Abstract
It has not been completely elucidated whether the liver injury induced by the hepatitis C virus (HCV) is due to direct cytopathic damage or to an immune-mediated response against HCV-infected hepatocytes. In this work, we have determined the percentage of HCV-infected hepatocytes, the histological activity index, and the viremia levels in chronically HCV-infected patients with different grades of liver injury to investigate any possible correlation between them. For that purpose, liver biopsies from 27 patients with HCV chronic hepatitis were analyzed by in situ hybridization. This technique revealed that the percentage of infected hepatocytes ranged from 0.04% to 83.6%. Regarding the viremia levels, HCV RNA concentration ranged from 1.8 × 103 to 1.4 × 106 genome copies/ml. A significant correlation (r = 0.54; P = 0.003) between the percentage of infected hepatocytes and the viremia levels was found. In contrast, no correlation was observed between the percentage of HCV-infected hepatocytes or the viremia levels and the histological activity index. In conclusion, we have shown that the HCV viremia reflects the extent of the infection in the liver and that the liver injury in chronic HCV infection is not directly related to either the number of infected hepatocytes or the serum HCV RNA concentration.
Chronic hepatitis C virus (HCV) infection is characterized by a wide spectrum of liver damage, ranging from minimal lesions to liver cirrhosis. The pathogenesis of HCV-induced liver injury has been attributed to either direct cytopathic damage 1-3 or to an immune-mediated injury against HCV-encoded proteins. 4-5 In an attempt to clarify this issue, several authors have analyzed the relation between serum HCV viremia or the viral load in the liver and the stage of liver disease with contradictory results. 6-12 However, none of these articles analyzed the relationship between the number of HCV-infected hepatocytes and the liver damage, which may be a better way to study this point.
On the other hand, whether serum HCV viremia reflects the viral load in liver is still controversial. 10,13 These differences may be due to the fact that the methods used to quantify the HCV-RNA levels in liver depend on a prior extraction of the total RNA from the hepatocytes so that the quantification of the viral load in the liver will depend on the efficiency of the extraction procedure. In a previous report, 14 we demonstrated using non-isotopic in situ hybridization and digital image analysis of the hybridization signals that serum HCV viremia is related to the percentage of infected hepatocytes in HCV-infected patients with chronic active hepatitis.
In the present work we have determined the percentage of HCV-infected hepatocytes in liver biopsies by in situ hybridization, the histological activity index, and the serum viremia levels in chronically HCV-infected patients with different grades of liver injury in an attempt to reveal any possible correlation or interdependence between them.
Patients and Methods
Paraffin-embedded liver biopsies from 27 anti-HCV-positive and serum HCV-RNA-positive patients (17 male, 10 female) with histologically proven chronic hepatitis and abnormal alanine aminotransferase (ALT) levels (mean, 174.88 ± 124.88 IU/L; range, 41–538 IU/L) were analyzed by in situ hybridization. None of the patients had hepatitis B surface antigen (HBsAg), anti-hepatitis D virus antibodies (anti-HDV), or anti-hepatitis A virus IgM antibodies (IgM anti-HAV). The histological diagnoses of the patients were minimal chronic hepatitis with no fibrosis in 1 patient, mild chronic hepatitis in 21 patients (2 with no fibrosis, 6 with mild fibrosis, 6 with moderate fibrosis, 7 with severe fibrosis) and moderate chronic hepatitis in 5 patients (1 with moderate fibrosis, 2 with severe fibrosis, 2 with cirrhosis).
None of the patients was receiving antiviral or immunosuppressive therapy when the liver biopsy was obtained. A serum sample obtained at the same time as the liver biopsy and properly stored at −20°C was available from each patient.
Viral Markers in Serum
HbsAg, anti-HDV, and IgM anti-HAV were tested by commercial enzyme immunoassays (Abbott Laboratories, North Chicago, IL). Anti-HCV was tested by a third-generation enzyme-linked immunosorbent assay test (Ortho Diagnostic Systems, Raritan, NJ) and confirmed by third-generation recombinant immunoblotting assay (RIBA III, Ortho).
HCV-RNA presence and levels were determined by using the Amplicor Monitor Test Kit (HCV Monitor quantitative assay, Roche Diagnostic Systems, Basel, Switzerland) following the instructions supplied by the manufacturer.
HCV-RNA Genotyping
HCV-RNA genotyping in the patients was performed as described by Navas et al. 15
In Situ Hybridization
Paraffin-embedded liver sections of the patients were in situ hybridized using a 340-bp cDNA corresponding to the 5′ noncoding region of the HCV genome as probe.
The in situ hybridization technique and the specific controls used were performed as described by Gosálvez et al. 14 In the in situ hybridization experiments, slides from two patients with chronic autoimmune hepatitis, two patients with chronic hepatitis B, and six patients with chronic alcoholic hepatitis, all of them without HCV markers, were used as negative controls.
Image capture of in situ hybridized liver biopsies were performed under a Leika DMB microscope. Images were acquired using a CCD camera (DIC-N, World Precision Instruments, Cambridge, UK) and for quantification of the infection low magnification (×20) objectives were used. Single frames of each biopsy were used to construct a whole liver biopsy using the Adobe Photoshop 4.0 software (Adobe Systems, San Jose, CA). The areas of infection visualized with in situ hybridization were localized by automatic segmentation protocols using VISILOG 5.0 Image analysis software (NOESIS Vision, Inc., Quebec, PQ). The percentage of infection in each patient was derived from the same analyzed surface.
Histological Diagnosis
Two consecutive slides from each liver biopsy were stained with hematoxylin-eosin for histological diagnosis. The histological activity index was estimated according to Knodell et al 16 and the histological diagnosis made according to Desmet et al. 17 The histological diagnoses were performed under code by two independent pathologists.
Liver Function Test
Liver function tests were analyzed by standard methods (SMAC-20, Technicon, New York, NY).
Statistical Analysis
The Spearman correlation was used for correlations. All statistical analyses were performed using the SPSS package (release 6.0 for Windows, SPSS, Chicago, IL).
Results
HCV-RNA in Serum
Genotyping of the HCV RNA isolated from serum and amplified by reverse transcription-polymerase chain reaction revealed that all patients were infected by the HCV 1b genotype.
With respect to serum viremia levels, the HCV RNA concentration in the patients included in the study ranged from 1.8 × 10 3 to 1.4 × 10 6 genome copies/ml (Table 1) ▶ .
Table 1.
Percentage of HCV-Infected Hepatocytes, Serum Viremia Levels, and Histological Activity Index (HAI) in the Patients Included in the Study
| Patient no. | % Infected hepatocytes | Serum viral load (genome copies/ml) | HAI* |
|---|---|---|---|
| 1 | 0.04% | 5 × 103 | (3,2,1,3) |
| 2 | 0.52% | 3.2 × 104 | (3,1,2,2) |
| 3 | 0.60% | 1 × 104 | (1,2,1,0) |
| 4 | 1% | 1.4 × 104 | (3,3,3,3) |
| 5 | 1.12% | 5 × 104 | (3,2,2,2) |
| 6 | 1.16% | 4.7 × 103 | (4,3,1,2) |
| 7 | 1.88% | 1.8 × 104 | (3,2,2,3) |
| 8 | 2.6% | 8.3 × 105 | (3,3,2,4) |
| 9 | 2.64% | 1.3 × 105 | (3,1,3,2) |
| 10 | 3.16% | 1.8 × 104 | (2,1,1,1) |
| 11 | 3.16% | 8.9 × 104 | (2,0,1,0) |
| 12 | 3.44% | 1.7 × 104 | (2,1,2,1) |
| 13 | 4.24% | 2.3 × 105 | (3,1,2,3) |
| 14 | 4.48% | 3.1 × 105 | (3,1,1,1) |
| 15 | 4.6% | 3 × 105 | (3,3,2,4) |
| 16 | 4.92% | 9.6 × 104 | (4,2,2,3) |
| 17 | 5.32% | 9.1 × 104 | (3,2,1,3) |
| 18 | 5.32% | 3.3 × 104 | (4,1,2,3) |
| 19 | 7.08% | 3 × 105 | (2,1,1,2) |
| 20 | 9.20% | 2.5 × 105 | (3,1,2,2) |
| 21 | 10.08% | 1.8 × 105 | (3,2,2,3) |
| 22 | 11.32% | 1 × 105 | (4,2,1,1) |
| 23 | 11.52% | 1.8 × 103 | (2,1,1,0) |
| 24 | 20.6% | 2.5 × 105 | (1,1,2,1) |
| 25 | 22.64% | 8 × 104 | (4,1,2,1) |
| 26 | 41.60% | 1.2 × 106 | (3,4,2,3) |
| 27 | 83.6% | 1.4 × 106 | (4,3,3,2) |
*Numbers refer to periportal necrosis, intralobular degeneration, portal inflammation, and fibrosis.
No statistical correlation was found between serum viremia levels and the total histological activity index (r = 0.25, P = 0.19) or its individual components: periportal necrosis (r = 0.14, P = 0.47), intralobular degeneration (r = 0.12, P = 0.52), portal inflammation (r = 0.31, P = 0.11), and fibrosis (r = 0.28, P = 0.15). Furthermore, no correlation was observed between the serum HCV-RNA concentration and the biochemical liver function test, ALT (r = 0.13, P = 0.48), aspartate aminotransferase-AST (r = 0.22, P = 0.26), and γ glutamiltranspeptidase-GGTP (r = 0.34, P = 0.08).
In Situ Hybridization
In situ hybridization using the HCV-derived probe showed positive signals in the hepatocytes from the liver biopsies of all anti-HCV-positive, serum HCV-RNA-positive patients analyzed. Positive hepatocytes were randomly distributed throughout the liver biopsies with no clustering in any specific part of the liver samples.
No hybridization signals were observed in other cell types within the liver (Figure 1) ▶ . Furthermore, no positive signals were detected in the liver biopsies from the 10 anti-HCV-negative patients used as negative controls (Figure 1) ▶ .
Figure 1.
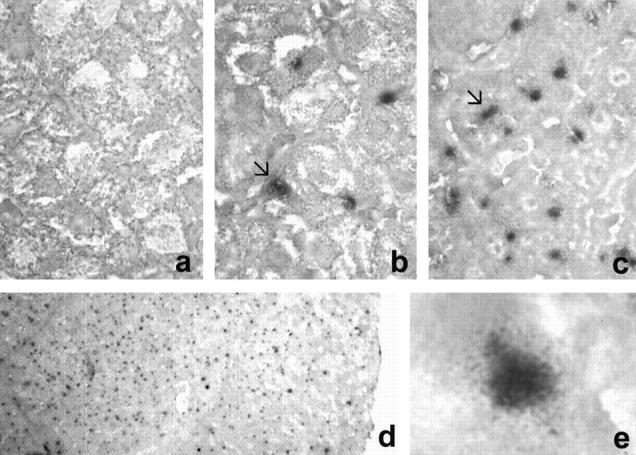
In situ hybridization and mapping of HCV virus in different patients. a: negative control with no traces of infected hepatocytes. Distribution of infected cells (arrow) in patients with 1.8 × 10 4 genome copies/ml (b) and with 1.2 × 10 6 genome copies/ml (c) in serum. d: General view of a single frame capture to show the distribution of the infection. e: Visualization of the infection in a single hepatocyte.
Regarding the localization of the hybridization signals, there were mainly detected in the cytoplasm of the hepatocytes, although nuclear staining was also observed in scattered hepatocytes.
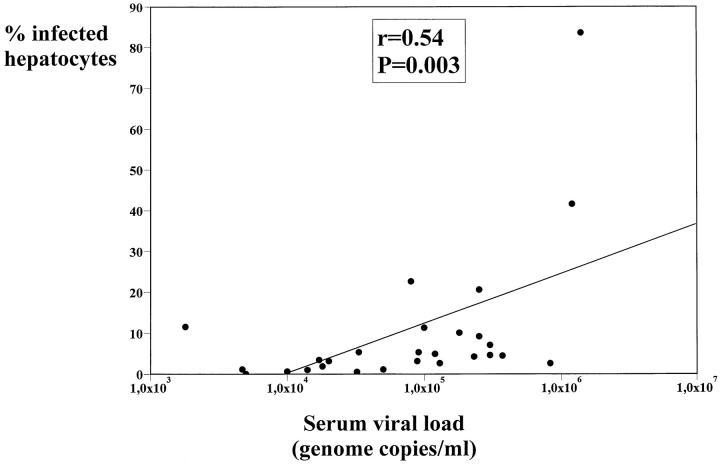
The percentage of infected hepatocytes ranged from 0.04% to 83.6% (Table 1) ▶ . A statistically significant correlation (r = 0.54; P = 0.003) between the percentage of infected hepatocytes and the serum viremia levels was found (Figure 2) ▶ .
Figure 2.
Regression analysis of correlation between the percentage of infected hepatocytes and the serum viral load (genome copies/ml).
In contrast, no correlation was observed between the percentage of HCV-infected hepatocytes and the histological activity index (r = 0.08; P = 0.67) or its individual components: periportal necrosis (r = 0.17, P = 0.38), intralobular degeneration (r = −0.06, P = 0.75), portal inflammation (r = 0.09, P = 0.62), and fibrosis (r = 0.116, P = 0.56). Figure 3 ▶ shows an example of two patients with different percentages of infected hepatocytes and serum viremia levels but with similar histological features. Furthermore, there is no correlation between the percentage of infected hepatocytes and the ALT (r = −0.06, P = 0.72), AST (r = −0.03, P = 0.85), and GGTP (r = 0.068, P = 0.73) levels.
Figure 3.
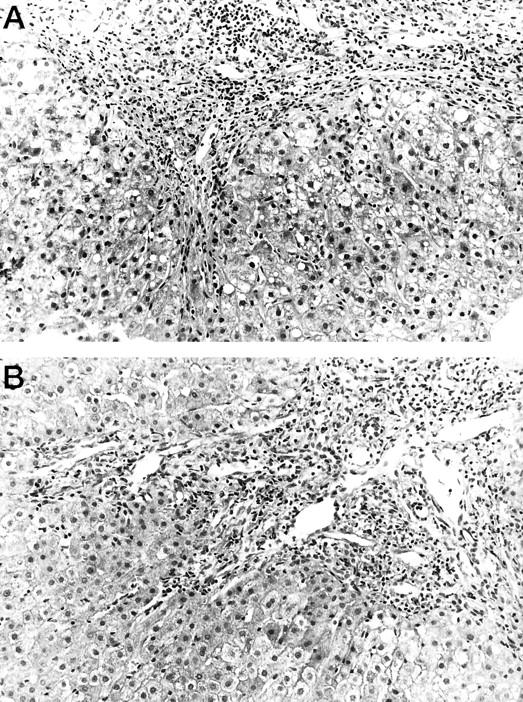
Fibrous and inflammatory portal expansion in two cases of chronic hepatitis C with very similar histological picture and different percentage of infected hepatocytes and viral load in serum. The patient in A had 1.8% infected hepatocytes and a viral load in serum of 1.8 × 10 4 copies genome/ml in serum. The patient in B had 41.6% infected hepatocytes and a viral load in serum of 1.2 × 10 6 genome copies/ml. These two patients’ in situ hybridization results are shown in Figure 1, b and c ▶ , respectively.
Discussion
The liver pathogenesis of chronic HCV infection remains unclear. It has been attributed to either a direct cytopathic effect of the virus 1-3 or to the host immune response against infected hepatocytes. 4,5 Several authors have tried to correlate the viral load in the serum or liver with the degree of liver injury, with conflicting results. 6-12 Furthermore, whether the viral load in the liver correlates with the serum viremia levels is still in controversy. 10,13 In the present study we have used in situ hybridization to determine the number of HCV-infected hepatocytes and we have correlated the results with the individual components of the histological activity index and with the serum viremia levels in patients with chronic HCV hepatitis.
We found that the serum viremia levels correlate with the percentage of HCV-infected hepatocytes in patients whose liver damage ranged from minimal hepatitis to liver cirrhosis. This finding confirms that the serum HCV viremia reflects the extent of HCV infection in the liver, 14 and shows that this correlation does not depend on the stage or grade of the liver injury. However, it should be noted that the correlation between percentage of labeled hepatocytes and serum viremia is not very strong. If the three patients with a high level of viremia are removed from the analysis, then the difference in percentage of labeled hepatocytes between high and low viremia is only about twofold (4–8%), whereas viremia varies tenfold.
The correlation between viremia and percentage of infected hepatocytes also suggests that HCV replication in extrahepatic tissues, such as peripheral blood mononuclear cells 18 or bone marrow cells, 19 does not significantly contribute to the HCV RNA levels in serum.
On the other hand, we did not find any relation between the number of HCV-infected hepatocytes and any of the parameters of the histological activity index (periportal necrosis, intralobular degeneration, portal inflammation, and fibrosis) described by Knodell et al. 16 Furthermore, no correlation between the number of HCV-infected hepatocytes and the liver function test (ALT, AST, and GGTP) was observed. Considered as a whole, all these data suggest that HCV infection of the hepatocytes does not per se contribute significantly to the liver injury.
Our findings agree with previous reports in which the intrahepatic HCV RNA concentration measured by different methods (dot blot polymerase chain reaction or bDNA assay) was correlated with the liver injury. 11,12 However, a problem exists when methods that depend on the extraction of the total RNA from the liver are used, because serum contamination lead to an inadequate measurement of intrahepatic HCV-RNA levels. In contrast, in situ hybridization provides a direct measurement of the number of infected hepatocytes and avoids this problem.
On the other hand, when the serum HCV RNA levels were correlated with the components of the histological activity index and with the liver enzyme levels, no correlation was found.
The data on the detection of HCV RNA in liver and in serum could imply that the liver injury in the chronic HCV infection is not directly related with the levels of viral replication and may be mediated by the host immune response against HCV-infected hepatocytes. The fact that there are HCV chronic carriers with normal or nearly normal liver histology 20 supports this hypothesis.
It should be pointed out that all of the patients included in this study were infected with the HCV 1b genotype and that the mechanism of liver injury induced by other HCV genotypes could be different. However, it has been shown that there are no differences in histological damage between patients infected by different HCV genotypes, 11,21 indicating that the same mechanism of liver damage is provoked by the different HCV genotypes.
In conclusion, in this report we have shown that the serum HCV viremia reflects the extent of the infection in the liver and that the liver injury in chronic HCV infection is not directly related to the number of infected hepatocytes or to the HCV RNA concentration in serum.
Acknowledgments
We thank Mercedes Casqueiro, Teresa Cuellar, Jose Luis Ramiro, and Mario Fernández for their cooperation in this work.
Footnotes
Address reprint requests to Dr. Vicente Carreño, Department of Hepatology, Fundación Jiménez Díaz, Avda Reyes Católicos 2, 28040, Madrid, Spain.
E. R. I. is recipient of a grant from the Fundación para el Estudio de las Hepatitis Virales. S. L. is a research fellow of the Fundación Conchita Rábago.
References
- 1.Shindo M, Di Bisceglie AM, Biswas R, Mihalik K, Feinstone SM: Hepatitis C virus replication during acute infection in the chimpanzee. J Infect Dis 1992, 166:424-427 [DOI] [PubMed] [Google Scholar]
- 2.Kato N, Yokosuka O, Hosoda K, Ito Y, Onto M, Omata M: Quantification of hepatitis C virus by competitive reverse transcription-polymerase chain reaction: increase of the virus in advanced liver disease. Hepatology 1993, 18:16-20 [PubMed] [Google Scholar]
- 3.Yuki N, Hayashi N, Kamada T: HCV viraemia and liver injury in symptom-free blood donors [letter]. Lancet 1993, 342:444. [DOI] [PubMed] [Google Scholar]
- 4.Hu KQ, Yu CH, Vierling JM: Direct detection of circulating hepatitis C virus RNA using probes from the 5′ untranslated region. J Clin Invest 1992, 89:2040-2045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Di Bisceglie AM, Hoofnagle JH, Krawczynski K: Changes in hepatitis C virus antigen in liver with antiviral therapy. Gastroenterology 1993, 105:858-862 [DOI] [PubMed] [Google Scholar]
- 6.Lau JY, Davis GL, Kniffen J, Quian KP, Urdea MS, Chan CS, Mizokani M, Neuwald PD, Wilber JC: Significance of serum hepatitis C virus RNA levels in chronic hepatitis C. Lancet 1993, 341:1501-1504 [DOI] [PubMed] [Google Scholar]
- 7.Hagiwara H, Hayashi N, Mita E, Naito M, Kasahara A, Fusamoto H, Kamada T: Quantitation of hepatitis C virus RNA in serum of asymptomatic blood donors and patients with type C chronic liver disease. Hepatology 1993, 17:545-550 [DOI] [PubMed] [Google Scholar]
- 8.Naito M, Hayashi N, Hagiwara H, Hiramatsu N, Kashahara A, Fusamoto H, Kamada T: Serum hepatitis C virus RNA quantity and histological features of hepatitis C virus carriers with persistently normal ALT levels. Hepatology 1994, 19:871-875 [PubMed] [Google Scholar]
- 9.Gretch D, Corey L, Wilson J, dela Rosa C, Willson R, Carithers R, Jr, Busch M, Hart J, Sayers M, Han J: Assessment of hepatitis C virus RNA levels by quantitative competitive RNA polymerase chain reaction: high-titer viremia correlates with advanced stage of disease. J Infect Dis 1994, 169:1219-1225 [DOI] [PubMed] [Google Scholar]
- 10.Coelho-Little E, Jeffers LJ, Bartholomew M, Reddy KR, Schiff ER, Dailey PJ: Correlation of HCV-RNA levels in serum and liver of patients with chronic hepatitis C [letter]. J Hepatol 1995, 22:248. [DOI] [PubMed] [Google Scholar]
- 11.McGuinness PH, Bishop GA, Painter DM, Chan R, McCaughan GW: Intrahepatic hepatitis C RNA levels do not correlate with degree of liver injury in patients with chronic hepatitis C. Hepatology 1996, 23:676-687 [DOI] [PubMed] [Google Scholar]
- 12.De Moliner L, Pontisso P, De Salvo GL, Cavaletto L, Chemello L, Alberti A: Serum and liver HCV-RNA levels in patients with chronic hepatitis C: correlation with clinical and histological features. Gut 1998, 42:856-860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Terrault NA, Dailey PJ, Ferrell L, Collins ML, Wilber JC, Urdea MS, Bhandari BN, Wright TL: Hepatitis C virus: quantitation and distribution in liver. J Med Virol 1997, 51:217-224 [PubMed] [Google Scholar]
- 14.Gosálvez J, Rodríguez-Iñigo E, Ramiro-Díaz JL, Bartolomé J, Tomás JF, Oliva H, Carreño V: Relative quantification and mapping of hepatitis C virus by in situ hybridization and digital image analysis. Hepatology 1998, 27:1428-1434 [DOI] [PubMed] [Google Scholar]
- 15.Navas S, Castillo I, Martín J, Quiroga JA, Bartolomé J, Carreño V: Concordance of hepatitis C virus typing methods based on restriction fragments length polymorphism analysis in 5′ non-coding region and NS4 serotyping, but not in core PCR or in a line probe assay. J Clin Microbiol 1997, 21:317-321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Knodell RG, Ishak KG, Black WL, Chen TS, Craig R, Kaplowitz N, Kiernan TW, Wollman J: Formulation and application of a numerical scoring system for assessing histological activity in asymptomatic chronic active hepatitis. Hepatology 1981, 1:431-435 [DOI] [PubMed] [Google Scholar]
- 17.Desmet VJ, Gerber M, Hoofnagle JH, Manns M, Scheuer PJ: Classification of chronic hepatitis: diagnosis, grading and staging. Hepatology 1994, 19:1513-1520 [PubMed] [Google Scholar]
- 18.Moldvay J, Deny P, Pol S, Brechot C, Lamas E: Detection of hepatitis C virus RNA in peripheral blood mononuclear cells of infected patients by in situ hybridization. Blood 1994, 83:269-273 [PubMed] [Google Scholar]
- 19.Sansonno D, Iacobeli AR, Cornacchiulo V, Iodice G, Dammacco F: Detection of hepatitis C virus (HCV) proteins by immunofluorescence and HCV RNA genomic sequences by non-isotopic in situ hybridization in bone marrow and peripheral blood mononuclear cells of chronically HCV-infected patients. Clin Exp Immunol 1996, 103:414-421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hoofnagle JH: Hepatitis C: The clinical spectrum of disease. Hepatology 1997, 26(Suppl):15S-20S [DOI] [PubMed] [Google Scholar]
- 21.Yamada M, Kakumu S, Yoshioka K, Higashi Y, Tanaka K, Ishikawa T, Takayanagi M: Hepatitis C virus genotypes are not responsible for development of serious liver disease. Dig Dis Sci 1994, 39:234-239 [DOI] [PubMed] [Google Scholar]