Abstract
To assess an unequivocal diagnosis of mantle cell lymphoma (MCL), we have developed a fluorescence in situ hybridization (FISH) assay, enabling the demonstration of t(11;14)(q13;q32) directly on pathological samples. We have first selected CCND1 and IGH probes encompassing the breakpoint regions on both chromosomes. Then, we have defined experimental conditions enabling us to obtain bright clear-cut signals in all of the samples, independently of the initial fixation conditions. We have analyzed single-cell suspensions from 26 formalin-fixed, paraffin-embedded MCL samples with this set of probes. In all cases, we have found a fusion signal (ie, a t(11;14)(q13;q32) translocation) in 14% to 99% of cells (median, 87%). So far, IGH-CCND1 fusions have been detected in all of the 51 MCL patients that we have analyzed by FISH (either on paraffin-embedded tumor samples or on peripheral blood samples). Regarding the low sensitivity of other techniques used to diagnose t(11;14)(q13;q32) (ie, 70% to 75% for cytogenetics and 50% to 60% for polymerase chain reaction), our FISH assay is by far the most sensitive technique. Moreover, because of the quality of the fluorescent signals and the rapidity of the experiment, this technique is widely applicable, even in routine cytogenetics or pathology laboratories. As MCL patients are usually refractory to standard therapy, an unambiguous diagnosis is needed to propose adapted therapeutic strategies, and this highly sensitive assay may be of great value for accurate diagnosis in difficult cases.
Classification of non-Hodgkin’s lymphoma (NHL) is essentially based on morphological features of malignant cells. A more recent classification, ie, the revised European-American classification of lymphoid neoplasms, 1 is probably the most accurate, taking into account morphological and immunological criteria, leading to the definition of several NHL subsets. One of this newly identified subgroup is the so-called mantle cell lymphoma (MCL), defined by CD5-positive/CD23-negative/sIgMD-positive lymphoid cells. 1-5 These cells are the malignant counterpart of naive B cells located in the mantle zone of secondary follicles. Besides these biological features, MCL is characterized by a male predominance, frequent advanced clinical stage at diagnosis, and poor prognosis. 5-8 This clinical aggressivity contrasts with the intermediate pathological grade, and reliable sensitive diagnostic tools would be highly useful for an unequivocal identification of this NHL subset.
Cytogenetic analyses have revealed that MCL is closely associated with the t(11;14)(q13;q32). 9-11 This translocation juxtaposes Ig heavy chain gene (IGH) sequences with the BCL-1 locus, leading to up-regulation of the CCND1 gene and consequently to an overexpression of cyclin D1. 12-15 However, whereas overexpression of cyclin D1 is thought to be present in 100% of patients with MCL, t(11;14)(q13;q32) is found in 70% to 75% of the patients only. 10,11 This discrepancy may be related to the low mitotic index of malignant cells and to the poor morphology of metaphase spreads. Moreover, cytogenetic analysis is a time-consuming technique, especially for the analysis of lymphoma samples. Other molecular techniques have also their own limitations, essentially because of the scattering of 11q13 breakpoints. Thus, polymerase chain reaction (PCR) techniques are positive in only 50% to 60% of cases. 12,16,17 Fluorescence in situ hybridization (FISH) may circumvent these difficulties, and we have previously demonstrated that this technique could be successfully applied on MCL leukemic phase samples. 18 However, this approach is applicable only to bone marrow or peripheral blood specimens, or to specimens prepared for cytogenetic analysis, and so cannot be used in patients with suspected MCL but without significant marrow involvement. To circumvent this limitation, we have developed and tested a dual-color FISH assay that could be performed directly on formalin-fixed, paraffin-embedded specimens.
Patients, Materials, and Methods
Patients
Twenty-six patients diagnosed as MCL (pan-B+, CD5+, IgMD+, CD23−) in our institution and for whom a paraffin-embedded sample was available were analyzed. The main clinical features of these patients are described in Table 1 ▶ . Briefly, they were 18 males and 8 females, with a median age of 65 (range, 40 to 78) years. Five patients with a peripheral blood involvement have been previously reported. 18 Twenty-four of these patients had been analyzed by immunohistochemistry, using a cyclin D1 monoclonal antibody (Immunotech, Marseille, France). All but one (patient 2) were positive. Routine cytogenetic or molecular analyses were not performed for any of these patients.
Table 1.
Patient Characteristics
| Patient | Sex | Age | IHC results* | % cells with t(11;14) | 11q13 probe split† |
|---|---|---|---|---|---|
| 1 | M | 56 | + | 95 | Y |
| 2 | F | 63 | − | 14 | Y |
| 3 | M | 70 | + | 99 | Y |
| 4 | M | 61 | + | 98 | Y |
| 5 | M | 54 | Not tested | 98 | Y |
| 6 | M | 67 | + | 92 | Y |
| 7 | M | 77 | + | 76 | Y |
| 8 | M | 61 | + | 52 | Y |
| 9 | M | 40 | + | 98 | Y |
| 10 | M | 64 | + | 91 | N |
| 11 | F | 68 | + | 95 | Y |
| 12 | M | 46 | + | 21 | N |
| 13 | F | 72 | + | 87 | N |
| 14 | F | 77 | + | 43 | Y |
| 15 | M | 62 | + | 76 | Y |
| 16 | M | 57 | + | 92 | Y |
| 17 | M | 78 | + | 68 | N |
| 18 | M | 65 | + | 76 | Y |
| 19 | M | 65 | Not tested | 63 | Y |
| 20 | M | 41 | + | 66 | Y |
| 21 | F | 70 | + | 92 | Y |
| 22 | F | 45 | + | 79 | Y |
| 23 | M | 71 | + | 99 | Y |
| 24 | M | 69 | + | 95 | Y |
| 25 | F | 68 | + | 87 | Y |
| 26 | F | 73 | + | 74 | Y |
M, male; F, female.
*Results of immunohistochemistry (IHC) using a cyclin D1 monoclonal antibody. +, positivity; −, absence of signal with this antibody.
†Y indicates a split of the 11q13 probe, whereas N indicates absence of detectable split.
Fluorescence in Situ Hybridization
The 14q32 (BAC 158A2) and 11q13 (cyclin D1 probe, Vysis, Downers Grove, IL) probes have been previously reported. 18 Briefly, the 14q32 probe maps to the JH and first constant regions of the IGH gene and was directly labeled with fluorescein isothiocyanate (green). The 11q13 probe was purchased from Vysis and was labeled with SpectrumOrange (orange). FISH was performed according to routine protocols. Briefly, 60 ng of 158A2 probe and 1 μl of the Vysis probe were mixed with 2 μg of Cot-1 DNA (Life Technologies, Gaithersburg, MD) in 10 μl of Hybrisol VII (Oncor, Gaithersburg, MD), denatured at 73°C for 10 minutes, and dropped on slides previously denatured at 73°C for 5 minutes. After overnight hybridization at 37°C, slides were washed in 2X SSC at 73°C for 5 minutes and rinsed in 2X SSC/0.1% Triton. Nuclei were then counterstained with 4′,6-diamidino-2-phenylindole in antifade, and at least 200 nuclei were examined using an epifluorescence microscope (Axioplan 2, Zeiss, Iena, Germany) equipped with a cooled CCD camera (Photometrics, Tucson, AZ) and appropriate filters specific for fluorescein isothiocyanate, SpectrumOrange, and DAPI (Chroma, Brattleboro, VT). Images were then captured using the SmartCapture VP software (Vysis).
Sample Preparation
In a first attempt, we performed FISH on thin sections (3 to 5 μm). Despite a good hybridization efficiency, analysis was limited by the small size and the high density of cells. Consequently, determination of the exact location of FISH signals within each nucleus was extremely difficult, and the method was abandoned. To perform a cell-by-cell analysis, we decided to include a tissue disaggregation step to obtain cell suspension. After dewaxing in xylene, 30-μm-thick slices were incubated for 30 minutes to 2 hours in 0.5% to 5% pepsin, pH 1.5, at 37°C. After a rapid wash in PBS, cells were incubated for 30 minutes in 0.075 mol/L KCl and fixed in methanol/acetic acid (3/1 v/v) and dropped on slides.
Results
Fluorescence in Situ Hybridization Analysis
We have previously shown that the t(11;14)(q13;q32) translocation leads to a split of the 11q13 probe in most patients (25/28) analyzed on bone marrow or peripheral blood samples. In this major configuration, t(11;14)(q13;q32) was assessed in case of the presence of a YGRR configuration (Figure 1A) ▶ : one yellow signal (fusion on the derivative chromosome 14), one green signal (normal 14), and two red signals (on both normal and derivative chromosome 11). Another possible configuration was YGR (Figure 1B) ▶ : in this case, the red signal on the derivative chromosome 11 is not observed, either because the size of the probe that hybridizes on this der(11) is too small or because the breakpoint occurs upstream from the probe without any split. In contrast, on chromosome 14, breakpoints are clusterized in the JH region, and in t(11;14)(q13;q32)-positive cases, almost the whole 158A2 probe remains on the der(14).
Figure 1.
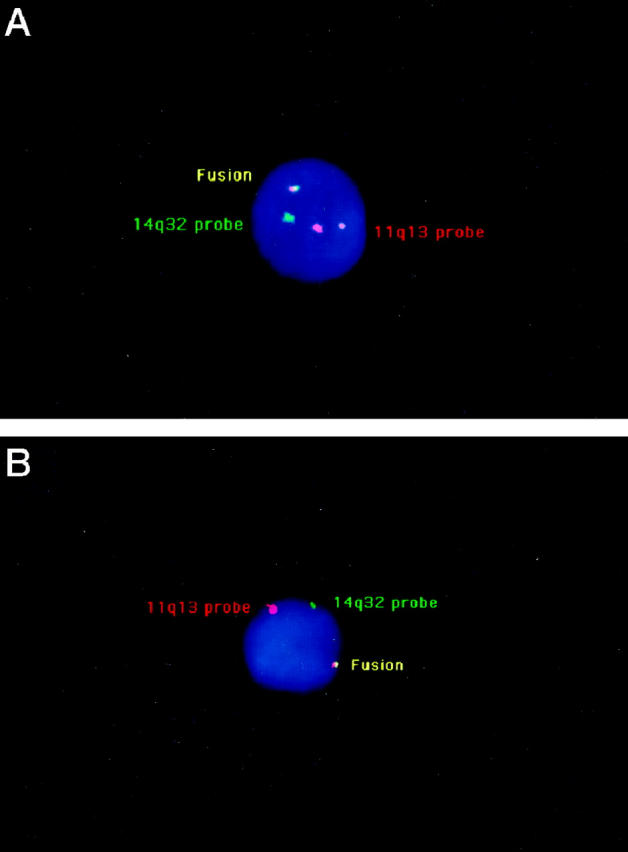
A: A malignant cell with IGH-CCND1 fusion and a split of the 11q13 probe (major configuration). The two red/orange signals correspond to the 11q13 probe, hybridized on the normal and derivative chromosomes 11, respectively. The green signal corresponds to the normal chromosome 14, whereas the yellow signal corresponds to the fusion of the 11q13 and 14q32 probes on the derivative chromosome 14. B: A malignant cell with an IGH-CCND1 fusion, but lacking a split of the 11q13 probe (minor configuration). Only one red/orange signal corresponding to the normal chromosome 11 is observed.
Different tissue digestion conditions were tested to obtain reproducible FISH results. We have tested different pepsin concentrations and different incubation times. The best results were obtained with incubation in 0.5% pepsin for 30 minutes. In these conditions, we achieved a good FISH efficiency in almost all of the cases. To determine the t(11;14)(q13;q32) positivity cut-off with these conditions, we analyzed other NHL subset specimens prepared in the same way (ie, from formalin-fixed, paraffin-embedded cells). We selected six B-cell diffuse large-cell lymphoma and six Burkitt’s lymphoma samples and blindly scored 300 nuclei in each case (total of 3600 nuclei). A nucleus was defined as t(11;14)(q13;q32) positive if a yellow fusion signal was observed, independently of the presence of a split of the 11q13 probe. In these controls, 2% to 5.7% (mean, 3.11; SD = 1.01%) of nuclei displayed a co-localization of a green and an orange signal. Thus, the cut-off level (mean + 3SD) for the detection of the t(11;14)(q13;q32) was set at 7%.
A t(11;14) Is Found by FISH in 26/26 Patients
In all 26 MCL samples investigated, more than 10% of nuclei displayed a fusion between 11q13 and 14q32 sequences. The percentage of cells with at least one co-localization was variable, ranging from 14% to 99% (median, 87%). In two patients (patients 2 and 12), we have observed IGH-CCND1 fusion only in a minor cell subset (14% and 21%, respectively). This apparent low tumor cell infiltration was in agreement with a marked lymphocytic reaction within the sample in both cases. The patient with 14% t(11;14)(q13;q32)-positive cells was the only case with negative immunohistochemistry with the cyclin D1 antibody. A split of the 11q13 probe (with a residual signal on the derivative chromosome 11) was observed in 22/26 cases.
Discussion
Mantle cell lymphoma is a recently identified subset of NHL, characterized by a male predominance, a high incidence of disseminated disease at diagnosis, and a poor outcome, even with intensive polychemotherapy regimens. 4-8 Incidence of this NHL subtype is variable in published series, but it probably represents 5% to 7% of all NHL cases. A recent retrospective study identified MCL in 121/1919 consecutive cases (6.8%). 4 Because of the poor prognosis of this NHL subgroup, its accurate identification is extremely important to propose specific intensive therapeutic strategies. Thus, detection of the molecular genetic hallmark of this NHL subtype would definitely help to propose the accurate diagnosis.
So far, several approaches have been reported, none of them fulfilling the requested criteria for an unequivocal diagnosis. The t(11;14)(q13;q32) is found in most but not all MCL cases, and up to 30% of patients with a characteristic MCL lack this translocation. 10,11 Likewise, molecular detection of the t(11;14)(q13;q32), using either Southern blotting or PCR, is hampered by the scattering of 11q13 breakpoints. As the functional consequence of the translocation is an overexpression of CCND1, the detection of high levels of RNA and/or protein would represent the ideal test. However, so far, no monoclonal antibody fulfills the specificity and sensitivity criteria for an undoubtful diagnosis. Currently, Northern blotting represents the gold standard for detection of CCND1 overexpression. However, this technique requires large amounts of RNA and is not adapted to routine diagnosis.
Several attempts to use FISH for identification of the t(11;14)(q13;q32) have been recently reported. 18-23 Even though the first reports were limited by a low sensitivity, the most recent studies, including ours, 18 based on dual-color FISH techniques, achieved this goal and demonstrated that virtually 100% of MCL patients displayed an IGH-CCND1 fusion. However, these series analyzed either patients for whom metaphase spreads were available or patients with a high percentage of malignant cells within bone marrow and/or peripheral blood. These studies had the ability to demonstrate the feasibility and reliability of this technique but were limited to a subset of MCL patients. Because NHL cytogenetic analyses are not performed in most routine cytogenetics laboratories, and as not all patients have a massive bone marrow involvement, many patients still escape from this evaluation. To analyze 100% of patients with a MCL diagnosis, we developed a FISH assay that could be performed directly on formalin-fixed, paraffin-embedded specimens.
An ideal experiment would be to perform FISH directly on thin sections, to combine morphological and genetic analyses. Even though we were successful in obtaining efficient hybridization, analysis was hampered by the small size and the high density of malignant cells, preventing us from an accurate cell-by-cell analysis. To circumvent this problem, we set up tissue digestion conditions, enabling the obtainment of a cell suspension while preserving a good cell morphology. Then, we progressively improved experimental conditions to obtain a high hybridization efficiency, even in formalin-fixed, paraffin-embedded cells. We were able to define FISH conditions applicable to all cases. Thus, we have detected IGH-CCND1 fusion in all 26 analyzed cases. In our previous study on peripheral blood or bone marrow samples, we have shown that IGH-CCND1 fusion was observed in 17/17 MCL patients. 18 Since this publication, we had the opportunity of analyzing 13 more patients with MCL and peripheral blood involvement. All of these 13 patients displayed an IGH-CCND1 fusion (unpublished data). So far, the combination of our analyses either on primary tumor or peripheral blood/bone marrow samples shows that an IGH-CCND1 fusion is observed in 51/51 patients with MCL. As cytogenetics was not performed in these cases, a direct correlation between IGH-CCND1 fusion and t(11;14)(q13;32) cannot be definitively assessed. However, this FISH assay enables the detection of every IGH-CCND1 rearrangement and thus is probably more sensitive than cytogenetics for the detection of cryptic rearrangements.
Thus, we think that we achieved the final goal of a highly sensitive, simple, and rapid technique, usable in 100% of patients, enabling an unambiguous MCL diagnosis. Moreover, as we used one commercially available probe and as the second probe gave bright clear signals without immunological amplification steps, we think that this set of probes could be used in most (if not all) routine cytogenetics (and probably pathology) laboratories.
Footnotes
Address reprint requests to Dr. Hervé Avet-Loiseau, Laboratoire d’Hématologie, Institut de Biologie, 9 quai Moncousu, 44093 Nantes Cédex 1, France. E-mail: havetloiseau@chu-nantes.fr.
J.-Y. Li is a grant recipient from the Conseil Régional des Pays de Loire.
References
- 1.Harris NL, Jaffe ES, Stein H, Banks PM, Chan JKC, Cleary ML, Delsol G, De Wolf-Peeters C, Falini B, Gatter KC, Grogan TM, Isaacson PG, Knowles DM, Mason DY, Muller-Hermelink HK, Pileri SA, Piris MA, Ralfkiaer E, Warnke RA: A revised European-American classification of lymphoid neoplasms: a proposal from the International Lymphoma Study Group. Blood 1994, 84:1361-1392 [PubMed] [Google Scholar]
- 2.Banks PM, Chan J, Cleary ML, Delsol G, de Wolf-Peeters C, Gatter K, Grogan TM, Harris NL, Isaacson PG, Jaffe ES, Mason D, Pileri S, Ralfkiaer E, Stein H, Warnke R: Mantle cell lymphoma: a proposal for unification of morphologic, immunologic, and molecular data. Am J Surg Pathol 1992, 16:637-642 [DOI] [PubMed] [Google Scholar]
- 3.Weisenburger DD, Armitage JO: Mantle cell lymphoma: an entity comes of age. Blood 1996, 87:4483-4494 [PubMed] [Google Scholar]
- 4.Samaha H, Dumontet C, Ketterer N, Moullet I, Thieblemont C, Bouafia F, Callet-Bauchu E, Felman P, Berger F, Salles G, Coiffier B: Mantle cell lymphoma: a retrospective study of 121 cases. Leukemia 1998, 12:1281-1287 [DOI] [PubMed] [Google Scholar]
- 5.Coiffier B, Hiddemann W, Stein H: Mantle cell lymphoma: a therapeutic dilemma. Ann Oncol 1995, 6:208-211 [DOI] [PubMed] [Google Scholar]
- 6.Fisher RI, Dahlberg S, Nathwani BN, Banks PM, Miller TP, Grogan TM: A clinical analysis of two indolent lymphoma entities: mantle cell lymphoma and marginal zone lymphoma (including mucosa-associated lymphoid tissue and monocytoid B cell categories): a Southwest Oncology Group study. Blood 1995, 85:1075-1082 [PubMed] [Google Scholar]
- 7.Teodorovic I, Pittaluga S, Kluin-Nelemans JC, Meerwaldt JH, Hagenbeek A, van Glabbeke M, Somers R, Bijnen L, Noordijk EM, De Wolfe-Peeters C: Efficacy of four different regimens in 64 mantle-cell lymphoma cases: clinicopathologic comparison with 498 other non-Hodgkin’s lymphoma subtypes. J Clin Oncol 1995, 13:1819-1828 [DOI] [PubMed] [Google Scholar]
- 8.Zucca E, Roggero E, Pinotti G, Pedrinis E, Cappella C, Venco A, Cavalli F: Patterns of survival in mantle cell lymphoma. Ann Oncol 1995, 6:257-262 [DOI] [PubMed] [Google Scholar]
- 9.Rimokh R, Berger F, Cornillet P, Wahbi K, Rouault JP, Ffrench M, Bryon PA, Gadoux M, Gentilhomme O, Germain D, Magaud JP: Break in the bcl-1 locus is closely associated with intermediate lymphocytic lymphoma subtype. Genes Chromosomes & Cancer 1990, 2:223-229 [DOI] [PubMed] [Google Scholar]
- 10.Leroux D, Le Marc’hadour F, Gressin R, Jacob MC, Keddari E, Monteil M, Caillot P, Jalbert P, Sotto JJ: Non-Hodgkin’s lymphomas with t(11;14)(q13;q32): A subset of mantle/intermediate lymphocytic lymphoma? Br J Haematol 1991, 77:346-353 [DOI] [PubMed] [Google Scholar]
- 11.Vandenberghe E, de Wolf-Peeters C, Wlodarska I, Stul M, Louwagie A, Verhoef G, Thomas J, Criel A, Cassiman JJ, Mecucci C, van den Berghe H: Chromosome 11q rearrangements in B non-Hodgkin’s lymphoma. Br J Haematol 1992, 81:212-217 [DOI] [PubMed] [Google Scholar]
- 12.de Boer CJ, Loyson S, Kluin PM, Kluin-Nelemans HC, Schuuring E, van Krieken JHJM: Multiple breakpoints within the BCL-1 locus in B-cell lymphoma: rearrangements of the cyclin D1 gene. Cancer Res 1993, 53:4148-4152 [PubMed] [Google Scholar]
- 13.Rimokh R, Berger F, Delsol G, Chanin C, Bertheas MF, French M, Garoscio M, Felman P, Coiffier B, Bryon PA, Rochet M, Gentilhomme O, Germain D, Magaud JP: Rearrangement and overexpression of the bcl-1/PRAD-1 gene in intermediate lymphocytic lymphomas and in t(11q13)-bearing leukemias. Blood 1993, 81:3063-3067 [PubMed] [Google Scholar]
- 14.de Boer CJ, van Krieken JHJM, Kluin-Nelemans HC, Kluin PM, Schuuring E: Cyclin D1 messenger RNA overexpression as a marker for mantle cell lymphoma. Oncogene 1995, 10:1833-1840 [PubMed] [Google Scholar]
- 15.de Boer CJ, Schuuring E, Dreef E, Peters G, Bartek J, Kluin PM, van Krieken JHJM: Cyclin D1 protein analysis in the diagnosis of mantle cell lymphoma. Blood 1995, 86:2715-2723 [PubMed] [Google Scholar]
- 16.Rimokh R, Berger F, Delsol G, Digonnet I, Rouault JP, Tigaud JD, Gadoux M, Coiffier B, Bryon PA, Magaud JP: Detection of the chromosomal translocation t(11;14) by polymerase chain reaction in mantle cell lymphomas. Blood 1994, 83:1871-1875 [PubMed] [Google Scholar]
- 17.Luthra R, Hai S, Pugh WC: Polymerase chain reaction detection of the t(11;14) translocation involving the bcl-1 major translocation cluster in mantle cell lymphoma. Diagn Mol Pathol 1995, 4:4-7 [DOI] [PubMed] [Google Scholar]
- 18.Avet-Loiseau H, Garand R, Gaillard F, Daviet A, Mellerin MP, Robillard N, Bouyge I, Arcot S, Batzer M, Talmant P, Harousseau JL, Milpied N, Bataille R: Detection of the t(11;14) using interphase molecular cytogenetics mantle cell lymphoma and atypical chronic lymphocytic leukemia. Genes Chromosomes & Cancer 1998, 23:175-182 [DOI] [PubMed] [Google Scholar]
- 19.Coignet LJA, Schuuring E, Kibbelar RE, Raap TK, Kleiverda KK, Bertheas MF, Wiegant J, Beverstock G, Kluin PM: Detection of 11q13 rearrangements in hematologic neoplasias by double-color fluorescence in situ hybridization. Blood 1996, 87:1512-1519 [PubMed] [Google Scholar]
- 20.Monteil M, Callanan M, Dascalescu C, Sotto JJ, Leroux D: Molecular diagnosis of t(11;14) in mantle cell lymphoma using two-colour interphase fluorescence in situ hybridization. Br J Haematol 1996, 93:656-660 [DOI] [PubMed] [Google Scholar]
- 21.Vaandrager JW, Schuuring E, Zwikstra E, de Boer CJ, Kleiverda KK, van Krieken JHJM, Kluin-Nelemans HC, van Ommen GJB, Raap AK, Kluin PM: Direct visualization of dispersed 11q13 chromosomal translocations in mantle cell lymphoma by muticolor DNA fiber fluorescence in situ hybridization. Blood 1996, 88:1177-1182 [PubMed] [Google Scholar]
- 22.Cuneo A, Bigoni R, Negrini M, Bullrich F, Veronese ML, Roberti MG, Bardi A, Rigolin GM, Cavazzini P, Croce CM, Castoldi G: Cytogenetic and interphase cytogenetic characterization of atypical chronic lymphocytic leukemia carrying BCL1 translocation. Cancer Res 1997, 57:1144-1150 [PubMed] [Google Scholar]
- 23.Siebert R, Matthiesen P, Harder S, Zhang Y, Borowski A, Zuhlke-Jenisch R, Plendl H, Metzke S, Joos S, Zucca E, Weber-Matthiesen K, Roggero E, Grote W, Schlegelberger B: Application of interphase cytogenetics for the detection of t(11;14)(q13;q32) in mantle cell lymphomas. Ann Oncol 1998, 9:519-526 [DOI] [PubMed] [Google Scholar]


