Abstract
Splenic marginal zone lymphoma (SMZL) has been recognized as an entity defined on the basis of its morphological, phenotypic, and clinical characteristic features. Nevertheless, no characteristic genetic alterations have been described to date for this entity, thus making an exact diagnosis of SMZL difficult in some cases. As initial studies showed that chromosome region 7q22-32 is deleted in some of these cases, we analyzed a larger group of SMZL and other lymphoproliferative disorders that may partially overlap with it. To better define the frequency of 7q deletion in SMZL and further identify the deleted region, polymerase chain reaction analysis of 13 microsatellite loci spanning 7q21-7q36 was performed on 20 SMZL and 26 non-SMZL tissue samples. The frequency of allelic loss in SMZL (8/20; 40%) was higher than that observed in other B-cell lymphoproliferative syndromes (2/26; 7.7%). This difference was statistically significant (P < 0.05). The most frequently deleted microsatellite was D7S487 (5/11; 45% of informative cases). Surrounding this microsatellite the smallest common deleted region of 5cM has been identified, defined between D7S685 and D7S514. By comparative multiplex polymerase chain reaction analysis, we detected a homozygous deletion in the D7S685 (7q31.3) marker in one case. These results suggest that 7q31-q32 loss may be used as a genetic marker of this neoplasia, in conjunction with other morphologic, phenotypic, and clinical features. A correlation between 7q allelic loss and tumoral progression (death secondary to the tumor or large cell transformation) in SMZL showed a borderline statistical significance. The observation of a homozygous deletion in this chromosomal region may indicate that there is a tumor suppressor gene involved in the pathogenesis of this lymphoproliferative neoplasia.
Although specific pathological and phenotypic traits are gradually being recognized for splenic marginal zone lymphomas (SMZL), 1-3 the distinction in comparison other lymphoproliferative disorders is frequently blurred by the lack of specific molecular markers for this neoplasia. Cytogenetic analysis of SMZL cases has showed diverse abnormalities at chromosomes 1, 3, 7, and 8, 4-5 chromosome 7 being the most frequently altered (3/19 cases). 4 Some SMZL cases showed del 7q as the only cytogenetic abnormality, this chromosome loss being found in 7q22 as well as in 7q32. 4 7q abnormalities have been previously published in studies of other chronic lymphoproliferative disorders by cytogenetic techniques. 6-10 Most reports center on tumors that may at least partially overlap SMZL, such as immunocytoma and splenic lymphoma with circulating villous lymphocytes (SLVL). In these series the most common deletions identified were also 7q22 and 7q32. 6-10
Some of the uncertainties in genetic studies of SMZL arise from the fact that cases with this diagnosis have been lumped in larger groups together with other lymphoproliferative disorders, as there is no requirement to confirm the diagnosis by study of a splenectomy specimen. 8-10 Here we have chosen a more conservative approach, exclusively studying cases in which the diagnosis of SMZL has been performed after histological study of a splenectomy specimen. At the same time, we have refined the molecular techniques used. Thus previous studies on SMZL were performed using mainly standard cytogenetic techniques, which do not detect the entire spectrum of chromosomal changes, including microdeletions. In this study loss of heterozygosity (LOH) analysis was used because it is a sensitive molecular method to screen for changes involving allele loss. 11-12
To clarify the frequency of 7q deletion in SMZL and define the deleted region, we analyzed 7q LOH specifically on 7q22-32 using a panel of highly polymorphic markers on 7q21-qter, in a series of SMZL cases defined on the basis of the splenic histology, and in a control series of lymphoproliferative processes whose morphology may mimic SMZL, such as B-chronic lymphocytic leukemia (B-CLL), mucosa-associated lymphoid tissue (MALT) lymphoma, follicular lymphoma (FL), and mantle cell lymphoma (MCL).
Materials and Methods
Tissue Samples
Twenty SMZL cases were included in this study. They were consecutive cases obtained from the routine and consultation files of the Pathology Laboratory of the Virgen de la Salud Hospital (Toledo, Spain). These cases were diagnosed on the basis of splenic morphology according to the criteria established by Isaacson 3,13 and Mollejo. 2,14 The only criterion for a case to be included in the series was the availability of control DNA extracted from oral swabs.
A control series of 26 lymphoproliferative disorders, including 7 B-CLL, 4 MCL, 9 MALT, and 6 FL cases, was also included. All of them were consecutive routine cases in which control DNA could be extracted either from oral swabs or microdissected nontumoral areas in the surgical specimens.
In all of the SMZL cases, a clinical follow-up was performed over a median range of 37 months. Large cell transformation was diagnosed if the patient presented a histology of large B-cell lymphoma in a different location after the initial diagnosis. Deaths secondary to the tumor were classified as attributable either to the tumor itself or to the consequences of the treatment.
Thirteen of the SMZL cases were previously analyzed by conventional cytogenetic means. Chromosome analysis was carried out on the spleen (six patients), lymph node (one case), and peripheral blood (six patients) by standard procedure. The cytogenetic findings of some cases (S1, S4, S74 and S149) have already been reported. 4
DNA Extraction
Tumoral DNA was extracted from fresh frozen tissue, paraffin-embedded tissue, or peripheral blood lymphocytes in non-SMZL cases with high tumoral lymphocyte counts. Fresh frozen tissue was treated with sodium dodecyl sulfate buffer and proteinase K, followed by phenol-chloroform purification and precipitation with ethanol, according to standard procedures.
Paraffin-embedded tissue sections were dewaxed in xylene and hydrated with ethanol. The samples were air-dried, incubated with buffer (100 mmol/L Tris, pH 8.5; 500 mmol/L KCl; 15 mmol/L Cl2Mg; 0.5% Tween 20) and heated for 7 minutes in a microwave oven. Peripheral blood lymphocytes were isolated by Histopaque gradient (Sigma, St. Louis, MO) and DNA was extracted using a standard procedure.
Normal DNA was obtained from oral swabs in 20 SMZL and 22 non-SMZL cases, or from microdissected nontumoral areas in surgical specimens in 4 cases of non-SMZL. Oral swabs were incubated for 2 hours in double distilled H2O at room temperature, centrifuged, and treated with sodium dodecyl sulfate buffer and proteinase K (200 μg/ml) at 37°C for 6 hours.
The quality of the extracted DNA was assessed using primers for the p53 gene, exon 8 (249 pb).
Microsatellite Analysis
Fourteen pairs of microsatellite markers on chromosome 7 were used (1 for the short arm and 13 for the long arm), all of which were obtained from Research Genetics (Huntsville, AL). The markers used in this study were D7S460, D7S492, D7S518, D7S471, D7S466, D7S486, D7S522, D7S2847, D7S480, D7S685, D7S487, D7S514, D7S530, and D7S550 (Table 1) ▶ . All microsatellite markers were previously published in studies of LOH in 7q, where they were found to be frequently deleted. 15-18 The location and linear order of the microsatellites used are based on the map release by Généthon. 19 A tetranucleotide repeat polymorphism in chromosome 7p (D7S460) was also tested. 20 The markers analyzed for non-SMZL cases were the most frequently deleted and informative for SMZL cases (D7S518, D7S466, D7S522, D7S480, D7S487, and D7S530).
Table 1.
Summary of Markers, Map Positions, and Frequency of 7q Allelic Loss in 20 Cases of SMZL
| Marker | Map Position | No. of cases studied (informative cases) | No. of cases with LOH (%) |
|---|---|---|---|
| D7S460 | 7p | 18 (14) | 0 |
| D7S492 | 7q21.1 | 19 (14) | 0 |
| D7S518 | 7q22.1 | 19 (16) | 0 |
| D7S471 | 7q22-31.1 | 18 (11) | 1 (8%) |
| D7S466 | 7q22-31.1 | 18 (12) | 4 (33%) |
| D7S486 | 7q31.1 | 20 (12) | 4 (33%) |
| D7S522 | 7q31.1 | 20 (12) | 3 (25%) |
| D7S2847 | 7q31.3 | 20 (18) | 5 (27%) |
| D7S480 | 7q31.3 | 19 (17) | 6 (35%) |
| D7S685 | 7q31.3 | 14 (10) | 3 (30%) |
| D7S487 | 7q31.3 | 19 (11) | 5 (45%) |
| D7S514 | 7q31.3-32 | 20 (13) | 4 (30%) |
| D7S530 | 7q32.2 | 19 (10) | 3 (33%) |
| D7S550 | 7q36 | 15 (12) | 2 (16%) |
Each microsatellite repeat was amplified using polymerase chain reaction (PCR). PCR reactions were performed in a final volume of 20 μl containing 50–100 ng of DNA template, 10 pmol of each primer, 200 mmol/L dNTPs, 1.5 mmol/L Cl2Mg, 1 μCi of [α-32P] dCTP, and 1 U of Taq polymerase (Boehringer Mannheim, Mannheim, Germany). Twenty-five cycles were performed, each consisting of 30 seconds at 94°C, with annealing temperatures at 57°C and 30 seconds at 72°C, using a Perkin Elmer 2400 GeneAmp PCR system (Norwalk, CT). The products of PCR reactions were mixed with an equal volume of formamide loading buffer (95% formamide, 20 mmol/L EDTA, 0.05% bromophenol blue, 0.05% xylencyanol), then denatured at 95°C for 5 minutes and cooled on ice. Approximately 2 μl of each sample were loaded onto 6% denaturing polyacrylamide gel containing 7 mol/L urea. Gels were fixed in 10% acetic acid, air-dried, and exposed to X-ray films.
Allelic Loss Determination
Cases were considered to be informative when heterozygosity was detected in normal tissue samples. LOH was determined visually by two different examiners, as complete loss of one allele or as quantified by densitometric analysis (1-D Analysis and Hand Scanner Settings, Biomed Instruments, Zeineh Programs (Fullerton, CA). By densitometric analysis a tumor was scored as LOH when the allelic ratio of the tumor tissue compared to the normal control tissue was >3. Allelic imbalance was considered to be present when this allelic ratio was >1.5 and <3. 15,21-24 Homozygous alleles in the normal tissue samples were considered uninformative.
In an attempt to distinguish whether allelic imbalance was due to gain or loss, comparative multiplex PCR was performed. 15,25,26 Microsatellite markers (D2S1360 and D7S550) without allelic imbalance were selected as internal controls. The intensity of the control alleles was compared with the intensity of the locus showing allelic imbalance by means of visual inspection and densitometric image analysis.
All cases of allelic loss were confirmed by three separate experiments with two different examiners.
Statistical Analysis
A χ 2 test was used to compare the frequency of tumoral progression between cases with and without 7q allelic losses. Differences were considered significant when P < 0.05.
Results
Allelic Loss 7q in SMZL
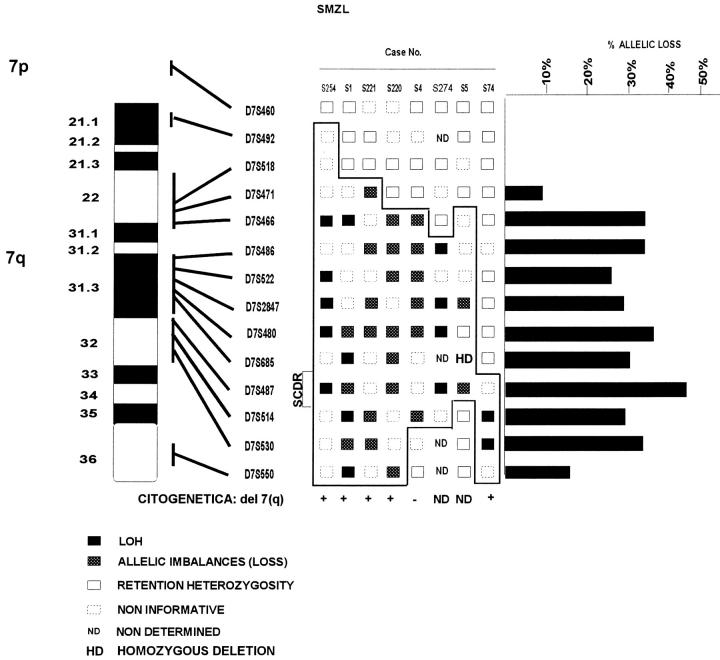
A set of 13 microsatellite markers spanning the region from 7q21 to 7q36 (Figure 1) ▶ were used to perform allelic loss analysis in 20 SMZL and 26 non-SMZL cases. Figure 1 ▶ summarizes the results obtained, and representative cases are illustrated in Figure 2 ▶ . As is shown in the deletion map in Figure 1 ▶ , all markers analyzed were highly informative (Table 1) ▶ .
Figure 1.
Summary of allelic loss data for 7q in SMZL. Graphic representation of the 13 microsatellite markers in 7q21-qter, giving the approximate position of each locus. Only cases showing LOH or allelic imbalance are illustrated. The results of the cytogenetic study are also included. Smallest common deletion region (SCDR) in the eight cases with allelic loss is shown, and spans about 5cM, from markers D7S685 to D7S514. A histogram shows the frequency of allelic loss for each of these markers in the informative samples.
Figure 2.
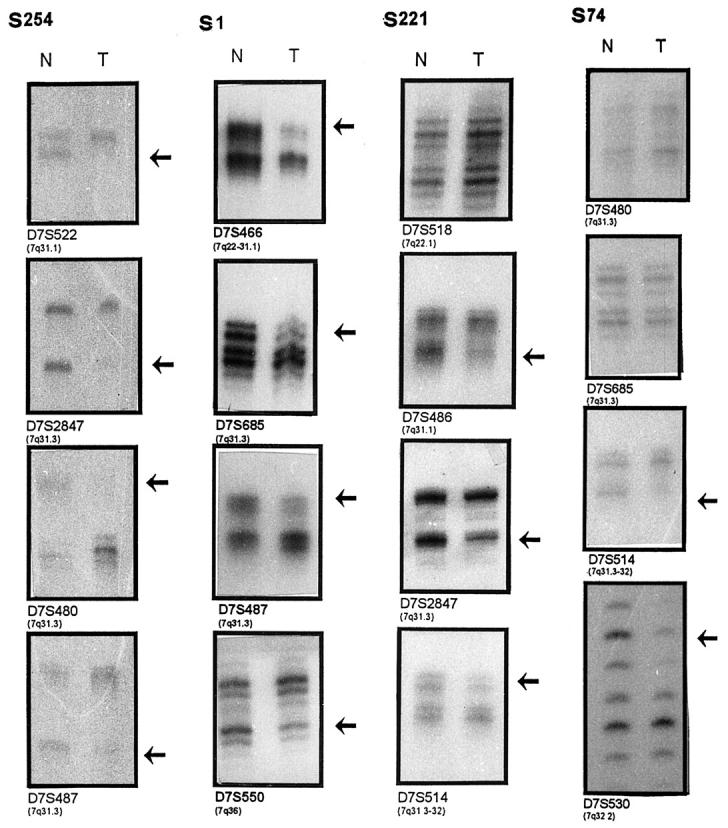
Representative examples of allelic loss in chromosome 7q. The presence of allelic losses in tumor (T) in comparison with matched normal (N) DNA is shown by arrows. Case S254 shows allelic loss in all informative loci. Case S1 shows loss of alleles for the loci mapping the 7q31-qter region (band intensity ratios of 3.1, 3.5, 1.6, and 2.7, respectively). Case S221 shows loss of alleles for loci D7S486, D7S2847, and D7S514 (band intensity ratios are 2.1, 2.8, 1.9 respectively) and retention of alleles for loci D7S518 mapped to 7q22. Case S74 shows loss of alleles for loci mapped to 7q32 (D7S514 and D7S530) and retention of alleles for loci D7S685 and D7S480, situated at the 7q31 band.
Allelic losses were detected in 8 of 20 (40%) SMZL. In the analysis of allelic loss we differentiated between allelic imbalances and LOH according to previous works. 15,21-24 In three tumors (S254, S274, and S74) LOH involved all the markers with allelic loss (Figures 1 and 2) ▶ ▶ . In addition, one tumor (S1) showed either LOH or allelic imbalances, whereas the other four tumors displayed only allelic imbalances (Figures 1 and 2) ▶ ▶ . To determine whether the observed allelic imbalances were gains or losses, cytogenetic analysis was used to confirm the existence of allelic loss in all three cases with allelic imbalances (S1, S220, and S221) (Figure 1) ▶ . In cases S4 and S5, comparative multiplex PCR analysis was performed, confirming the existence of allelic loss (Figure 3) ▶ . All cases were considered to demonstrate genuine LOH according to the criteria of previous works. 15,21-24
Figure 3.
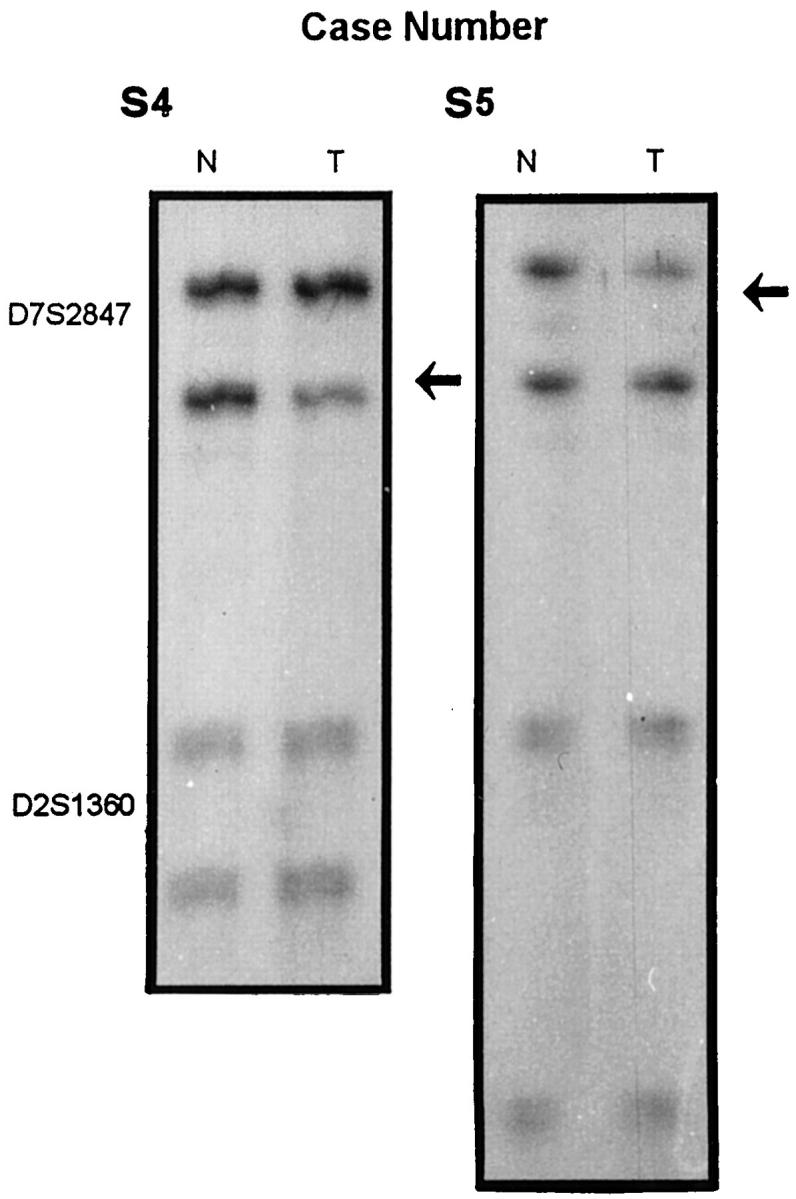
Example of comparative multiplex PCR in cases S4 and S5. An internal control marker (D2S1360) shows the amount of tumor (T) and normal (N) DNA amplified. Case S5 shows a loss of the upper band, whereas case S4 has a loss of the lower band, as shown by arrows.
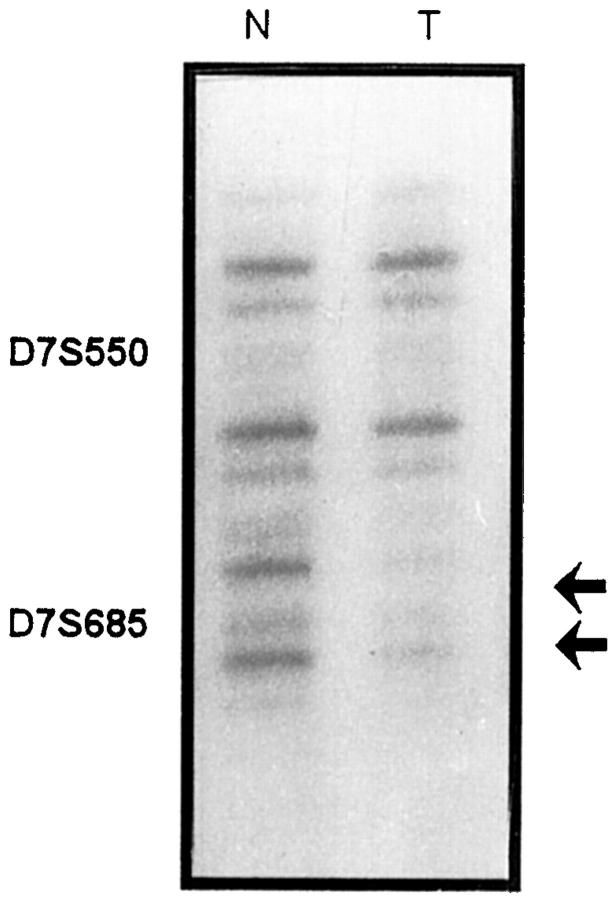
A 7q31.3-q32 homozygous deletion was observed in one case (S5). Multiplex PCR with primers for both D7S685 and D7S550 show that when the D7S550 alleles in the normal and tumor DNA lanes were of equal intensity, the signal for D7S685 in the tumor lane was absent or very weak (Figure 4) ▶ .
Figure 4.
Case number S5, showing homozygous deletion of microsatellite D7S685 (arrows). Autoradiography of multiplex PCR products showed that primer sets D7S550 and D7S685 amplified products from the normal (N) DNA, whereas only D7S550 amplified a product from the corresponding tumor (T) DNA. (The signal for D7S685 in the tumor lane was absent or very weak)
The results of the cytogenetic studies performed in a subset of these cases are shown in Figure 1 ▶ . Deletion 7q detectable by karyotype was seen in five cases, all of which were confirmed in this LOH analysis. Additionally, one case (S4) without cytogenetic 7q alterations showed LOH after molecular study (Figure 1) ▶ .
The frequency of allelic loss at each locus is shown Table 1 ▶ . The locus that showed the highest percentage of LOH was D7S487 (5 of 11 informative cases, 45%). The lowest incidence of allelic loss was found in microsatellites D7S518 and D7S471 (0% and 8%, respectively), spanning the 7q22 band.
Five of the eight cases with allelic loss showed LOH in all the informative loci of band 7q31-32, as analyzed here. In contrast with this finding, case S74 showed just a small deletion, thereby making it possible to delimit the smallest commonly deleted region (SCDR) (Figures 1 and 2) ▶ ▶ . The localization of the SCDR was inferred from the pattern of allelic loss in these tumors, defined by flanking markers D7S685 and D7S514, as demonstrated by tumor samples S74 and S5, respectively (Figure 1) ▶ . This region is located in bands 7q31-32.
7q Allelic Loss in Non-SMZL
Two of 26 (7.7%) non-SMZL cases showed allelic loss. Positive 7q LOH cases were 1/4 MCL and 1/5 FL. When comparing the frequency of 7q allelic loss between SMZL and non-SMZL cases, a statistically significant difference was found (P < 0.008; Fisher’s Exact Test) (Table 2) ▶ .
Table 2.
Frequency of Allelic Loss at 7q in the Different Types of Lymphoma
| Diagnosis | Allelic loss at 7q(%) |
|---|---|
| non-SMZL | 2 /26 (7.7%) |
| SMZL | 8 /20 (40%) |
P < 0.008
Correlation between del(7q) and Tumoral Progression in SMZL
To explore the relationship between tumoral aggressiveness and 7q LOH, we selected a group of cases with morphological or clinical evidence of tumoral progression, and compared their frequency of 7q LOH to that in other cases. Four of eight (50%) SMZL cases with 7q LOH showed either death attributable to the tumor or large cell transformation, whereas a similar clinical aggressiveness was found only in 1/12 cases without 7q LOH. This had a borderline statistical significance (P < 0.05779, Fisher’s Exact Test) (Table 3) ▶ .
Table 3.
Correlation between Features at Diagnosis, Neoplastic Progression, and 7q Allelic Loss in SMZL Cases
| 7q allelic loss | P | ||
|---|---|---|---|
| Yes | No | ||
| Progression | |||
| Large cell transformation | 1 /8 | 0 /12 | 0.05779 |
| Death secondary to the tumor | 3 /8 | 1 /12 |
Discussion
In this study we used 13 dinucleotide repeats spanning the 7q21-q36 band to deduce the frequency of deletion and draw a deletion map for this LOH. In these tumors we detected 8/20 (40%) cases of SMZL with allelic losses, as opposed to a frequency of 2/26 (7.7%) in non-SMZL B-cell lymphoproliferative disorders. Cytogenetic studies and comparative multiplex PCR excluded the possibility that the allelic imbalance found here in some cases could be attributable to allelic gains. These results indicate that 7q31-32 allelic loss is a characteristic finding in SMZL, which, in conjunction with other features of the tumor, may be used in differential diagnosis of SMZL versus other types of lymphoma.
Chromosome 7 abnormalities in non-Hodgkin’s lymphomas (NHLs) have been described in different studies using cytogenetic, fluorescent in situ hybridization and comparative genomic hybridization techniques. 4-8,27-32 Although in some studies no significant incidence of 7q loss in NHLs was found, 27,30,31 other studies have shown increased frequency in some specific lymphoproliferative processes. 8,9,32 Thus, two consecutive studies performed by the same group 9,32 showed that, in spite of an overall 7q loss incidence of 3.6% in NHLs, 26–31% of the cases with a diagnosis of SLVL display different abnormalities of chromosome 7 including del(7)(q22-32), del(7)(q34-36), and t(7q22). Other comprehensive studies on NHLs confirm that the overall incidence of 7q loss is low in the group of NHLs as a whole, 24/558 cases (4.3%). 8 This frequency is higher when only small lymphocytic lymphoma is considered.
The data so far described are therefore consistent with those reported here, because the histological types where a higher incidence of 7q loss has been found are those which overlap SMZL. Different reports agree that the splenic histology of SLVL cases is similar to that described for SMZL, 6 although some differences between these two groups have also been stated to exist, such as the reported presence of t(11;14) in a subset of SLVL cases (this has never been found in SMZL) and the occasional lack of peripheral blood involvement in cases of SMZL. 4,6 The selection of cases used here, taking splenic histology into account together with the other features of the tumors, made it possible to restrict the cases included to a relatively homogeneous histological type, thus avoiding any bias arising from the selection of the sample.
The relevance of this genetic abnormality in the pathogenesis of SMZL is underlined by the fact that in different descriptions it appears as a single cytogenetic alteration. 4,7,8,10 The exact region of genetic loss seems to be closer to microsatellite D7S487, where the highest incidence of LOH (45%) has been identified. This region is situated between the D7S685 and D7S514 markers, which define the smallest commonly deleted region observed by Hernandez and colleagues in a previous study of a group of NHL. 33
The data reported here confirm and expand initial observations made by Sole et al 4 using cytogenetic and fluorescence in situ hybridization techniques pointing to 7q alterations as a significant cytogenetic finding in SMZL defined on the basis of splenic histology. It now appears after LOH analysis that the incidence of this genetic loss is more frequent than initially expected and that, additionally, it seems to be associated with a more aggressive course.
Loss of this chromosome region is not restricted to lymphoproliferative disorders, 8-10 but has also been described in other solid tumor and myeloid disorders. 15-18,23,26,34-37 This recurrent abnormality suggests that these regions contain a novel tumor suppressor gene that has yet to be identified. In several studies of other types of tumors it has been observed that genetic alterations at 7q31 may participate in tumor progression. 15,34 The existence in one of these cases of a biallelic deletion in the D7S685 locus, as detected by the use of comparative multiplex PCR analysis, offers additional evidence in favor of the existence of a tumor suppressor gene in this location.
To summarize, this study shows that 7q31-q32 loss is a relatively specific genetic marker of SMZL, which may be used in the differential diagnosis of this entity in conjunction with other clinical, morphological, and phenotypic traits of the neoplasia. The data obtained here seem to show, additionally, that the loss of this genetic region causes these tumors to display more aggressive behavior and thereby supplying data that is potentially useful in the treatment of these patients. Further genetic studies could make progress towards the identification of a tumour suppressor gene that may be located in this area.
Acknowledgments
We thank Dr. Jose Fernandez Piqueras of Laboratory Human Molecular Genetic, Department of Biology (University Autonoma, Madrid, Spain) for his critical review of the manuscript and technical advice, and Drs. T. Flores, L. Bernardo, M. Morente, and P. Gonzalvo from the pathology departments at hospitals in Salamanca, Gerona, Guadalajara, and Asturias, Spain, for kindly providing the cases included in this series.
Footnotes
Address reprint requests to Dr. Miguel A. Piris, Dept. of Pathology, Hospital Virgen de la Salud, Av. Barber No. 30, 45004 Toledo, Spain. E-Mail: mpiris@cht.es.
Supported by a grant (96/1382) from the Fondo de Investigaciones Sanitarias, Ministerio de Sanidad y Consumo, Spain.
References
- 1.Harris NL, Jaffe NS, Stein H, Banks PM, Chan JKC, Cleary M, Delsol G, de Wolf-Peters C, Falini B, Gatter KC, Grogan TM, Isaacson PG, Knowles DM, Masson SY, Muller-Hermelink HK, Pileri SA, Piris MA, Ralfkiaer E, Warnke RA: A revised European-American classification of lymphoid neoplasms: a proposal from the International Lymphoma Study Group. Blood 1994, 84:1361-1392 [PubMed] [Google Scholar]
- 2.Mollejo M, Menarguez J, LLoret E, Sanchez A, Campo E, Algara P, Cristobal E, Sanchez E, Piris MA: Splenic marginal zone lymphoma, a distinctive type of low-grade B-cell lymphoma: a clinicopathological study of 13 cases. Am J Surg Pathol 1995, 19:1146-1157 [PubMed] [Google Scholar]
- 3.Isaacson PG, Piris MA: Splenic marginal zone lymphoma. Adv Anat Pathol 1997, 4:191-201 [Google Scholar]
- 4.Sole F, Woessner S, Florensa L, Espinet B, Mollejo M, Martinez P, Piris MA: Frequent involvement of chromosomes 1, 3, 7 and 8 in splenic marginal zone B-cell lymphoma. Br J Haematology 1998, 98:446-449 [DOI] [PubMed] [Google Scholar]
- 5.Dierlam D, Pittaluga S, Wlodarska I, Stul M, Thomas J, Boogaerts M, Michaux L, Driessen A, Mecucci C, Cassiman J-J, De Wolf-Peeters, Van den Berghe H: Marginal zone B-cell lymphomas of different sites share similar cytogenetic an morphologic features. Blood 1996, 87:299–307 [PubMed]
- 6.Oscier DG, Matutes E, Gardiner A, Glide S, Mould S, Brito-Babapulle V, Ellis J, Catovsky D: Cytogenetic studies in splenic lymphoma with villous lymphocytes. Br J Haematology 1993, 85:487-491 [DOI] [PubMed] [Google Scholar]
- 7.Sole F, Woessner L, Florensa L, Montero S, Asensio A, Besses C, Sans-Sabafren J: A new chromosomal anomaly associated with mature B-cell chronic lymphoproliferative disorders: del(7)(q32). Cancer Genet Cytogenet 1993, 65:170-172 [DOI] [PubMed] [Google Scholar]
- 8.Offit K, Louie DC, Parsa NZ, Noy A, Chaganti RSK: Del (7) (q32) is associated with a subset of small lymphocytic lymphoma with plasmacytoid features. Blood 1995, 86:2365-2370 [PubMed] [Google Scholar]
- 9.Oscier DG, Gardiner A, Mould S: Structural abnormalities of chromosome 7q in chronic lymphoproliferative disorders. Cancer Genet Cytogenet 1996, 92:24-27 [DOI] [PubMed] [Google Scholar]
- 10.Hernandez JM, Mecucci C, Michaux L, Criel A, Stul M, Meeus P, Wlodarska I, Van Orshoven A, Cassiman J-J, De Wolf-Peeters C, Van Den Berghe H: Del(7q) in chronic B-cell lymphoid malignancies. Cancer Genet Cytogenet 1997, 93:147-151 [DOI] [PubMed] [Google Scholar]
- 11.Vogelstein B, Fearon ER, Kern SE, Hamilton SR, Preisinger AC, Nakamura Y, White R: Allelotype of colorectal carcinomas. Science 1989, 244:207-211 [DOI] [PubMed] [Google Scholar]
- 12.Luis DN, Deimling A, Seisinger BR: A (CA)n dinucleotide repeat assay for evaluating loss of allelic heterozygosity in small and archival human brain tumor specimens. Am J Pathol 1992, 141:778-782 [PMC free article] [PubMed] [Google Scholar]
- 13.Isaacson PG, Matutes E, Burke M, Catovsky D: The histopathology of splenic marginal lymphoma with villous lymphocytes. Blood 1994, 84:3828-3834 [PubMed] [Google Scholar]
- 14.Mollejo M, Lloret E, Menarguez J, Piris MA: Lymph node involvement by splenic marginal zone lymphoma morphological and immunohistochemical features. Am J Surg Pathol 1997, 21:772-780 [DOI] [PubMed] [Google Scholar]
- 15.Takahashi A, Shan AL, Ritland SR, Delacey KA, Bostwick DG, Lieber MM, Thibodeau SN, Jenkins RB: Frequent of loss of heterozygosity at 7q31.1 in primary prostate cancer is associated with tumour aggressiveness and progression. Cancer Res 1995, 55:4114-4119 [PubMed] [Google Scholar]
- 16.Ishwad CS, Ferrell RE, Davare J, Meloni AM, Sandberg AA, Surti U: Molecular and cytogenetic analysis of chromosome 7 in uterine leiomyomas. Genes Chromosomes Cancer 1995, 14:51-55 [DOI] [PubMed] [Google Scholar]
- 17.Edelson MI, Scherer SW, Tsui LC, Welch WR, Bell DA, Berkowitz RS, Mok SC: Identification of a 1300 kilobase deletion unit on chromosome 7q31.3 in invasive epithelial ovarian carcinomas. Oncogene 1997, 14:2979-2984 [DOI] [PubMed] [Google Scholar]
- 18.Zenklusen JC, Weitzel JN, Ball HG, Conti CJ: Allelic loss at 7q31.1 in human primary ovarian carcinomas suggests the existence of a tumour suppresser gene. Oncogene 1995, 11:359-363 [PubMed] [Google Scholar]
- 19.Gyapay G, Morissette J, Vignal A, Dib C, Fizames C, Millasseau P, Marc S, Bernardi G, Lathrop M, Weissenbach J: The 1993–94 Généthon human genetic linkage map. Nat Genet 1994, 7:246-339 [DOI] [PubMed] [Google Scholar]
- 20.Hudson TJ, Engelstein M, Lee MK, Ho EC, Rubenfield MJ, Adams CP, Housman DE, Dracopoli NC: Isolation and chromosomal assignment of 100 highly informative human simple sequence repeat polymorphisms. Genomics 1992, 13:622-629 [DOI] [PubMed] [Google Scholar]
- 21.Santos J, Perez de Castro I, Herranz M, Pellicer A, Fernandez-Piqueras J: Allelic losses on chromosome 4 suggest the existence of a candidate tumour suppresser gene region of about 0.6 cm in γ-radiation-induced mouse primary thymic lymphomas. Oncogene 1996, 12:669-676 [PubMed] [Google Scholar]
- 22.Hegi ME, Devereux TR, Dietrich WH, Cochran CJ, Lander ES, Foley JF, Maronpot RR, Anderson MW, Wiseman RW: Allelotype analysis of mouse lung carcinomas reveals frequent allelic losses on chromosome 4 and an association between allelic imbalances on chromosome 6 and K-ras activation. Cancer Res 1994, 54:6257-6264 [PubMed] [Google Scholar]
- 23.Achille A, Biasi MO, Zamboni G, Bogina G, Magalini AR, Pederzoli P, Perucho M, Scarpa A: Chromosome 7q allelic losses in pancreatic carcinoma. Cancer Res 1996, 56:3808-3813 [PubMed] [Google Scholar]
- 24.Haddad R, Held WA: Genomic imprinting and Igf2 influence liver tumorigenesis and loss of heterozygosity in SV40 T antigen transgenic mice. Cancer Res 1997, 57:4615-4623 [PubMed] [Google Scholar]
- 25.Cairns P, Tokino K, Eby Y, Sidransky D: Homozygous deletions of 9p21 in primary human bladder tumors detected by comparative multiplex polymerase chain reaction. Cancer Res 1994, 54:1422-1424 [PubMed] [Google Scholar]
- 26.Shridhar V, Sun QC, Miller OJ, Kalemkerian GP, Petros J, Smith DI: Loss of heterozygosity on the long arm of human chromosome 7 in sporadic renal cell carcinomas. Oncogene 1997, 15:2727-2733 [DOI] [PubMed] [Google Scholar]
- 27.Wotherspoon AC, Pan L, Diss TC, Isaacson PG: Cytogenetic study of B-cell lymphoma of mucosa-associated lymphoid tissue. Cancer Genet Cytogenet 1992, 58:35-38 [DOI] [PubMed] [Google Scholar]
- 28.Wotherspoon AC, Finn TM, Isaacson PG: Trisomy 3 in low-grade B-cell lymphomas of mucosa-associated lymphoid tissue. Blood 1995, 8:2000-2004 [PubMed] [Google Scholar]
- 29.Brynes RK, Almaguer PD, Leathery KE, McCourty A, Arber DA, Medeiros J, Nathwani BN: Numerical cytogenetic abnormalities of chromosomes 3, 7 and 12 in marginal zone B-cell lymphomas. Mod Pathol 1996, 9:995-1000 [PubMed] [Google Scholar]
- 30.Dierlam J, Rosenberg C, Stul M, Pittaluga S, Wlodarska I, Michaux L, Dehaen, Verhoef G, Thomas J, de Kelver W, Bakker-Schut T, Cassiman JJ, Raap AK, De Wolf-Peeters C, Van den Berghe H, Hagemeijer A: Characteristic pattern of chromosomal gains and losses in marginal zone B cell lymphoma detected by comparative genomic hybridization. Leukaemia 1997, 11:747–758 [DOI] [PubMed]
- 31.Avet-Loiseau H, Vigier M, Moreau A, Mellerin MP, Gaillard F, Harousseau JL, Bataille R, Milpied N: Comparative genomic hybridization detects genomic abnormalities in 80% of follicular lymphomas. Br J Haematology 1997, 97:119-122 [DOI] [PubMed] [Google Scholar]
- 32.Gillingham R, Oscier D: Genetic analysis of splenic lymphoma with villous lymphocytes using comparative genomic hybridization and fluorescent in situ. Br J Haematology 1998, 101:45 [Google Scholar]
- 33.Hernandez JM, Schoenmakers EFPM, Dal Chin P, Michaux L, Van den Ven WJM, Van den Berghe H: Molecular delineation of the commonly deleted segment in mature B-cell lymphoid neoplasias with deletion of 7q. Genes Chromosomes Cancer 1997, 18:147-150 [PubMed] [Google Scholar]
- 34.Velloso ERP, Michaux L, Ferrant A, Hernandez JM, Meeu P, Dierlamm J, Criel A, Louwagie A, Verhoef G, Boogaerts M, Michaux J-L, Bosly A, Mecucci C, Van den Bergue H: Deletions of the long arm of chromosome 7 in myeloid disorders: loss of band 7q32 implies worst prognosis. Br J Haematology 1996, 92:574-581 [DOI] [PubMed] [Google Scholar]
- 35.Le Beau MM, Esinosa R, Davis EM, Eisenbart JD, Larson RA, Green ED: Cytogenetic and molecular delineation of a region of chromosome 7 commonly deleted in malignant myeloid disorders. Blood 1996, 88:1930-1935 [PubMed] [Google Scholar]
- 36.Kiuru-Kuhlefelt S, Kristo P, Ruutu T, Knuutila S, Kere J: Evidence for two molecular steps in the pathogenesis of myeloid disorders associated with deletion of chromosome 7 long arm. Leukaemia 1997, 11:2097-2104 [DOI] [PubMed] [Google Scholar]
- 37.Fisher K, Frohling S, Scherer SW, McAllister, Brown J, Scholl C, Stilgenbauer S, Tsui L-C, Lichter P, Dohner H: Molecular cytogenetic delineation of deletions and translocations involving chromosome band 7q22 in myeloid leukemias. Blood 1997, 89:2036-2041 [PubMed] [Google Scholar]