Abstract
Identification of specific and primary chromosomal alterations during the course of neoplastic development is an essential part of defining the genetic basis of cancer. We have developed a transgenic mouse model for liver neoplasia in which chromosomal lesions associated with both the initial stages of the neoplastic process and the acquisition of malignancy can be analyzed. Here we analyze chromosomal alterations in 11 hepatocellular carcinomas from the c-myc/TGF-α double-transgenic mice by fluorescent in situ hybridization with whole chromosome probes, single-copy genes, and 4′-6-diamidino-2-phenylindole (DAPI-) and G-banded chromosomes and report nonrandom cytogenetic alterations associated with the tumor development. All tumors were aneuploid and exhibited nonrandom structural and numerical alterations. A balanced translocation t(5:6)(G1;F2) was identified by two-color fluorescent in situ hybridization in all tumors, and, using a genomic probe, the c-myc transgene was localized near the breakpoint on derivative chromosome der 6. Partial or complete loss of chromosome 4 was observed in all tumors with nonrandom breakage in band C2. Deletions of chromosome 1 were observed in 80% of the tumors, with the most frequent deletion at the border of bands C4 and C5. An entire copy of chromosome 7 was lost in 80% of the tumors cells. Eighty-five percent of the tumor cells had lost one copy of chromosome 12, and the most common breakpoint on chromosome 12 occurred at band D3 (28%). A copy of chromosome 14 was lost in 72%, and band 14E1 was deleted in 32% of the tumor cells. The X chromosome was lost in the majority of the tumor cells. The most frequent deletion on the X chromosome involved band F1. We have previously shown that breakages of chromosomes 1, 6, 7, and 12 were observed before the appearance of morphologically distinct neoplastic liver lesions in this transgenic mouse model. Thus breakpoints on chromosome 4, 9, 14, and X appear to be later events in this model of liver neoplasia. This is the first study to demonstrate that specific sites of chromosomal breakage observed during a period of chromosomal instability in early stages of carcinogenesis are later involved in stable rearrangements in solid tumors. The identification of the 5;6 translocation in all of the tumors has a special significance, being the first balanced translocation reported in human and mouse hepatocellular carcinoma and having the breakpoint near a tumor susceptibility gene and myc transgene site of integration. Moreover, its early occurrence indicates that this is a primary and relevant alteration to the initiation of the neoplastic process. In addition, the concordance between the breakpoints observed during the early dysplastic stage of hepatocarcinogenesis and the stable deletions of chromosomes 1, 4, 6, 7, 9, and 12 in the tumors provides evidence for preferential site of genetic changes in hepatocarcinogenesis.
Murine liver neoplasia is a multistage phenomenon that may be induced either by chemicals 1 or by targeting overexpression or deletion of selective genes. 2-8 The evolution of neoplastic development in the c-myc/TGF-α mice has been described in detail. 9 The early appearance of dysplastic lesions is followed by formation of foci and nodules, and by 8 months of age 100% of the mice have developed numerous hepatocellular carcinomas. 9 This sequential process of neoplastic development in the c-myc/TGF-α transgenic mice is highly reproducible and therefore offers an attractive model to analyze both cellular and genetic alterations during hepatocarcinogenesis. For example, recent work from our laboratory has demonstrated that disruption of the pRb/E2F pathway and inhibition of apoptosis are significant oncogenic events in the development of liver tumors in the c-myc/TGF-α mice. 10 However, the characterization of the genetic lesions that promote the sequential process of carcinogenesis in the liver is still to be accomplished.
In this study, we have used the c-myc/TGF-α transgenic mouse model of liver carcinogenesis to characterize the time course of cytogenetic alterations during the neoplastic process and to target chromosomal regions for identification of genes that may be important in the development of hepatocellular carcinoma. Our previous results have demonstrated that the ploidy and karyotype of the 3-week-old c-myc/TGF-α double-transgenic hepatocyte is statistically the same as that observed in normal control mice. However, at 10 weeks, the c-myc/TGF-α liver is dysplastic with focal lesions, and 80% of the hepatocytes are aneuploid and 32% have chromosomal breakage. 9,11 The importance of these chromosomal regions in liver neoplasia is further indicated by alteration of the corresponding genetic regions in human hepatocellular carcinoma. 12-21
To evaluate the possible significance of the specific chromosomal alterations seen during the early dysplastic stage of liver tumorigenesis in the c-myc/TGF-α mice we have now analyzed chromosomal alterations in hepatocellular carcinomas from these mice. Here we show that the early sites of breakage on mouse chromosomes 1, 6, 7, and 12 that are associated with genomic instability and correspond with regions of tumor susceptibility 11 are later involved in stable rearrangements and deletions in mouse liver tumors.
Materials and Methods
Construction of Fusion Genes and Generation of Transgenic Mice
The development of the double-transgenic mouse model using (CD I × B6CBA)F1 mice was described by Murakami et al. 8 The TGF-α transgene was constructed by Jhappan et al, 6 and the myc transgene by Murakami et al. 8 The screening for the transgene was performed by Southern blot analysis of tail DNA. 8 The mice were maintained on 50 mmol/L ZnCl2 drinking water from 3 weeks of age to maximize induction of the TGF-α transgene expression. Animals were treated according to National Institutes of Health guidelines.
Perfusion and Chromosome Preparation from Hepatocytes
The cytogenetic aberrations in tumors prepared from c-myc/TGF-α mice at 30 weeks of age as well as age-matched controls has been analyzed using G-banded karyotypes and chromosome paints. The livers of Fl control mice (CDI × B6CBA) were examined at 30 weeks of age. Five control animals were anesthetized with avertine, and the livers were perfused with a collagenase solution as described previously. 22 A section of the liver was tied off with silk thread during the initial wash with Hanks’ balanced salt solution, and the liver piece was placed in formalin for pathological analysis. After the collagenase digestion, hepatocytes were separated from the littoral cells by a Percoll (Sigma Chemical Co., St. Louis, MO) isodensity centrifugation and immediately plated in a 75-cm collagen-type-I-coated flask (Vitrogen 100, Celtrix Laboratories, Santa Barbara, CA) at a density of 5 × 10 6 cells in 15 ml of Dulbecco’s modified Eagle’s medium (DMEM)/F12 medium containing 10% serum and supplemented with 18 mmol/L HEPES, 5 mmol/L sodium pyruvate, 1 mmol/L NaHCO3, 1 mg/ml galactose, 30 μg/ml proline, 100 U/ml penicillin, 100 μg/ml streptomycin, and ITS liquid media supplement (Collaborative Research, Bedford, MA) containing 5 μg/ml insulin and 5 μg/ml transferrin. 11 The medium was changed 2 hours later, and 10 ng/ml murine epidermal growth factor (Gibco/BRL, Gaithersburg, MD) was added. Forty-four hours after plating, colchicine was added to the medium to a final concentration of 0.05 μg/ml. After another 2- to 3-hour incubation, the hepatocytes were removed from the flask with 0.25% trypsin solution and harvested for chromosome analysis by hypotonic treatment (0.075 mol/L KCL) for 9.5 minutes. The hepatocytes were then further treated with 3:1 (v/v) acetic methanol fixative, and slides were prepared as described. 11 Twenty G-banded metaphase spreads of good morphology were analyzed. The karyotypic analysis of the early lesions and the tumors indicated specific breakage on chromosomes 1, 4, 5, 7, 9, 12, 14, and X (Vector Laboratories, Burlingame, CA). To confirm the karyotypic results, chromosomal paints for the significantly altered chromosomes were hybridized to metaphase spreads. Twenty metaphase spreads of good morphology were photographed and analyzed. The c-myc transgene and the chromosome painting probes were labeled with biotin or digoxigenin. The genomic c-myc clone used to localize the transgene and the endogenous c-myc was a generous gift from Dr. Frederic J. Mushinski, Laboratory of Genetics, National Cancer Institute, Bethesda, MD. The conditions for fluorescent in situ hybridization (FISH), the detection of the signal, and the digital imaging were performed as described previously. 22,23 Lesions were mapped to specific bands using the karyotypes and ideograms according to Cowell. 24
Isolation and Chromosome Preparation from Tumor Cells
The hepatocellular carcinomas (HCCs) were isolated from 30-week-old c-myc/TGF-α mice. Eleven primary tumors were analyzed by FISH with chromosome probes and 4′-6-diamidino-2-phenylindole (DAPI)-banded chromosomes and three tumors by banding alone. Cells from tumors > 1 cm were prepared for transplantation into nude mice to confirm the tumorigenicity of each sample. The minced tumor tissue was digested at 37°C in a solution of 0.035% type IV collagenase (Gibco/BRL) and 0.015% type I collagenase (Sigma). After a 15-minute digestion, the supernatant was removed, and the cells washed in plating media. The cells were pelleted at 700 rpm, resuspended, and washed in fresh medium. The washed and resuspended tumor cells were then plated on a 35-mm plastic culture dish and allowed to attach for 5 minutes to preferentially select out contaminating fibroblasts. The unattached tumor cells were then plated on collagen-coated plates (Vitrogen 100, Celtrix Laboratories) at the same density and culture conditions as used for primary hepatocytes. This procedure was repeated three times for each tumor cell population. The metaphase spreads of the tumor cells were harvested and analyzed according to the same protocol used for primary hepatocytes isolated from control animals. The data were analyzed as the percentage and SD of cells with a specific aberration. The incidence of chromosome damage in tumors was compared with the expected incidence in the hepatocytes from five F1 age-matched controls using standard χ 2 analysis.
Results
Specific Chromosome Breakage in Diploid Hepatocytes Isolated from 10-Week-Old c-myc/TGF-α Mice
The hepatocytes isolated from 10-week-old c-myc/TGF-α mice were 9% diploid. The total breakage in the diploid hepatocytes was the same as the cells with higher chromosome number. The diploid population was confirmed to have less specific breakage than the tetraploid and octaploid cells. 11 Significant breakage was observed at chromosome regions 1C4/C5, 5G1, 6F2, 7D3, and 12D3 (Table 1) ▶ . Although elevated breakage of chromosomes 4 and 9 was observed previously in the total hepatocytes isolated from the 10-week-old c-myc/TGF-α mice, no significant breakage of chromosome 4 or 9 was observed in the diploid population.
Table 1.
Percentage of Diploid Cells Demonstrating Nonrandom Chromosomal Lesions in Total Liver Isolated from 10-Week-Old c-myc/TGF-α Mice and Diploid Cell Population
| Chromosome number and band region | % of cells with altered band region (± SD) |
|---|---|
| 1 C4/C5 | 17.0 ± 5.0 |
| 4 C2, C6 | <1.0 |
| 5 G1 | 8.0 ± 2.0 |
| 6 F2 | 7.0 ± 2.0 |
| 7 D3 | 10.0 ± 3.0 |
| 9 B5 | <1.0 |
| 12 D3 | 10.0 ± 3.0 |
Values are expressed as mean percentage of cells with alterations in specific chromosome bands. The standard deviation is indicated to the right of the percent. At least 50 cells from each of five animals were examined. Hepatocytes from 10-week-old c-myc/TGF-α mice and (CD1 × B6CBA)F1 mice were isolated, cultured, and prepared for chromosome analysis as described in Materials and Methods. At least 50 banded metaphase spreads were analyzed for each animal. Lesions for each chromosome were mapped using the ideograms of Cowell. 24
Cytogenetic Alterations in Mouse Liver Tumors Isolated from 30-Week-Old c-myc/TGF-α Mice
Deletions on chromosome 1, 4, 5, 6, 7, 9, 12, 14, and X were observed in all of the tumors examined (Table 2) ▶ . Loss of all or part of chromosome 1 was observed in 80% of the tumor cells (Tables 2 and 4 ▶ ▶ ; Figure 1 ▶ ) and in all of the tumors. The most frequent breakpoint was observed on chromosome 1 at the border of bands C4/C5 (Table 3) ▶ . The whole or part of chromosome 4 was lost in all of the tumors examined (Tables 2 and 4 ▶ ▶ ; Figure 2a ▶ ), with the most frequent breakpoint at band C2-terminal (Figures 2a and 3) ▶ ▶ . The minimal deletion of chromosome 4 at band C6 was observed in 23 ± 9% of the cells (Table 3) ▶ . An entire copy of chromosome 7 was lost in 86.0 ± 10% of the tumors cells (Table 2) ▶ , and 70% of the cells also had significant deletions of bands B5 and D3 (Figures 1, 2, and 3 ▶ ▶ ▶ ; Table 3 ▶ ). The minimal region of deletion at 7D3 was observed in 40 ± 18% of the neoplastic cells (Table 3) ▶ . The terminus of chromosome 6 was broken at 6F2-ter in all of the tumor cells (Table 3 ▶ ; Figure 3 ▶ ). In addition to the break, all of the cells had a rearrangement of chromosome 6F2-ter (Tables 3 and 4 ▶ ▶ ; Figure 3 ▶ ). Sixty-six percent of the cells had lost a copy of chromosome 9. Eleven percent of the tumor cells had deletions at band 9E1 (Table 3 ▶ ; Figure 3 ▶ ). Eighty-six percent of the cells had only one normal copy of chromosome 12 (Table 2 ▶ ; Figure 1 ▶ ). The most frequent breakpoint and the minimal deletion on chromosome 12 occurred at band D3 (28 ± 15%; Table 3 ▶ ; Figures 2b and 3 ▶ ▶ ). An entire copy of chromosome 14 was lost in 72 ± 8% of the tumor cells (Tables 2 and 4) ▶ ▶ . A deletion on 14qE1 was observed in 33 ± 15% of the cells (Table 3 ▶ ; Figure 3 ▶ ). The whole or part of the X chromosome was lost in 98% of the tumor cells (Tables 2 and 4 ▶ ▶ ; Figure 1 ▶ ), and 50 ± 25% of the cells had deletions of a segment of the X chromosome. Loss of material from band X D1-ter was found in 13 ± 8% and X F1 in 25 ± 11% of the cells (Table 3 ▶ ; Figure 3 ▶ ).
Table 2.
Percentage of Cells with Chromosomal Alterations Isolated from 32-Week-Old Mice
| Chromosome number | |||||||
|---|---|---|---|---|---|---|---|
| 1- | 4- | 7- | 9- | 12q | 14- | X- | |
| Control | <1 | 6.0 ± 3.0 | <1.0 | 6.0 ± 3.0 | <1.0 | <1.0 | <1.0 |
| Tumors | 80.0 ± 26 | 98.0 ± 3 | 86.0 ± 10 | 75.0 ± 13 | 86.0 ± 7 | 72.0 ± 8 | 98.0 ± 20 |
Eleven hepatocellular carcinomas isolated from the c-myc/TGF-α mice and hepatocytes from five age-matched (Cd 1 × B6CBA)F1 mice were cultured as described in Materials and Methods, and metaphase preparations were made from the first two divisions. The metaphase spreads from 11 tumors and hepatocytes from five age-matched controls were hybridized with chromosomal paints for chromosomes 1, 4, 5, 7, 8, 9, 11, and 12 and the X chromosome. At least 20 metaphase spreads were analyzed for each chromosome paint. The inverted DAPI from each hybridized metaphase spread was analyzed to determine the band region that was altered. A minimum of 20 G-banded metaphase spreads were karyotyped for each tumor and control. The data are expressed as the mean percent ± SD of cells with a specific aberration.
Table 4.
Karyotype of Tumor Cells Isolated from 30-Week-Old Mice
| Tumor number | Karyotype |
|---|---|
| 241D | −1, 1C4 ter-, −3, 4Cr ter-,−5,−6, T (5:6) (G1 ter/F2), 7dup(7D1/D8),+7,−9, 12D1 ter-,−12,−X, XF1 ter-; N-50 |
| 220A | 1C4ter-,−3,−4, −5,−6, T (5:6) (G1 ter/F2) 7Dter-,−7,−9, T(9:12) (E1 ter/D1), −12, 14 E1 ter-,−X, XD1ter-, N=60 |
| 249C | 1C4/C5 ter-, 4C6, 6F1ter-, T (5:6) (G1 ter/F2), 7 dup (7B to D1), T(7D1/9E1),+8,−12, 14E1ter-,−X, N=65 |
| 261.1 | 1C4./C5 ter-,−4, 4/C5 ter-m 5G1ter-,−6, 6 F1 ter-, T (5:6) (G1 ter/F2)−7, 7 D1 ter-,−12, 12 D3ter-,−14,−17, XF1 ter-, N= |
| 221 | +1, 1 C4/C5 ter-,+2,−4, 4 C6 ter-, 5 F1 ter-,−6, 6F2 ter-,T (5:6) (G1 ter/F2),−7, 7 B5 ter-,−9, 9E1 ter-,−10,−12, 12D3 ter-, 14 E1 ter-,−14,−19, −X, X F1 ter- N=60 |
| 267 T1 | −1,−4,−5,−6, 6F6ter-, T (5:6) (G1 ter/F2),−7, 7D3ter-,−9, 9E1ter-,−12,−14, 14B1ter-,−X, XD1ter-, N=55 |
| 267 T2 | −1, T (1:14) (C4/B1ter),−2,−4,−5, −6, 6F2 ter-, T (5:6) (G1 ter/F2), T (6F1 ter/5G1),−7, 7 B5 ter-, −9, 9E1 ter-,−10,−12, 12 D1 ter-,+13,−14, 14 B1 ter-,−X, XD1 ter-, N=60 |
| 267 T3 | 1 C4/C5 ter-, T (2:3) (),−4, 5 G1 ter-,−6, T (5:6) (G1 ter/F2),−7, 7 B1 ter-,−9, 9 E1 ter-,−12, 12 D3 ter-, 14 E1 ter-,−14,−X, X F1 ter- N=60 |
| 223 | −1,−2, 1 C5 ter-, 1 C6 ter-,−2,−4, 4 C6 ter-, 5 D1 ter-,−5,−6, 6F1 ter-, T (5:6) (G1 ter/F2), 7 D ter-, T 7D1/XD1ter),−9, 9 E1 ter-,−12, Iso-12 (D3 ter-) T(12A1-D1/12A1-D1),+13,−14, 14 B1 ter-,−X, Iso-X (D1 ter-)I B=56 |
| 263 | −1,−4, 4 C6 ter-,−6, 6 F2 ter-, T (5:6) (G1 ter/F2), 7 D3 ter-,+7,−9,−12, 12 D3 ter-,−14, 14 E1 ter-,−X X D1 ter- N=65 |
| 241 | −1,−4, 4C2 ter-, 5 G1ter-,−6, 6 F2 ter-, T (5:6) (G1 ter/F2),−9, 9A5 ter-,−12, 12 D3 ter-,−14, 14 B1 ter-, −X, XD1ter- N=55 |
Figure 1.
A band karyotype of a metaphase from a 30-week-old c-myc/TGF-α double-transgenic mouse liver tumor demonstrating deletion of chromosome 1 band C4 and E, loss of chromosome 2, deletion and loss of chromosome 4, deletion of chromosome 7, a deletion of 17, and loss of the X chromosome. Chromosome 12 has a deletion of band D1 and a fusion of the deleted copy at the centromere resulting in a metacentric chromosome. The major breakpoints on chromosomes 1, 4, 5, 7, 9, and 12 and the loss of the X chromosome are indicated by arrows. A balanced translocation 5;6 identified by FISH is only partially detected by G-banding as a terminal deletion of 5, but the translocated material on 6 is not discernible.
Table 3.
Frequency of Chromosomal Breakpoints in Tumor Cells Isolated from 30-Week-Old Mice
| Chromosome number | % of cells with altered band (± SD) |
|---|---|
| 1 band C4/C5 | 40.0 ± 13.0 |
| 4 band C2 | 30.0 ± 10.0 |
| 5 band G1 | 98.0 ± 10.0 |
| 6 band F2 | 98.0 ± 20.0 |
| 7 band B5; band D3 | 31.0 ± 8.0; 40.0 ± 18.0 |
| 9 band E1 | 11.0 ± 5.0 |
| 12 band D3 | 27.0 ± 15.0 |
| 14 band E1 | 32.0 ± 8.0 |
| X band D1; band F1 | 13.0 ± 8.0; 25.0 ± 10.0 |
Fourteen hepatocellular carcinomas isolated from the c-myc/TGF-α mice and hepatocytes from five age-matched (CD 1 × B6CBA)F1 mice were cultured as described in Materials and Methods, and metaphase preparations were made from the first two divisions. The metaphase spreads from 11 tumors were hybridized with chromosomal paints for chromosomes 1, 4, 5, 6, 7, 8, 9, 11, and 12 and the X chromosome. Twenty metaphase spreads were analyzed for each chromosome paint. Twenty G-banded metaphase spreads were karyotyped for each tumor and control. The data are expressed as a percentage of cells with the specific band alteration.
Figure 2.
FISH analysis of the c-myc/TGF-α tumors. a: Metaphase spreads after hybridization with biotinylated probes for chromosome 4 demonstrating deletion and translocation of chromosome 4 at band C2 indicated by the arrow. b: An example of a deletion of chromosome 12 at region D3 and a centromeric fusion of two deleted chromosomes demonstrated by painting as indicated by the arrow. c: The photograph is a partial metaphase hybridized with a genomic c-myc probe. Fluorescent signals were observed on chromosome 15 at the normal c-myc gene location (indicated by arrows) and at the terminus of chromosome 5, which was translocated to another chromosome 6 containing the translocated 5 material and the c-myc transgene sequences. The details of the hybridization are described in Materials and Methods. d: The slide was destained and rehybridized with chromosome 5 probe to confirm the localization of the c-myc transgene to 5G1-ter. e: The balanced translocation of t(5;6)(G1:F1) was identified by two-color FISH with digoxigenin (red) and biotin-labeled (yellow) probes for chromosomes 5 and 6, respectively. The localization of the c-myc transgene was therefore to 5G1-ter translocated to 6G-1. The arrows on the right indicate a derivative chromosome 5 in red with a yellow translocation from chromosome 6 as a yellow band. The arrows on the left indicate a derivative chromosome 6 in yellow with the translocation of 5 as a red band.
Figure 3.
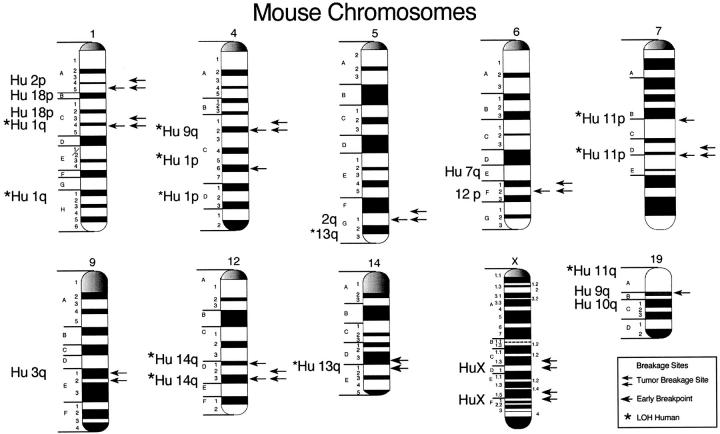
Ideograms of chromosomes 1, 4, 5, 6, 7, 9, 12, 14, X, and 19. The single arrows indicate regions that were found to have a statistically significant number of chromosome breaks in 10-week-old c-myc/TGF-α double-transgenic mouse livers. The double arrows indicate regions that were found to have statistically significant chromosome breakage in metaphase spreads isolated from hepatocellular carcinoma prepared from 30-week-old double-transgenic c-myc/TGF-α mice. The homologous region of the human chromosome is indicated at the left of the figure. A starred human homologous region indicates loss of heterozygosity in human liver tumors.
Balanced Translocation of Chromosomes 5 and 6 in the c-myc/TGF-α Transgenic Mouse Model
The site of the c-myc transgene integration was localized by FISH on derivative chromosome 6 carrying the translocated segment 5G1-ter (Figure 2, c and d) ▶ . The breakpoint at 5G1 was proximal to the c-myc transgene (Tables 2 and 4 ▶ ▶ ; Figure 2d ▶ ). The localization of the endogenous c-myc on chromosome 15 served as an internal control (Figure 2c) ▶ . The balanced translocation between chromosome 5G1-ter to 6F1 and 6F2-ter to 5F1 was identified by two-color FISH with probes for chromosomes 5 and 6 in all of the tumors examined (Figure 2e) ▶ . This balanced translocation was not discernible on G-banded karyotypes due to the small size of the translocated chromosome segments (Figure 1) ▶ . The normal copy of chromosomes 5 and 6 was lost in 80% of the cells examined and significantly lost in all of the tumors.
Comparison of Specific Chromosomal Breakpoints in Dysplastic Liver Lesions at 10 Weeks and in HCCs from c-myc/TGF-α Transgenic Mice
Figure 3 ▶ is an ideogram comparing the chromosomal breakpoints that were previously reported in early lesions observed in 10-week-old c-myc/TGF-α mouse livers to the breakpoints observed in the tumors. The most frequent breakpoints that were observed in tumor metaphase spreads on chromosomes 1, 4, 6, 7, 9, and 12 were the same band regions observed in early liver lesions. Alterations of mouse chromosome 14 and X are seen only in tumor cells.
Transplantation of Mouse Liver Tumors into Nude Mice
All of the cell lines established from the liver tumors from the c-myc/TGF-α mice formed tumors when transplanted onto nude mice. The histological pattern of the tumors were that of well to intermediate differentiated HCC (data not shown).
Discussion
We have established a time course of cytogenetic alterations in the transgenic mouse model of hepatocarcinogenesis. The breakage of chromosomes 1, 5, 6, 7, and 12 in diploid hepatic cells was observed before the appearance of morphologically distinct liver lesions in the transgenic mouse model. The breakpoints of these chromosomes occurred in loci that are associated with susceptibility to liver tumorigenesis. Breakage of chromosomes 4, 9, 14, and X seem to be later events in this model of liver neoplasia. The correspondence between the stable deletions of chromosomes 1, 5, 6, 7, and 12 and regions of breakage identified at an early stage of hepatocarcinogenesis suggest the importance of these linkage groups in liver neoplasia.
The region of chromosome 1 that was frequently deleted in the c-myc/TGF-α tumors has been reported to have a locus that confers tumor resistance in many strains of mice. A tumor resistance gene in the B6C3Fl mouse near the midpoint of the chromosome has been reported by Bennett. 25 Lee and colleagues 26 identified a tumor resistance gene on chromosome 1 in the DBA/2J mice. The distal region of chromosome 1 has genetic linkage groups that confer susceptibility in B6C3F1, C3H, and C57BR/cd mice. 25-27 Deletions of the band regions C5 and C4 have been observed in liver tumors from B6C3F1. 1 The analysis of congenic lines has demonstrated that the C4/C5 locus has genes that influence the susceptibility to the induction of lung and intestinal tumors. 28,29 The tryosine kinase Abll, which is related to the abelson proto-oncogene, 30 and RXRγ, the retinoid X receptor-γ, 31 a kinetechore protein, 32 as well as a microtubule-associated protein 33 mapped to this locus. The Scfr1 gene is an important predictor of the size of the stem cell population of the hematopoietic cells and is located in the fragile site of mouse chromosome 1. 34 The breakage of the mouse chromosome 1 C4/C5 region was observed in the early liver lesions of the c-myc/TGF-α mice 11 and was the most significant breakage observed in the diploid cells. The diploid population is the subset of the liver cells that may give rise to liver tumors as has been proposed in the rat liver model. 35,36 The deletion of the linkage group that contains two spindle-associated proteins as well as a gene regulating the size of the stem cell compartment was concurrent with the onset of aneuploidy and expansion of a liver cell population in this mouse liver model of carcinogenesis. The deletion of the corresponding region of the human chromosome, 1q32–41, has been observed in human liver neoplasia. 37
The balanced translocation of chromosomes 5 and 6 in the c-myc/TGF-α tumors is the first rearrangement of this type to be described in either human or mouse liver tumors. As the c-myc transgene is near the breakpoint of this translocation, it is tempting to speculate that the insertion of the transgene has triggered the rearrangement. Balanced translocations commonly lead to activation of the proto-oncogene or to the formation of new oncogenic chimeric genes. Both oncogene products and gene fusion proteins are often transcriptional factors. Thus, the disruption of the transcriptional control might be a critical and etiologically relevant alteration in the development of certain forms of cancers. 38 Also, the consistency of this alteration indicates that the linkage groups on 5G1 and 6F2 may be important in the etiology of the liver tumors. The loss of the normal chromosomes 5 and 6 indicates a possible selection for the translocation. The translocation near the c-myc transgene did not result in the up-regulation of c-myc in the tumors. 9 The site of rearrangement of chromosome 5 is near a tumor susceptibility gene at the Zp3 locus 26 that has been identified to cause a twofold increase in tumor incidence in DBA mice. Loss of the distal region of chromosome 5 has been observed in mouse liver tumors. 1 The Zp3 locus corresponds to human 13q12–13 proximal to the retinoblastoma gene. 39 Human 13p12–13 is frequently altered in hepatocellular carcinoma. 40,41 The region on the 6F2 band is associated with increased sensitivity to lung tumors, and loss of chromosome 6 has been reported in mouse liver tumor. 41 The syntenic region of human chromosome 12 is deleted in hepatocellular carcinoma. 42-45
The region of mouse chromosome 7 that is deleted in the liver tumors corresponds to rat Iq4.l and human 11p15.5 and 11p13. Rearrangements of the homologous region on rat 1q4l and human 11p13 and 11p15 have been reported in hepatocellular carcinoma. 13,14,45,46 The loss of human chromosome 11p has also been observed in Wilms tumor and hepatoblastoma. 45-47 Garibaldi et al 48 have identified a tumor susceptibility gene near the imprinted locus of insulin-like growth factor II, H19, and p57 as well as the H-ras gene on mouse chromosome 7. Although activated H-ras has been observed in mouse liver tumors, 49,50 Ohgaki et al 51 did not detect a mutated H-ras in the c-myc/TGF-α mice. Loss of the normal H-ras allele has been reported when one copy of H-ras is mutated. 52,53 The maternal copy of H19 on chromosome 7 is lost in SV40-T transgenic mouse liver tumors. 54 Microcell fusion with human chromosome 11 will inhibit the tumor phenotype of tumor cells. 55
The breakage observed in the c-myc/TGF-α mouse on chromosome 12 is in the region of the chromosome designated by Garibaldi et al 48 to have a tumor resistance gene. The proximal region of chromosome 12 has a gene that confers sensitivity to liver tumors in DBA/2J mice. 26 Loss of heterozygosity of the same band region of mouse 12 has been reported in hepatocellular carcinoma isolated from C57BL/6JBY mice, 1 and furthermore, the same linkage group is lost in rat liver tumors. 47 The homologous region on human 14q32 is frequently deleted in hepatocellular carcinoma. 15,16
Loss of the distal portion of mouse chromosome 9 was a common event in the c-myc/TGF-α liver tumors. Deletion of the same region has been observed in chemically induced liver tumors isolated from B6C3F1 mice. 1 The distal portion of mouse chromosome 9 is homologous to human 3p. A candidate tumor suppressor gene in this region is the FHIT gene. 56 Although deletion of 3p has not been reported in human liver cancer, the FHIT gene is one of the most frequently reported deletions in human cancer. The transforming growth factor β II receptor (TGF-β receptor II) is mapped to mouse chromosome 9, 57 and human 3p 58 is another candidate tumor susceptibility gene. Not only is the expression and binding of TGF-β II receptor down-regulated in human liver tumors, 59 80% of the c-myc/TGF-α tumors demonstrate reduced expression. 10
The loss of chromosomes 4, 9, X, and 14 appeared to be a later event in the c-myc/TGF-α tumors. Another hepatocyte tumor resistance locus has been reported on chromosome 4 near the break site observed in the hepatocellular carcinomas isolated from the c-myc/TGF-α mice. 10,60 Deletions of chromosome 4 have been observed in transformed liver epithelial cells at the D4WSMI locus. 44,60,61 Loss of heterozygosity of mouse chromosome 4 has been observed in C3H/MSM mouse hepatocellular carcinoma. 27,44 The fragile region of chromosome 4 is homologous to human chromosome 1p32–41 and 9p21. Deletions of human 1p have been reported in human hepatocellular carcinoma and breast, lung, and colon carcinoma. 62,63 Mouse chromosome 4 and human chromosome 1 suppress the malignant phenotype in cell fusions of normal and tumor cells. 62,63 The loss of heterozygosity of chromosome 4 is associated with spontaneous immortalization. 44,60-65 The cyclin-dependent kinase inhibitors CDKN2/p16, p15, and p19 are deleted in murine liver and lung carcinoma. 66,67 Another candidate susceptibility gene, Mom-1, which is known to modulate the expression of the intestinal tumor phenotype in the APC mice has been mapped to the breakpoint on mouse 4. 68 Phospholipase A2, which resides at 66.6 cM, has been implicated as a candidate gene for Mom-1. The expression of phospholipase A2 has been shown to be a prognostic indicator in human breast and liver cancer. 69-74 The midpoint of chromosome 4 has a number of candidate tumor suppressor genes that are lost in the later stages of carcinogenesis.
Deletion of chromosome 14 was a common deletion in the tumor cells; however, this deletion was not observed in the early lesions isolated from the c-myc/TGF-α mice. Loss of heterozygosity of mouse chromosome 14 has been associated with the later stages of mouse neoplasia. 75 The retinoblastoma gene, 39,76 the urokinase plasminogen activator, and retinoic receptor-related gene are candidate susceptibility genes that are located in the deleted region of chromosome 14. Although significant loss of the syntenic region of the human chromosome 13q has been found in human hepatocellular carcinoma, no significant mutation of Rb has been associated with human liver neoplasia. 39,77 This corresponding region on human 13q is often deleted without a mutation of retinoblastoma gene in chronic lymphocytic leukemia and mammary and lung as well as human liver neoplasia. 78-80
The loss of the X chromosome in 98% of the cells indicates the importance of this region in the c-myc/TGF-α tumor model. Other murine and rat models of carcinogenesis have also reported the loss of the X chromosome in liver tumors. 47 The expression of connexin 32 genes mapped to the X chromosome has been known to be suppressed in neoplastic hepatocytes and other tumor cell populations. 81 The suppression of the tumor phenotype of neoplastic cells after transfection with connexin 32 suggests that this gene can act as a tumor suppressor gene. 82
Specific breakage indicates genetic regions that are important to the development of hepatocellular carcinoma. The regions on chromosomes 1, 4, 5, 6, 7, 9, 12, 14, and X occurred at high frequency and have been observed to have tumor susceptibility genes as well as loss of heterozygosity in the c-myc/TGF-α mouse liver tumors. The breakpoints on mouse chromosomes 1, 4, 5, 6, 7, 9, 12, 14, and X correspond to human 1q, 1p, 3p, 13q, 11p15.5 and 11p12–13, 11p13, 12p, 14q32, and X. 81,82 Human 1q, 1p, 11p, 12p, 13q, and 14q are also rearranged in human liver tumors. The breakpoints on chromosomes 1, 4, 5, 6, 7, 12, and 14 correspond to tumor susceptibility genes in mouse and loss of heterozygosity in mouse as well as in human.
Although many investigations have determined specific gene changes in hepatic tumors, this study demonstrates that specific chromosomal breakpoints observed during the early stages of mouse liver carcinogenesis are later involved in stable rearrangement in the tumors. The results of this study further indicate that the breakage and deletion of chromosomes 1, 5, 6, 7, and 12 may be early events, and the deletion of chromosomes 4, 12, 14, and X are later events in this liver tumor model. The alteration of the same genetic linkage groups in mouse and human liver tumors indicates that these regions of the genome are critical in the etiology of the neoplastic process. Due to the highly conserved genetic linkage groups between human and mouse, 81-83 further characterization of these breakpoints may provide critical information on tumor susceptibility genes that are important in the early development of human hepatocellular carcinoma.
Footnotes
Address reprint requests to Dr. Snorri S. Thorgeirsson, National Cancer Institute, Building 37, Room 3C28, 37 Convent Drive, MSC4255, Bethesda, MD 20892-4255. E-mail: snorri_thorgeirsson@nih.gov.
References
- 1.Davies LM, Caspary WJ, Sakallah SE, Maronpot R, Wiseman R, Barrett JC, Elliott R, Hozier JC: Loss of heterozygosity in spontaneous and chemically induced tumors of the B6C3F1 mouse. Carcinogenesis (Lond) 1994, 15:1637-1645 [DOI] [PubMed] [Google Scholar]
- 2.Adams J, Cory S: Transgenic models of tumor development. Science 1991, 254:1161-1166 [DOI] [PubMed] [Google Scholar]
- 3.Hanahan D: Dissecting multistep tumorigenesis in transgenic mice. Annu Rev Genet 1988, 22:479-519 [DOI] [PubMed] [Google Scholar]
- 4.Merlino G: Transgenic mice: designing genes for molecular models. ed 3 Arias I Boyer J Fausto N Jakoby W Schater D eds. The Liver: Biology and Pathobiology, 1994, :pp 1579-1590 Raven Press, New York [Google Scholar]
- 5.Sandgren EP, Quaife CJ, Pinkert CA, Palmiter RD, Brinster RL: Oncogene-induced liver neoplasia in transgenic mice. Oncogene 1989, 4:7l5-724 [PubMed] [Google Scholar]
- 6.Jhappan Ch, Stahle CN, Harkins RN, Fausto N, Smith GH, Merlino GT: TGF-α over-expression in transgenic mice induces liver neoplasia and abnormal development of the mammary gland and pancreas. Cell 1990, 61:1137-1146 [DOI] [PubMed] [Google Scholar]
- 7.Sandgren EP, Luetteke NC, Qiu TH, Palmiter RD, Brinster RL, Lee DC: Transforming growth factor α dramatically enhances oncogene induced carcinogenesis in transgenic mouse pancreas and liver. Mol Cell Biol 1993, 13:320-330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Murakami H, Sanderson N, Nagy P, Marino P, Merlino GT, Thorgeirsson SS: Transgenic mouse model for synergistic effects of nuclear oncogenes and growth factors in tumorigenesis: interaction of c-myc and transforming growth factor α in hepatic oncogenesis. Cancer Res 1993, 53:1719-1723 [PubMed] [Google Scholar]
- 9.Santoni-Rugiu E, Nagy P, Jensen MR, Factor V, Thorgeirsson SS: Evolution of neoplastic development in the liver of transgenic mice co-expressing c-myc and transforming growth factor-α. Am J Pathol 1996, 149:401-428 [PMC free article] [PubMed] [Google Scholar]
- 10.Santoni-Rugiu E, Jensen MR, Thorgeirsson SS: Disruption of the pRb/E2F pathway and inhibition of apoptosis are major oncogenic events in liver constitutively expressing c-myc and transforming growth factor α. Cancer Res 1998, 58:123-134 [PubMed] [Google Scholar]
- 11.Sargent LM, Sanderson ND, Thorgeirsson SS: Ploidy and karyotypic alterations associated with early events in the development of hepatocarcinogenesis in transgenic mice harboring c-myc and transforming growth factor α transgenes. Cancer Res 1996, 56:2137-2142 [PubMed] [Google Scholar]
- 12.Yeh S-H, Chen P-J, Lai M-Y, Wang C-C, Chen D-S: Frequent genetic alterations at the distal region of chromosome 1p in human hepatocellular carcinomas. Cancer Res 1994, 54:4188-4192 [PubMed] [Google Scholar]
- 13.Wang HP, Rogler C: Deletions in human chromosome arms 11p and 13q in primary hepatocellular carcinomas. Cytogenet Cell Genet 1988, 48:72-78 [DOI] [PubMed] [Google Scholar]
- 14.Buetow KH, Murray JC, Israel JL, London WT, Smith M, Kew M, Blanquet V, Brechot C, Redeker A, Govindarajah S: Loss of heterozygosity suggests tumor suppressor gene responsible for primary hepatocellular carcinoma. Genetics 1989, 86:8852-8856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tsuda H, Zhang W, Shimasato Y: Allele loss on chromosome 16 associated with progression of human hepatocellular carcinoma. Proc Natl Acad Sci USA 1990, 87:6791-6794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Emi M, Fujiwara Y, Nakajima T, Tsuchiya E, Tsuda H, Hirohashi S, Maeda Y, Tsuruta K, Miyaki M, Nakamura Y: Frequent loss of heterozygosity for loci on chromosome 8p in hepatocellular carcinoma, colorectal cancer and lung cancer. Cancer Res 1992, 52:5368-5372 [PubMed] [Google Scholar]
- 17.Zhang W, Hirohashi S, Tsuda H, Shimasato Y, Yokata J, Terada M, Sugimura T: Frequent loss of heterozygosity on chromosome 16 and 4 in human hepatocellular carcinoma. Jpn J Cancer Res 1990, 81:108-111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fujimori M, Tokino T, Hino O, Kitagawa T, Imamura T, Okamoto E, Mitsunobu M, Ishikawa T, Nakagama H, Yagura M, Matsubara K, Nakamura Y: Allelotype study of primary hepatocellular carcinoma. Cancer Res 1991, 51:89-93 [PubMed] [Google Scholar]
- 19.Simon D, Knowles BB, Weith A: Abnormalities of chromosome 1 and loss of heterozygosity on 1p in primary hepatomas. Oncogene 1991, 6:765-770 [PubMed] [Google Scholar]
- 20.Walker GJ, Hayward NK, Falvery S, Cooksley WGE: Loss of somatic heterozygosity in hepatocellular carcinoma. Cancer Res 1991, 51:4367-4370 [PubMed] [Google Scholar]
- 21.Fujimoto Y, Hampton LL, Wirth PJ, Wang NJ, Xie JP, Thorgeirsson SS: Alterations of tumor suppressor genes and allelic losses in human hepatocellular carcinomas in China. Cancer Res 1994, 54:281-285 [PubMed] [Google Scholar]
- 22.Zimonjic DB, Popescu NC, Matsui T, Ito M, Chihara K: Localization of the human cholecystokinin-B/gastrin receptor gene (CCKBR) to chromosome 11p15.5-p15.4 by fluorescence in situ hybridization. Cytogenet Cell Genet 1994, 65:184-185 [DOI] [PubMed] [Google Scholar]
- 23.Zimonjic DB, Rezanka L, Di Paolo JA, Popescu NC: Refined localization of the erbB-3 proto-oncogene by direct visualization of FISH signals on LUT-inverted and contrast-enhanced digital images of DAPI-banded chromosomes. Cancer Genet Cytogenet 1995, 80:100-102 [DOI] [PubMed] [Google Scholar]
- 24.Cowell JK: A photographic representation of the variability in the G-banded structure of the chromosomes in the mouse karyotype. Chromosoma 1984, 89:294-320 [DOI] [PubMed] [Google Scholar]
- 25.Bennett ML, Drinkwater NR: Localization of the Htm gene to the distal region of mouse chromosome one. Proc Am Assoc Cancer Res 1993, 34:144 [Google Scholar]
- 26.Lee G-H, Bennett LM, Carabao RA, Drinkwater NR: Identification of hepatocarcinogen-resistance genes in DBA/2 mice. Genetics 1995, 139:387-395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Drinkwater NR: Genetic control of hepatocarcinogenesis in C3H mice. Drug Metab Rev 1994, 26:201-208 [DOI] [PubMed] [Google Scholar]
- 28.Van Wezel T, Stassen AP, Moen CJ, Hart AA, van der Vvalk MA, Demant P: Gene interaction and single gene effects in colon tumour susceptibility in mice. Nature Genet 1996, 14:468-470 [DOI] [PubMed] [Google Scholar]
- 29.Herzog CR, Chen B, Wang Y, Schut HAJ, You M: Loss of heterozygosity on chromosomes 1, 11, 12 and 14 in hybrid mouse lung adenocarcinomas. Mol Carcinog 1996, 16:83-90 [DOI] [PubMed] [Google Scholar]
- 30.Seldin MF, Kruh GD: Mapping of ABII within a conserved linkage group on distal mouse chromosome 1 syntenic with human chromosome using an interspecific cross. Genomics 1989, 4:221-223 [DOI] [PubMed] [Google Scholar]
- 31.Hoopes CW, Taketo M, Ozato K, Liu Q, Howard TA, Linney E, Seldin MF: Mapping of the mouse Rxr loci encoding nuclear retinoid X receptors RXR α, RXR β, and RXR γ. Genomics 1992, 14:611-617 [DOI] [PubMed] [Google Scholar]
- 32.Testa JR, Zhou J-Y, Bell DW, Yen TJ: Chromosomal localization of the genes encoding the kinetochore proteins CENPE and CENPF to human chromosomes 4q24–25 and 1q32–q41, respectively by fluorescence in situ hybridization. Genomics 1994, 23:691-693 [DOI] [PubMed] [Google Scholar]
- 33.Lafuse WP, Brown D, Zwilling BS: Assignment of the microtubule-associated protein 2 gene to mouse chromosome 1. Mammalian Genome 1992, 3:48-51 [DOI] [PubMed] [Google Scholar]
- 34.Mulleer-Sieburg CE, Riblet R: Genetic control of the frequency of hematopoietic stem cells in mice: mapping of a candidate locus to chromosome 1. J Exp Med 1996, 183:141-150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sargent LM, Xu HY, Sattler GL, Meisner LF, Pitot HC: Ploidy and karyotype of hepatocytes isolated from enzyme-altered foci in two different protocols of multistage hepatocarcinogenesis in the rat. Carcinogenesis 1989, 10:387-391 [DOI] [PubMed] [Google Scholar]
- 36.Saeter G, Schwarze PE, Nesland JM, Juul N, Pettersen EO, Seglen PO: The polyploidizing growth of normal rat liver is replaced by divisional, diploid growth in hepatocellular nodules and carcinomas. Carcinogenesis 1988, 9:939-945 [DOI] [PubMed] [Google Scholar]
- 37.Lowickhik A, Schneider NR, Tonk V, Ansari MQ, Timmons CF: Report of a complex karyotype in recurrent metastatic fibrolamellar hepatocellular carcinoma and a review of hepatocellular carcinoma cytogenetics. Cancer Genet Cytogenet 1995, 88:170-174 [DOI] [PubMed] [Google Scholar]
- 38.Rabbits TH: Chromosomal translocations in human cancers. Nature 1994, 372:143-149 [DOI] [PubMed] [Google Scholar]
- 39.Kuroki T, Fujiwara Y, Nakamori S, Imaoka S, Kanematsu T, Nakamura Y: Evidence for the presence of two tumor-suppressor genes for hepatocellular carcinoma on chromosome 13q. Br J Cancer 1995, 72:383-385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Katagiri T, Yusuke N, Miki Y: Mutations in the BRCA2 gene in hepatocellular carcinomas. Cancer Res 1996, 56:4575-4577 [PubMed] [Google Scholar]
- 41.Hegi ME, Devereux TR, Dietrich WF, Cochran CJ, Lander ES, Foley JF, Maronpot RR, Anderson MW, Wiseman RW: Allelotype analysis of mouse lung carcinomas reveals frequent allelic losses on chromosome 4 and an association between allelic imbalances on chromosome 6 and K-ras activation. Cancer Res 1994, 54:6257-6264 [PubMed] [Google Scholar]
- 42.Aihara T, Noguchi S, Sasaki Y, Nakano H, Monden M, Imaoka S: Clonal analysis of precancerous lesion of hepatocellular carcinoma. Gastroenterology 1996, 111:455-461 [DOI] [PubMed] [Google Scholar]
- 43.Osawa N, Sakamoto M, Saito T, Kobayashi M, Hirohashi S: Numerical chromosome aberrations in hepatocellular carcinoma detected by fluorescence in situ hybridization. J Hepatol 1996, 25:655-662 [DOI] [PubMed] [Google Scholar]
- 44.Lee G-H, Ogawa K, Nishimori H, Drinkwater NR: Most liver epithelial cell lines from C3B6F1 mice exhibit parentally-biased loss of heterozygosity at the LCI (liver cell immortalization) locus on chromosome 4. Oncogene 1995, 11:2281-2287 [PubMed] [Google Scholar]
- 45.Byrne JA, Smith PJ: The 11p15.5 ribonucleotide reductase M1 subunit locus is not imprinted in Wilms’ tumor and hepatoblastoma. Hum Genet 1993, 91:275-277 [DOI] [PubMed] [Google Scholar]
- 46.Sargent LM, Dragan YP, Sattler G, Xu Y-H, Wiley JE, Pitot HC: Specific chromosomal changes in albumin simian virus 40 T antigen transgenic rat liver neoplasms. Cancer Res 1997, 57:3451-3456 [PubMed] [Google Scholar]
- 47.Albrecht S, von Schweinitz D, Waha A, Kraus JA, Vondeimling A, Pietsch T: Loss of maternal alleles on chromosome arm 11p in hepatoblastoma. Cancer Res 1994, 54:5041-5044 [PubMed] [Google Scholar]
- 48.Garibaldi M, Manenti G, Canaan F, Falvella FS, Pierotti MA, Porta GD, Binelli G, Dragani TA: Chromosome mapping of murine susceptibility loci to liver carcinogenesis. Cancer Res 1993, 53:209-211 [PubMed] [Google Scholar]
- 49.Reynolds SH, Stowers SJ, Patterson RM, Maronpot RR: Activated oncogenes in B6C3F1 mouse liver tumors: implications for risk assessment. Science 1987, 237:1309-1316 [DOI] [PubMed] [Google Scholar]
- 50.Lee GH: Detection of activated c-H-ras oncogene in hepatocellular carcinomas developing in transgenic mice harboring albumin promoter-regulated simian virus 40 gene. Carcinogenesis 1990, 11:1145-1148 [DOI] [PubMed] [Google Scholar]
- 51.Ohgaki H, Sanderson ND, Ton P, Thorgeirsson SS: Molecular analyses of liver tumors in c-myc transgenic mice and c-myc and TGF-α double transgenic mice. Cancer Lett 1996, 106:43-49 [DOI] [PubMed] [Google Scholar]
- 52.Bremner R, Balmain A: Genetic changes in skin tumor progression: correlation between presence of a mutant ras gene and loss of heterozygosity on mouse chromosome 7. Cell 1990, 61:407-417 [DOI] [PubMed] [Google Scholar]
- 53.Held WA, Pazik J, Giancola J, O’Brien G, Kerns K, Gobey M, Mels R, Kenny L, Rustum Y: Genetic analysis of liver tumorigenesis in SV40 T antigen transgenic mice implies a role for imprinted genes. Cancer Res 1994, 54:6489-6495 [PubMed] [Google Scholar]
- 54.Casola S, Ungaro P, Pedone PV, Lassaro D: Loss of heterozygosity of imprinted genes in SV40 t/T antigen induced hepatocellular carcinomas. Oncogene 1995, 11:711-721 [PubMed] [Google Scholar]
- 55.Weissman BE, Saxon PJ, Pasquale SR, Jones GR, Geiser AG, Standbridge EJ: Introduction of a normal human chromosome 11 into Wilms’ tumor cell line controls its tumorigenic expression. Science 1987, 236:175-180 [DOI] [PubMed] [Google Scholar]
- 56.Bhla M, Inoue H, Contticellis MG, Kastury K, Baffa R, Palazzo J, Siprashvili Zl, Mori M, McCue P, Druck T, Croce CM, Huebner K: The FHIT gene spanning the chromosome 3p14.2 fragile site, and renal carcinoma-associated t(34;8) breakpoint, is abnormal in digestive tract cancers. Cell 1996, 84:587-597 [DOI] [PubMed] [Google Scholar]
- 57.Bonyadi M, Cui W, Nagase H, Akhurst RJ: The TGF β type II receptor, Tgfb2, maps to distal mouse chromosome 9. Genomics 1996, 33:328-329 [DOI] [PubMed] [Google Scholar]
- 58.Mathew S, Murty VV, Cheifetz S, George D, Massague J, Chaganti RR: Transforming growth factor receptor gene TGFB2 maps to human chromosome band 3p22. Genomics 1994, 20:114-115 [DOI] [PubMed] [Google Scholar]
- 59.Sue SR, Chari RS, Kong FM, Mills JJ, Fine RL, Jirtle RL, Meyers WC: Transforming growth factor-β receptors and mannose 6-phosphate/insulin-like growth factor II receptor expression in human hepatocellular carcinoma. Ann Surg 1995, 222:17-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Miyasaka K, Ohtake K, Nomura K, Kanda H, Kominami R, Miyashita N, Kitagawa T: Frequent loss of heterozygosity on chromosome 4 in diethylnitrosamine-induced C3H/MSM mouse hepatocellular carcinomas in culture. Mol Carcinog 1995, 13:37-43 [DOI] [PubMed] [Google Scholar]
- 61.Potter M, Mushinski EB, Wax JS, Hartley J, Mock BA: Identification of two genes on chromosome 4 that determine resistance to plasmacytoma induction in mice. Cancer Res 1994, 54:969-975 [PubMed] [Google Scholar]
- 62.Genuardi M, Tsihiara H, Anderson DE, Saunders GF: Distal deletion of chromosome 1p in ductal carcinoma of the breast. Am J Hum Genet 1989, 45:73-82 [PMC free article] [PubMed] [Google Scholar]
- 63.Bieche I, Champene MH, Matifas F, Cropp CS: Two distinct regions involved in 1p deletion in human primary breast cancer. Cancer Res 1993, 53:1990-1994 [PubMed] [Google Scholar]
- 64.Jonasson J, Povey S, Harris H: The analysis of malignancy by cell fusion. VII. Cytogenetic analysis of hybrids between malignant and diploid cells and of tumours derived from them. J Cell Sci 1977, 24:217-254 [DOI] [PubMed] [Google Scholar]
- 65.Tanaka K, Yanoshita R, Konishi M, Oshimura M: Suppression of tumourigenicity in human colon carcinoma cells by introduction of normal chromosome 1p36 region. Oncogene 1993, 8:2253-2258 [PubMed] [Google Scholar]
- 66.Obata M, Lee GH, Kanda H, Kitagawa T, Ogawa K: Loss of heterozygosity at loci on chromosome 4, a common genetic event during the spontaneous immortalization of mouse embryonic fibroblasts. Mol Carcinog 1998, 19:17-24 [PubMed] [Google Scholar]
- 67.Belinsky SA, Swafford DS, Middleton SK, Kennedy CH, Tesfaigzi J: Deletion and differential expression of p16INK4a in mouse lung tumors. Carcinogenesis (Lond) 1997, 18:115-120 [DOI] [PubMed] [Google Scholar]
- 68.Maneti G, Garibaldi M, Fiorino A, Zanesi N, Pierotti MA, Dragani TA: Genetic mapping of lung cancer modifier loci specifically affecting tumor initiation and progression. Cancer Res 1997, 57:4164-4166 [PubMed] [Google Scholar]
- 69.Dietrich WF, Lander ES, Smith JS, Moser AR, Gould KA, Luongo C, Borenstein N, Dove W: Genetic identification of Mom-1, a major modifier locus affecting Min-induced intestinal neoplasia in the nude mouse. Cell 1993, 75:631-639 [DOI] [PubMed] [Google Scholar]
- 70.Miyasaka K, Fukui T, Kitagawa T: Allelotype analysis in mouse hepatocellular carcinomas: frequent homozygous deletion of mouse homolog of p16/CDKN2 gene on chromosome 4 in culture. Biol Pharmaceut Bull 1996, 19:683-691 [DOI] [PubMed] [Google Scholar]
- 71.Wever B, Shen F, Prajada N, Yeh YA, Yang H, Herenyiova M, Look KY: Increased signal transduction activity and down-regulation in human cancer cells. Anticancer Res 1996, 16:3271-3281 [PubMed] [Google Scholar]
- 72.Yamashita S, Ogawa M, Sakamato K, Abe T, Arakawa H, Yamashita J: Elevation of serum group II phospholipase A2 levels in patients with advanced cancer. Clin Chem Acta 1994, 228:91-99 [DOI] [PubMed] [Google Scholar]
- 73.Yamashita J, Ogawa M, Sakai K: Prognostic significance of three novel biologic factors in clinical trial of adjuvant therapy for node-negative breast cancer. Surgery 1995, 117:601-608 [DOI] [PubMed] [Google Scholar]
- 74.Yamashita S, Yamashita J, Ogawa M: Overexpression of group II phospholipase A2 in human breast cancer tissues is closely associated with their malignant potency. Br J Cancer 1994, 69:1166-1170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pathak S, Dave BJ, Gadhia PK: Mouse chromosome 14 is altered in different metastatic murine neoplasias. Cancer Genet Cytogenet 1995, 56:172-173 [DOI] [PubMed] [Google Scholar]
- 76.Rajput B, Marshal A, Killary AM, Lalley PA, Naylor SL, Belin D, Rickles RJ, Strickland S: Chromosomal assignments of genes for tissue plasminogen activator and urokinase in mouse. Somat Cell Mol Genet 1987, 13:581-586 [DOI] [PubMed] [Google Scholar]
- 77.Zhang X, Murakami Y, Sachse R, Yashima K, Hirohashi S, Hu SX, Benedict WF, Sekiya T: Deletions of chromosome 13q, mutations in retinoblastoma 1, and retinoblastoma protein state in human hepatocellular carcinoma. Cancer Res 1994, 54:4177-4182 [PubMed] [Google Scholar]
- 78.Karhu R, Knuutila S, Kallioniemi OP, Stiltonen S, Aine R, Vilpo J: Frequent loss of the 11q14–24 region in chronic lymphocytic leukemia: a study by comparative genomic hybridization. Genes Chromosomes & Cancer 1997, 19:286-290 [DOI] [PubMed] [Google Scholar]
- 79.Sato T, Tanigami A, Yamakawa K: Allelotype of breast cancer: cumulative allele losses promote tumor progression in primary breast cancer. Cancer Res 1990, 50:7184-7189 [PubMed] [Google Scholar]
- 80.Tamura K, Zhang X, Murakami Y, Hirohashi S, Xu HJ, Hu SX, Benedict WF, Sekiya T: Deletion of three distinct regions on chromosome 13q in human non-small cell lung cancer. Int J Cancer 1997, 74:45-49 [DOI] [PubMed] [Google Scholar]
- 81.Copeland NG, Jenkins NA, Gilbert DJ, Eppig JT: A genetic linkage map of the mouse: current applications and future prospects. Science 1993, 262:57-66 [DOI] [PubMed] [Google Scholar]
- 82.Yamasaki H, Mesnil M, Omori Y, Mironov N, Krutovskikh V: Intercellular communication and carcinogenesis. Mutat Res 1995, 333:181-188 [DOI] [PubMed] [Google Scholar]
- 83.Searle AG, Peter J, Lyon MF, Evans EP, Edwards JH, Buckle VJ: Chromosome maps of man and mouse, III. Genomics 1987, 1:3-18 [DOI] [PubMed] [Google Scholar]