Abstract
The widely used, very-low-copy BAC (bacterial artificial chromosome) vectors are the mainstay of present genomic research. The principal advantage of BACs is the high stability of inserted clones, but an important disadvantage is the low yield of DNA, both for vectors alone and when carrying genomic inserts. We describe here a novel class of single-copy/high-copy (SC/HC) pBAC/oriV vectors that retain all the advantages of low-copy BAC vectors, but are endowed with a conditional and tightly controlled oriV/TrfA amplification system that allows: (1) a yield of ∼100 copies of the vector per host cell when conditionally induced with l-arabinose, and (2) analogous DNA amplification (only upon induction and with copy number depending on the insert size) of pBAC/oriV clones carrying >100-kb inserts. Amplifiable clones and libraries facilitate high-throughput DNA sequencing and other applications requiring HC plasmid DNA. To turn on DNA amplification, which is driven by the oriV origin of replication, we used copy-up mutations in the gene trfA whose expression was very tightly controlled by the araC–ParaBAD promoter/regulator system. This system is inducible by l-arabinose, and could be further regulated by glucose and fucose. Amplification of DNA upon induction with l-arabinose and its modulation by glucose are robust and reliable. Furthermore, we discovered that addition of 0.2% d-glucose to the growth medium helped toward the objective of obtaining a real SC state for all BAC systems, thus enhancing the stability of their maintenance, which became equivalent to cloning into the host chromosome.
[The following individuals kindly provided reagents, samples or unpublished information as indicated in the paper: F.R. Blattner D. Helinski, S. Valla, F. Schomburg, C. Small, R. Bogden, C. Gaskins, M.P. Mayer, D.C. Schwartz, and O. Azzam.]
The progress in genomic research in the present decade relied on use of the very-low-copy bacterial artificial chromosome (BAC) vectors developed by Shizuya et al. (1992). The BAC vectors were extensively used for preparation of DNA libraries, for physical mapping, and for large-scale DNA sequencing efforts. BAC libraries from fungi (Diaz-Perez et al. 1996), plants (Woo et al. 1994; Mozo et al. 1998), mammals (Cai et al. 1995; Schibler et al. 1998), and human DNA (Kim et al. 1996; Asakawa et al. 1997) have been constructed. The main advantage of using BACs for genomic library construction was the stability of the large, very low or single-copy (SC) clones. However, the SC state of vectors and clones is also of great disadvantage, because of very low levels of DNA recovery, and consequently, reduced purity of DNA with respect to host DNA.
Our objective was to construct vectors that retain the advantages of the SC stability of the BAC clones during the maintenance phase while acquiring elements for a conditional in vivo amplification of BAC vectors and clones. Such amplification would be turned on only when high yields of DNA are required, as when preparing vector DNA for library construction or cloned DNA for sequencing. Ideally, such high yields will lower the dependence on the DNA purification. To achieve such a goal, we constructed the SC pBAC/oriV vector whose conditional, high-copy (HC) origin of DNA replication oriV is completely inactive in the commonly used hosts, because they do not produce the TrfA replication protein upon which replication at oriV depends. To supply the TrfA protein, we constructed special hosts, in which synthesis of copy-up TrfA mutant protein is very tightly controlled by the ParaBAD (PBAD) promoter and AraC protein. Thus, we created a system permitting conditional amplification of BAC plasmids (without or with) inserts consisting of the pBAC/oriV vector and a host supplying (only upon induction) a copy-up mutant of TrfA protein. In such a system, the pBAC/oriV clone is maintained at the SC level, but when the synthesis of the TrfA protein is induced, DNA is amplified up to 100-fold.
In our earlier preliminary studies, the trfA gene was under control of the inducible Ptet promoter (Hradecna et al. 1998; Szybalski et al. 1999), but in the present study we used the araC–PBAD expression system, which offers additional advantages.
RESULTS
Derivatives of the pBeloBAC 11 Vector Allowing “on Command” Amplification of BAC Clones
For construction of stable genomic libraries, Shizuya et al. (1992) developed the pBAC108L vector, whose replication mode was that of the Escherichia coli F factor. This pBAC108L vector is maintained at 1–2 copies per host chromosome, and its replication is stringently controlled at the level of initiation. Replication initiates at oriS (ori2), which consists of (1) four directly repeated sequences of 19 bp (iterons), (2) an AT-rich region, and (3) binding sites for the host DnaA protein. The RepE protein (251 residues, 29 kD), when in the monomeric form, mediates assembly of a replication complex at oriS. The dimeric form of RepE binds to the inverted repeats of the repE operator exerting autogenous repression (Komori et al. 1999). Similarly to the F plasmid, the stability of the pBAC's maintenance is ensured by the partition system consisting of ParA, ParB, and ParC elements (Mori et al. 1986).
The pBAC108L vector, however, was of limited use, because clones had to be identified by colony hybridization. To ease detection of clones, Kim et al. (1996) constructed the pBeloBAC11 vector carrying the lacZα fragment for blue/white color screening based on the α-complementation of β-galactosidase (Ullmann et al. 1967; Ullmann 1992). Libraries of DNA fragments up to 300 kb prepared in this vector were shown to be rather stable, and chimeric forms were rarely found (Boysen et al. 1997). Yet another BAC derivative, the SacB-based positive-selection vector with MCS was constructed (Frengen et al. 1999) and used for constructing libraries from human, primate, canine, and murine genomes (Osoegawa et al. 1998). However, the amounts of DNA that are generated from such BAC-derived clones are usually suboptimal, especially for genetic manipulations or sequencing, because the plasmids are present at only 1–2 copies per chromosome. Moreover, preparation of large quantities of the pBeloBAC11 vector DNA, essential for library construction, is laborious and time consuming.
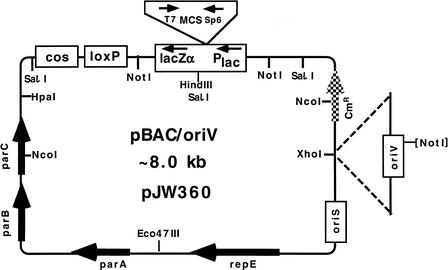
To overcome the above-discussed shortcomings, but retain all the advantages of conventional low-copy BAC systems, we have reengineered both the pBeloBAC11 vector and the DH10B host so as to allow “on command” amplification of the vector alone or carrying the genomic or cDNA fragments. As a suitable system to turn on the DNA amplification on command, we chose the oriV/TrfA replicon system of the broad-host-range RK2 plasmid (Perri and Helinski 1993). The oriV origin of replication consists of eight 17-bp direct repeats (iterons) that bind a monomeric form of the initiation protein TrfA (Toukdarian et al. 1996). As a source of oriV, we used the EcoRI–BamHI fragment of the pSV16 plasmid (Haugan et al. 1992), which was modified to eliminate the NotI site (see Methods). The modified oriV was cloned into the PolIk-blunted (PolIk, Klenow fragment of Pol I DNA polymerase) XhoI site of pBeloBAC11, creating pBAC/oriV (Fig. 1). We have also cloned oriV into other sites of the BAC (see legend to Fig. 1). The resulting pBAC/oriV vector retains all features of pBeloBac11, including (1) stability of SC clones; (2) the MCS within the lacZα to detect cloned inserts by α-complementation; (3) two NotI sites flanking the MCS for excision of cloned inserts; (4) two phage promoters (T7 and SP6) reading into MCS to generate RNA probes for blotting procedures and containing sequences complementary to generally used sequencing primers; (5) the cosN site of phage λ, enabling packaging into phage λ particles, specific labeling of λ cohesive ends used for restriction mapping, and in vitro linearization by λ terminase; and (6) a loxP site for linearization and/or introduction of additional DNA fragments via the Cre–loxP system of phage P1 (used in our novel in vivo method to retrofit existing BAC library clones with oriV; Wild et al. 2001a).
Figure 1.
The pBAC/oriV vector permitting its single-copy (SC) maintenance and, alternatively, its conditional, tightly regulated DNA amplification. This new derivative of the pBeloBAC11 vector preserves most of its original specific features, including the plasmid F-derived SC maintenance system based on the oriS–repE–parABC genes (see Kim et al. 1996), but was equipped with a second origin of DNA replication, oriV, from the broad-host-range plasmid RK2 (Stalker et al. 1981). We cloned the NotI-less oriV at the HpaI or XhoI sites, but for reasons not fully anticipated, the TrfA–oriV-directed DNA amplification (see Figs. 2 and 3) was the highest for oriV in the XhoI site. Four derivatives of pBAC/oriV have been constructed: (1) The pBeloBAC11/SceI/oriV (pJW408 = pBAC/oriV/SceI) vector with the I-SceI recognition site at the HpaI site of pBAC/oriV. (2) The pTrueBlue-BAC2/oriV (pJW406) vector, with oriV at the XhoI site. This vector features dark-blue colonies (darker than for original BACs and similarly dark as for pIndigoBAC/oriV; see below), thus allowing more accurate blue/white colony screening. It is based on pTrueBlue-BAC2 (Genomics One 1999 Catalog), which contains four additional cloning sites, as compared with pBeloBAC11, all within the specially constructed lacZα segment (Slilaty and Lebel 1998). (3) The pTrueBlue-BAC2/oriV/SceI (pJW419) vector with the I-SceI recognition site cloned into the Eco47III site of pTrueBlue-BAC2/oriV. (4) The pIndigoBAC/oriV (pJW550) vector with oriV at the XhoI site. This vector features enhanced, dark-blue-color colony screening and is based on pIndigoBAC-5 (Epicentre 2001 Catalog). Details of the construction of pBAC/oriV and their derivatives are described in Methods. [NotI] Inactivated NotI site.
As the optical mapping became a tool for construction of restriction mega-maps (Giacalone et al. 2000), it also became very desirable to have a very rare and reliable restriction site on the vector for efficient and convenient linearization of clones. To meet this goal, we used the very rare restriction site, I-SceI, for the intron homing endonuclease (Monteilhet et al. 1990) and constructed pBAC/oriV/SceI by cloning the NotI-less oriV fragment (see legend to Fig. 1 and Methods section) into the PolIk-blunted XhoI site of pBeloBAC11/SceI, provided by the F.R. Blattner laboratory (University of Wisconsin).
Color screening of clones based on insertional inactivation (between ATG and codon 7) of the lacZα that encodes the α-peptide of β-galactosidase is widely used for detecting recombinant clones. However, this screening method often leads to false results, both positives (white colonies that do not contain the insert) and negatives (blue colonies that contain the insert). To provide high-accuracy color screening, Slilaty and Lebel (1998) reengineered the lacZα fragment so as to ensure detection of DNA insertions within the region that encodes amino acids 11–36 of β-galactosidase. The DNA containing this TrueBlue gene fragment was cloned into the pBeloBAC11 vector creating TrueBlue-BAC2, which claims to offer 100% accuracy in blue/white screening together with six unique cloning sites capable of accepting DNA fragments generated by >70 different restriction enzymes or obtained by shearing or sonication (Genomics One Catalogue 1999). To obtain amplifiable derivatives, we have cloned the modified oriV (see Methods) into the PolIk-blunted XhoI site of pTrueBlue-BAC2, creating the pTrueBlue-BAC2/oriV vector (see legend 2 to Fig. 1 and Methods). Furthermore, we introduced a recognition site for the I-SceI mega-nuclease (see legend 3 to Fig. 1) into pTrueBlue-BAC2/oriV creating the pTrueBlue-BAC2/oriV/SceI vector (see legend to Fig. 1). When compared with other lacZα plasmids, the pTrueBlue-BAC2/oriV vector offers another advantage by providing a much darker blue color of colonies when enough IPTG inducer (40 μg/mL) is used. Because colonies of Indigo derivative of pBeloBAC11 (Epicentre Catalogue 2000) develop a similar deep blue color, we have also cloned oriV into this derivative and obtained the pIndigoBAC-5/oriV vector (see (4) in legend to Fig. 1).
Modified E. coli DH10B Hosts for On-Command Amplification of pBAC/oriV
We have chosen the oriV/TrfA replication system of the RK2 plasmid for ”at-wish” amplification of modified pBAC vectors because this replication system is simple and requires only one RK2-encoded protein, TrfA. Replication at oriV depends on the RK2-encoded replication protein TrfA that binds specifically to direct repeats (iterons) at oriV. Specific copy-up mutations in trfA increase the copy number from 3- to 20-fold, as compared with the wild-type TrfA. We tested several trfA copy-up mutations (Durland et al. 1990; Haugan et al. 1992) for their effect on replication of BAC vectors that contained oriV (see below, “Effect of Various Copy-up Mutations in the trfA Gene on the Amplification of Large DNA Inserts” and Fig. 6). To tightly control expression of the trfA gene mutants, we used the regulatory system of the arabinose operon consisting of araC (encoding the AraC regulatory protein) and the PBAD promoter (Guzman et al. 1995). The ara expression cassette was cloned into the integration vector containing the attP site from λ (for site-specific recombination), and the pBR322 ori, the latter flanked by two NotI sites, for its convenient removal (M. Koob, pers. comm.). The resulting pJW344 plasmid served for cloning the trfA copy-up mutant genes and for inserting them into the host genome (see Fig. 2, Table 1 below, and Methods for details). In the presence of inducer, L-arabinose, transcription of trfA mutants from the PBAD promoter is turned on; whereas in the absence of inducer, the trfA expression is undetectable.
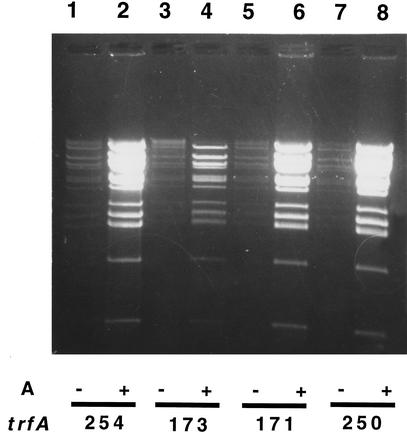
Figure 6.
Effect of copy-up mutations in the trfA gene on DNA amplification. All host strains carry the same pBAC/oriV plasmid with a 108-kb DNA insert (pCG275). Growth conditions and the induction of TrfA synthesis by L-arabinose (A) are described in Methods. DNA samples prepared by phenol extraction and ethanol precipitation were digested with SmaI and run on a 0.6% agarose gel. (Lanes 1,3,5,7) LB medium (LB); (lanes 2,4,6,8) LB + 0.01% A. (Lanes 1,2) Strain JW439 containing the trfA254 mutation; (lanes 3,4) strain JW499 containing the trfA173 mutation; (lanes 5,6) strain JW500 containing the trfA171 mutation; (lanes 7,8) strain JW501 containing the trfA250 mutation. Amplifications (lanes 2,6,8) of plasmid + 108-kb insert are estimated to be up to 30-fold.
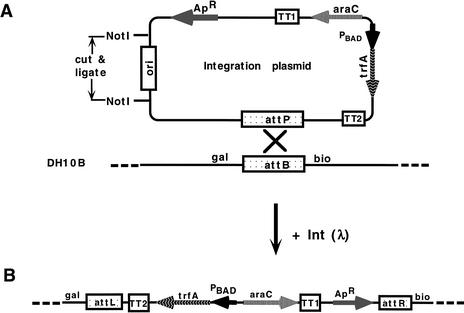
Figure 2.
Construction of four DH10B-based hosts carrying a tightly regulated trfA gene that supplies, but only upon induction, the TrfA replication protein. (A) A representation of an integration plasmid and of a fragment of the host genome with the attB site for site-specific recombination. Four integration plasmids carrying four various trfA copy-up mutations have been constructed (see Table 1), as described in Methods. Each integration plasmid carries a cassette consisting of araC–PBAD fused to the specific trfA gene copy-up mutant. All integration plasmids have (1) an easily removable NotI-flanked ori of plasmid pBR322, and (2) the attPλ site for site-specific integration into attB of the DH10B host genome, as shown below the plasmid drawing. (B) A diagram of the genomic segment of the host upon recombination of the trfA-integration plasmid. Such hosts permit conditional, tightly regulated synthesis of the TrfA protein. Experimental details on Int-mediated integration of the four plasmids into the DH10B host strains (Table 2) are described in Methods. TT1 represents the t1 and t2 terminators (both clockwise) from rrnB; TT2 represents the tL3 (clockwise) and tL1 (anticlockwise) terminators of phage λ.
Table 1.
List of Plasmids
| Plasmid | Pertinent features | Source/Reference |
|---|---|---|
| pBeloBAC11 | F plasmid-based very low-copy vector | Kim et al. (1996) |
| pBeloBAC11/SceI | pBeloBAC11 with the I–SceI recognition site inserted at the HpaI site | F. Blattner's lab |
| TrueBlue-BAC2 | Accurate color selection vector | Genomics One |
| pBAC/oriV | pBeloBAC11 with oriV inserted at the XhoI site | Fig. 1 and Fig. 3 |
| pCG273 | pBAC/oriV containing 140-kb insert of wheat DNA | C. Gaskins |
| pCG274 | pBAC/oriV containing 122-kb insert of wheat DNA | C. Gaskins |
| pCG275 | pBAc/oriV containing 108-kb insert of wheat DNA | C. Gaskins |
| pMPM123 | ColE1 ori, araC-PBAD, SpR, ApR | Mayer (1995) |
| p67 | pBAC/oriV containing 40-kb insert of trout DNA | C. Small and B. Bogden |
| p69 | pBAc/oriV containing 80-kb insert of trout DNA | C. Small and B. Bogden |
| pJW22 | pBR322 with its ori flanked by two NotI sites (formerly pANTS) | M. Koob, unpubl. (see Hasan et al. 1994) |
| pJW344 | pJW22 containing araC-PBAD | This paper |
| pJW349 | pJW344 containing araC-PBAD-trfA203 cassette | Fig. 2 |
| pJW360 | pBAC/oriV | Fig. 1 |
| pJW378 | pBAC/oriV containing 20 kb of Arabidopsis thaliana DNA inserted between two NotI sites | Fig. 4A; Fig. 5 |
| pJW406 | pTrueBlue-BAC2/oriV | Fig. 1, legend |
| pJW408 | pBeloBAC11/SceI/oriV | Fig. 1, legend |
| pJW419 | pTrueBlue-BAC2/oriV/SceI | Fig. 1, legend |
| pJW424 | pJW344 containing araCPBADtrfA254 cassette | Fig. 6 |
| pJW457 | pJW344 containing araCPBADtrfA250 cassette | Fig. 6 |
| pJW458 | pJW344 containing araCPBADtrfA173 cassette | Fig. 6 |
| pJW459 | pJW344 containing araCPBADtrfA171 cassette | Fig. 6 |
| pJW487 | pBAC containing 100 kb of Arabidopsis thaliana DNA inserted between two NotI sites and retrofitted with oriV | This paper |
| pJW511 | pBAC containing 77 kb of rice DNA retrofitted with oriV | This paper |
| pJW547 | pIndigoBAC-5; phosphorylated and recircularized from the linear epicenter pIndigoBAC-5 (HindIII) | This paper; Methods |
| pJW550 | pIndigoBAC/oriV | Fig. 1, legend |
Construction and Maintenance of Genomic Libraries
Methods for constructing libraries in pBAC/oriV should not differ from those in regular BACs (Cai et al. 1995; Diaz-Perez et al. 1996; Asakawa et al. 1997), other than simplifying preparation of large quantities of very pure pBAC/oriV vector. The resulting clones, however, could be transformed either into the standard DH10B, which would not allow DNA amplification, or into the DH10B/trfA derivatives, like JW366, that only on command, that is, when the inducer L-arabinose is added, could provide TrfA function that promotes DNA amplification. We believe that preparation of libraries directly in the latter JW366-like hosts is much more efficient than first preparing a DH10B-based library as an intermediate, and then transferring the clones into hosts with the inducible TrfA function. Such a two-step procedure is unwarranted, because libraries constructed directly in JW366-like hosts (in L-arabinose-free media) are as stable as when using oriV-less BACs or trfA-less hosts.
More recently, we also constructed pBAC/oriV derivatives that, in addition, carry the inducible trfA cassette; such plasmids can be amplified in the plain DH10B hosts. We found such pBAC/oriV/trfA plasmids perfectly stable. We used them for expression vectors (Wild et al. 2001b), but we did not evaluate them as yet for library constructions. The examples above illustrate the great flexibility of our oriV/trfA cloning and amplification systems.
Stability and Amplification of the pBAC/oriV Vector and Clones
Various stability controls (Fig. 3, lanes 1–7) together with the dramatic (50- to 100-fold) amplification of the pBAC/oriV vector are shown in Figure 3. The latter occurs only when the TrfA function is provided by the host (lane 8). There was no amplification when hosts do not provide the TrfA function, for example, DH10B (Fig. 3, lanes 1,2), or when the plasmid does not carry the oriV site (pBeloBAC11; lanes 3–5), even in the presence of the L-arabinose inducer (lane 5).
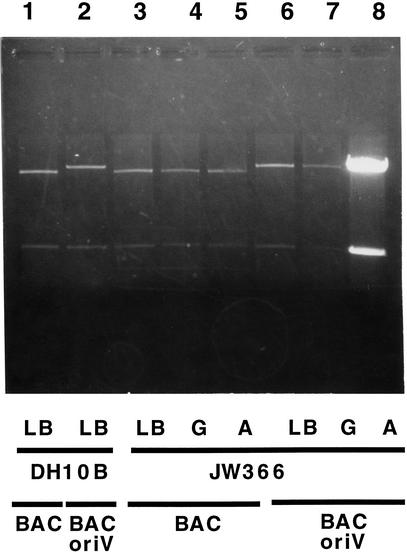
Figure 3.
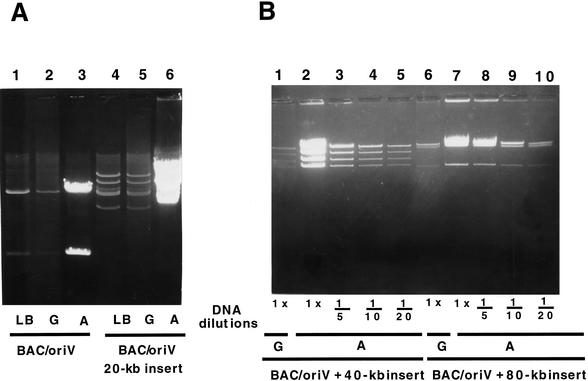
Maintenance and amplification of the pBAC/oriV vector: effects of the host, glucose, and L-arabinose-induced synthesis of the TrfA protein. The DH10B host and its derivative JW366, containing the araC–PBAD–trfA203 cassette at the λ attB site (Table 2), were transformed either with pBeloBAC11 (BAC) or pBAC/oriV (see Table 1). Transformants were grown in the Luria-Bertani medium (LB), LB + 0.2% D-glucose (G) or LB + 0.01% L-arabinose (A). After 5 h of growth, a 4.5-mL volume of each culture was centrifuged and the DNA was prepared using Wizard columns (Promega). All lanes (0.8% agarose gel) show two NcoI fragments of plasmid pBeloBAC11, either without (lanes 1, 3–5) or with inserted oriV (pBAC/oriV in lanes 2, 6–8). Successful DNA amplification is seen only in lane 8, whereas lanes 1–7 represent various controls. (Lane 1) pBeloBAC11 in the DH10B host grown in LB; (lane 2) pBAC/oriV in the DH10B host grown in LB; (lanes 3–5) pBeloBAC11 in the JW366 host grown in LB, LB + 0.2% G or LB + 0.01% A, respectively; (lanes 6–8) pBAC/oriV in the JW366 host grown in LB, LB + 0.2% G or LB + 0.01% A, respectively. Whereas 0.2% G reduces the plasmid number to one per cell (lane 7 vs. 6), induction with A amplifies DNA up to 100-fold (lane 8 vs. 6). The induced high-copy (HC) replication of pBAC/oriV provides an ample amount of vector DNA for construction of libraries.
When testing various media for optimal DNA amplification, we have discovered that glucose, at 0.2%, reduces the number of BAC copies (Fig. 3, cf. lane 4 with 3, lane 7 with 6). This makes the 0.2% glucose a rather important novel tool for the maintenance of BACs in an SC state, because it prevents any undesirable rearrangements that might occur as a result of recombination between two or more plasmids present within a single host cell.
Amplification of the pBAC/oriV Clones
The DNA restriction pattern remains unchanged over many months of maintenance of the large clones and also after the entire cycle of their amplification (Figs. 4–7), as confirmed by DNA sequencing of selected clones. Amplification of the pBAC/oriV vector (Fig. 4A, lane 1 or 2 vs. 3) and the same vector carrying a 20-kb insert (Fig. 4A, lane 4 and 5 vs. 6) each in the same host, are shown in Figure 4A. Again, one can see the dramatic DNA amplification after induction of the TrfA function by L-arabinose (Fig. 4A, lanes 3,6), and the glucose-dependent reduction of copy number of uninduced pBAC/oriV plasmids, from about two to one per cell (Fig. 4A, lane 2 vs. 1, lane 5 vs. 4). We compared the extent of amplification (as assessed by a series of dilutions; Fig. 4B) in clones carrying foreign DNA inserts of 40 kb (Fig. 4B, lanes 1–5), and 80 kb (Fig. 4B, lanes 6–10). In the DH10B host expressing the trfA203 gene (JW366), the 40- and 80-kb clones were amplified ∼40-fold and ∼20-fold, respectively. This host (JW366) was best suited for the amplification of clones up to 50 kb. The effect of other trfA mutations, more suitable for amplification of larger clones, is described below and is illustrated in Figures 6 and 7.
Figure 4.
Maintenance and amplification of pBAC/oriV that carries DNA inserts of various length. (A) Comparison of amplification of pBAC/oriV vector, with or without a 20-kb insert. After 5 h of growth, cells from the 4.5-mL volume of the culture were collected, and DNA was phenol-extracted, precipitated with 70% ethanol, digested with NcoI (lanes 1–3) or SalI (lanes 4–6), and run on an 0.8% agarose gel. (Lanes 1–3) Strain JW371 carrying pBAC/oriV grown in the LB medium (LB), LB + 0.2% D-glucose (G) or LB + 0.01% L-arabinose (A), respectively (two bands are analogous to those in Fig. 3); (lanes 4–6) strain JW378 carrying pBAC/oriV with the 20-kb insert grown, respectively, in LB, LB + 0.2% G or LB + 0.01% A. (B) Assessment of amplification by diluting of the amplified DNA of pBAC/oriV clones containing 40-kb or 80-kb inserts. Growth conditions, DNA analysis after SalI digestion, and abbreviations are as described for A and in Methods. Numbers below the lanes indicate the fold of DNA dilution prior to SalI digestion (results were similar for dilutions made after digestions and are not shown here). (Lane 1) Uninduced strain JW389 carrying pBAC/oriV with the 40-kb insert grown in LB + G; (lanes 2–5) induced strain JW389 grown in LB + A; (lane 6) uninduced strain JW390 carrying pBAC/oriV with the 80-kb insert grown in LB + G; (lanes 7–10) induced strain JW390 grown in LB + A. The DNA in lanes 1, 2, 6, and 7 is undiluted. In lanes 3–5 and 8–10, the DNA was diluted, as specified below the lanes.
Figure 7.
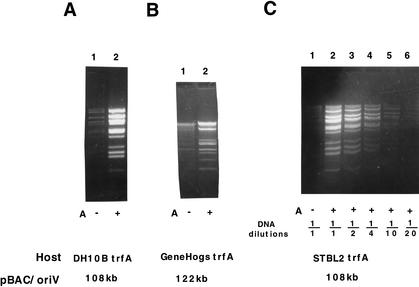
Amplification of the DNA of pBAC/oriV clones carrying (A,C) 108-kb or (B) 122-kb inserts of foreign DNA, when propagated in the DH10B host and in two commercial hosts, GeneHogs (Invitrogen) and Stbl2 (Life Technologies, presently Invitrogen). All three host strains contain the araC–PBAD–trfA254 cassette at their attBλ site. Growth conditions and induction of TrfA synthesis by L-arabinose (A) are described in Methods. DNA samples prepared by phenol extraction and ethanol precipitation were digested with SalI (A,C) or with SmaI (B) and run on a 0.6% agarose gel. (A) Amplification of pBAC/oriV carrying a 108-kb insert (pCG275) in the JW427 host (trfA254; see JW439 in Table 2). (Lane 1) LB medium (LB); (lane 2) LB + 0.01% A. (B) Amplification of pBAC/oriV carrying a 122-kb insert (pCG274) in JW480 (GeneHogs trfA254). (Lane 1) LB; (lane 2) LB + 0.01% A. (C) Amplification of pBAC/oriV carrying a 108-kb insert (pCG275) in JW526 (Stbl2 trfA254). (Lane 1) LB; (lanes 2–6) LB + 0.01% A. (Lanes 3–6) DNA preparations were diluted 1/2, 1/4, 1/10, and 1/20, respectively. Comparison of lanes 1 and 6 indicates an ∼30-fold amplification of the plasmid with the 108-kb DNA insert.
Regulation of Amplification of the pBAC/oriV Clones by L-Arabinose, D-Fucose, and D-Glucose
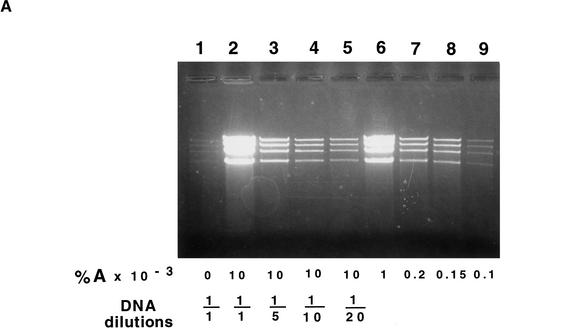
The extent of DNA amplification could be regulated either by varying concentrations of L-arabinose (Fig. 5A) or by modulating induction by other sugars (Fig. 5B,C). The optimal amplification was achieved at 0.01% of the L-arabinose (Fig. 5A, lane 2). However, lowering the concentration to 0.001% reduced the induction only slightly (Fig. 5A, lane 6). Comparison of DNA dilutions (Fig. 5A, lanes 1–5) indicates that the extent of amplification was 50- to 100-fold. Less than 0.001% of L-arabinose resulted in progressively lower DNA amplification (Fig. 5A, lanes 7–9).
Figure 5.
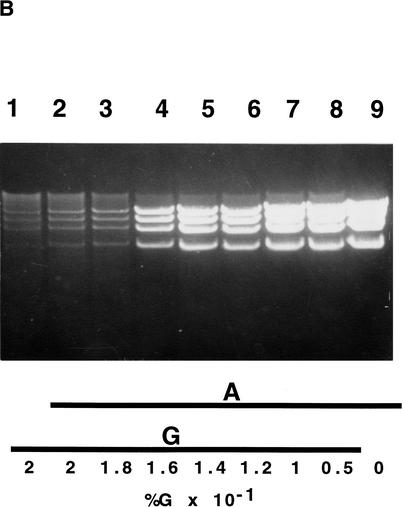
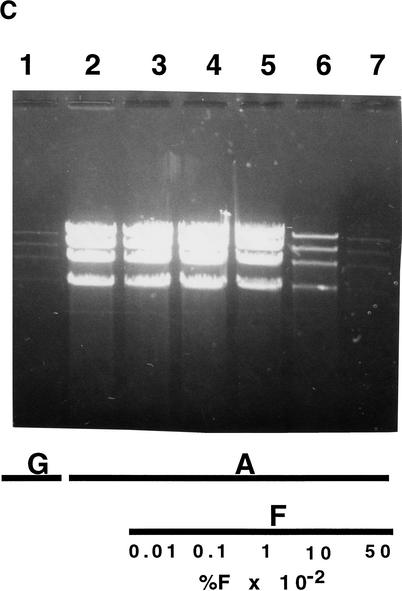
Effects of L-arabinose, D-glucose, and D-fucose on the amplification of the pBAC/oriV with a 20-kb insert. (A) Induction by L-arabinose (A). Strain JW378 (pBAC/oriV + 20-kb insert) was grown in LB medium (LB) supplemented with various concentrations of A. Induction, DNA extraction, and digestion were performed as described in Methods and in the legend to Figure 4A (lanes 4–6). (Lane 1) No inducer present in LB; (lanes 2–5) LB + 0.01% A. (Lane 2) An undiluted DNA sample was run; (lanes 3–5) DNA samples were diluted 5-, 10-, or 20-fold, respectively, prior to the SalI digestion; (lanes 6–9) LB supplemented with 0.001%, 0.0002%, 0.00015%, or 0.0001% A, respectively. By comparing the DNA bands in lanes 1 and 5, we estimate that DNA amplification was ∼80-fold. (B) Inhibition of amplification by D-glucose (G). Strain, experimental design, and abbreviations are as in description of A. (Lane 1) No A added, LB + 0.2% G; (lanes 2–8) LB was supplemented with 0.01% A and with 0.2, 0.18, 0.16, 0.14, 0.12, 0.1, or 0.05% G, respectively; (lane 9) LB supplemented only with 0.01% A. (C) Inhibition of amplification by D-fucose (F). Strain, experimental panel design, and abbreviations are as in description of A. (Lane 1) LB + 0.2% G only; (lanes 2–7) LB + 0.01% A, supplemented with none, 0.0001%, 0.001%, 0.01%, 0.1%, or 0.5% F, respectively.
The inhibition of the L-arabinose-induced DNA amplification by D-glucose or D-fucose is shown in Figure 5, B and C, respectively. Glucose showed a very sharp transition between noninhibitory (0.1%) and very inhibitory (0.18–0.2%) concentrations (Fig. 5B). Significantly, at concentrations just below 0.1%, glucose enhanced the growth of the host and the final yield of the amplified DNA. Fucose blocked quite effectively the induction of DNA replication at concentrations above 0.01% (Fig. 5C, lanes 6,7). Modulation of DNA replication was mainly applicable for expression vectors that were based on the pBAC/oriV SC/HC plasmids (Wild et al. 2001b).
Effect of Various Copy-Up Mutations in the trfA Gene on the Amplification of Large DNA Inserts
Because the trfA203-expressing host was most effective in amplification of clones smaller than 50 kb, we evaluated several other copy-up mutants of trfA for their ability to amplify larger DNA inserts. As shown in Figure 6, three of the trfA copy-up mutants were somewhat more effective than trfA173 in the amplification of the pBAC/oriV 108-kb clone. Amplification of this 108-kb clone host was less effective in the trfA203-bearing host than that shown in lane 4. On the other hand the amplification of smaller (20- to 40-kb) clones was better in hosts carrying the trfA203 mutation than in some other hosts listed in Figure 6 (systematic comparative results are not shown here).
BAC amplification in the commercial derivatives of DH10B, into which we have integrated cassettes expressing various trfA copy-up mutations, is shown in Figure 7. The availability of such commercial hosts is of advantage for various high-throughput uses, especially for large genome sequencing. Moreover, commercially available competent cells of such hosts are more efficiently transformed by electroporation (see below) and thus are more suitable for high-throughput constructions of libraries.
Efficiency of Transformation
Efficient transformation is crucial for cloning large (>30-kb) DNA fragments. The DNA of such clones is introduced into cells by electroporation, the efficiency of which depends both on the DNA used for electroporation (its quality and size) and on the recipient bacteria. We have investigated here the effects of oriV presence and its various locations on the plasmid, of plasmid and insert size, of trfA mutations in the host, and of the method of host cell preparation. We first compared plasmid pBeloBAC11 and its pBAC/oriV derivative using as recipients strains DH10B and JW463, the latter carrying the araC–PBAD–trfA250 cassette at attλ. In all four experiments, the number of transformations obtained (CmR colonies), that is, 1–5 × 106 CFU/μg DNA, was similar. To be sure that in transformation experiments we were using equivalent amounts of pBeloBAC11 or pBAC/oriV (pJW360) DNA, each plasmid was digested with NcoI and various DNA aliquots were run on the gel to estimate DNA concentrations. When evaluating the commercially available Electrocomp GeneHogs and Electrocomp GeneHogs(trfA) (constructed by us and listed in Invitrogen 2001 Catalog, p. 53, and 2002 Catalog No. T 1060-01) as recipients for electroporation of pBeloBAC11 and pBAC/oriV DNA, we found that the number of transformants was 1 or 2 orders of magnitude higher compared with regular DH10B and JW463 hosts.
Libraries constructed in pBeloBAC11 usually contain DNA inserts of 100–200 kb. Therefore, it was important to test the efficiency of transformation using pBAC/oriV carrying large DNA inserts. To obtain such clones, we retrofitted existing pBeloBAC11 clones with oriV (using an in vivo procedure, outlined by Wild et al. 2001a). The resulting plasmids, pJW487 (pBAC/oriV with a 100-kb Arabidopsis thaliana DNA insert) and pJW511 (pBAC/oriV with a 77-kb rice DNA insert), were used to transform Electrocomp GeneHogs, DH10B, and JW463 cells. The number of transformants obtained with either plasmid was 0.5–1 × 105 CFU/μg DNA when GeneHogs were used and 1 × 104 CFU/μg DNA when electroporating into DH10B and JW463 competent cells. These results confirm that the presence of either the oriV or the trfA cassette had no effect on the efficiency of transformation, even under conditions when such efficiency coordinately decreased owing to the large size of the plasmid, or increased when using the commercially prepared recipient cells.
DISCUSSION
The present study describes important and useful improvements to the conventional low-copy BAC vectors that are a major workhorse of large genomes projects.
The most important feature of BAC clones is their stability resulting from their very low copy number. We showed here that the copy number can be lowered even further by the addition of glucose (0.2%) to the growth medium, both for conventional BACs and our pBAC/oriV. The real SC state should improve stability of maintenance of BAC libraries by reducing the opportunity for intracellular recombination between clones.
For practical applications like library construction and sequencing, ample amounts of pure DNA are required. Therefore, we developed the conditional oriV–TrfA DNA amplification system, which permits easy and prompt 30- to 100-fold increases in the amount of DNA for (1) preparing the pBAC/oriV vectors for constructing libraries, or (2) high-throughput DNA sequencing of clones. Obviously, clones that have undergone the amplification process should not be used for clone maintenance, but only for biochemical procedures, including sequencing or gene expression. Furthermore, our system offers great advantages for the purification of pBAC/oriV clones free of host DNA, usually a rather laborious procedure. The large size of BAC clones and the probability that their DNA would become sheared along with the contaminating host genomic DNA make DNA purification of traditional BACs even more difficult. Amplification of BACs using our oriV–TrfA system enriches the BAC clones 30- to 100-fold, thus enhancing by a similar factor the purity of BAC DNA.
Further improvements include the following: (1) Incorporation of the I-SceI site into pBAC/oriV derivatives creates clones more suitable for the optical mapping (Giacalone et al. 2000). Our vectors pBAC/oriV/SceI and pTrueBlue-BAC2/oriV/SceI were designed for that purpose. (2) Introduction of the lacZα derivatives into pBAC/oriV allowing a deeper blue color development (pIndigoBAC-5/oriV), together with more reliable blue/white screen for successful cloning (pTrueBlue-BAC2/oriV), and developing methods for the positive selection of clones, prepared either by restriction enzyme digestion or random sheer, that will soon be available for construction of libraries based on our pBAC/oriV vectors. (3) Both the pBAC/oriV vectors (see Fig. 3) and clones (Wild et al. 2001a) are as stable as the commonly used BACs. They should be even more stable when maintained as a single copy, in the presence of 0.2% glucose. (4) Our hosts, which contain the tightly regulated trfA cassette, are as easily transformed as the trfA-less parental hosts. Our trfA hosts are now commercially available in highly electro-competent forms (e.g., strains Stbl2–trfA, Life Technologies; GeneHogs–trfA, Invitrogen), and TransforMax EPI300 (Epicentre). (5) The extent of the L-arabinose-induced DNA amplification can be easily modulated by the appropriate concentrations of glucose (with a sharp transition in the 0.1%–0.2% range). This is illustrated in Figure 5B. (6) Our system is specially designed for high-throughput sequencing of both ends of cloned fragments. At present, this is probably the most effective approach for sequencing of large genomes.
An addition to the improvements listed above is the reduction in plasmid size as mediated by the Flp/FRT excision system (see Szybalski et al. 1999). In this mode of amplification, only the cloned fragment and oriV are excised and replicated.
We are in the process of preparing genomic libraries, using our pBAC/oriV vectors to evaluate the stability of clones and the effectiveness of DNA amplification. Moreover, several libraries have been constructed by commercial laboratories, including Research Genetics (at present Invitrogen), using our pBAC/oriV–TrfA systems, and we were informed that, if anything, they are superior to regular BAC libraries as far as the ease of construction and maintenance are concerned (M. Ragland, pers. comm.). The amplification of clones yielded ample amounts of DNA, similar to the results reported here, and the fidelity of DNA sequencing was high, as expected.
The plasmids and hosts are available from the authors, unless they are or will become available from commercial or other sources.
METHODS
Bacterial Strains and Media
The E. coli strains used in this study were mostly derivatives of strain DH10B (see Table 2), widely used for the preparation of genomic libraries. Bacterial cultures were routinely grown with shaking in Luria-Bertani broth (LB medium) at 37°C, unless different temperatures are indicated. Antibiotics were added at following concentrations: ampicillin (Ap), 50 μg/mL; chloramphenicol (Cm), 12.5 μg/mL; and spectinomycin (Sp), 30 μg/mL.
Table 2.
List of Strains
| E. coli strains | Pertinent genotype |
|---|---|
| DH10Ba | F− mcrA Δ(mrr-hsdRMS-mcrBC) φ80dlacZΔM15ΔlacX74 endA1 recA1 deoR Δ(ara,leu)7697 araD139 galU galK nupG rpsL |
| JW366 | DH10B, with pJW349 (trfA203) integrated at the attB site; see Figs. 2 and 3 |
| JW371 | JW366 [pBAC/oriV] |
| JW378 | JW366 [pBAC/oriV+20-kb insert (pJW378)] |
| JW389 | JW366 [pBAC/oriV+40-kb insert (p67)] |
| JW390 | JW366 [pBAC/oriV+80-kb insert (p69)] |
| JW427 | DH10B with pJW424 (trfA254) integrated at the attB site; see Table 1 and Fig. 2 |
| JW439 | JW427 [pBAC/oriV+108-kb insert (pCG275)] |
| JW450 | JW427 [pBAC/oriV+122-kb insert (pCG274)] |
| JW461 | DH10B with pJW458 (trfA173) integrated at the attB site; see Table 1 and Fig. 2 |
| JW462 | DH10B with pJW459 (trfA171) integrated at the attB site; see Table 1 and Fig. 2 |
| JW463 | DH10B with pJW457 (trfA250) integrated at the attB site; see Table 1 and Fig. 2 |
| JW467 | JW461 [pJW378] |
| JW468 | JW462 [pJW378] |
| JW471 | JW461 [pBAC/oriV+140-kb insert (pCG273)] |
| JW472 | JW462 [pBAC/oriV+140-kb insert (pCG273)] |
| JW480b | GenHogs (Invitrogen) with pJW424 (trfA254) integrated at the attB site, see Table 1 and Fig. 2 |
| JW499 | JW461 [pBAC/oriV+108-kb insert (pCG275)] |
| JW500 | JW462 [pBAC/oriV+108-kb insert (pCG275)] |
| JW501 | JW463 [pBAC/oriV+108-kb insert (pCG275)] |
| JW526c | Stbl2 cells (Life Technologies/Invitrogen) with pJW424 (trfA254) integrated at the attB site; see Table 1 and Fig. 2 |
| JW550 | JW463[pIndigoBAC/oriV] |
The E. coli DH10B strain is described in Invitrogen Catalog 2002, p. 229.
The GeneHogs strain is described in Invitrogen Catalog 2002, p. 230.
The Stbl2 strain is the gal+ lon− parental strain of Stbl4 described in Invitrogen Catalog 2002, p. 234.
Brackets indicate plasmid, as carried in the host strain. Plasmid designations (see Table 1) or mutant numbers are in parentheses. Δ( ), deletion (followed by deleted genes, in parentheses, and number).
DNA Manipulations
Standard media, buffers, and DNA techniques were used (Sambrook et al. 1989). DNA from small plasmids was purified using the Wizard DNA Purification System (Promega), whereas DNA from plasmids carrying inserts >20 kb was extracted by alkaline lysis, followed by the phenol–chloroform treatment and precipitation with 70% ethanol. We routinely prepared DNA from 4.5 mL of overnight culture. Purified DNA was resuspended in 40–50 μL of TE buffer.
Construction of pBAC/oriV Derivatives
As a source of oriV, we cloned the EcoRI–BamHI fragment of the pSV16 plasmid (Durland et al. 1990) into the same sites of plasmid pUC19, resulting in pJW32. The 617-bp oriV sequence contains the NotI site preceding the eight iterons required for replication (Stalker et al. 1981). We have shown that truncation of 92 bp from the 5′ end of this oriV fragment does not affect replication directed by oriV when cloned into the XhoI site of pBeloBAC11. Therefore, pJW32 was digested with NotI + BamHI, and the 0.5-kb fragment was gel-purified, blunted with PolIk (Klenow fragment of DNA polymerase Pol I), and ligated to pBeloBAC11 (that was digested with XhoI, blunted with PolIk, and dephosphorylated with alkaline phosphatase). The resulting plasmid, pBAC/oriV, is shown in Figure 1. To obtain pBAC/oriV/SceI, the oriV-containing fragment was prepared as described above and cloned into the XhoI site of pBeloBAC11/SceI (gift from F.R. Blattner's laboratory, University of Wisconsin), as described above for pBAC/oriV (see legend to Fig. 1). The pTrueBlue-BAC2/oriV vector (pJW406; Table 1) was constructed in a similar manner (see legend to Fig. 1) by cloning the oriV-containing fragment into the XhoI-digested pTrueBlue-BAC2 plasmid (1999 Catalogue of Genomics One). To obtain the TruBlue-BAC2/oriV/SceI vector (pJW419), a 0.5-kb EcoRI–SalI fragment containing the recognition sequence for I-SceI was prepared from pSCM522 (Monteilhet et al. 1990), blunted with PolIk, gel-purified, and ligated to EcoN47III-digested and dephosphorylated pTrueBlue-BAC2/oriV (JW406), resulting in pTrueBlue-BAC2/oriV/SceI (JW419). To construct pIndigoBAC/oriV, a commercially available linearized plasmid, pIndigoBAC-5 (HindIII-Cloning Ready from Epicentre Technologies), was phosphorylated and religated to reconstruct circular pIndigoBAC-5. This vector was digested with ScaI + StuI, and the smaller fragment (1.75 kb) was replaced with the oriV-containing ScaI–StuI fragment (2.2 kb) from pJW360 (see Table 1).
Construction of Plasmids That Deliver TrfA
To secure very tight regulation of TrfA synthesis, we chose the regulatory system of the ara operon. A cassette containing the regulatory gene araC, the PBAD promoter, and the SpR gene was obtained from pMPM123 (Mayer 1995) as a 4-kb KpnI–SacI fragment. Upon blunting with PolIk, the fragment was ligated to the integration vector pJW22 (Table 1), which was digested with EcoRI + HindIII, blunted with PolIk, and dephosphorylated with alkaline phosphatase. The resulting pJW344 plasmid was used for cloning of the trfA mutant genes. The trfA203 mutation (Haugan et al. 1995) was retrieved as a 1.2-kb EcoRI–PstI fragment from pRD110-34, blunted by PolIk, and ligated to pJW344, which was digested with XbaI, blunted, and dephosphorylated, creating pJW349. The 1-kb EcoRI–PstI fragment containing a trfA254 copy-up mutation (Durland et al. 1990) was blunted and cloned into the HincII site within the MCS of pUC19. The XbaI–HindIII fragment of this clone was ligated with pJW344 (digested with the same enzymes), resulting in pJW424. Plasmids pJW457, pJW458, and pJW459 were constructed by cloning EcoRI–PstI fragments carrying trfA250 (Durland et al. 1990), trfA173, and trfA171 (Haugan et al. 1995), respectively, into pJW344 (digested with the same two enzymes). All four integration plasmids were used to create trfA-expressing hosts, as described in the next section.
Site-Specific Recombination into the attB Site in the Host Genome
The integration plasmids listed in Table 1 were inserted into the attB site in the E. coli genome by site-specific recombination, using the Int/att system of phage λ. The Int-producing pINT-ts plasmid (Hasan et al. 1994), in which the CmR marker replaced ApR, carries a gene encoding a heat-sensitive mutant of the pSC101 Rep protein; therefore, these ts plasmids are easy to eliminate at elevated temperatures. Preparation of competent cells carrying pINT-ts (ensuring delivery of Int) was as described by Hasan et al. (1994). The DNA fragment destined for integration was cloned into the pJW22 carrying the phage λ attP site, the ApR gene, MCS, and pBR322 ori, the latter flanked by two NotI sites. After digestion with NotI, the fragment containing cloned DNA, but missing the pBR322 ori, was gel-purified, recircularized using NotI–NotI ligation, and transformed at 30°C into competent DH10B cells already carrying pINT-ts (Hasan et al. 1994). Transformants were grown in LB + Ap for 2–3 h, then transferred to 37°C for overnight growth. Serial dilutions of these cultures were plated on LB + Ap plates and incubated at 42°C. Single colonies were tested for growth on LB + Ap and LB + Ap + Cm at 42°C. Colonies unable to grow on LB + Ap + Cm must have lost the pINT-ts CmR plasmid and were selected as trfA integrants.
Induction of TrfA Synthesis
Overnight cultures grown in LB supplemented with the appropriate antibiotics were used to inoculate fresh cultures that were grown in the same medium to A590 = 0.2–0.3; then L-arabinose (A) inducer was added to the final concentration of 0.01%. Parallel uninduced cultures were grown in LB or LB + 0.2% D-glucose (G); because G reduces the pBAC/oriV copy number to one. Cultures were grown for an additional 4–5 h before the cells were harvested and the DNA was extracted.
Additional methods are described in the legends of the individual figures.
Acknowledgments
We thank F.R. Blattner (University of Wisconsin) for providing the pBeloBAC11/SceI plasmid; D. Helinski's (UCSD) and S. Valla's (University of Trondheim) laboratories for the plasmids carrying the trfA gene with copy-up mutations and plasmids with oriV; M.P. Mayer for the pMTM123 plasmid carrying araC–ParaBAD; F. Schomburg from R.M. Amasino's lab (University of Wisconsin) for the T1024 and T20K24 clones of Arabidopsis thaliana DNA; C. Small and R. Bogden from Genomex for the pBAC/oriV clones carrying inserts of trout DNA; C. Gaskins from Invitrogen for pBeloBAC11 with inserts of wheat DNA; and O. Azzam for Messing's (Rutgers University) clone of rice DNA. We thank Barbara Hunter-Sandor for the expert editing.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL szybalski@oncology.wisc.edu; FAX (608) 262-2824.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.130502.
REFERENCES
- Asakawa S, Abe I, Kudoh Y, Kishi N, Wang Y, Kubota R, Kudoh J, Kawasaki K, Minoshima S, Shimizu N. Human BAC library: Construction and rapid screening. Gene. 1997;191:69–79. doi: 10.1016/s0378-1119(97)00044-9. [DOI] [PubMed] [Google Scholar]
- Boysen C, Simon MI, Hood L. Analysis of the 1.1-Mb human α/δ T-cell receptor locus with bacterial artificial chromosome clones. Genome Res. 1997;7:330–338. doi: 10.1101/gr.7.4.330. [DOI] [PubMed] [Google Scholar]
- Cai L, Taylor JF, Wing RA, Gallagher DS, Woo SS, Davis SK. Construction and characterization of a bovine bacterial artificial chromosome library. Genomics. 1995;29:413–425. doi: 10.1006/geno.1995.9986. [DOI] [PubMed] [Google Scholar]
- Diaz-Perez SV, Crouch VW, Orbach MJ. Construction and characterization of Magnaporthe grisea bacterial artificial chromosome library. Fungal Genet Biol. 1996;20:280–288. doi: 10.1006/fgbi.1996.0042. [DOI] [PubMed] [Google Scholar]
- Durland RH, Toukdarian A, Fang F, Helinski DR. Mutations in the trfA replication gene of the broad-host-range plasmid RK2 result in elevated plasmid copy numbers. J Bacteriol. 1990;172:3859–3867. doi: 10.1128/jb.172.7.3859-3867.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frengen E, Weichenhan D, Zhao B, Osoegawa K, van Geel M, de Jong PJ. A modular, positive selection bacterial artificial chromosome vector with multiple cloning sites. Genomics. 1999;58:250–253. doi: 10.1006/geno.1998.5693. [DOI] [PubMed] [Google Scholar]
- Giacalone J, Delobette S, Gibaja V, Ni L, Skiadas Y, Qi R, Edington J, Lai Z, Gebauer D, Zhao H, et al. Optical mapping of BAC clones from the human Y chromosome DAZ locus. Genome Res. 2000;10:1421–1429. doi: 10.1101/gr.112100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzman L-M, Belin D, Carson MJ, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasan N, Koob M, Szybalski W. Escherichia coli genome targeting, I. Cre-lox-mediated in vitro generation of ori− plasmids and their in vivo chromosomal integration and retrieval. Gene. 1994;150:51–56. doi: 10.1016/0378-1119(94)90856-7. [DOI] [PubMed] [Google Scholar]
- Haugan K, Karunakaran P, Blatny JM, Valla S. The phenotypes of temperature-sensitive mini-RK2 replicons carrying mutations in the replication control gene trfA are suppressed nonspecifically by intragenic cop mutations. J Bacteriol. 1992;174:7026–7032. doi: 10.1128/jb.174.21.7026-7032.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haugan K, Karunakaran P, Tondervik A, Valla S. The host range of RK2 minimal replicon copy-up mutants is limited by species-specific differences in the maximum tolerable copy number. Plasmid. 1995;33:27–39. doi: 10.1006/plas.1995.1004. [DOI] [PubMed] [Google Scholar]
- Hradecna Z, Wild J, Szybalski W. Conditionally amplifiable inserts in pBAC vectors. Microbial Comp Genom. 1998;3:58. [Google Scholar]
- Kim U-J, Birren BW, Slepak T, Mancino V, Boysen C, Kang H-L, Simon MI, Shizuya H. Construction and characterization of a human bacterial artificial chromosome library. Genomics. 1996;34:213–218. doi: 10.1006/geno.1996.0268. [DOI] [PubMed] [Google Scholar]
- Komori H, Matsunaga F, Higuchi Y, Ishiai M, Wada C, Miki K. Crystal structure of a prokaryotic replication initiator protein bound to DNA at 2.6 Å resolution. EMBO J. 1999;18:4597–4607. doi: 10.1093/emboj/18.17.4597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer MP. A new set of useful cloning and expression vectors derived from pBlueScript. Gene. 1995;163:41–46. doi: 10.1016/0378-1119(95)00389-n. [DOI] [PubMed] [Google Scholar]
- Monteilhet G, Perrin A, Thierry A, Colleaux L, Dujon B. Purification and characterization of the in vitro activity of I-SceI, a novel and highly specific endonuclease encoded by a group I intron. Nucleic Acids Res. 1990;18:1407–1413. doi: 10.1093/nar/18.6.1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori H, Kondo A, Ohshima A, Ogura T, Hiraga S. Structure and function of the F plasmid genes essential for partitioning. J Mol Biol. 1986;192:1–15. doi: 10.1016/0022-2836(86)90459-6. [DOI] [PubMed] [Google Scholar]
- Mozo T, Fischer S, Shizuya H, Altmann T. Construction and characterization of the IGF Arabidopsis BAC library. Mol Gen Genet. 1998;258:562–570. doi: 10.1007/s004380050769. [DOI] [PubMed] [Google Scholar]
- Osoegawa K, Woon PY, Zhao B, Frengen E, Tateno M, Catanase JJ, de Jong PJ. An improved approach for construction of bacterial artificial chromosome libraries. Genomics. 1998;52:1–8. doi: 10.1006/geno.1998.5423. [DOI] [PubMed] [Google Scholar]
- Perri S, Helinski DR. DNA sequence requirements for interaction of the RK2 replication initiation protein with plasmid origin repeats. J Biol Chem. 1993;268:3662–3669. [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: A laboratory manual. 2nd ed. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Schibler L, Vaiman D, Oustry A, Guinec N, Dangy-Caye AL, Billault A, Cribiu EP. Construction and extensive characterization of a goat bacterial artificial chromosome library with threefold genome coverage. Mamm Genome. 1998;9:119–124. doi: 10.1007/s003359900701. [DOI] [PubMed] [Google Scholar]
- Shizuya H, Birren B, Kim U-J, Mancino V, Slepak T, Tachiiri Y, Simon M. Cloning and stable maintenance of 300-kilobase-pair fragments of human DNA in Escherichia coli using an F-factor-based vector. Proc Natl Acad Sci. 1992;89:8794–8797. doi: 10.1073/pnas.89.18.8794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slilaty SN, Lebel S. Accurate insertional inactivation of lacZα: Construction of pTrueBlue and M13TrueBlue cloning vectors. Gene. 1998;213:83–91. doi: 10.1016/s0378-1119(98)00209-1. [DOI] [PubMed] [Google Scholar]
- Stalker DM, Thomas CM, Helinski DR. Nucleotide sequence of the region of the origin of replication of the broad host range plasmid RK2. Mol Gen Genet. 1981;181:8–12. doi: 10.1007/BF00338997. [DOI] [PubMed] [Google Scholar]
- Szybalski, W., Wild, J., and Hradecna, Z. 1999. Conditionally amplifiable BAC vector. US Patent No. 5,874,259 [revised].
- Toukdarian AE, Helinski DR, Perri S. The plasmid RK2 initiation protein binds to the origin of replication as a monomer. J Biol Chem. 1996;271:7072–7078. doi: 10.1074/jbc.271.12.7072. [DOI] [PubMed] [Google Scholar]
- Ullmann A. Complementation in β-galactosidase: From protein structure to genetic engineering. BioEssays. 1992;14:201–205. doi: 10.1002/bies.950140311. [DOI] [PubMed] [Google Scholar]
- Ullmann A, Jacob F, Monod J. Characterization by in vitro complementation of a peptide corresponding to an operator-proximal segment of the β-galactosidase structural gene of Escherichia coli. J Mol Biol. 1967;24:339–343. doi: 10.1016/0022-2836(67)90341-5. [DOI] [PubMed] [Google Scholar]
- Wild J, Hradecna Z, Szybalski W. The 2001 Molecular Genetics of Bacteria & Phages Meeting. 2001a. Easy conversion of single-copy BAC libraries into conditionally high-copy pBAC/oriV clones. Ju1y 31–August 5, 2001163. Madison, WI. [Google Scholar]
- ————— Single-copy/high-copy (SC/HC) pBAC/oriV novel vectors for genomics and gene expression. Plasmid. 2001b;45:142–143. [Google Scholar]
- Woo S-S, Jiang J, Gill BS, Paterson AH, Wing RA. Construction and characterization of a bacterial artificial chromosome library of Sorghum bicolor. Nucleic Acids Res. 1994;22:4922–4931. doi: 10.1093/nar/22.23.4922. [DOI] [PMC free article] [PubMed] [Google Scholar]