065199. [DOI] [PubMed] [Google Scholar]
- Galtier N, Gouy M. Inferring pattern and process: Maximum-likelihood implementation of a nonhomogeneous model of DNA sequence evolution for phylogenetic analysis. Mol Biol Evol. 1998;15:871–879. doi: 10.1093/oxfordjournals.molbev.a025991. [DOI] [PubMed] [Google Scholar]
- Hurst LD, Williams EJB. Covariation of GC content and the silent site substitution rate in rodents: Implications for methodology and for the evolution of isochores. Gene. 2000;261:107–114. doi: 10.1016/s0378-1119(00)00489-3. [DOI] [PubMed] [Google Scholar]
- Jukes TH, Cantor CR. Evolution of protein molecules. In: Munro HN, editor. Mammalian protein metabolism. New York: Academic Press; 1969. pp. 21–123. [Google Scholar]
- Kimura M. The neutral theory of evolution. Cambridge: Cambridge University Press; 1983. [Google Scholar]
- Kumar S, Subramanian S. Mutation rates in mammalian genomes. Proc Natl Acad Sci. 2002;99:803–808. doi: 10.1073/pnas.022629899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Lercher MJ, Williams EJB, Hurst LD. Local similarity in evolutionary rates extends over whole chromosomes in human-rodent and mouse-rat comparisons: Implications for understanding the mechanistic basis of the male mutation bias. Mol Biol Evol. 2001;18:2032–2039. doi: 10.1093/oxfordjournals.molbev.a003744. [DOI] [PubMed] [Google Scholar]
- Matassi G, Sharp PM, Gautier C. Chromosomal location effects on gene sequence evolution in mammals. Curr Biol. 1999;9:786–791. doi: 10.1016/s0960-9822(99)80361-3. [DOI] [PubMed] [Google Scholar]
- Meisler MH. Evolutionarily conserved noncoding DNA in the human genome: How much and what for? Genome Res. 2001;11:1617–1618. doi: 10.1101/gr.211401. [DOI] [PubMed] [Google Scholar]
- Morgenstern B. DIALIGN 2: Improvement of the segment-to-segment approach to multiple sequence alignment. Bioinformatics. 1999;15:211–218. doi: 10.1093/bioinformatics/15.3.211. [DOI] [PubMed] [Google Scholar]
- Nachman MW, Crowell SL. Estimate of the mutation rate per nucleotide in humans. Genetics. 2000;156:297–304. doi: 10.1093/genetics/156.1.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennacchio LA, Rubin EM. Genomic strategies to identify mammalian regulatory sequences. Nat Rev Genet. 2001;2:100–109. doi: 10.1038/35052548. [DOI] [PubMed] [Google Scholar]
- Piganeau G, Mouchiroud D, Duret L, Gautier C. Expected relationship between the silent substitution rate and the GC content: Implications for the evolution of isochores. J Mol Evol. 2002;54:129–133. doi: 10.1007/s00239-001-0011-3. [DOI] [PubMed] [Google Scholar]
- Sachidanandam R, Weissman D, Schmidt SC, Kakol JM, Stein LD, Marth G, Sherry S, Mullikin JC, Mortimore BJ, Willey DL, et al. A map of human genome sequence variation containing 1.42 million single nucleotide polymorphisms. Nature. 2001;409:928–933. doi: 10.1038/35057149. [DOI] [PubMed] [Google Scholar]
- Smith NGC, Eyre-Walker A. Synonymous codon bias is not caused by mutation bias in G+C-rich genes in humans. Mol Biol Evol. 2001;18:892–896. doi: 10.1093/oxfordjournals.molbev.a003899. [DOI] [PubMed] [Google Scholar]
- Smith NGC, Hurst LD. Molecular evolution of an imprinted gene: Repeatability of patterns of evolution within the mammalian insulin-like growth factor type II receptor. Genetics. 1998a;150:823–833. doi: 10.1093/genetics/150.2.823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— Sensitivity of patterns of molecular evolution to alterations in methodology: A critique of Hughes and Yeager. J Mol Evol. 1998b;47:493–500. doi: 10.1007/pl00013151. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. ClustalW: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acid Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams EJB, Hurst LD. The proteins of linked genes evolve at similar rates. Nature. 2000;407:900–903. doi: 10.1038/35038066. [DOI] [PubMed] [Google Scholar]
- Wolfe KH, Sharp PM, Li WH. Mutation rates differ among regions of the mammalian genome. Nature. 1989;337:283–285. doi: 10.1038/337283a0. [DOI] [PubMed] [Google Scholar]