Abstract
Evolutionarily conserved noncoding genomic sequences represent a potentially rich source for the discovery of gene regulatory regions. However, detecting and visualizing compositionally similar cis-element clusters in the context of conserved sequences is challenging. We have explored potential solutions and developed an algorithm and visualization method that combines the results of conserved sequence analyses (BLASTZ) with those of transcription factor binding site analyses (MatInspector) (http://trafac.chmcc.org). We define hits as the density of co-occurring cis-element transcription factor (TF)-binding sites measured within a 200-bp moving average window through phylogenetically conserved regions. The results are depicted as a Regulogram, in which the hit count is plotted as a function of position within each of the two genomic regions of the aligned orthologs. Within a high-scoring region, the relative arrangement of shared cis-elements within compositionally similar TF-binding site clusters is depicted in a Trafacgram. On the basis of analyses of several training data sets, the approach also allows for the detection of similarities in composition and relative arrangement of cis-element clusters within nonorthologous genes, promoters, and enhancers that exhibit coordinate regulatory properties. Known functional regulatory regions of nonorthologous and less-conserved orthologous genes frequently showed cis-element shuffling, demonstrating that compositional similarity can be more sensitive than sequence similarity. These results show that combining sequence similarity with cis-element compositional similarity provides a powerful aid for the identification of potential control regions.
In higher multicellular organisms, cell-type-specific nuclear machinery has an uncanny ability to direct precise patterns of gene expression through the recognition of arrays of cis-elements specified by primary DNA sequence lying in the context of higher-order chromatin structure. Computationally, however, we have little ability to identify cis-regulatory regions from primary sequence and even less ability to predict cellular compartments into which expression is specified. Improved ability to do so will advance our understanding of eukaryotic gene regulatory mechanisms, facilitate improved annotation of the genome, and provide insight into the potential effects of sequence polymorphisms on gene expression patterns. Moreover, improved understanding of cis-element clusters present in coordinately regulated gene groups may allow for the prediction of gene regulatory network behaviors during development, homeostasis, and disease. To exploit the potential power of conserved cis-element clusters to contribute to our understanding of eukaryotic gene regulation, it is critical to create database resources from multiple sequence analysis methods on the basis of both phylogenetic conservation and known binding-site matches that can be mined for patterns that correlate with experimentally gathered expression profile data. As shown by many transgenic analyses, promoter regions alone frequently are not able to direct in vivo gene expression patterns that correspond to that of their gene of origin, and regulatory regions may occur many kilobases upstream or downstream of a gene as well as within the introns. Moreover, observed in vivo gene regulatory patterns can also be the result of the interplay of multiple cis-regulatory regions spread out over several hundreds of kilobases. Therefore, it is critical to substantially reduce the search space for functional cis-element clusters prior to whole-genome examination. We have therefore sought to do this in the context phylogeny and compositional similarity within a sequence window of 200 bp that is appropriate for local cis-element interactions.
Attempts to find putative TF-binding sites in regulatory DNA sequences began with a database of TF-binding-site sequences (Ghosh 1990) and a program developed to scan a DNA sequence against that database (Prestridge 1991). Since then, several programs have been developed to analyze DNA sequences against TF-binding-site sequence databases.
Software tools that allow the recognition of individual TF-binding sites invariably give the user an unacceptably large list of putative TF-binding sites, irrespective of the methods or databanks used. In most cases, many of these may be false positive sites. This is most likely a result of attempting to recognize binding sites independently of their context (Trifonov 1996). Functional context might include the presence of other binding sites, relative position and helical alignment to other binding sites (Fickett 1996), DNA structural characteristics (Benham 1996; Karas et al. 1996) such as curvature (Shpigelman et al. 1993; Ponomarenko et al. 1997), or other local or distant sequence characteristics.
Seeking out phylogenetic footprints, the clusters of invariant or slowly changing positions in the aligned sequences of related but divergent organisms, has now become a standard approach to examine those DNA segments flanking and interrupting the coding regions. Phylogenetic footprints have been defined as noncoding sequence motifs that show 100% conservation in several species over a region of six or more contiguous base pairs (Gumucio et al. 1996). Exploring and analyzing these footprints for regulatory elements has been fruitful. For instance, one of the first tissue-specific enhancers identified, the immunoglobulin κ enhancer, was at first distinguished as a highly conserved region within an intron (Emorine et al. 1983). Further, analysis of noncoding regions with high percent identity has determined that they are also frequently conserved in other mammals and unique within the human genome, which are two common features of long-range regulatory elements. Hardison et al. (1997) and Oeltjen et al. (1997) have also shown this fact in their studies on BTK (Bruton's Tyrosine Kinase) and HBB (β-Hemoglobin) loci, respectively.
Approaches that cluster together sites with some biologically intuitive connection between the predicted TF-binding sites and the function of the gene often reduce the output to a manageable size even though individual putative TF-binding sites are too abundant to analyze in full. This would also bring to the forefront reasonable hypotheses for testing. To test the significance of a given cluster of sites, one may calculate the probability of finding k number of sites within a space of x number of nucleotides, taking sequence heterogeneity into account (Wagner 1998).
Our present study was based on the postulate that highly conserved sequences are usually invariably involved in important functions. There has already been ample evidence to prove this (Gumucio et al. 1996; Hardison et al. 1997; Oeltjen et al. 1997; Brickner et al. 1999; Loots et al. 2000).
TraFaC is a Web-based application for analysis and display of a pair of DNA sequences with an emphasis on the detection of conserved TF-binding sites. A number of programs are used to analyze the sequences and identify various genomic features (for example, exons, repeats, conserved regions, TF-binding sites). Repeat elements are masked out using RepeatMasker and the sequences are aligned using the PipMaker-BLASTZ algorithm (Schwartz et al. 2000). MatInspector Professional (Quandt et al. 1995) or Match (BioBase) is run to scan the sequences for TF-binding sites. TraFaC then integrates analysis results from these applications and generates graphical outputs, the Regulogram and Trafacgram.
RESULTS
Accessing and Using TraFaC
TraFaC is accessed through a Web browser (http://trafac.chmcc.org), which is the main page with links to log in and instructions. The basic version allows access and visualization of the data stored in the database. Registered users can upload and view the results for sequences relevant to their interest or can compare their sequences with the existing sequences in the database through an advanced menu. However, a fundamental problem with distributed user input is the issue of quality control of the input data.
Through the cis-element clusters within BLASTZ alignments link, users will be able to see the sequence alignment, conservation data, and the number of TF-binding sites, which we refer to as hits, occurring in the conserved regions. The user has the option of either seeing the results as an image/table or a cis-element hit-density graph (Regulogram).
For comparisons of unrelated heterologous or coexpressed genes or genes, which do not yet have an ortholog available, the cis-elements shared between any gene pairs link should be opted.
A typical TraFaC result consists of the following two types of outputs, each of which can be visualized in the browser: Regulogram, a graphical representation (line graph) of the number of common binding sites (hits) in the context of sequence similarity between two sequences (Figs. 1A, 2); and Trafacgram, graphical representation of shared TF-binding sites between two sequences (Figs. 1B, 3).
Figure 1.
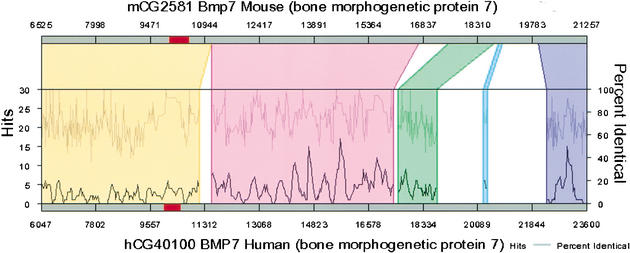
(A,B) Regulogram depiction of shared cis-elements between two sequences in the context of their sequence similarity. The two sequences are represented as horizontal bars. The colored segments on these bars are exons. The regions of alignment are represented as different colored quadrilaterals that relate one sequence to another. Within each shaded block, the percent sequence similarity and the number of TF-binding sites are represented as two separate line graphs. The percent similarity is the average sequence conservation as determined by the BLASTZ algorithm and the shared cis-element hits are determined by an algorithm that uses a 200-bp moving window that looks through the cis-elements that are present within the conserved sequence block. Numbers are nucleotide positions. Regulogram can be clicked to zoom in or view the TF-binding sites that are in common between the two sequences at the click-point coordinate.
Figure 2.
Regulogram of BMP7 with the first intronic region zoomed in. There are several clusters of shared TF-binding sites (hits) within this highly conserved region.
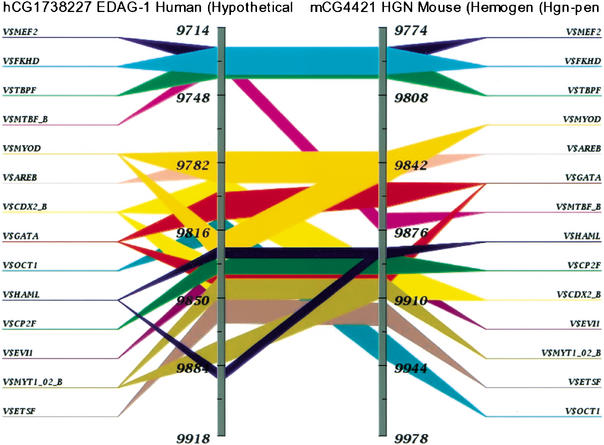
Figure 3.
TraFaC image of the promoter region of mouse hemogen and human homolog, EDAG. This region revealed a strong conservation of consensus TF-binding sites in relatively the same order of occurrence. The two gray vertical bars are the two genes (human EDAG and mouse HGN) that are compared. The numbers represent the nucleotide positions with respect to the sequences used. The TF-binding sites occurring in both the genes are highlighted as various colored bars drawn across the two genes. The image can be clicked to zoom in on a site of interest.
Known Regulatory Regions
To validate TraFaC analyses, we examined several genes with known experimentally demarcated regulatory regions. The genomic sequences were searched for shared binding sites in the phylogenetically conserved regions. Choosing the phylogenetic footprints alone reduced the total sequence space to be searched by ∼75% (Table 1). Additionally, looking for binding sites within these footprints in a moving window of 200 bp further minimized the overall sequence data to be analyzed (Table 2). A hit count was generated for each of these blocks of similar regions.
Table 1.
Reduction of total sequence search space for regulatory region identification by restriction to phylogenetically conserved sequences between orthologous gene pairs.
| Gene | Genomic sequences | Total sequence length (bp) (a) | Sum of conserved regions (bp) (b) | % Sequence to be searched for shared TF-binding sites.(b/a) | |
|---|---|---|---|---|---|
| Human (bp) | Mouse (bp) | ||||
| ADA | 36741 | 29807 | 66548 | 16668 | 25 |
| APEX | 22527 | 21963 | 44490 | 5982 | 13 |
| XRCC1 | 37785 | 37349 | 75134 | 11260 | 15 |
| ERCC2 | 54336 | 32595 | 86931 | 14257 | 16 |
| CD4 | 39512 | 43508 | 83020 | 18828 | 23 |
| PAX6 | 378625 | 400000 | 778625 | 187432 | 24 |
| ATM | 162429 | 116461 | 268890 | 58828 | 21 |
| MYO7A | 106974 | 75825 | 182799 | 50247 | 27 |
Examples of genes in which sequence search space was reduced 70% to 85%.
Table 2.
Reduction of total cis-element search space for identification of conserved TF-binding site clusters by restriction to phylogenetically conserved sequence regions.
| Gene | Number of TF binding sites in genomic sequences | Total number of TF binding sites of the two sequences (a) | TF binding sites in common within BLASTZ aligned regions (b) | % Reduction based on no. shared cis-elements within BLASTZ-aligned sequence blocks (100—b/a%) | |
|---|---|---|---|---|---|
| Human | Mouse | ||||
| ADA | 2225 | 1740 | 3965 | 798 | 80 |
| APEX | 1850 | 1721 | 3571 | 458 | 87 |
| XRCC1 | 2276 | 2209 | 4485 | 469 | 90 |
| ERCC2 | 1806 | 1051 | 2857 | 299 | 90 |
| CD4 | 2609 | 2609 | 5218 | 910 | 83 |
| PAX6 | 29398 | 27321 | 56719 | 12444 | 78 |
| ATM | 12653 | 8753 | 21406 | 4093 | 81 |
| MYO7A | 7492 | 5146 | 12638 | 3116 | 75 |
Examples of genes in which cis-element matrix hits are reduced 75% to 90%.
ADA
ADA is a key enzyme of purine catabolism that deaminates adenosine and deoxyadenosine. It is expressed at low levels ubiquitously, but at elevated levels in cortical thymocytes and the small bowel epithelial cells. Its deficiency in humans results in a failure to develop T-cells, causing severe combined immunodeficiency (SCID). Brickner et al. (1999) examined homology in noncoding regulatory regions of the human and murine ADA genes and correlated regions of homology with critical ADA T-cell regulatory regions in the first intron of the human ADA gene (Aronow et al. 1989, 1992). Recently, it has been shown that there are additional regions within the first and second introns of the human ADA gene that separately facilitate T-cell enhancer activation (Aronow et al. 1995) and control GI expression with both a duodenal enhancer and spatio-temporal modifiers of GI expression (Dusing et al. 2000).
A BLASTZ sequence alignment of human and mouse ADA genomic sequences (with ∼4.0 kb upstream and >1.0 kb downstream) revealed 17 blocks with >50% sequence similarity (indicated as different color stretches on the Regulogram, extending from one genomic sequence to another). Within each of these sequence similarity regions, TraFaC identified the shared cis-elements. Relatively bigger peaks are seen in the promoter region and also in the first and second intronic regions. On analyzing the individual constituent TF-binding sites of these regions with peaks, we observed that they have those elements that were reported previously to be responsible for the regulation of ADA expression in T-cells (first intron) and duodenum (second intron) (Fig. 1A,B).
APEX
A comparison of human and murine sequences depicted sequence conservation in the noncoding regions apart from the coding regions. The tallest peak corresponding to the shared cis-elements between human and mouse was observed in the minimal promoter region. Analysis of the minimal promoter region revealed conservation of consensus TF-binding sites for CREB, USF, PBX, and 2 CCAAT boxes (Harrison et al. 1997). Interestingly, Harrison et al. (1997) reported that the mouse APEX lack the USF sites in the promoter, whereas TraFaC analyses revealed presence of USF sites in mouse APEX also.
XRCC1
Comparison of the upstream region of XRCC1 in human and baboon (∼1600 bp upstream), and mouse (∼1200 bp upstream) revealed a significantly conserved 220-bp region. When we analyzed this region for potential cis-elements, we found that whereas human and baboon share a cluster of TF-binding sites (MTF1, HEAT, STAT, and CREB), the human and mouse shared only a single binding site for SMAD3. Interestingly, within this region, mouse and baboon had two entirely different sets of TF-binding sites in common (NFKB and SP1F). Another relatively less-conserved segment revealed TF-binding sites RFX1, a transcriptional represso, and CREB, SP1, and NF. Yu et al. (2001), in their study on splicing variant of ERCC1, another DNA repair gene, reported the presence of a binding site for RFX1 in a 42-bp segment in the human ERCC1 5′ UTR region. Loss of this segment was associated with increased ERCC1 mRNA expression in ovarian cancer specimens.
ERCC2
A comparison of the human and mouse sequences (Lamerdin et al. 1996) revealed sequence conservation for only ∼200 bp upstream of the first exon. Analysis of this region did not reveal a strong conservation of TF-binding sites.
CD4
The comparison of human and mouse CD4 genes showed high-scoring matches within the noncoding part, especially the first intron. This region has been shown to harbor important regulatory elements, the enhancer and silencer regions (Siu et al. 1994; Sarafova and Siu 1999). TraFaC revealed significant conserved TF-binding sites in both of these known regulatory regions shown as multiple peaks on the Regulogram.
Comparison of Orthologous Promoters, Enhancers, and Silencers
Known tissue-specific promoters (mainly from EPD; http://www.epd.isb-sib.ch), enhancers, and silencers were analyzed with TraFaC. Comparison of human and mouse promoters usually revealed a very high rate of conservation with respect to the TF-binding sites. For instance, strong conservation of cis-elements was observed between IL2 promoters of human and mouse. A similar result was also seen when promoters of liver-specific genes (albumin, AFP) of human and mouse were compared.
Significant representation of TF-binding sites was also frequently seen within intronic and downstream regions. For instance, the PAX6 gene in human, mouse, and fugu fish reveals strong sequence and TF-binding sites conservation in the seventh intronic region. These regions have been reported as being important regulatory regions for the development of the eye. Similarly, the fourth intronic region is also highly conserved.
All three upstream enhancers of the AFP gene could be identified as highly conserved, cis-element dense regions indicated by several peaks in the Regulogram image.
Sequence comparison of the human CD4 first intron with the CD4 silencer of African green monkey revealed high-sequence similarity. This region, extending over ∼300 bp also showed conservation of TF-binding sites with that of the first intronic region of the mouse. However, upon further analysis, we observed that humans and primates both share consensus TF-binding sites for CDX2, STAT, and SEF1 in addition to HAML, ETSF, MYT, GATA, NFKB, and EVI, which are also found in the mouse CD4 silencer.
Tissue-Specific Genes: Shared Clusters of Cis-Elements
The promoter regions of liver-specific genes, albumin, α fetoprotein, and transthyretin in mouse revealed common binding sites for the TFs HNF1, OCT1, GATA, CRBP, CLOX, and AREB. Albumin and AFP also shared additional consensus binding sites for PDX1, HNF4, HOXF, MYOF, and EVI1.
Coordinately Regulated Genes: Shared Clusters of Cis Elements
Another potential use of TraFaC is in identifying consensus-shared TF-binding sites within genes that exhibit coordinate regulation.
Shared Regulatory Elements in Coordinately Expressed Gastrointestinal Genes
Using TraFaC, we compared 4-kb upstream regions of human FABP2 and CUBN genes, both of which have been shown to have an increased expression in the small intestine of mouse (Bates et al. 2002). A segment of 300 bp, ∼1300 bp upstream of human FABP2 and CUBN genes, not only revealed a sequence similarity of >90%, but also strong conservation of cis-elements, viz., SP1F, SREB, EBOR, PAX4, AREB, RORA, EBOX, LYMF, and IKRS.
Shared Regulatory Elements in Coordinately Expressed Brain Genes
Comparing the genes PRPH and GFRA3 both highly expressed in dorsal root ganglion (Orozco et al. 2001; J. Zhang et al., in prep.), we observed that the proximal upstream regions of PRPH and GFRA3 genes share ETSF and NRSF transcription factor-binding sites. These results are in agreement with previous reports of implication of ETSF and NRSF site binding proteins in transcriptional regulation of PRPH (Chang et al. 1996).
Shared Regulatory Elements in Genes Expressed in Intestines and Lungs
We used TraFaC to analyze the expression of the gene intelectin. This gene, normally expressed in intestinal epithelium, was expressed in lungs following an exposure to ragweed (J. Santeliz and W. Karp, unpubl.). When we compared the upstream regions of mouse ITLN with mouse CLCA3 (chloride channel, calcium activated, family member), we found that these two genes share TF-binding sites of HMYO, CDX2, MYT1, MEF, EVI1, and OCT1. We also compared the same region of mouse ITLN to the CLCA family members of human, namely, CLCA1, CLCA2, and CLCA3. They all revealed TF-binding sites for OCT1, HMYO, GATA, CDX2, and EVI1. We then proceeded to compare the ITLN gene with human SFTPA1 and SFTPA2, each of which has been reported to be expressed in the intestine, in addition to their role in local host defense and the regulation of inflammation in lung (Lin et al. 2001). The downstream region of SFTPA1 and 3′-UTR of SFTPA2 shared TF-binding sites for CDX2, GATA, FKHD, and HOX.
Novel Regulatory Regions
An important use of TraFaC is in the identification of potential novel regulatory regions. Whereas any computationally identified regulatory region will require experimental validation, TraFaC analysis can serve as an excellent filter. The Regulogram feature facilitates searches for clusters of conserved cis-elements in phylogenetically conserved regions.
BMP7
The first intronic region of human and mouse BMP7 (Bone Morphogenetic Protein 7) genes is strikingly well conserved in at least nine segments. The first segment (∼6000 bp with an overall similarity of 65%) had several gapless local alignment regions (seven regions with >100 bp and a similarity ranging from 77% to 95%). Each of these phylogenetic footprints was a repository for clusters of binding sites with conserved order and extent (Fig. 2). On the basis of the high sequence similarity and also highly conserved cis-element clusters, we hypothesize that these regions may have a role in modulating BMP7 expression.
CDKN1B
A comparison of the mouse promoter and 5′ UTR of CDKN1B to its human counterpart revealed several regions of conserved noncoding segments. The promoter region had a sequence similarity of >90% and showed strong cis-element conservation. An additional conserved region was observed 3.5 kb upstream from the start site in mouse genes and ∼3 kb upstream in human genes. The 3′ downstream region also exhibited strong sequence and cis-element conservation. There were two regions. The first one was ∼250 bp and 600 bp downstream of 3′, the second was much farther removed.
PAX6
Plaza et al. (1999) reported an upstream neuroretina-specific enhancer in the human PAX6 gene. Using TraFaC, we could identify another region of high-sequence conservation in the fourth intronic regions of human and mouse PAX6. When we analyzed this region, it showed strong conservation of TF-binding sites. In addition, apart from human and mouse, this region is also highly conserved phylogenetically among fugu and quail, and is also proven as a neuroretina-specific enhancer region in fugu, quail, and mouse. Hence, this region in human is most likely to be another important regulatory segment apart from the already-known upstream neuroretina-specific enhancer in human PAX6 gene.
Hemogen
Hemogen (Hemopoietic gene) in mouse is sequentially expressed in active hematopoietic sites and down-regulated in the process of blood cell differentiation. The human homolog is EDAG, which is also specifically expressed in hematopoietic cells. EDAG maps to chromosome 9q22, a region containing the breakpoints of several hematopietic neoplasms (Yang et al. 2001).
We compared the immediate upstream regions (∼400-bp segment) of the mouse Hgn and human EDAG genes and observed high-sequence similarity. This region also revealed a strong conservation of consensus TF-binding sites. The shared cis-elements included MEF2, FKHD, MYOD, GATA1, AML1, CP2, ETS, AREB6, and EVI among others (Fig. 3). It has been proven that the transcription factor CP2 is crucial in hemoglobin synthesis during erythroid terminal differentiation in vitro (Chae et al. 1999; Solis et al. 2001). They also reported that the GATA1 and CP2 transcriptional-binding elements are functionally important for erythroid-specific heme biosynthesis. AML and ETS families of TFs are also shown to play roles in myeloid cell differentiation. AREB6 and EVI-1 are known to be haematopoietic transcriptional repressors (Turner and Crossley 1999). The mouse hemogen showed a consensus-binding site for PU.1 (∼157 bp upstream of the transcriptional start site), an Ets family member, and is considered as one of the master TFs identified in regulating development of both granulocytes and monocytes (Nagamura-Inoue et al. 2001). We did not find this site in the human homolog, EDAG, in the immediate upstream region.
We also observed another region of conserved cis-elements in the 3′ region (∼470 bp downstream to the fourth and last exon of human EDAG and ∼1400 bp downstream to the fifth and last exon of mouse Hgn). The shared consensus-binding sites included STAT, GATA, EVI, NFAT, IRFF, STAT, GATA, and EVI have been shown to be functionally important in myeloid cell differentiation (Turner and Crossley 1999; Nagamura-Inoue et al. 2001; Solis et al. 2001).
On the basis of these findings, and the strong conservation of cis-elements in this region between mouse and human, we hypothesize that these two regions might be involved in important regulatory functions.
DISCUSSION
Extraction of relevant biological information from the inundation of accumulating genomic sequence data is still a formidable challenge. This task is particularly arduous in the case of large genomes with large components of noncoding sequences, much of which may be functionless. The amount of experimental work that would be required to systematically analyze the noncoding sequences simply exceeds known research methodologies. Hence, there is a critical need for computational tools that identify potential regulatory regions with which researchers could focus their experiments. Any improvement in the prescreening process of regulatory region identification by sequence analysis is therefore highly desirable and welcome.
Phylogenetic comparison of homologous sequences has been the most promising approach for the identification of new unknown regulatory elements. There has been an increasingly convergent view on the organization of gene regulatory regions and also an emerging paradigm for their computational characterization. Bucher (1999) summarized it as follows: The elementary units of transcriptional regulatory regions are transcription factor-binding sites (Mitchell and Tjian 1989); control regions, such as promoters, enhancers, silencers, locus control regions, and so on, are modular; the regulatory output of a control region depends on the specific combination of its elements, as well as on the order and orientation in which they occur, and genes are typically controlled by several control regions located upstream or downstream from the transcription initiation site.
The TF-binding recognition sites are usually 6–8 nucleotides long, and limited variants of this short string will bind the protein with high affinity. However, Dembo et al. (1994) suggested that runs of 16 consecutive identical nucleotides could be expected to occur strictly by chance when comparing two 100-kb sequences. This raises the possibility that single isolated TF-binding sites will be lost in the background noise of spurious matches. Also, biological variation between two species can confound the approach on the basis of similarity and conservation. For instance, homologous human and murine TFs may have somewhat different specificity, and in some cases, humans may use a different set of TFs than mice to regulate a homologous gene in a different way. This search for conserved noncoding sequences is further complicated by the difference in patterns of evolution at various loci (Koop 1995). Therefore, how can regulatory factors have gene-specific effects on the rate of transcription? The strongest answer lies in the fact that TFs never act in isolation (Courey 2001), and transcriptional activity of any gene is governed by a combination of TFs acting in clusters of localized domains.
Known Regulatory Regions
Analysis of known regulatory regions proved that most of these regions lie in evolutionarily conserved regions. However, the sequence similarity alone is not always the right criterion to identify regulatory regions. For instance, the thymic enhancer region of the ADA gene is not a highly conserved region between human and mouse. However, this region is constitutionally similar with respect to the cis-elements. Hence, when looking for regulatory regions, it is highly imperative to look in those regions that also do not have relatively high-sequence similarity. Therefore, we found it difficult to set a cut-off limit for similarity score. Nevertheless, the BLASTZ algorithm of PipMaker, which we used to align the genomic sequences, uses 50% as the cut-off limit. The hits or the shared cis-elements, therefore, are limited to those regions that have 50% or more over all local sequence similarity. The window size for calculating hits was set as a minimum of 200 bp.
Presently, a limiting factor for TraFaC is that it identifies known binding sites, basically the entries in the Transfac library. We have seen cases in which there was high-sequence conservation in the upstream promoter regions, but not significant conservation with respect to the cis-elements. Even though sequence similarity doesn't necessarily always have to be reflected in TF-binding sites conservation, we felt these regions might harbor sites that have not yet been discovered. A motif search among all such sites might be a useful exercise.
The DNA repair genes APEX, ERCC2, and XRCC1 revealed smaller regulatory regions and little conservation among human and mouse with respect to cis-elements. A probable explanation for this phenomenon is that the excision repair genes may be expressed at about the same level in all cells, given the universal need for excision repair of the DNA. Thus, these genes may be under relatively simple control manifested in the TraFaC comparative analysis as a limited number of conserved cis-regulatory elements. Analyzing regulatory regions of genes with similar function would help in laying groundwork in determining potential roles for positive or negative, synergistic or antagonistic regulatory elements in various physiological functions like cell cycle control, DNA repair pathways, etc.
Coordinately Expressed Genes: Common TF-Binding Sites
Following the advent of DNA microarray technology, which can measure mRNA expression levels of different genes, obtaining clusters of similarly expressed genes has become relatively less cumbersome. Such genes with a similar expression profile may be assumed to result, at least partly, because of similar transcription machinery. In other words, upstream promoter regions of these genes might contain the binding sites for the same TFs. As is known, in most cases, TFs do not function in isolation, but rather as organized functional groups called modules (Yuh et al. 1998; Werner 1999) and regulate transcription as synergistic or antagonistic pairs (Fickett 1996; Aronone and Davidson 1997; Yuh et al. 1998). Prior reports indicate that the combined presence of a set of TF-binding sites is strong evidence for similar gene regulation. Kel et al. (1999) showed this in their study on activated T-cells. Michelson (2002) opined that a more efficient approach to the identification of coexpressed genes and their associated regulatory elements would accelerate the field of identification of regulatory regions greatly.
Even though the results for yeast have been promising in linking the gene expression data to regulatory segments in the genomic sequences, it remains one of the great challenges while analyzing the mammalian expression data. Iyer et al. (2001), following the array of noncoding DNA of yeast, identified the genes regulated by the cell cycle TFs, SBF, and MBF. Ren et al. (2000) reported similar results for Gal4 and Ste12. Similarly, Livesey et al. (2000) have identified the response element configuration and genes responsive to the mouse homeobox gene Crx (cone-rod homeobox).
Tissue-Specific Genes: Common TF-Binding Sites
The unique expression of genes appears more tissue specific than the transcription factors that regulate them. Earlier studies (Cordle et al. 1991) led to the concept that many regulatory modules contain cis-acting regions that repress transcription in nonexpressing cells. What is yet to be understood is whether these effects represent repressors or are due to incompatible TF interactions. Subtle differences in abundance or timing, absence of one critical factor, or addition of one new factor could redirect a regulatory module. Finally, there may be a higher level of cell-specific chromosome mechanism regulating the accessibility of genes to transcription machinery. We also observed that high-scoring regions frequently showed cis-element shuffling, particularly in nonorthologous genes with similar expression profiles. This was evident in the comparison, interspecies and intraspecies, between liver-specific genes like AFP, TAT, and albumin, and in between IL3, IL4, and IL5.
Novel Regulatory Regions
The regulatory regions in higher multicellular organisms may occur upstream or downstream of a gene or even within the introns; sometimes they are also spread out over several hundreds of kilobases. Hence, in investigations of regulatory regions, any approach that substantially reduces the size of the sequence space to be searched can be very valuable. Searching for regulatory regions in phylogenetic footprints using TraFaC reduces the sequence space to be searched by ∼75% (Table 1). The various software tools that allow the recognition of individual TF-binding sites invariably give the user an unacceptably large list of putative TF-binding sites, irrespective of the methods or databanks used. In most cases, many of these may be false positive sites. This is most likely a result of attempting to recognize binding sites independently of their context. However, when the occurrence of cis-elements in conserved regions alone was taken into account, there was a considerable reduction in the overall number of cis-elements (75%–90%, Table 2). When additional parameters, such as the presence of other binding sites, window size of 200 bp, and relative position and spread of each element was taken account, the data to be analyzed became easily manageable.
In hypothesizing regions of highly conserved cis-element clusters as novel regulatory regions, we took into account the following three criteria: sequence similarity, cis-element conservation (order and extent – a 200-bp window was set as minimum), and the individual constituent TF-binding sites – whether these are reported previously to be involved in regulation of other genes belonging to the same family or genes having the same function.
CisMols
We are in the process of identifying and storing clusters of cis-elements, which we refer to as CisMols (Cis regulatory modules), pertaining to a defined set of genes. The classifying criteria can be a gene expression pattern (coordinately expressed genes), gene ontology functionalities, a phenotype, or simply a tissue specificity.
CONCLUSION
The need for computational tools that identify potential regulatory elements with which researchers could focus their experiments is of paramount importance. In the present study, an attempt has been made to identify conserved cis-elements/TF-binding sites by adopting a comparative genomic analysis of human and murine DNA sequences. Phylogenetically conserved noncoding regions tend to be good indicators of regions of gene regulatory functions (ADA, APEX, XRCC1, ERCC2, CD4, and several other orthologous promoters from the EPD database). However, small introns and high conservation at the protein level diminish the power of noncoding region sequence homology. We also observed that combinations of TF-binding sites in the same relative order and distance can be reliable indicators of potential novel regulatory regions. This was further strengthened when we analyzed the individual cis-elements making up these shared clusters. These shared elements were either implicated previously to be involved in similar function in other annotated genes or were similar to the other members of these gene families or groups (first intron of BMP7, upstream and downstream regions of CDKN1B, PAX6, Hemogen). Genes with coordinate expression frequently tend to share similar TF-binding sites. This was apparent when we analyzed genes with a coordinate expression in the gastrointestinal tract (FABP2 and CUBN), dorsal root ganglion of brain (PRPH and GFRA3), and intestine and lungs (ITLN and CLCA3).
The search for clusters of cis-regulatory elements in sequence databases is still a difficult task. TraFaC programs can help reduce the signal-to-noise ratio considerably, especially when comparing genomic orthologs, because by looking for conserved cis-elements in the context of sequence similarity, the overall sequence space to be searched for the identification of the regulatory keys is reduced considerably. However, an added advantage is that it also aids in identifying constitutionally similar cis-element clusters in the absence of sequence similarity. Finally, even though TraFaC and other software with similar functionality can be used for identifying potential novel regulatory regions, the challenge remains, however, in the transfer of useful biological knowledge to these regions. Computational methods only provide useful suggestions of regulatory regions and functions, however, only a biologist can truly assign function to a regulatory region by use of these results, and the ultimate confirmation of these assignments must be based on experimental evidence.
METHODS
Complete genomic sequences were extracted from the GenBank (GenBank http://www.ncbi.nlm.nih.gov) and Celera human and mouse databases (Celera http://www.celera.com). The repeat elements were masked using the RepeatMasker program (http://ftp.genome.washington.edu/RM/RepeatMasker.html; Smit and Green 1999) prior to computational alignment of the two genomic sequences using Advanced PipMaker (http://bio.cse.psu.edu). PipMaker's BLASTZ computes local alignments using dynamic programming. These alignments are then post-processed by a tool that arranges for each nucleotide in human to map to, at most, one nucleotide in mouse, possibly by dividing the original BLASTZ alignments into several smaller ones. Every local alignment is a series of gap-free alignments and gaps. The positions of gap-free portions of alignments are plotted with respect to the human sequence along the x-axis, and the percent identity is plotted along the y-axis (from 50% to 100%). The programs Match (http://www.biobase.de) or MatInspector Professional Version 4.3, 2000 (http://www.genomatix.de) were used to locate putative TF-binding sites in a DNA sequence. Both of these programs utilize the TRANSFAC database (TRANSFAC is the database on eukaryotic TFs, their genomic binding sites, and DNA-binding profiles; Wingender et al. 2000; http://transfac.gbf.de) to identify matches in DNA sequences. The output consists of a table indicating a list of putative binding sites.
Thus, for a set of orthologous genes, the following files were obtained and saved to upload to TraFaC, sequence files in Fasta format; Exons file; RepeatMasker output file; BLASTZ, concise and PIP (pdf file) output files from PipMaker; list of putative binding sites for both the sequences (output from MatInspector/Match).
Basic User Version
Unregistered users can only access the data available. There are two approaches to visualize the data. The approach depends on the purpose of analysis, viz., to compare a homologous pair of genes or just compare any gene with any other gene (heterologous genes) or a known regulatory sequence like promoters or enhancers in the TraFaC database.
Through the cis-element clusters within BLASTZ alignments' link, users will be able to see the sequence alignment, conservation data, and the number of TF-binding sites, which we refer to as hits, occurring in the conserved regions. The user has the option of seeing either the results as an image/table or a graph (Regulogram).
For comparing unrelated heterologous or coexpressed genes or genes, which do not have an ortholog available yet, the cis-elements shared between any gene pairs' link should be opted.
Advanced User Version
Registered users have the privilege of uploading sequences relevant to their interest and can compare their data with other genes in the database. The user is required to submit the following input files. Again, the number or type of input files to be submitted here depends on the purpose TraFaC is intended to be used for. To visualize a complete picture of sequence similarity with TF homologies and exon annotations, the following files have to be submitted. The BLASTZ alignment file and the alignment summary file from the PipMaker. These files are referred to as text and concise files in the output of PipMaker. A list of TF-binding sites for each of the sequences. The programs MatInspector or Match can be used to generate this file. The sequence file in a FASTA format, the Exons file, and the RepeatMask file are optional. The instructions page on the Web site provides a more detailed description.
WEB SITE REFERENCES
http://www.celera.com; Celera.
http://www.epd.isb-sib.ch; EPD.
http://www.biobase.de; Match.
http://www.genomatix.de; MatInspector.
http://www/ncbi.nlm.nih.gov; GenBank.
http://bio.cse.psu.edu; PipMaker.
http://ftp.genome.washington.edu/RM/RepeatMasker.html; RepeatMasker.
http://trafac.chmcc.org; TraFaC.
http://transfac.gsf.de); Transfac.
Acknowledgments
This work was supported in part by a grant from the Howard Hughes Medical Institute to the University of Cincinnati for the Development of Bioinformatics Core Resources and from the NIEHS U01 ES11038 Mouse Centers Genomics Consortium.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL bruce.aronow@chmcc.org; FAX (513) 636–2056.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.255002.
REFERENCES
- Aronone MI, Davidson EH. The hard wiring of development: Organization and function of genomic regulatory sequences. Development. 1997;124:1857–1864. doi: 10.1242/dev.124.10.1851. [DOI] [PubMed] [Google Scholar]
- Aronow B, Lattier D, Silbiger R, Dusing M, Hutton J, Jones G, Stock J, McNeish J, Potter S, Witte D, et al. Evidence for a complex regulatory array in the first intron of the human adenosine deaminase gene. Genes & Dev. 1989;3:1384–1400. doi: 10.1101/gad.3.9.1384. [DOI] [PubMed] [Google Scholar]
- Aronow B, Silbiger RN, Dusing MR, Stock JL, Yager KL, Potter, Hutton J, Wiginton DA. Functional analysis of the human adenosine deaminase gene thymic regulatory region and its ability to generate position-independent transgene expression. Mol Cell Biol. 1992;12:4170–4185. doi: 10.1128/mcb.12.9.4170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aronow BJ, Ebert CA, Valerius MT, Potter SS, Wiginton DA, Witte DP, Hutton JJ. Dissecting a locus control region: Facilitation of enhancer function by extended enhancer-flanking sequences. Mol Cell Biol. 1995;15:1123–1135. doi: 10.1128/mcb.15.2.1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates MD, Erwin CR, Sanford LP, Wiginton D, Bezerra JA, Schatzman LC, Jegga AG, Ley-Ebert C, Williams SS, Steinbrecher KA, et al. Novel genes and functional relationships in the adult mouse gastrointestinal tract identified by microarray analysis. Gastroenterology. 2002;122:1467–1482. doi: 10.1053/gast.2002.32975. [DOI] [PubMed] [Google Scholar]
- Benham CJ. Computation of DNA structural variability - a new predictor of DNA regulatory regions. Comp App Biosci. 1996;12:375–381. doi: 10.1093/bioinformatics/12.5.375. [DOI] [PubMed] [Google Scholar]
- Brickner AG, Koop BF, Aronow BJ, Wiginton DA. Genomic sequence comparison of the human and mouse adenosine deaminase gene regions. Mamm Genome. 1999;10:95–101. doi: 10.1007/s003359900951. [DOI] [PubMed] [Google Scholar]
- Bucher P. Regulatory elements and expression profiles. Curr Opin Struct Biol. 1999;9:400–407. doi: 10.1016/S0959-440X(99)80054-2. [DOI] [PubMed] [Google Scholar]
- Chae JH, Lee YH, Kim CG. Transcription factor CP2 is crucial in hemoglobin synthesis during erythroid terminal differentiation in vitro. Biochem Biophys Res Comm. 1999;263:580–583. doi: 10.1006/bbrc.1999.1408. [DOI] [PubMed] [Google Scholar]
- Chang L, Thompson MA. Activity of the distal positive element of the peripherin gene is dependent on proteins binding to an Ets-like recognition site and a novel inverted repeat site. J Biol Chem. 1996;271:6467–6475. doi: 10.1074/jbc.271.11.6467. [DOI] [PubMed] [Google Scholar]
- Cordle SR, Whelan J, Henderson E, Masuoka H, Weil PA, Stein R. Insulin gene expression in nonexpressing cells appears to be regulated by multiple distinct negative-acting control elements. Mol Cell Biol. 1991;11:2881–2886. doi: 10.1128/mcb.11.5.2881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courey AJ. Regulatory transcription factors and cis-regulatory regions. In: Locker J, editor. Transcription factors. Oxford, UK: Academic Press; 2001. pp. 17–34. [Google Scholar]
- Dembo A, Karlin S, Zeitouni O. Limit distribution of maximal non-aligned two-sequence segmental score. Ann Probability. 1994;22:2022–2039. [Google Scholar]
- Dusing MR, Brickner AG, Lowe SY, Cohen MB, Wiginton DA. A duodenum-specific enhancer regulates expression along three axes in the small intestine. Am J Physiol Gastrointest Liver Physiol. 2000;279:G1080–G1093. doi: 10.1152/ajpgi.2000.279.5.G1080. [DOI] [PubMed] [Google Scholar]
- Emorine L, Kuehl M, Weir L, Leder P, Max EE. A conserved sequence in the immunoglobulin JK-CK intron: Possible enhancer element. Nature. 1983;304:447–449. doi: 10.1038/304447a0. [DOI] [PubMed] [Google Scholar]
- Fickett JW. Coordinate positioning of MEF2 and myogenin binding sites. Gene. 1996;172:GC19–GC32. doi: 10.1016/0378-1119(95)00888-8. [DOI] [PubMed] [Google Scholar]
- Ghosh D. A relational database of transcriptional factors. Nucleic Acids Res. 1990;18:1749–1756. doi: 10.1093/nar/18.7.1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gumucio D, Shelton D, Zhu W, Millinoff D, Gray T, Bock JH, Slightom JL, Goodman M. Evolutionary strategies for the elucidation of cis and trans factors that regulate the developmental switching programs of the β-like globin genes. Mol Phylogenet Evol. 1996;5:18–32. doi: 10.1006/mpev.1996.0004. [DOI] [PubMed] [Google Scholar]
- Hardison R, Slightom JL, Gumucio DL, Goodman M, Stojanovic N, Miller W. Locus control regions of mammalian β-globin gene clusters: Combining phylogenetic analyses and experimental results to gain functional insights. Gene. 1997;205:73–94. doi: 10.1016/s0378-1119(97)00474-5. [DOI] [PubMed] [Google Scholar]
- Harrison L, Ascione AG, Takiguchi Y, Wilson DM, Chen DJ, Demple B. Comparison of the promoters of the mouse (APEX) and human (APE) apurinic endonuclease genes. Mutat Res. 1997;385:159–172. doi: 10.1016/s0921-8777(97)00053-0. [DOI] [PubMed] [Google Scholar]
- Iyer VR, Horak CE, Scafe CS, Botstein D, Snyder M, Brown PO. Genomic binding sites of the yeast cell-cycle transcription factors SBF and MBF. Nature. 2001;409:533–538. doi: 10.1038/35054095. [DOI] [PubMed] [Google Scholar]
- Karas H, Knuppel R, Schulz W, Sklenar H, Wingender E. Combining structural analysis of DNA with search routines for the detection of transcription regulatory elements. Comput Appl Biosci. 1996;12:441–446. doi: 10.1093/bioinformatics/12.5.441. [DOI] [PubMed] [Google Scholar]
- Kel A, Kel-Margoulis O, Babenko V, Wingender E. Recognition of NFATp/AP-1 composite elements within genes induced upon the activation of immune cells. J Mol Biol. 1999;288:353–376. doi: 10.1006/jmbi.1999.2684. [DOI] [PubMed] [Google Scholar]
- Koop BF. Human and rodent sequence comparisons: A mosaic model of genomic evolution. Trends Genet. 1995;11:367–371. doi: 10.1016/s0168-9525(00)89108-8. [DOI] [PubMed] [Google Scholar]
- Lamerdin JE, Stilwagen SA, Ramirez MH, Stubbs L, Carrano AV. Sequence analysis of the ERCC2 gene regions in human, mouse, and hamster reveal three linked genes. Genomics. 1996;34:399–409. doi: 10.1006/geno.1996.0303. [DOI] [PubMed] [Google Scholar]
- Lin Z, deMello D, Phelps DS, Koltun WA, Page M, Floros J. Both human SP-A1 and Sp-A2 genes are expressed in small and large intestine. Pediatr Pathol Mol Med. 2001;20:367–386. [PubMed] [Google Scholar]
- Livesey FJ, Furukawa T, Steffen MA, Church GM, Cepko CL. Microarray analysis of the transcriptional network controlled by the photoreceptor homeobox gene Crx. Curr Biol. 2000;10:301–310. doi: 10.1016/s0960-9822(00)00379-1. [DOI] [PubMed] [Google Scholar]
- Loots GG, Locksley RM, Blankespoor CM, Wang ZE, Miller W, Rubin EM, Frazer KA. Identification of a coordinate regulator of interleukins 4, 13, and 5 by cross-species sequence comparisons. Science. 2000;288:136–140. doi: 10.1126/science.288.5463.136. [DOI] [PubMed] [Google Scholar]
- Michelson AM. Deciphering genetic regulatory codes: A challenge for functional genomics. Proc Natl Acad Sci. 2002;99:546–548. doi: 10.1073/pnas.032685999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell PJ, Tjian R. Transcriptional regulation in mammalian cells by sequence-specific DNA binding proteins. Science. 1989;245:371–378. doi: 10.1126/science.2667136. [DOI] [PubMed] [Google Scholar]
- Nagamura-Inoue T, Tamura T, Ozato K. Transcription factors that regulate growth and differentiation of myeloid cells. Int Rev Immunol. 2001;20:83–105. doi: 10.3109/08830180109056724. [DOI] [PubMed] [Google Scholar]
- Oeltjen JC, Malley TM, Muzny DM, Miller W, Gibbs RA, Belmont JW. Large-scale comparative sequence analysis of the human and murine Bruton's tyrosine kinase loci reveals conserved regulatory domains. Genome Res. 1997;7:315–329. doi: 10.1101/gr.7.4.315. [DOI] [PubMed] [Google Scholar]
- Orozco OE, Walus L, Sah DW, Pepinsky RB, Sanicola M. GFRα3 is expressed predominantly in nociceptive sensory neurons. Eur J Neurosci. 2001;13:2177–2182. doi: 10.1046/j.0953-816x.2001.01596.x. [DOI] [PubMed] [Google Scholar]
- Plaza S, Saule S, Dozier C. High conservation of cis-regulatory elements between quail and human for the Pax-6 gene. Dev Genes Evol. 1999;209:165–173. doi: 10.1007/s004270050240. [DOI] [PubMed] [Google Scholar]
- Ponomarenko, M.P., Ponomarenko, J.V., Kel, A.E., and Kolchanov, N.A. 1997. Search for DNA conformational features for functional sites. Investigation of the TATA box. (Eds. R.B. Altman, K. Dunker, L. Hunter, T.E. Klein). Proc. 1997 Pacific Symp. Biocomput., pp. 340–351. World Scientific Publishing Company, Singapore. [PubMed]
- Prestridge DS. SIGNAL SCAN: A computer program that scans DNA sequences for eukaryotic transcriptional elements. Comput Appl Biosc. 1991;7:203–206. doi: 10.1093/bioinformatics/7.2.203. [DOI] [PubMed] [Google Scholar]
- Quandt K, Frech K, Karas H, Wingender E, Werner T. MatInd and MatInspector: New fast and versatile tools for detection of consensus matches in nucleotide sequence data. Nucleic Acids Res. 1995;23:4878–4884. doi: 10.1093/nar/23.23.4878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren B, Robert F, Wyrick JJ, Aparicio O, Jennings EG, Simon I, Zeitlinger J, Schreiber J, Hannett N, Kanin E, et al. Genome-wide location and function of DNA binding proteins. Science. 2000;290:2306–2309. doi: 10.1126/science.290.5500.2306. [DOI] [PubMed] [Google Scholar]
- Sarafova SD, Siu G. Control of CD4 gene expression: Connecting signals to outcomes in T cell development. Brazilian J Med Biol Res. 1999;32:785–803. doi: 10.1590/s0100-879x1999000700001. [DOI] [PubMed] [Google Scholar]
- Schwartz S, Zhang Z, Frazer KA, Smit A, Riemer C, Bouck J, Gibbs R, Hardison R, Miller W. PipMaker: A Web server for aligning two genomic DNA sequences. Genome Res. 2000;10:577–586. doi: 10.1101/gr.10.4.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shpigelman ES, Trifonov EN, Bolshoy A. CURVATURE: Software for the analysis of curved DNA. Comput Appl Biosci. 1993;9:435–440. doi: 10.1093/bioinformatics/9.4.435. [DOI] [PubMed] [Google Scholar]
- Siu G, Wurster AL, Duncan DD, Soliman TM, Hedrick SM. A transcriptional silencer controls the developmental expression of the CD4 gene. EMBO J. 1994;13:3570–3579. doi: 10.1002/j.1460-2075.1994.tb06664.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solis C, Aizencang GI, Astrin KH, Bishop DF, Desnick RJ. Uroporphyrinogen III synthase erythroid promoter mutations in adjacent GATA1 and CP2 elements cause congenital erythropoietic porphyria. J Clin Invest. 2001;107:753–762. doi: 10.1172/JCI10642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trifonov EN. Interfering contexts of regulatory sequence elements. Comput Appl Biosci. 1996;12:423–429. doi: 10.1093/bioinformatics/12.5.423. [DOI] [PubMed] [Google Scholar]
- Turner J, Crossley M. Basic Kruppel-like factor functions within a network of interacting haematopoietic transcription factors. Int J Biochem Cell Biol. 1999;10:1169–1174. doi: 10.1016/s1357-2725(99)00067-9. [DOI] [PubMed] [Google Scholar]
- Wagner, A. 1998. A computational “genome walk” technique to identify regular interactions in gene networks. Pacific Symposium on Biocomputation, pp. 264–278. [PubMed]
- Werner T. Models for prediction and recognition of eukaryotic promoters. Mamm Genome. 1999;10:168–175. doi: 10.1007/s003359900963. [DOI] [PubMed] [Google Scholar]
- Wingender E, Chen X, Hehl R, Karas H, Liebich I, Matys V, Meinhardt T, Pruss M, Reuter I, Schacherer F. TRANSFAC: An integrated system for gene expression regulation. Nucleic Acids Res. 2000;28:316–319. doi: 10.1093/nar/28.1.316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang LV, Nicholson RH, Kaplan J, Galy A, Li L. Hemogen is a novel nuclear factor specifically expressed in mouse hematopoietic development and its human homologue EDAG maps to chromosome 9q22, a region containing breakpoints of hematological neoplasms. Mechan Dev. 2001;104:105–111. doi: 10.1016/s0925-4773(01)00376-8. [DOI] [PubMed] [Google Scholar]
- Yu JJ, Thronton K, Guo Y, Kotz H, Reed E. An ERCC1 splicing variant involving the 5′-UTR of the mRNA may have a transcriptional modulatory function. Oncogene. 2001;20:7694–7698. doi: 10.1038/sj.onc.1204977. [DOI] [PubMed] [Google Scholar]
- Yuh CH, Bolouri H, Davidson EH. Genomic cis-regulatory logic: Experimental and computational analysis of a sea urchin gene. Science. 1998;279:1896–1902. doi: 10.1126/science.279.5358.1896. [DOI] [PubMed] [Google Scholar]