Abstract
We assessed the frequency of muscle fibers showing histochemical phosphorylase activity in 27 muscle biopsies from 25 unrelated patients with McArdle’s disease and studied by immunohistochemistry and in situ hybridization whether the muscle-specific isoform was expressed. Positive phosphorylase fibers were observed in 19% of our series of biopsies. We show that the enzyme isoform expressed in regenerating fibers differs according to the genotype of patients: the muscle-specific isoform is transcribed and translated in patients with none of the described mutations in at least one allele of the myophosphorylase gene, whereas it is neither transcribed nor translated in patients with identified mutations in both alleles.
McArdle’s disease (myophosphorylase deficiency) is the most common glycogenosis affecting skeletal muscle. 1 The biochemical hallmark of the disease is impaired muscle glycogenolysis due to lack of muscle glycogen phosphorylase (M-GP) activity. M-GP is the muscle-specific isoform of the enzyme, with two others being the liver- and the brain-specific forms. The three isoforms are coded by separate genes, 2 and their expression is developmentally regulated. 3
The molecular basis of McArdle’s disease has been linked to several mutations, either nonsense or missense, in the PYGM gene, which codes for M-GP. 4,5 The most common mutation is the nonsense mutation at codon 49 (R49X), accounting for up to 60% of the alleles. Many patients, however, defy even an extensive search for mutations. 6,7
Muscle biopsies of McArdle’s patients are characterized by subsarcolemmal storage of normal glycogen and by absent stain with the histochemical phosphorylase reaction. In some patients, however, it is possible to find scattered fibers with positive phosphorylase staining. 8,9 The identity of the glycogen phosphorylase isoform expressed in those phosphorylase-positive fibers has been a matter of debate. After the initial report describing the reappearance of phosphorylase activity in myophosphorylase-deficient patients, 8 subsequent studies supported the assumption that those fibers, similarly to cultured immature myotubes, express non-muscle-specific (ie, fetal) isoforms. 10,11 Later studies on cultured muscle in which differentiation was more advanced due to different culture techniques or innervation showed that M-GP was indeed expressed, albeit in small proportion, in both control and patients’ myotubes in vitro. 12,13
In this paper we assessed the frequency of phosphorylase-positive fibers in a consecutive series of 27 muscle biopsies of 25 McArdle’s patients. We found phosphorylase-positive fibers in 19% of our muscle biopsies. Virtually all positive fibers are regenerating fibers. We show that the isoform expressed in phosphorylase-positive fibers differs according to the genetic background of the patient: in patients with no detected mutation in at least one allele of the PYGM gene, the muscle-specific isoform is transcribed and translated, whereas it is neither transcribed nor translated in patients with mutations identified in both alleles of the gene.
Materials and Methods
Patients
A total of 27 muscle biopsies of 25 patients with Mc-Ardle’s disease were screened for scattered phosphorylase positivity. The biopsies showing at least one positive fiber were further studied by immunohistochemistry and in situ hybridization.
All patients presented clinical and laboratory findings typical of McArdle’s disease, with muscle glycogen storage and severely reduced glycogen phosphorylase activity. Clinical, biochemical, and molecular data for patients whose biopsy showed phosphorylse positive fibers are summarized in Table 1 ▶ .
Table 1.
Clinical, Biochemical, and Molecular Summary of Patients Showing Fibers with Histochemical Phosphorylase Activity in Muscle Biopsy
| Patient | Age at biopsy | Serum CK (U/l) | Myoglo- binuria | Biopsied muscle | GP activity | GP protein | M-GP mRNA | Mutation | HC+ fibers (%) |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 28 | 4000 | Yes | Triceps | 0 | No | + | − /49 | 5.2 |
| 2 | 69 | 740 | No | Quad. | 1% | No | + | − /49 | 0.3 |
| 3 | 59 | 200 | No | Biceps | 3% | No | ++ | − /396 | 0.2 |
| 4 | 51 | 3100 | Yes | Quad. | 0 | No | + | 49 /204 | 6.9 |
| 5 | 17 | 2200 | Yes | Triceps | 0 | No | − | 49 /49 | 0.1 |
Glycogen phosphorylase activity (assessed by spectrophotometric assay on muscle homogenate) is expressed as % of normal control. Number of fibers showing positivity at the phosphorylase histochemical stain (HC+) is expressed as % of total number of fibers in the biopsy slice.
Cell Culture
Cultured myoblasts were expanded from muscle explants obtained from patient and control biopsies as previously described. 13 Myoblasts were allowed to fuse in myotubes, and selected dishes were innervated with rat fetal spinal cord explants as described. 13 Aneural and innervated cultured muscle from patients and control were harvested, and the total RNA was extracted as described below.
Molecular Characterization
Total RNA and DNA were extracted from muscle biopsies or cultured muscle with the TRI-zol reagent according to the manufacturer’s instructions (Gibco BRL, Gaithersburg, MD). Northern analysis for M-GP mRNA was carried out as described. 13 The following mutations known to be associated with McArdle’s disease were searched in all patients with described methods 4,5 : R49X, G204S, L291P, L396P, K542T, E654K, del708, 1778del67 (splice junction mutation at the 5′ end of intron 14), delG510, A to G transition in the translation initiation codon, frameshift mutation in exon 1, R575X, G685R, and delA753.
In addition, in those patients in whom none of the above mutations could be detected, we directly sequenced the entire PYGM cDNA.
Histochemistry (HC) and Immunohistochemistry (IHC)
Serial cryostat sections (6 μm thick) were placed on slices pretreated with 0.25% gelatin, 0.025% chrome alum (Merck, Darmstadt, Germany), and processed separately. One section was used for histochemical phosphorylase stain. As marker of muscle regeneration, we used a monoclonal antibody against fetal myosin heavy chain (Novocastra, Newcastle, UK), diluted 1:100 in PBS and incubated for 1 hour at room temperature. The expression of M-GP was studied using sheep anti-M-GP antiserum. 13 To block nonspecific staining, sections were incubated with 5% bovine serum albumin (BSA) in PBS for 1 hour. Anti-M-GP antiserum diluted 1:25 in 1% BSA/PBS was incubated overnight at 4°C and for one additional hour at room temperature on a shaker. The specific labeling was developed by an immunofluorescence method using appropriate biotinylated antibodies (Amersham, Little Chalfont, UK) for 30 minutes at 1:200 dilution and streptavidin-Texas red (Amersham) for 30 minutes at 1:100 dilution.
M-GP Ribonucleic Probes
A 232-bp stretch of the 3′ end of the M-GP cDNA (bp 3243 to 3475) in which the divergence among muscle- and non-muscle-specific isoforms was maximal was subcloned in the SmaI/ApaI site of Bluescript KS+. Orientation was checked by direct sequencing. RNA probes were synthesized using [35S]UTP (1300 Ci/mmol) using T3 (antisense) and T4 (sense) polymerase following the supplier’s instructions.
In Situ Hybridization (ISH)
ISH was performed as described 14 on 10-μm cryostat muscle sections. Transverse sections were fixed 30 minutes at room temperature in 4% paraformaldehyde, treated for 7.5 minutes with proteinase K (20 μg/ml) in Tris/HCl (50 mmol/L), pH 7.2, and EDTA (5 mmol/L), acetylated in 0.5% acetic anhydride in 0.1 mol/L triethanolamine, pH 8.0, for 10 minutes, and dehydrated. The probe (10 8 cpm/ml) was applied directly to dried sections in hybridization buffer (50% deionized formamide, 0.3 mol/L NaCl, 20 mmol/L Tris/HCl, pH 7.4, 5 mmol/L EDTA, 10 mmol/L NaPO4, pH 8.0, 10% dextran sulfate, 1X Denhardt’s, 50 μg/ml yeast RNA). Hybridization was carried out for 16 hours at 52°C in a humid chamber. The slides were washed 30 minutes at 42°C in 5X SSC (1X SSC contains 0.15 mol/L NaCl, 0.015 mol/L sodium citrate), 10 mmol/L dithiothreitol (DTT); 20 minutes at 65°C in 50% formamide, 2X SSC, 10 mmol/L DTT; 30 minutes at 37°C in 0.5 mol/L NaCl, 0.05 mmol/L NaP04, pH 7.0, 20 μg/ml RNAse; 15 minutes at 37°C in 1X SSC; and 15 minutes at 37°C in 0.1X SSC. The slices were rapidly dehydrated, processed for autoradiography using Kodak NBT-2 emulsion, exposed for 10 days at 4°C, and developed according to the manufacturer’s directions.
Results
Five biopsies from five unrelated patients showed scattered stained fibers after phosphorylase HC. In three biopsies, stained fibers were extremely rare (one to seven per slice), whereas in the other two, the stained fibers accounted for 5% to 7% of the total. Recent myoglobinuria was reported for the patients showing a high number of stained fibers but not in the others. All five patients had typical McArdle’s disease and were comparable for age, duration of symptoms, and severity to the other 21 patients with no phosphorylase HC-positive fibers. Phosphorylase activity by spectrophotometric assay was below 3% of normal control in two and absent in three. Immunoblot with sheep anti-M-GP antiserum 13 was negative in all. Molecular characterization of the patients showed normal M-GP mRNA in patient 3, reduced transcript of normal size in patients 1, 2, and 4, and no mRNA in patient 5. Genotype of the patients was as follows: one allele mutated in patients 1, 2 (R49X), and 3 (L396P), R49X/G204S in patient 4, and R49X/R49X in patient 5 (Table 1) ▶ .
There was complete overlapping between fibers positive with HC for phosphorylase and fibers positive with IHC for fetal myosin (Figure 1) ▶ , confirming their regenerating status. In one case (patient 3), however, we observed four faintly stained, normal-sized fibers, which were IHC fetal myosin negative.
Figure 1.
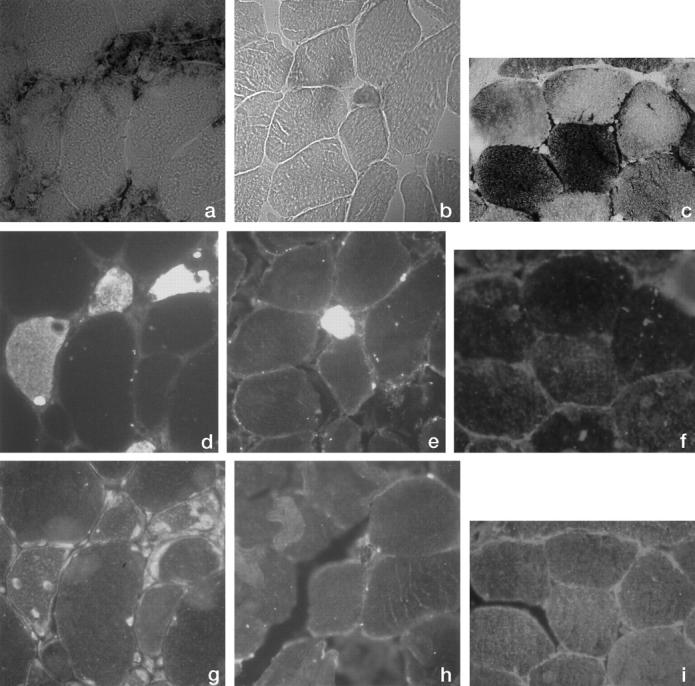
Photomicrographs (×100) of serial sections from muscle biopsy of patient 1 (a, d, and g), patient 5 (b, e, and h), and normal control (c, f, and i). a to c: Histochemistry for glycogen phosphorylase; d to f: Immunohistochemistry for fetal myosin; g to i: Immunohistochemistry for muscle-specific glycogen phosphorylase.
Regenerating phosphorylase-positive fibers reacted specifically with the anti-M-GP antiserum in patients 1 and 2 (Figure 1g) ▶ , indicating in these patients localized re-expression of the defective enzyme.
In situ hybridization with the M-GP riboprobe showed diffuse subsarcolemmal positivity in control, scattered patchy positivity in the patients with IHC M-GP-positive fibers, and no specific signal in the patients with regenerating IHC M-GP-negative fibers (Figure 2 ▶ ; Table 2 ▶ ). M-GP mRNA accumulation therefore positively correlates with M-GP expression.
Figure 2.
Photomicrographs (×25) of sections from muscle biopsy of patient 1 (a), patient 5 (b), and normal control (c) after in situ hybridization with myophosphorylase-specific riboprobe.
Table 2.
Results of Phosphorylase HC, IHC with Anti-Fetal Myosin Antibodies, IHC with Anti-Myophosphorylase Antiserum, and in Situ Hybridization with M-GP-Specific Riboprobe in 5 Biopsies with HC-Positive Fibers
| Patient No. | Phosphorylase HC | Fetal myosin IHC | M-GP IHC | M-GP ISH |
|---|---|---|---|---|
| 1 | +++ | +++ | ++ | ++ |
| 2 | + | + | + | + |
| 3 | + | − | − | − |
| 4 | +++ | +++ | − | − |
| 5 | + | + | − | − |
M-GP cDNA obtained by reverse transcription polymerase chain reaction of total RNA purified from control and patients’ cultured muscle was studied by restriction fragment length polymorphism and direct sequencing. There was complete identity between M-GP mRNA sequence of the adult and the cultured muscle of any given patient or control.
Discussion
In this work we show that in a large series of patients with biochemically defined myophosphorylase deficiency, histochemical phosphorylase activity is detected in less than 20% of cases, and it is limited to fetal-myosin-positive fibers.
The relatively low frequency of patients with HC-positive fibers in our series is in agreement with previous reports, 6,9 in which only single or very few patients were presented. Recent myonecrosis, as suggested in our cases by high serum creatine kinase and reported myoglobinuria, correlates with the presence of regenerating phosphorylase-positive fibers.
Phosphorylase expression is mostly restricted to regenerating fibers, as was shown by morphological criteria in previous studies. 8,9 However, few normal-sized fibers that were fetal myosin negative showed faint phosphorylase positivity in one patient. In those fibers, the regeneration process could be just completed, and consequently, the expression of fetal myosin and possibly M-GP could have been already shut down.
The results of IHC and ISH indicate that regenerating fibers in patients in whom at least one allele of the PYGM gene does not carry an identified mutation express M-GP. This finding is in agreement with previous in vitro studies showing re-expression of the defective enzyme in aneural 12 and innervated 13 cultured muscle of patients with McArdle’s disease. In aneural and innervated cultured muscle from myophosphorylase-deficient patients, the deleterious mutations identified in the adult muscle were retained; therefore, we can exclude gene reversion as the explanation for the observed phenomenon. The similarity between in vitro and in vivo behavior of regenerating muscle cells from myophosphorylase-deficient patients suggests that the same conclusion could be applied to our in vivo observations.
The present work provides the first evidence that re-expression of M-GP can occur also in vivo, and correlates this finding with the molecular basis of the defect. Patients with mutation identified in both alleles, and with absent or grossly reduced M-GP mRNA, did not express M-GP, despite the presence of regenerating fibers (almost 7% in patient 4).
IHC and ISH could identify in patients 1 and 2 M-GP as the GP isoform expressed. The same was not possible for the other patients. From what is known of GP gene regulation we can hypothesize that the observed GP activity in IHC M-GP-negative fibers is due to a fetal isoform. In vivo regenerating rat muscle grafts do not express non-M-GP, in contrast to primary rat skeletal muscle explants in vitro. 15 In vivo regenerating muscle from non-myophosphorylase-deficient patients (eg, Duchenne muscular dystrophy patients) shows intense staining for M-GP IHC in fetal-myosin-positive fibers (data not shown), suggesting that M-GP is the prevalent isoform expressed in regenerating human muscle in vivo.
The mechanism leading to the activation of M-GP expression in regenerating fibers and to its suppression in mature fibers is not known. Possible explanations are: 1) activation in regenerating cells of a PYGM pseudogene normally silent, 2) mRNA editing with correction, in regenerating fibers, of deleterious mutations, as described for transcripts with GAGAG motifs in neurons, 16 and 3) use of an alternative promoter or enhancer in poorly differentiated muscle fibers, leading to reversion in the dividing cells of a transcriptional or translational block preventing expression in mature myotubes.
The striking sequence homology among the genes coding for the three GP isozymes suggests that they have evolved through duplication and translocation events, 17 although no PYGM-related sequences have been spotted in mapping studies covering the whole genome. 18 Several GAGAG islands are present in the second half of the coding sequence of the PYGM gene, although none is the site of a known mutation, 4,5 and none were found to be mutated in our patients after direct sequencing. There are conflicting reports on the presence in rat and human muscle of alternative spliced M-GP transcripts 15,19 ; alternative splicing and transcript processing is a common feature in muscle-specific isoforms of glycolytic enzymes such as aldolase A, phosphofructokinase, and phosphorylase kinase-α. 20-22 This last hypothesis would imply that in some patients with McArdle’s disease the defect is due to a reversible impairment of PYGM transcription or translation. If confirmed, this mechanism could be exploited for a novel form of gene therapy, which would take advantage of the self-healing potential of the diseased gene itself.
Acknowledgments
We thank Prof. S. Schiaffino for his valuable advice and Prof. C. Angelini for providing access to his very rich muscle tissue bank.
Footnotes
Address reprint requests to Dr. Andrea Martinuzzi, Scientific Institute “Eugenio Medea”, Conegliano Research Center, via Costa Alta 37, 31015 Conegliano, Italy. E-mail: andree.martinuzzi@cn.luf.it.
Supported by Telethon of Italy grant 855 to A. Martinuzzi.
References
- 1.Di Mauro S, Tsujino S: Nonlysosomal glycogenoses. ed 2 Engel AG Banker BQ eds. Myology, 1994, :pp 1554-1576 McGraw-Hill, New York [Google Scholar]
- 2.Newgard CB, Littman DR, van Genderen C, Smith M, Fletterick RJ: Human brain glycogen phosphorylase: cloning, sequence analysis, chromosomal mapping, tissue expression and comparison with the human liver and muscle isozymes. J Biol Chem 1988, 83:8132-8136 [PubMed] [Google Scholar]
- 3.Newgard CB, Norkiewicz B, Hughes SD, Frenkel RA, Coats WS, Martiniuk F, Johnston JM: Developmental expression of glycogenolytic enzymes in rabbit tissues: possible relationship to fetal lung maturation. Biochim Biophys Acta 1991, 1090:333-342 [DOI] [PubMed] [Google Scholar]
- 4.Tsujino S, Shanske S, Nonaka I, DiMauro S: The molecular genetic basis of myophosphorylase deficiency (McArdle’s disease). Muscle Nerve 1995, 18(suppl 3):S23-S27 [DOI] [PubMed] [Google Scholar]
- 5.Vorgerd M, Kubisch C, Burwinkel B, Reichmann H, Mortier W, Tottenborn B, Pongratz D, Lindemuth R, Tagenthoff M, Malin JP, Kilimann MW: Mutation analysis in myophosphorylase deficiency (McArdle’s disease). Ann Neurol 1998, 43:326-331 [DOI] [PubMed] [Google Scholar]
- 6.Martinuzzi A, Tsujino S, Vergani L, Schievano G, Cadaldini M, Bartoloni L, Fanin M, Siciliano G, Shanske S, DiMauro S, Angelini C: Molecular characterization of myophosphorylase deficiency in a group of patients from Northern Italy. J Neurol Sci 1995, 137:14-19 [DOI] [PubMed] [Google Scholar]
- 7.Andreu AL, Bruno C, Gamez J, Shanske S, Cervera C, Navarro C, Arbos MA, Tamburino L, Schwartz S, DiMauro S: Molecular genetic analysis of McArdle’s disease in Spanish patients. Neurology 1998, 51:260-262 [DOI] [PubMed] [Google Scholar]
- 8.Roelofs RI, Engel WK, Chauvin PB: Histochemical demonstration of phosphorylase activity in regenerating skeletal muscle fibres from myophosphorylase deficient patients. Science 1972, 177:795-797 [DOI] [PubMed] [Google Scholar]
- 9.Mitsumoto H: McArdle disease: phosphorylase activity in regenerating muscle fibers. Neurology 1979, 29:258-262 [DOI] [PubMed] [Google Scholar]
- 10.Sato K, Imai I, Hatayama I, Roelofs RI: Characterization of glycogen phosphorylase isoenzymes present in cultured skeletal muscle from patients with McArdle’s disease. Biochem Biophys Res Commun 1977, 78:663-668 [DOI] [PubMed] [Google Scholar]
- 11.DiMauro S, Arnold S, Miranda A, Rowland LP: McArdle disease: the mystery of reappearing phosphorylase activity in muscle culture: a fetal isoenzyme. Ann Neurol 1978, 3:60-66 [DOI] [PubMed] [Google Scholar]
- 12.Meienhofer MC, Askanas V, Proux-Daegelen D, Dreyfus JC, Engel WK: Muscle-type phosphorylase activity present in muscle cells cultured from three patients with myophosphorylase deficiency. Arch Neurol 1977, 34:779-781 [DOI] [PubMed] [Google Scholar]
- 13.Martinuzzi A, Vergani L, Carrozzo R, Fanin M, Bartoloni L, Angelini C, Askanas V, Engel WK: Expression of muscle-type phosphorylase in innervated and aneural cultured muscle of patients with myophosphorylase deficiency. J Clin Invest 1993, 92:1774-1780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sassoon DA, Garner I, Buckingham M: Transcripts of α-skeletal actins are early markers for myogenesis in the mouse embryo. Development 1988, 104:155-164 [DOI] [PubMed] [Google Scholar]
- 15.Gorin F, Ignacio P, Gelinas R, Carlsen R: Abnormal expression of glycogen phosphorylase genes in regenerated muscle. Am J Physiol 1989, 257:C495-C503 [DOI] [PubMed] [Google Scholar]
- 16.van Leeuwen FW, de Kleijn DPV, van den Hurk HH, Neubauer A, Sonnemans MAF, Sluijs JA, Koycu Ramdjielal RDJ, Salehi A, Martens GJM, Grosveld FG, Burbach JPH, Hol EM: Frameshift mutants of β amyloid precursor protein and ubiquitin-B in Alzheimer’s and Down patients. Science 1998, 279:242-247 [DOI] [PubMed] [Google Scholar]
- 17.Browner MF, Fletterick RJ: Phosphorylase: a biological transducer. Trends Biochem Sci 1992, 17:66-71 [DOI] [PubMed] [Google Scholar]
- 18.Lebo RV, Anderson LA, Di Mauro S, Lynch E, Hwang P, Fletterick RJ: Rare McArdle disease locus polymorphic site on 11q13 contains CpG sequence. Hum Genet 1990, 86:17-24 [DOI] [PubMed] [Google Scholar]
- 19.Lockyer JM, Mc Cracken JB, Jr: Identification of a tissue specific regulatory element within the human muscle glycogen phosphorylase gene. J Biol Chem 1991, 266:20262-20269 [PubMed] [Google Scholar]
- 20.Hori K, Mukai T, Joh K, Arai Y, Sakakibara M, Yatsuki H: Stucture and expression of human and rat aldolase isozyme genes: multiple mRNA species of aldolase A produced from a single gene. Isozymes Curr Top Biol Med Res 1987, 14:153-175 [PubMed] [Google Scholar]
- 21.Nakajima H, Kono N, Yamasaki T, Hotta K, Kuwajima M, Noguchi T, Tanaka T, Tarui S: Tissue specificity in expression and alternative RNA splicing of human phosphofructokinase-M and -L genes. Biochem Biophys Res Commun 1990, 173:1317-1321 [DOI] [PubMed] [Google Scholar]
- 22.Wullrich A, Hamacher C, Schneider A, Kilimann MW: The multiphosphorylation domain of the phosphorylase kinase αM and αL subunits is a hotspot of differential mRNA processing and molecular evolution. J Biol Chem 1993, 268:23208-23214 [PubMed] [Google Scholar]



