Abstract
We originally isolated the HIP/PAP gene in a differential screen of a human hepatocellular carcinoma cDNA library. This gene is expressed at high levels in 25% of primary liver cancers but not in nontumorous liver. HIP/PAP belongs to the family of C-type lectins and acts as an adhesion molecule for hepatocytes. In normal adult human tissues, HIP/PAP expression is found in pancreas (exocrine and endocrine cells) and small intestine (Paneth and neuroendocrine cells). In order to gain insight into the possible role of HIP/PAP in vivo, we have investigated the pattern of HIP/PAP expression in the developing postimplantation mouse embryo by in situ hybridization. Detailed analysis of developing mouse embryos revealed that HIP/PAP gene exhibits a restricted expression pattern during development. Thus, HIP/PAP transcripts are first observed within the nervous system from day 14.5 onwards in trigeminal ganglia, dorsal root ganglia, and spinal cord where it appears to be an early specific marker of a subpopulation of motor neurons. At laster stages, HIP/PAP transcripts were detected in intestine and pancreas at day 16.5 but not in embryonic liver. This highly restricted expression pattern suggests that HIP/PAP might participate in neuronal as well as intestinal and pancreatic cell development.
Lectins are proteins that have the ability to recognize carbohydrate residues that may be part of cell surface, extracellular matrix, or soluble glycoproteins. In the past few years, a large number of animal lectins have been isolated and in many cases have been ascribed a role in biological recognition events such as cell-cell interactions, intracellular glycoprotein routing and phagocytosis. 1 Lectins may also play a role in cellular invasion and metastasis. 2 Most animal lectins can be grouped into two main classes, based on the nature of their carbohydrate recognition domain (CRD). 3 The first class comprises a family of small molecular mass proteins named galectins that specifically bind to beta-galactoside derivatives in a calcium-independent manner. The second class, the C-type lectins, comprises a large family of multifunctional proteins found in serum, extracellular matrix, and membranes, which require Ca2+ for carbohydrate binding. C-type lectins share a common sequence motif in their CRD consisting of 18 highly conserved amino acid residues.
By differential screening of a human hepatocellular carcinoma cDNA library, we previously identified a gene named hepatocarcinoma-intestine-pancreas (HIP) whose expression was elevated in 25% of human primary liver cancers and was not detected in nontumoral liver tissues. 4 By Northern analysis of a series of human adult tissues, we only found HIP expression in small intestine and pancreas. The HIP protein belongs to the C-type lectin family according to sequence homologies and its ability to bind lactose residues. 5 Recently, we have shown that this protein is able to promote adhesion of hepatocytes and to bind laminin, an extracellular matrix protein. 6 In contrast to the other C-type lectins that are multifunctional proteins, HIP is comprised of a single CRD linked to a signal peptide. Therefore, we previously proposed that HIP and related proteins belong to a new family of C-type lectins, 4 and this protein family is now classified as the free CRD lectins (group VII). 1 Several other proteins exhibiting the same structural features were isolated from pancreas and included in this group. In rat, three members of this lectin group are highly expressed during acute pancreatitis and called pancreatitis-associated protein (PAP1, PAP2, PAP3). Human and mouse cDNA encoding proteins homologous to rat PAP1 were isolated, 7,8 and sequence identity research showed that human HIP and PAP genes were identical. 9 We will thus refer to this gene as HIP/PAP. In addition, a rat putative growth factor peptide 23, identical to rat PAP1, is secreted by pituitary cells under the influence of growth-hormone-releasing hormone (GHRH). 10 Thus, the same protein has been identified using several independent approaches. Although several roles have been proposed such as bacterial agglutination, 11 cell adhesion, 6 cell proliferation, 10,4 and resistance to apoptosis, 12 the in vivo function of HIP/PAP remains unclear.
In this view, it is important to investigate whether overexpression of HIP/PAP during hepatocarcinogenesis results of the reexpression of a fetal liver marker in tumorous cells as shown for alphafetoprotein, 13 glutamin synthetase, 14 and insulin-like growth factor II. 15 The tissue specificity of HIP/PAP expression should be indeed interpretated taking into account the common embryonic origin of liver, pancreas, and intestine. In addition, the so-called oval liver cells, presumably of ductal origin, proliferate in rodents in response to various chemical carcinogens and are “pretumorous candidate cells” during liver carcinogenesis. They can differentiate into both pancreatic cells and hepatocytes 16 and in certain conditions give rise to intestinal metaplasia. 17 Conversely, rat pancreatic ductular cells can differentiate into hepatocytes following a copper-depletion regimen. 18,19 We have therefore investigated by in situ hybridization the cell types involved in overexpression of HIP/PAP in liver tumor. We have also examined the pattern of expression of HIP/PAP in both mouse adult tissues (liver, intestine, and pancreas) and in postimplantation mouse embryos. We show that HIP/PAP gene is not broadly expressed during development but instead, presents a remarkably restricted expression pattern. In particular, we provide evidence of its expression in developing nervous system in addition to pancreas and small intestine. In contrast, we never detected HIP/PAP mRNA in fetal liver. Our results are therefore consistent with a role for HIP/PAP in neuronal as well as intestinal and pancreatic cell development.
Materials and Methods
Plasmids and RNA Probes
Mouse HIP/PAP clone was obtained by reverse transcription (RT) of mouse small intestine RNA and amplification by polymerase chain reaction (PCR). The primers were derived from the mouse HIP/PAP sequence of Itoh and Terakoa 8 and numbered as in this publication. A 488-bp fragment was amplified by Taq polymerase using a sense (5′TGGATGCTGCTCTCCTGCCT3′, nt 37–57) and an antisense (5′TTGCCCTATGTCTGCAAATTT3′, nt 505–525) primers. Samples were heated for 5 minutes at 94°C, and 40 cycles of amplification were performed with denaturation at 94°C for 1 minute, annealing at 55°C for 1 minute, and extended at 72°C for 1 minute, and the final cycle was identical except for 10 minutes extension. Reaction product was digested with EcoR1 and HindIII, subcloned into SK +pBluescript vector, and sequenced using T7/T3 primers. The sequence is identical to that previous published sequence. 8 High specific activity RNA probes were synthetized using 35S-UTP (specific activity above 1000 Ci/mmol, Amersham, UK) and T3 RNA polymerase for antisense mouse and human HIP/PAP 5 probes and T7 RNA polymerase for antisense Galectin 1 20 and sense mouse HIP/PAP probes.
In Situ Hybridization
Embryo and adult tissues were fixed overnight in 4% (w/w) paraformaldehyde, dehydrated, and embedded in paraffin wax; 7-μm sections were cut and hybridized with 35S-labeled antisense RNA probes using the procedure of Wilkinson et al 21 with some modifications. Tissues and embryo sections were deparaffined in histoclear (twice for 10 minutes), rehydrated through a graded series of ethanol solutions (100 to 30%), rinsed in 0.85% saline solution (5 minutes), l× phosphate-buffered saline (PBS) (5 minutes), and postfixed in a freshly prepared solution of 4% paraformaldehyde in 1× PBS (20 minutes). Slides carrying sections were then rinsed in PBS (twice for 10 minutes) and treated with a fresh solution of proteinase K (20 μg/ml) in 50 mmol/L Tris Hcl, 5 mmol/L EDTA, pH 7.2) for 7.5 minutes. Slides were then rinsed in PBS (3 minutes), refixed in 4% paraformaldehyde solution in PBS, dipped in distilled water, and acetylated in 0.1 mol/L solution of triethanolamine and acetic anhydre (1:400 v/v) for 10 minutes. Slides were subsequently rinsed in 1× PBS and 0.85% saline solution (5 minutes each) and quickly dehydrated through a series of ethanol solutions (30 to 100%). Probes were applied directly to tissue sections (40 μl) at a final concentration of 100,000 cpm/μl in hybridization buffer (50% formamide, 0.3 mol/L NaCl, 10 mmol/L Tris HCL (pH 8), 5 mmol/L EDTA, 10 mmol/L NaPO4 (pH 8), 50 mmol/L dithiothreitol (DTT), 10% dextran sulfate, 1× Denhart’s, 0.5 mg/ml of total yeast RNA). Hybridization was carried out at 52°C for 16 hours in a humid chamber. Coverslips were gently floated off in a solution of 5× SSC, 10 mmol/L DTT at 50°C, and the slides were then rinsed at 60°C in a stringent washing solution of 50% formamide, 2× SSC, 20 mmol/L DTT, treated with RNAse A (20 μg/ml) for 30 minutes at 37°C. After washes at 55°C in 2× SSC and 0.1× SSC (15 minutes each) sides were dehydrated in a series of ethanol containing 0.3 mol/L ammonium acetate and dipped in Ilford K5 emulsion. Exposure times varied from 6 to 15 days.
Northern Blot
Human small intestine obtained at surgery was immediately frozen in liquid nitrogen and stored at −80°C until used. Total RNA was extracted using the hot phenol method. Human liver fetal RNA was kindly provided by Clare Selden (Hammersmith Hospital, London, UK). RNA samples were run on a denaturing formaldehyde gel and transferred onto Hybond-C-extra nirocellulose membranes (Amersham, UK). The probe used to hybridize the filter was a DNA fragment corresponding to the human HIP/PAP gene coding sequence cloned in pBluescript in SmaI and EcorI sites. 5 The insert was isolated before labeling. Filters were hybridized as previously described 15 and washed in 2× SSC, 0.1% (w/w) sodium dodecyl sulfate at 42°C for 30 minutes. Human glucose 6 phosphate dehydrogenase cDNA was used as reference probe to verify the quantity of RNA transferred to the filters.
Reverse Transcription (RT) and Polymerase Chain Reaction (PCR)
RNA was extracted from embryonic tissues using a Promega kit following the recommended procedure. For adult tissues, RNA was extracted according to TRIZOL (Life Technology) supplied instructions. Primer used for RT and PCR were derived from the mouse published sequence of Itoh and Terakoa. 8 To evaluate the amplifiability of RNA, RT was also performed with β-actin primer. One microgram of RNA was reverse transcribed at 37°C for 60 minutes in 25 μl of final volume with 10 pmol of primer, 10 U of RNasin, 1× buffer supplied with the enzyme, 40 mmol/L of the four deoxynucleotides, and 20 U of Moloney murine leukemia virus reverse transcriptase (Life Technologies; Gibco BRL). Two antisense primers were used in the same reaction, the first is specific of mouse HIP/PAP (5′GACATGTGAGGTGAAGTTGCC3′, nt 490–510), whereas the second is specific of β-actin (5′TTGGCCTTAGGGTTCAGGGGGG3′). Five microliters of cDNA were then amplified by two PCR assays performed in parallel, one for HIP/PAP and one for β-actin, in a final volume of 100 μL containing 2.5 U of Taq polymerase (Life Technologies; Gibco BRL), 1× supplied buffer, 1.5 mmol/L MgCl2, 0.2 mmol/L each deoxyribonucleotide. HIP/PAP primers were sense (5′TCCAACAGCCTGCTCCGTCATG3′, nt 13–33) and antisense (5′CAGAAGCTCTTGACAAGCTGCCAC3′, nt 457–477). β actin primers were antisense (5′TTGGCCTTAGGGTTCAGGGGGG3′) and sense (5′CGTGGGCCGCCCTAGGCACCA3′). Samples were subjected to 40 cycles of PCR amplification (1 minute at 94°C, 1 minute at 58°C, 1 minute at 72°C). PCR products were transferred to a nylon membrane and hybridized with specific 32P-labeled oligonucleotide probes: 5′AAGGACTCCTATGTGGGTGACG3′ for β actin and ′5′TCTACTGCCTTAGACCGTG3′ (nt 416–435) for HIP/PAP.
Neural tube was excised from 16.5 dpc embryos. RNA extraction and RT-PCR assays were performed as above. For nested PCR, two microliters of the amplification product obtained by HIP/PAP RT-PCR was reamplified with the inner primers (sense 5′ATGCTCTTATCTCAGGTTCAAG3′, nt 59–81) and antisense (5′TCTACTGCCTTAGACCGTG3′, nt 416–435) with the same protocol for a further 40 cycles. Nested PCR product was sequenced using the same primers.
All experiments were approved by local ethical committee.
Results
HIP/PAP Expression in Human HCC and Human Fetal Liver
We have shown previously by Northern blot that the HIP/PAP gene is not expressed in human adult liver, whereas it is abundantly expressed in primary liver tumors. In order to identify cells overexpressing HIP/PAP mRNHA in liver tumors, we performed an in situ hybridization analysis of three tumors that exhibit a strong expression of HIP/PAP transcripts. The probe used in this experiment was the original human HIP/PAP clone. 4,5 We examined three human hepatocellular carcinomas by in situ hybridization and in all cases we found that 10 to 50% of tumorous hepatocytes were intensely labeled, whereas no signal was detected in adjacent nontumorous areas (Figure 1A) ▶ . We also performed a Northern blot analysis of HIP/PAP mRNA expression in several human fetal liver samples from 9.6 to 21.5 weeks of pregnancy. The results shown in Figure 1B ▶ indicate that there is no detectable expression of HIP/PAP in fetal liver, whereas a strong expression of this gene is readily observed in human intestine.
Figure 1.
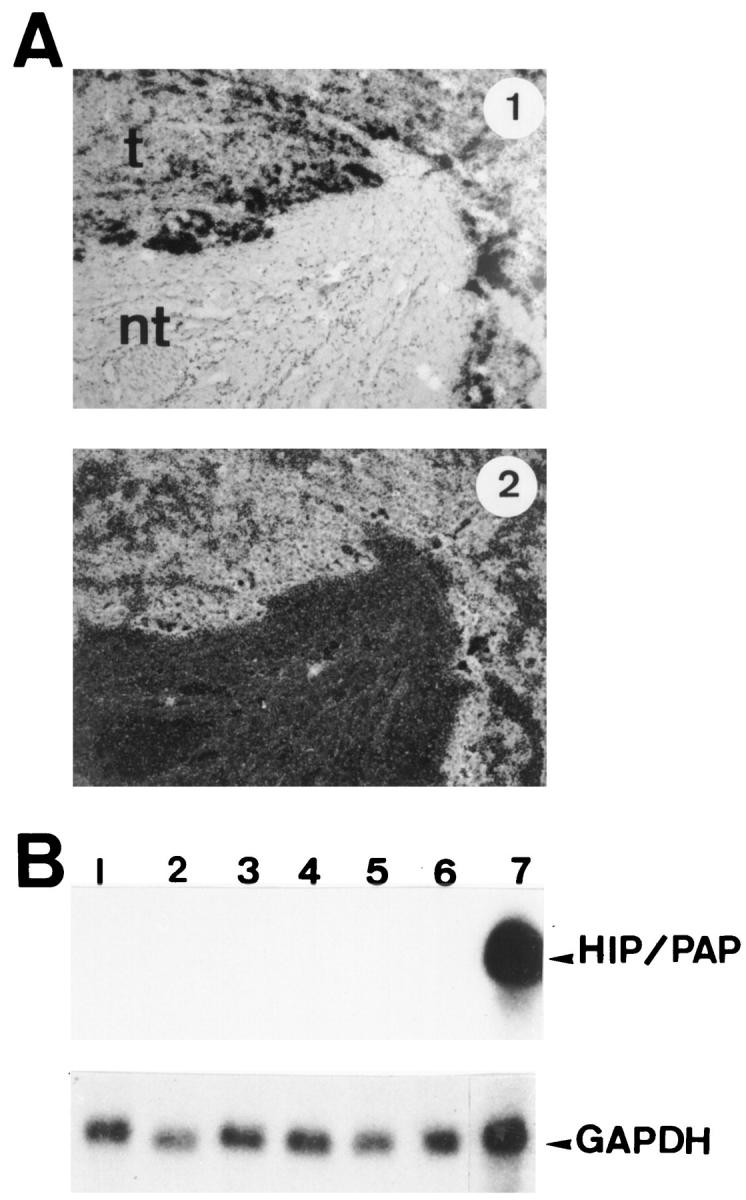
Expression of HIP/PAP mRNA in human tissues. A: In situ hybridization of 35S-labeled antisense human HIP/PAP probe to sections of human hepatocarcinoma (HCC) under bright field and dark field illumination: t, tumor; nt, nontumoral tissue. Original magnification, ×75. Exposure time is 2 days. B: Northern blot analysis of human fetal liver mRNA. The probe was the full human HIP/PAP cDNA. Lanes 1 to 6: 10 μg of total RNA from human fetal liver (7 days exposure); lane 1: 21.5 weeks old; lane 2: 14.6 weeks old; lane 3: 11.3 weeks old; lane 4: 9.6 weeks old; lane 5: 10 weeks old; lane 6: 9 weeks old. Lane 7: human adult ileum, 20 μg of total RNA (1 day exposure). The same filter was stripped and reprobed with human glyceraldehyde-3-phosphate dehydrogenase (GAPDH) probe. For GAPDH hybridization (lower panel) exposure time is 1 day.
Expression of HIP/PAP Gene in Intestine, Liver, and Pancreas
We have previously reported by Northern blot analysis that at least in human tissues HIP/PAP transcripts distribution presents some degree of tissue-specificity. Thus, we found HIP/PAP mRNA in small intestine and pancreas but not in liver, colon, brain, kidney, or lung. 4 In order to perform a more extensive and more sensitive analysis of HIP/PAP expression, we used in situ hybridization and RT-PCR methods on embryonic and adult mouse tissues. Primers used for RT-PCR were derived from the mouse cDNA HIP/PAP published sequence of Itoh and Terakoa. 8 For in situ hybridization, a cDNA clone was amplified from mouse small intestine by RT-PCR and subcloned in pBLuescript vector; its sequence was done and found identical to the mouse cDNA HIP/PAP published sequence. 8
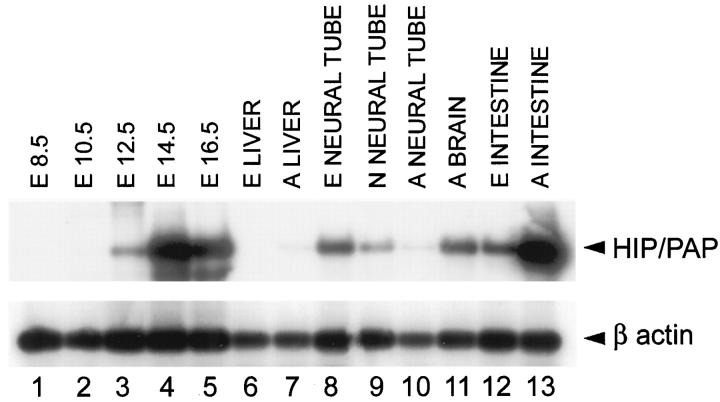
No expression was detected by RT-PCR in embryos at days 8.5 (E8.5) and 10.5 (E10.5) (Figure 2 ▶ , lanes 1 and 2). At E14.5 and E16.5, a strong signal was detected at the expected size of 440 bp (Figure 2 ▶ , lanes 4 and 5). At E12.5 a weak signal was detected in Southern blot autoradiography of the RT-PCR products (Figure 2 ▶ , lane 3). Thus, HIP/PAP mRNA expression is initiated at late stages of development.
Figure 2.
RT-PCR analysis of mouse HIP/PAP expression in embryonic and adult tissues. RNA extracted from total embryos (lanes 1 to 5) of 8.5 days old (E8.5), 10.5 days old (E10.5), 12.5 days old (E12.5), 14.5 days old (E14.5), and 16.5 days old (E16.5). RNA extracted from isolated mouse tissues (lanes 6 to 13) : liver from E16.5 embryo (E LIVER); adult liver (A LIVER); neural tube from E16.5 embryo (E NEURAL TUBE); neural tube from newborn mouse (N NEURAL TUBE); adult neural tube (A NEURAL TUBE); adult brain (A BRAIN); intestine from E16.5 embryo (E INTESTINE); adult small intestine (A INTESTINE). Reaction products were electrophoresed and an autoradiography is shown. Exposure times were 1 day for HIP/PAP and 1 hour for β actin.
In order to identify tissues expressing HIP/PAP RNA during mouse development, an in situ hybridization analysis was performed on embryos from E8.5 to E16.5 with an antisense HIP/PAP RNA probe. By this method, HIP/PAP transcripts were first detected at E14.5 (see below). We found no evidence for expression of HIP/PAP gene at E8.5, E10.5, and E12.5. It is likely that the lower sensitivity of in situ hybridization compared with RT-PCR accounts for the lack of detection of HIP/PAP at E12.5.
In embryonic intestine, a strong punctate signal appeared (Figure 3, A and B) ▶ at E16.5. At birth, HIP/PAP expression was localized to epithelial villi cells (Figure 3, C and D) ▶ . In adults, we observed a strong signal in three segments of small intestine: duodenum, jejunum, and ileum. The positive cells were identified as enterocytes (Figure 3, E and F) ▶ . Location of HIP/PAP expression in villi cells was previously described in rat adult intestine by immunocytochemistry. 22 To characterize the cell type expressing HIP/PAP in E16.5, sections adjacent to those hybridized with the HIP/PAP probe were stained with gremilius, a histological marker of goblet cells, and with PAS-blue alcyan, a marker of neuroendocrine cells. The comparison of neuroendocrine, goblet cells, and HIP/PAP expressing cells distribution suggested that the small number of cells expressing HIP/PAP might be neurodendocrine cells (data not shown). This result is consistent with our previous observation of HIP/PAP immunocytolocalisation in human intestine neuroendocrine cells. 6 It is interesting that unlike the mouse HIP/PAP gene, the human homolog is mainly expressed in Paneth cells. 6 HIP/PAP mRNA was also detected by RT-PCR assay in both E16.5 and adult intestine (Figure 2B ▶ , lane 12 and 13).
Figure 3.
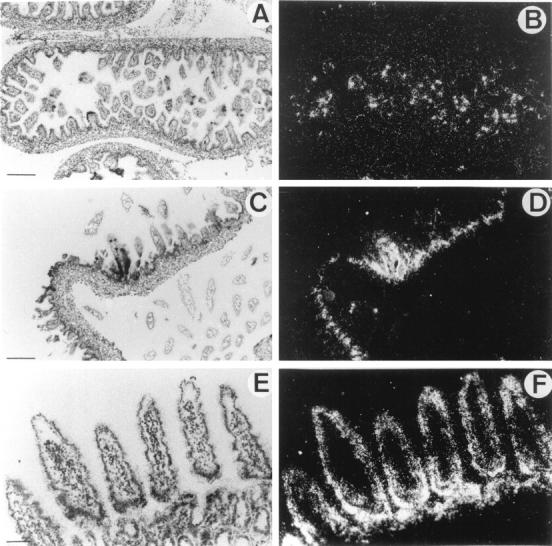
In situ hybridization of 35S-labeled antisense mouse HIP/PAP probe to mouse intestine sections under bright-field (A,C,E) and dark field (B,D,F) illumination. A and B: Longitudinal section through small intestine of E16.5 mouse embryo. C and D: Section of mouse newborn intestine. E and F: Section of mouse adult jejunum. Scale bar, 250 μm (A,B,C,D); 100 μm (E,F).
HIP/PAP transcripts were undetectable by in situ hybridization analysis in fetal liver at E10.5, E12.5, E14.5 (data not shown), and E16.5 (Figure 4, A and B) ▶ . Adult liver tissue was also devoid of HIP/PAP transcripts (Figure 4, C and D) ▶ . This is confirmed by the absence of detectable signal in fetal liver of E16.5 embryo by RT-PCR (Figure 2 ▶ , lane 6). By contrast, in adult liver, a weak signal was only detectable upon hybridization of PCR products with the HIP/PAP probe (Figure 2 ▶ , lane 7). As before, the RT-PCR method appears to be more sensitive than in situ hybridization.
Figure 4.
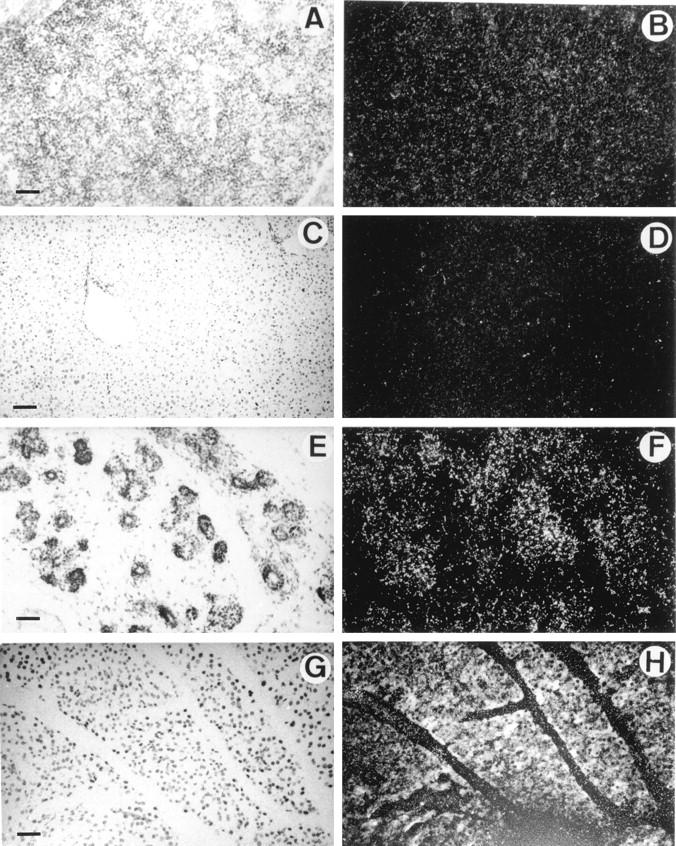
In situ hybridization of 35S-labeled antisense mouse HIP/PAP probe to sections of liver and pancreas under bright field (A,C,E,G) and dark field (B,D,F,H) illumination. A and B: Section through liver of a E16.5 mouse embryo; C and D: Section of mouse adult liver; E and F: Section through pancreas of a E16.5 mouse embryo; G and H: Section of mouse adult pancreas (10-day exposure time). Note that the white specks seen in B are caused by the refringent membrane of erythrocytes and not to silver grains. Scale bar, 250 μm (A,B,C,D); 100 μm (E,F,G,H).
A weak signal appeared in pancreas from E16.5 onward (Figure 4, E and F) ▶ . In adult pancreas, HIP/PAP was strongly expressed in acinar cells (Figure 4, G and H) ▶ . This observation is consistent with the results of a recent study that showed high levels of HIP/PAP transcripts in mouse pancreas by Northern blot. 23 With sense RNA HIP/PAP probe, no signal was detected in adult adjacent sections of adult small intestine and in longitudinal sections of E16.5 (data not shown). No HIP/PAP expression was observed in heart, smooth muscle, skeletal muscle, lung, kidney, skin, and bone from E10.5 to E16.5 (see Figure 3, 4, 5, and 6 ▶ ▶ ▶ ).
Figure 5.
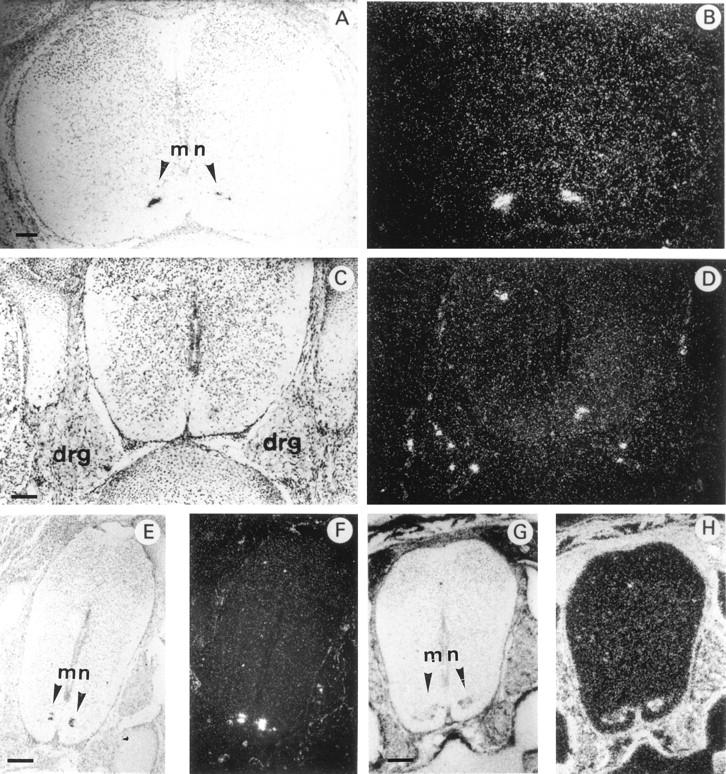
In situ hybridization of 35S-labeled antisense mouse HIP/PAP or mouse Galectin 1 probe to transverse sections through the spinal cord of E16.5 mouse embryo under bright field (A,C,E,G) and dark field (B,D,F,H) illumination. A and B: cervical region; C–H: lumbar region. For A–F the probe was mouse HIP/PAP, for G and H the probe was Galectin 1 cDNA (scale bars, 250 μm). mn, motor neuron. Note dorsal root ganglia (DRG) in C and D. DRG are aggregate of neuronal cell bodies that transmit somatic sensory information from periphery to spinal cord. Dorsal side towards the top, ventral side towards the bottom of the photographs.
HIP/PAP Expression in the Nervous System
HIP/PAP transcripts were first observed in the developing nervous system from E14.5 onwards in the neural tube and the spinal and trigeminal ganglia. On transverse sections, a strong hybridization signal was detected in the ventral horns of the spinal cord in a region that is occupied by motor neurons. This signal was restricted to a very small number of cells, distributes all along the anteroposterior axis in cervical (Figure 5, A and B) ▶ as well as lumbar regions (Figure 5, E and F) ▶ . To better characterize the cells expressing HIP/PAP mRNA, we hybridized adjacent sections with mouse galectin 1 probe, a gene expressed in many tissues but specific for motor neurons within the spinal cord. 20 Figure 5 ▶ shows a comparison of HIP/PAP and galectin 1 mRNA distribution in E16.5 spinal cord at the lumbosacral level. The cluster of cells expressing HIP/PAP (Figure 5, E and F) ▶ was included in the region positively stained for galectin 1 (Figure 5, G and H) ▶ but appeared to be restricted to a subpopulation of motor neurons. No HIP/PAP signal was found in adult spinal cord regions highly labeled with galectin 1 probe (data not shown). However, weak HIP/PAP signals were revealed by Southern blotting of RT-PCR products from newborn and adult spinal cord RNA (Figure 2 ▶ , lane 9 and 10). With E16.5 spinal cord, a strong RT-PCR signal was easily detectable without autoradiography (Figure 2 ▶ , lane 8). To definitively establish that HIP/PAP is expressed in the developing spinal cord, we performed a nested PCR from an aliquot of the RT-PCR product obtained using E16.5 neural tube RNA. As expected, the sequence of the nested PCR product was identical to that previously reported by Itoh and Terakoa. 8 Thus, we clearly demonstrate that HIP/PAP transcripts are present in the neurons of the late gestation mouse embryo.
Finally, HIP/PAP transcripts were also detected in some sensory neurons of the dorsal root ganglia (DRG) of E14.5 (data not shown) and E16.5 (Figure 5, C and D) ▶ . Figure 6 ▶ shows that in brain HIP/PAP gene was very restrictively expressed in neural crest derivatives such as trigeminal ganglia, whereas in other cerebral tissues such as cerebral hemispheres, midbrain, and pituitary gland no signal was detected (Figure 6, A and B) ▶ . Moreover, higher magnification of longitudinal section showed that, in trigeminal ganglia, only a few cells, most likely sensory neurons, were intensively labeled (Figure 6, E and F) ▶ .
Figure 6.
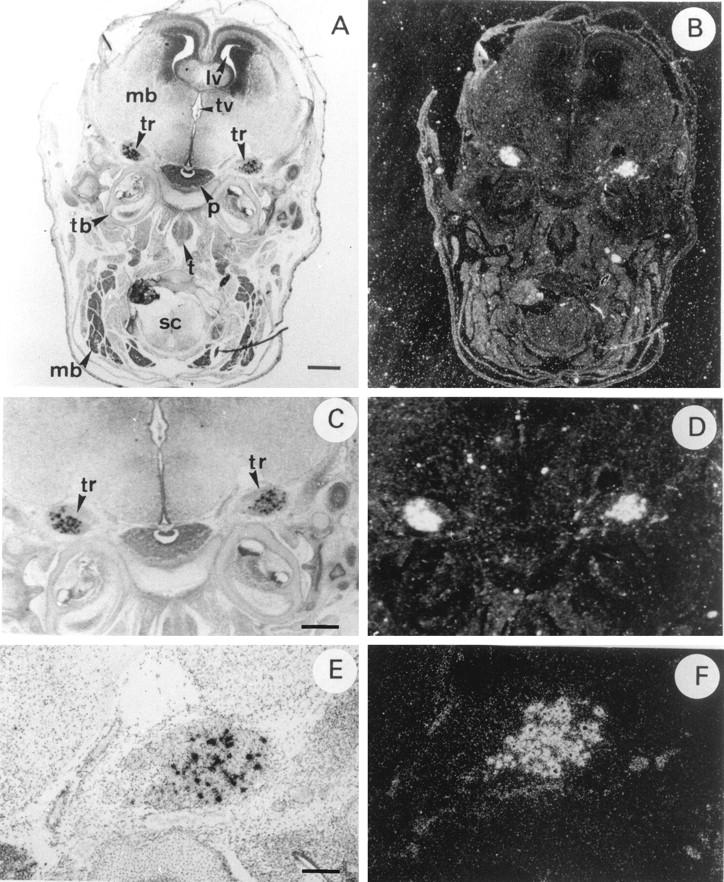
In situ hybridization of 35S-labeled antisense mouse HIP/PAP probe to sections of E16.5 mouse embryo central nervous system under bright field (A,C,E) and dark field (B,D,F) illumination. A and B: Coronal section of the mouse head of a E16.5; C and D: Detail of the same coronal section of the mouse brain at E16.5. Note trigeminal ganglia (tr) that are aggregate of neuronal cell bodies of the primary sensory neurons of V cranial nerve; E and F: Longitudinal section through mouse brain at E16.5. mb, midbrain; lv, lateral ventricule; tv, third ventricule; tr, trigeminal ganglia; p, pituitary gland; tb, temporal bone; t, tongue; sc, spinal cord; mb, muscle blocks. Scale bar, 800 μm (A,B); 400 μm (C,D); 100 μm (E,F).
Discussion
In this study, we have shown by combining in situ hybridization and RT-PCR analyses that HIP/PAP gene presents a restricted and unexpected expression pattern during postimplantation development of the mouse embryo. It is indeed striking that HIP/PAP transcripts are observed in the nervous system in the neural tube, trigeminal ganglia, and spinal ganglia from E14.5 onwards. At later stages (E16.5), HIP/PAP transcripts appear in pancreas and small intestine where a punctate signal indicated expression in only a small number of epithelial cells, possibly neuroendocrine cells. At birth, in intestine, the signal is stronger and expressing cells are spread all along the lower part of epithelium villi. In adult an intense signal is observed in all three segments of the small intestine epithelium. The same distribution has been described in rat adult intestine by immunocytochemistry, 22 although a Northern blot analysis failed to detect HIP/PAP RNA expression before weaning, which occurs around the third postnatal week. 24 This difference can be explained by the high sensitivity of in situ hybridization compared with Northern blot. In human intestine, we and others have recently reported that by immunohistochemistry localization, HIP/PAP is expressed in Paneth cells. 6,25 Although Paneth cells and enterocytes arise from the same stem cells located in crypts, this difference between HIP/PAP expression pattern in human and mouse intestine is surprising. In this respect it is important to note that the HIP/PAP gene family includes several members: three in rat, 26 three in mouse, 27,8 and only one in human. 4 Given the high stringency of the experimental conditions used for in situ hybridization analysis, we believe that the strong signal observed in intestine epithelial cells does correspond to mouse HIP/PAP expression and not to mouse-related genes. This was further demonstrated by our results on human intestine tissues. Indeed, by using the mouse cDNA HIP/PAP as probe, we detected by in situ hybridization a strong signal in human intestine cells localized in crypts, consistent with Paneth cells; thus, this result completely agrees with those obtained with the human probe (data not shown). It is worth noting that there are a few precedents for such differences between mouse and human gene expression patterns such as α1-antitrypsin, 28 α(1,2)-fucosyltransferase, 29 l -isoform of carnitine palmitoyltransferase (CPT). 30
The embryonic and adult expression profiles suggest a role for HIP/PAP in development and differentiation of intestinal epithelial cells. The epithelial cells lining the small intestine mucosa are renewed every 2 to 3 days in rodents. This renewal occurs by continuous division of a stem cell population located in intestinal crypt followed by migration and differentiation of daughter cells along the villus and final extrusion of senescent cells into the intestinal lumen. At E14.5 no HIP/PAP signal was observed in embryonic intestine at a time when the initial cytodifferentiation of the intestinal endoderm into an epithelial monolayer has not yet occurred, whereas a signal was detected at E16.5 when morphological waves of cytodifferentiation generate nascent villi. 31,32 The data are therefore consistent with the role for HIP/PAP in this process.
We have previously shown by Northern blot analysis that HIP/PAP gene was not expressed in human adult liver, although it was abundantly expressed in many primary liver tumors. In situ hybridization analysis of three tumors showed that 10 to 50% of tumorous hepatocytes are intensely labeled, whereas no signal was detected in nontumorous areas adjacent to tumors. HIP/PAP expression in tumors might have been the consequence of hepatocytes dedifferentiation and expression of a liver fetal marker. However, this now seems unlikely: no expression was detected by Northern blot in several human fetal liver samples age 9.6 to 21.5 weeks. Similarly, in mouse, we showed by in situ hybridization analysis that HIP/PAP transcripts are not expressed in fetal liver from E10.5 to E16.5. RT-PCR allowed us to confirm the absence of HIP/PAP transcript at an early stage, E8.5, at the beginning of hepatic determination 33 and at E10.5. Finally, at E16.5 fetal liver was isolated and no HIP/PAP expression was detected by RT-PCR. These results indicate that expression of HIP/PAP in hepatocellular carcinomas is not related to reexpression in tumorous hepatocytes of a gene normally expressed in fetus but instead is related to the liver carcinogenis.
One of the most striking results of our present study is the specific and intense expression of HIP/PAP in the embryonic nervous system. HIP/PAP is found in some cells of trigeminal ganglia, in DRG sensory neurons, and in the ventral horns of mouse spinal cord, a region occupied by motor neurons. In spinal cord, the signal is present in an area that also scores positive for galectin 1, a marker of motor neurons within this tissue. However, it is restricted to a small number of cells and seems distributed all along the ventral tube. Motor neurons located at different positions in the embryonic spinal cord project their axons to innervate distinct targets in the periphery, establishing a topography of motor connections. 34 Molecular specificities of these neurons was demonstrated by Tsushida et al 35 who showed that the expression pattern of four genes of the LIM homeobox gene family defines subclasses of motor neurons that segregate into different spinal cord columns and thus select distinct axonal pathways. It would be interesting to compare expression of HIP/PAP with that of the LIM genes. In any case our results indicate that HIP/PAP is a marker of a subset of the motor neurons. This observation provides an additional tool for understanding the mechanisms of neuronal differentiation.
In addition, HIP/PAP itself might play a role in neuron growth/guidance or survival. The importance of carbohydrates in neuronal migration during late embryonic life and their interaction with adhesion molecules such as neural cell adhesion molecules (N-CAM) is well established. 36 At least one lectin has already been demonstrated to play a role in axonal guidance because the Galectin-1 null mutation affects the outgrowth and/or guidance of a subset of olfactory neurons. 37 Lectins might mediate by cross-linking of carbohydrate ligands present on neurons and promote axonal adhesion of these neurons to laminin sugar chains. 38 Moreover laminin, which promotes neurite outgrowth in vitro is present in the pathways through which central and peripherical axons project to their cellular targets. The localization of HIP/PAP in motor neurons and DRG and the ability of this lectin to bind laminin 5 suggest that HIP/PAP might be a new lectin implicated in axonal guidance and/or growth. In this regard, it is also interesting that HIP/PAP (which is also called Reg-2) was recently isolated in a search for genes expressed in regenerating rat motor neurons and likely acts as a Schwann cell mitogen in vitro. 39
In summary, our data suggest that HIP/PAP might be involved during development in the differentiation processes of neuronal, pancreatic, and intestinal cells. Furthermore, its high expression in liver cancer and not in fetal liver also suggest that HIP/PAP might participate in the expansion of liver cells during liver carcinogenesis.
Acknowledgments
We thank F. Demaugre and R. Karess for critical reading the manuscript and M-A Ripoche for helpful discussions.
Footnotes
Address reprint request to C. Lasserre, INSERM U370, CHU Necker, 156 rue de Vaugirard, 75742 Paris Cedex 15 France. E-mail: lasserre@necker.fr.
Supported in part by Institut National de la Santé et de la Recherche Médicale, Association pour la Recherche Contre le Cancer, and Ligue Nationale Contre le Cancer.
References
- 1.Drickamer K, Taykor ME: Biology of animal lectins. Annu Rev Cell Biol 1993, 9:237-264 [DOI] [PubMed] [Google Scholar]
- 2.Ochieng J, Warfield P: Galectin-3 binding potentials of mouse tumor EHS and human placental laminins. Biochem Biophys Res Commun 1995, 217:402-406 [DOI] [PubMed] [Google Scholar]
- 3.Drickamer K: Two distinct classes of carbohydrate-recognition domain in animal lectins. J Biol Chem 1988, 263:9557-9560 [PubMed] [Google Scholar]
- 4.Lasserre C, Christa L, Simon M-T, Vernier P, Brechot C: A novel gene (HIP) activated in human primary liver cancer. Cancer Res 1992, 52:5089-5095 [PubMed] [Google Scholar]
- 5.Christa I, Felin M, Morali O, Simon M-T, Lasserre C, Brechot C, Seve A-P: The human HIP gene, overexpressed in primary liver cancer encodes a C-type carbohydrate binding protein with lactose binding activity. FEBS Lett 1994, 337:114-118 [DOI] [PubMed] [Google Scholar]
- 6.Christa L, Carnot F, Simon M-T, Levavasseur F, Stinnakre M-G, Lasserre C, Thepot D, Clement B, Devinoy e, Brechot C: HIP/PAP is an adhesive protein expressed in hepatocarcinoma, normal Paneth, and Pancreatic cells. Am J Physiol 1996, 271:G993-G1002 [DOI] [PubMed] [Google Scholar]
- 7.Orelle B, Keim V, Masciotra L, Dagorn J-C, Iovanna J-L: Human pancreatitis associated protein: messenger RNA cloning and expression in pancreatic diseases. J Clin Invest 1992, 90:2284-2291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Itoh T, Teraoka H: Cloning and tissue-specific expression of cDNAs for the human and mouse homologues of rat pancreatitis-associated protein (PAP). Biochim Biophys Acta 1993, 1172:184-186 [DOI] [PubMed] [Google Scholar]
- 9.Lasserre C, Simon M-T, Ishikawa H, Diriong S, Nguyen V, Christa L, Vernier P, Brechot C: Structural organization and chromosomal localization of a human gene (HIP/PAP) encoding a C-type lectin overexpressed in primary liver cancer. Eur J Biochem 1994, 224:29-38 [DOI] [PubMed] [Google Scholar]
- 10.Chakraborty C, Vrontakis M, Molnar P, Schroedter IC, Katsumata N, Murphy LJ, Shiu RPC, Friesen HG: Expression of pituitary peptide 23 in the rat uterus: regulation by estradiol. Mol Cell Endo 1995, 108:149-154 [DOI] [PubMed] [Google Scholar]
- 11.Iovanna JL, Orelle B, Keim V, Dagorn J-C: Messenger RNA sequence and expression of rat pancreatitis-associated protein, a lectin-related protein overexpressed during acute experimental pancreatitis. J Biol Chem 1991, 266:24664-24669 [PubMed] [Google Scholar]
- 12.Ortiz EM, Dusetti NJ, Vasseur S, Malka D, Bödeker H, Dagorn J-C, Iovanna JL: The pancreatitis-associated protein is induced by free radicals in AR4–2J cells and confers cell resistance to apoptosis. Gastroenterology 1998, 114:808-816 [DOI] [PubMed] [Google Scholar]
- 13.Petropoulos C, Andrews G, Tamaoki T, Fausto N: Alphafetoprotein and albumin mRNA levels in liver regeneration and carcinogenesis. J Biol Chem 1983, 258:4901-4906 [PubMed] [Google Scholar]
- 14.Christa L, Simon M-T, Finnois J-P, Gebhart R, Brechot C, Lasserre C: Overexpression of glutamin synthetase in human primary liver cancer. 1994, 106:1312–1320 [DOI] [PubMed]
- 15.Cariani E, Lasserre C, Seurin E, Hamelin B, Kemeny F, Franco D, Czech MP, Ullrich A, Bréchot C: Differential expression of insulin-like growth factor II mRNA in human primary liver cancers, benign liver tumors, and liver cirrhosid. Proc Natl Acad Sci USA 1988, 48:6844-6849 [PubMed] [Google Scholar]
- 16.Fausto N: Hepatocyte differentiation and liver progenitor cells. Curr Opin Cell Biol 1990, 2:1036-1042 [DOI] [PubMed] [Google Scholar]
- 17.Elmore LW, Sirica AE: Phenotypic characterization of metaplasic intestinal glands and ductular hepatocytes in cholangiofibrotic lesions rapidly induced in the caudate liver lobe of rats treated with furan. Cancer Res 1991, 51:5752-5759 [PubMed] [Google Scholar]
- 18.Dwivedi RS, Reddy JK, Rao MS, Yeldandi AV, Tan X, Dwivedi RS: Pancreatic hepatocytes: An in vivo model for cell lineage in pancreas of adult rat. Dig Dis Sci 1991, 36:502-509 [DOI] [PubMed] [Google Scholar]
- 19.Dabeva MD, Hwang SG, Vasa SR, Hurston E, Novikoff PM, Hixson DC, Gupta S, Shafritz DA: Differentiation of pancreatic epithelial progenitor cells into hepatocytes following transplantation into rat liver. Proc Natl Acad Sci 1997, 94:7356-7361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Poirier F, Timmons PM, Chan C-TJ, Guenet J-L, Rigby PW: Expression of the L14 lectin during mouse embryogenesis suggests multiple roles during pre- and post-implantation development. Development 1992, 115:143-155 [DOI] [PubMed] [Google Scholar]
- 21.Angerer LM, Angerer RC: In situ hybridization to cellular RNA with radiolabeled RNA probes. Edited by DG Wilkinson. Oxford, Oxford University Press, 1987, pp 15–32
- 22.Iovanna J-C, Keim V, Bosshard A, Orelle B, Frigerio J-M, Dusetti N, Dagorn J-C: PAP, a pancreatic secretory protein induced during acute pancreatitis, is expressed in rat intestine. Am J Physiol 1993, 265:G611-G618 [DOI] [PubMed] [Google Scholar]
- 23.Fu K, Sarras MP, Jr, De Lisle RC, Andrews GK: Regulation of mouse pancreatitis-associated protein-1 gene expression during caerulein-induced acute pancreatitis. Digestion 1996, 57:333-340 [DOI] [PubMed] [Google Scholar]
- 24.Sansonetti A, Romeo H, Berthezene P, Scacchi P, Dusetti N, Keim V, Dagorn J-C, Iovanna J-L: Developmental, nutritional and hormonal regulation of the pancreatitis-associated protein I and III gene expression in the rat small intestine. Scand J Gastroenterol 1995, 30:664-669 [DOI] [PubMed] [Google Scholar]
- 25.Masciotra L, Lechêne de la porte P, Frigerio J-M, Dusetti NJ, Dagorn J-C, Iovanna JL: Immunocytochemical localization of pancreatitis-associated protein in human small intestine. Dig Dis Sci 1995, 40:519-524 [DOI] [PubMed] [Google Scholar]
- 26.Frigerio J-M, Dusetti NJ, Garrido P, Dagorn J-C, Iovanna J-C: The pancreatitis-associated protein III (PAP III), a new member of the PAP gene family. Biochim Biophys Acta 1993, 1216:329-331 [DOI] [PubMed] [Google Scholar]
- 27.Narushima Y, Unno M, Nakagawara K-I, Mori M, Miyashita H, Suzuki Y, Nogushi N, Takasawa S, Kumagai T, Yonekura H, Okamato H: Structure, chromosomal localization and expression of mouse genes encoding type III Reg, RegIIIa, RegIIIb, RegIIIc. Gene 1997, 185:159-168 [DOI] [PubMed] [Google Scholar]
- 28.Koopman P, Povey S, Lovell-Badge H: Widespread expression of human a1-antitrypsin in transgenic mice revealed by in situ hybridization. Genes Dev 1989, 3:16-25 [DOI] [PubMed] [Google Scholar]
- 29.Piau J-P, Labarriere N, Dabouis G, Denis MG: Evidence for two distinct a(1,2)-fucosyltransferase genes differentially expressed throughout the rat colon. Biochem J 1994, 300:623-626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brown NF, Hill JK, Esser V, Kirkland JL, Corkey BE, Foster DW, McGarry JD: Mouse white adipocytes and 3T3–L1 cells display an anomalous pattern of carnitine palmitoyltransferase (CPT) I isoform expression during differentiation. Biochem J 1997, 327:225-231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Simon TC, Gordon JL: Intestinal epithelial cell differentiation: new insights from mice, flies and menatodes. Curr Opin Cell Biol 1995, 5:577-586 [DOI] [PubMed] [Google Scholar]
- 32.Bry L, Falk P, Huttner K, Ouellette A, Midtvedt T, Gordon JI: Paneth cell differentiation in the developing intestine of normal and transgenic mice. Proc Natl Acad Sci 1994, 91:10335-10339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gualdi R, Bossard P, Zheng M, Hamada Y, Coleman JR, Zaret KS: Hepatic specification of the gut endoderm in vitro: cell signaling and transcriptional control. Genes Dev 1996, 10:1670-1682 [DOI] [PubMed] [Google Scholar]
- 34.Lewin B: On neuronal specificity and the molecular basis of perception. Cell 1994, 79:935-943 [DOI] [PubMed] [Google Scholar]
- 35.Tsushida T, Ensini M, Morton SB, Baldassare M, Edlund T, Jessel TM, Pfaff SL: Topographic organization of embryonic motor neurons defined by expression of Lim homeobox genes. Cell 1994, 79:957-970 [DOI] [PubMed] [Google Scholar]
- 36.Hynes MA, Dodd J, Jessel TM: Carbohydrate recognition, cell interactions, and vertebrate neural development. Edited by RU Margolis, RK Margolis. New York, Plenum Publishers Corp., 1989, pp 337–365
- 37.Puche AC, Poirier F, hair M, Bartlett PF, Key B: Role of Galectin-1 in developing mouse olfactory system. Dev Biol 1996, 179:274-287 [DOI] [PubMed] [Google Scholar]
- 38.Mahanthappa NK, Cooper DNW, Barondes SH, Schwarting GA: Rat olfactory neurons can utilize the endogeneous lectin, L-14, in a novel adhesion mechanism. Development 1994, 120:1373-1384 [DOI] [PubMed] [Google Scholar]
- 39.Livesey FJ, O’Brien JA, Li M, Murphy LJ, Hunt SP: A Schwann cell mitogen accompanying regeneration of motor neurons. Nature 1997, 390:614-618 [DOI] [PubMed] [Google Scholar]