Abstract
The Notch family of cell surface receptors plays a key role in cell-fate determination and differentiation, functioning in a cell- and context-specific manner. In mammalian cells, Notch activation is generally thought to maintain stem cell potential and inhibit differentiation, thereby promoting carcinogenesis. However, in other contexts such as primary epithelial cells (keratinocytes), increased Notch activity causes exit from the cell cycle and/or commitment to differentiation. We now report that expression of the endogenous Notch1 gene is markedly reduced in a panel of cervical carcinoma cells whereas expression of Notch2 remains elevated, and Notch1 expression is similarly reduced or absent in invasive cervical cancers. Conversely, expression of activated Notch1 causes strong growth inhibition of HPV-positive, but not HPV-negative, cervical carcinoma cells, but exerts no such effects on other epithelial tumor cells. Increased Notch1 signaling, but not Notch2, causes a dramatic down-modulation of HPV-driven transcription of the E6/E7 viral genes, through suppression of AP-1 activity by up-regulation of the Fra-1 family member and decreased c-Fos expression. Thus, Notch1 exerts specific protective effects against HPV-induced transformation through suppression of E6/E7 expression, and down-modulation of Notch1 expression is likely to play an important role in late stages of HPV-induced carcinogenesis.
Keywords: Notch, p21WAF1/Cip1, Fra-1, c-Fos, keratinocyte transformation
The Notch gene family encodes evolutionarily conserved transmembrane receptors with a key role in cell-fate determination and differentiation. Notch activation is triggered by interactions with ligands of the Delta and Serrate/Jagged families, and results in release of the intracellular region of the receptor by proteolytic cleavage and nuclear translocation (Artavanis-Tsakonas et al. 1999; Mumm and Kopan 2000). The activated Notch intracellular region binds to a ubiquitous DNA-binding protein of the CSL family [Drosophila Suppressor of hairless, Su(H), or its mammalian homolog RBP-Jκ/CBF1], converting it from a repressor into an activator of transcription. This results in the induction of genes that antagonize differentiation in a number of cell types, of neuronal origin as well as mesenchymally derived (hematopoietic cells, myoblasts, etc.; Artavanis-Tsakonas et al. 1999; Mumm and Kopan 2000). However, in cell types of other lineages and/or in different contexts, increased Notch signaling can promote differentiation (Morrison et al. 2000; Schroeder and Just 2000; Shou et al. 2001). In primary keratinocytes, increased Notch activity promotes exit from the cell cycle and entry into differentiation (Lowell et al. 2000; Rangarajan et al. 2001b). In particular, in mouse keratinocytes, Notch signaling induces differentiation directly through two distinct mechanisms, one involving the RBP-Jκ-dependent up-regulation of p21WAF1/Cip1 expression and the other the RBP-Jκ-independent induction of terminal differentiation markers of the intermediate epidermal layers (Rangarajan et al. 2001b). In human primary keratinocytes, increased Notch1 activity is not sufficient to induce p21 expression and growth arrest (C. Talora and G.P. Dotto, unpubl.), but it promotes the commitment of self-renewing stem cells to transit amplifying populations that are still actively proliferating but only for a limited amount of time (Lowell et al. 2000).
The dual capability of Notch to suppress or induce differentiation can have important consequences for tumorigenesis. Development of human T-cell acute lymphoblastic leukemia (T-ALL) has been linked to a specific chromosomal translocation that generates an aberrant Notch1 protein, lacking most of the extracellular domain and therefore functioning as a constitutive active mutant (Ellisen et al. 1991). Forced expression of similarly activated forms of the Notch1 and/or Notch2 proteins in cultured cells or transgenic animals have been shown to have a transforming potential (Capobianco et al. 1997 and references therein), as it has the inappropriate expression of the endogenous Notch4 (wnt3) gene by integration of an activating retrovirus in mammary tumorigenesis (Uyttendaele et al. 1996). Based on these findings, and the observation that expression of activated Notch proteins can have protective effects against apoptosis (Miele and Osborne 1999), it has been postulated that antagonists of Notch signaling could provide new powerful anti-neoplastic agents (Jang et al. 2000). Although such an approach could be very useful, it would need to be validated for each individual tumor type and specific stages of the carcinogenesis process, given the already mentioned possibility that increased Notch signaling can also suppress growth and/or induce differentiation.
Cervical carcinomas are a major type of epithelial keratinocyte-derived tumors. Infection with human papillomaviruses (HPVs), more specifically the high-risk HPV16 and HPV18, is associated with most cervical cancers and is thought to have a causal link with the disease (zur Hausen 2000). In particular, the E6 and E7 oncoproteins of HPV16 or HPV18 perturb normal cell cycle control through interaction with a number of cellular proteins. Thus, E6 targets the p53 tumor suppressor protein for ubiquination and degradation (Scheffner et al. 1990), whereas E7 functionally inactivates p105-Rb (Dyson et al. 1989). Additionally, E6 can increase telomerase activity in normal primary cells (Klingelhutz et al. 1996), providing an additional mechanism for escape from cellular senescence, whereas E7 binds to other members of the p105-Rb family, p107 and p130 (Dyson et al. 1992), as well as the cyclin/CDK inhibitor p21WAF1/Cip1 (Funk et al. 1997; Jones et al. 1997), suppressing their function. Compelling evidence indicates that sustained expression of the E6 and E7 oncoproteins is required for maintenance of the transformed malignant phenotype (Scheffner et al. 1994; zur Hausen 2000). In fact, in cervical cancer cell lines like HeLa, the p53 and p105-Rb pathways are only dormant and can be reactivated by loss of E6 and E7 expression (Goodwin and DiMaio 2000). The consequent increase in p21WAF1/Cip1 expression in these cells can account, to a large extent, for growth suppression and senescence (Wells et al. 2000).
Clinically, the HPV E6/E7 genes are expressed at low levels in low-grade cervical intraepithelial neoplasias but are abundantly expressed in high-grade malignant lesions (zur Hausen 2000), consistent with deregulated expression of viral oncogenes being a key element in the later stages of neoplastic development. In fact, the HPV life cycle is intimately connected with the normal growth/differentiation program of cervical keratinocytes. The viruses persist in an episomal state for long periods of time, with balanced proliferation and maturation being limited to the upper differentiated layers of the stratified epithelium (McMurray et al. 2001). Neoplastic transformation has been linked to integration of the viral DNA into the host genome, together with disruption of the viral E2 open reading frame (Scheffner et al. 1994; zur Hausen 2000). E2 functions as a repressor of the viral upstream regulatory region (URR) promoter that drives transcription of the E6 and E7 oncogenes, so that loss of E2 appears to be a prerequisite for increased E6/E7 expression (Goodwin and DiMaio 2000; Wells et al. 2000). However, besides loss of E2 function, additional changes in the host genome are likely to be involved in deregulating E6/E7 expression in HPV-transformed malignant cells. In fact, expression of the E6/E7 genes can be down-modulated in HPV-immortalized keratinocytes by a number of cytokines involved in intercellular forms of growth regulation, whereas sensitivity to these cytokines is lost with malignant transformation (zur Hausen 2000). The cellular events responsible for deregulated HPV-driven gene expression in malignant cervical carcinoma cells have not been elucidated.
Previous immunohistochemical data have indicated that Notch1 expression is elevated in squamous metaplasia of the cervical columnar epithelium and in early HPV-induced lesions, that is, cervical intraepithelial neoplastic (CIN) lesions and well-differentiated superficial carcinomas of the cervix (Zagouras et al. 1995; Daniel et al. 1997; Gray et al. 1999). We report here that in more aggressive cervical cancers, as well as cells derived from these cancers, Notch1 expression is substantially and specifically reduced. Down-modulation of Notch1, but not Notch2, signaling is required for sustained HPV-E6/E7 expression and HPV-induced malignant transformation.
Results
Specific down-modulation of endogenous Notch1 expression in cervical cancer cells and tumors
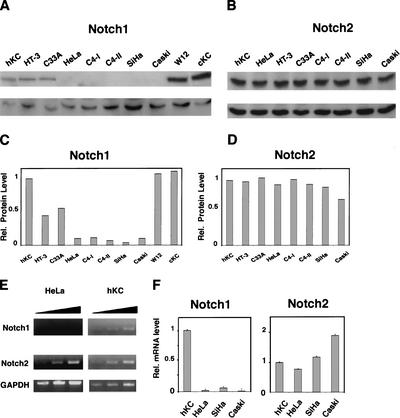
As Notch signaling promotes exit of keratinocytes from the cell cycle and/or commitment to differentiation (Lowell et al. 2000; Rangarajan et al. 2001b), this pathway may be down-modulated in cells derived from cervical carcinomas, a major type of keratinocyte-derived tumors. Immunoblotting of total cell extracts with antibodies against Notch1 allowed ready detection of this protein in primary human keratinocytes, HPV-negative cervical carcinoma cells (C33a), as well as a keratinocyte cell line with episomal HPV initiated from a low-grade cervical lesion (W12; Stanley et al. 1989). In contrast, Notch1 levels were undetectable or strongly reduced in all HPV-positive malignant cervical cancer cells that were tested (HeLa, C4-I, C4-II, SiHa, and Caski), except HT-3 cells, a cervical cell line that harbors HPV30 DNA and contains a dominant-negative p53 gene and a mutant p105Rb allele (Naeger et al. 1999; Fig. 1A,C). Unlike Notch1, the Notch2 protein was expressed in all cervical cancer cells to similar levels as in human primary keratinocytes (Fig. 1B,D). Notch3 and Notch4 expression was undetectable in all cells (data not shown).
Figure 1.
Specific down-modulation of endogenous Notch1 expression in cervical carcinoma cells. (A,B) Total cell extracts from primary human foreskin keratinocytes (hKC), a panel of cervical carcinoma cell lines, cervical keratinocytes (cKC), and a keratinocyte cell line with episomal HPV (W12) were analyzed by immunoblotting with antibodies against Notch1 (C-20) and Notch2 (C65) as indicated (upper panels). Immunoblotting with antibodies against Cdk2 was used as an equal loading control (lower panels). Similar results were obtained in at least two other experiments, including use of Tan-20 antibodies for detection of Notch1, and of anti-β actin antibodies for an equal loading control. (C,D) Densitometric scanning of the autoradiographs shown in A and B was used for quantification of the relative levels of Notch1 and Notch2 proteins in human primary keratinocytes versus the other cells. Cdk2 expression was used for internal value normalization. (E) Total RNA from human primary keratinocytes and HeLa cells was analyzed by RT-PCR with oligonucleotide primers specific for the 3′-untranslated regions of the Notch1 and Notch2 transcript and for GAPDH as indicated. Each sample was analyzed in three serial dilutions (1:1, 1:10, 1:100). (F) Total RNA from human primary keratinocytes and various carcinoma cells was analyzed by real-time RT-PCR with primers specific for the 3′-untranslated regions of the Notch1 and Notch2 transcripts as indicated.Values were normalized for β-actin expression and expressed as relative levels in reference to primary human keratinocytes. All RNAs were tested in triplicate samples, and the standard deviation is indicated.
Southern blot analysis with a Notch1-specific probe indicated that the copy number of this gene is not reduced in the HPV-positive cervical cancer cells (data not shown). Treatment of HeLa cells with proteasome inhibitors failed to elevate Notch1 protein levels (data not shown). To determine whether there is a block in mRNA expression, RNA derived from HeLa cells as well as from primary human keratinocytes was analyzed by semiquantitative RT-PCR using primers for the 3′ unique regions of the Notch1, Notch2, and GAPDH transcripts. Whereas Notch2 mRNA expression was similar in the two cells, that of Notch1 mRNA was drastically reduced in HeLa cells (Fig. 1E). This analysis was quantified and extended to other carcinoma cells by real-time PCR using the same set of oligonucleotide primers. As shown in Figure 1F, in HeLa cells endogenous levels of Notch1 mRNA were found to be >45-fold lower than in human primary keratinocytes, and a lesser but still significant reduction (greater than eightfold) was observed in all the other cervical carcinoma cells. In contrast to Notch1, Notch2 mRNA levels were approximately the same in all cells.
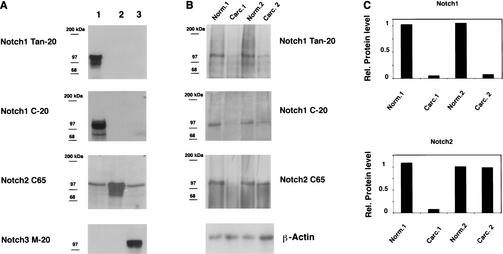
To examine expression of Notch1 and Notch2 in cervical carcinomas in vivo, we used a combined biochemical and immunohistochemical approach. Antibodies with tested specificity against these proteins (Fig. 2A) were used for immunoblotting of protein extracts derived from two surgically excised tumors with confirmed histological diagnosis (invasive moderately differentiated squamous cell carcinoma, Carc.1, and invasive poorly differentiated squamous cell carcinoma, Carc.2) versus surrounding normal regions of the same patients. This analysis revealed that levels of Notch1 were substantially reduced in both tumors (>10-fold), whereas levels of Notch2 were reduced in one case but not the other (Fig. 2B,C).
Figure 2.
Down-modulation of Notch1 protein expression in cervical carcinomas. (A) Total cell extracts derived from 293 cells transfected with plasmid expression vectors for Notch1 (lane 1), Notch2 (lane 2), or Notch3 (lane 3) were immunoblotted with the indicated antibodies as control of their specificity. (B) Extracts from surgically excised invasive cervical carcinomas (Carc.1 and Carc.2) and surrounding normal cervical tissues from the same patients (Norm.1 and Norm.2) were analyzed for levels of Notch1 and Notch2 protein expression by immunoblotting with the corresponding antibodies as indicated. Immunoblotting with antibodies against β-actin was used for an equal loading control. (C) Densitometric scanning of the autoradiographs shown in B was used for quantification of the relative levels of Notch1 and Notch2 proteins after normalization for levels of β-actin expression.
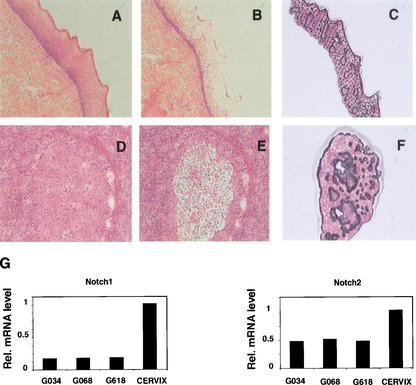
As an alternative approach, laser capture microscopy was used to quantify levels of Notch1 and Notch2 mRNA expression in the neoplastic areas of three independent cervical carcinomas versus normal epithelium. Total RNA prepared from the captured tissues was converted into cDNA followed by real-time PCR using the same set of oligonucleotide primers used for analysis of cultured cells. As shown in Figure 3, levels of Notch1 mRNA expression in the tumors were substantially lower (∼10-fold) than in the normal epithelium, but Notch2 mRNA levels were reduced to a more limited extent (∼twofold).
Figure 3.
Down-modulation of Notch1 mRNA expression in cervical carcinomas as assessed by laser capture microscopy and real-time RT-PCR. (A–F) Histological slides of normal cervical tissue (A–C) and an invasive cervical tumor (D–F) photographed before laser capture microscopy (A,D) and after removal of the normal (B) and neoplastic (E) epithelium. (C,F) Cap images of the corresponding captured epithelia. Hematoxylin and eosin, ×40. (G) Total RNAs from normal squamous epithelium and three invasive squamous carcinomas of the cervix microdissected by the above technique were analyzed in triplicate samples for levels of Notch1 and Notch2 mRNA expression by real-time RT-PCR as described in Materials and Methods.
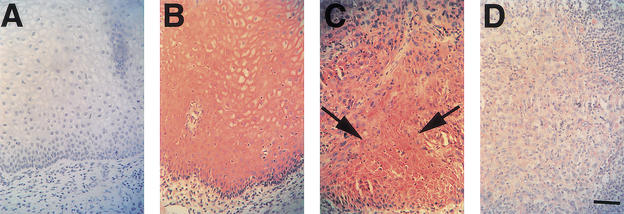
Finally, the same tumors examined above as well as 10 additional tumors were analyzed by immunohistochemistry. Strong staining for Notch1 expression was found in the stratified epithelium of the normal outer cervix (Fig. 4B), but no positive signal was detected in the simple columnar epithelium of the inner cervix (data not shown). As reported previously (Zagouras et al. 1995; Daniel et al. 1997; Gray et al. 1999), staining for Notch1 expression was elevated at the junction between normal stratified epithelium and adjacent areas of metaplastic/neoplastic transformation (Fig. 4C). In contrast, little or no Notch1 staining was found in the deeper and more invasive tumor areas (Fig. 4D), similar to what was seen by others (P. Zagouras, pers. comm.).
Figure 4.
Localization of endogenous Notch1 protein expression in normal squamous epithelium and invasive squamous carcinoma of the cervix. (A,B) Serial sections of normal squamous epithelium of the cervix stained with affinity-purified nonimmune antiserum (A) or Tan-20 antibodies against Notch1 (B). (C,D) Staining with Tan-20 antibodies of an area of cervical intraepithelial neoplastic lesion (C) or invasive cervical carcinoma (D) present in the same histological slide as the normal squamous epithelium shown in B. Note the positive staining for Notch1 expression in cells at the transition between normal and neoplastic epithelium (C, arrows). In contrast, little or no Notch1 staining is detectable in the invasive tumor area (D). Similarly, inconspicuous staining with Notch1 antibodies was also found in 10 other advanced cervical cancers that were analyzed. Bar, 80 μm.
Specific growth inhibition of cervical cancer cells by activated Notch1
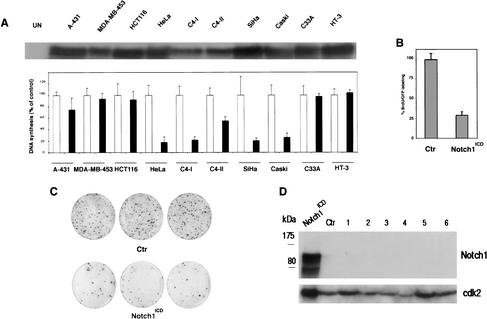
There may be a link between the specific down-modulation of Notch1 expression in cervical cancer cells and their deregulated growth. To assess whether increased Notch1 signaling can directly affect growth of these cells, we resorted to an adenovirus-mediated approach. Cervical cancer cell lines were infected along with other epithelial cancer cells with either a recombinant adenovirus expressing the intracellular constitutive active form of Notch1 (Ad-Notch1) or a control virus expressing GFP (Ad-GFP). All cells expressed comparable levels of adenovirally transduced Notch1 protein (Fig. 5A, upper panel). Cellular DNA synthesis was measured at 36 h after infection by a thymidine incorporation assay (Fig. 5A, lower panel). DNA synthesis in cells derived from epidermoid (A-431), breast (MDA-MB-453), and colon (HCT116) carcinomas was affected to a very limited extent by infection with the Ad-Notch1 virus versus the Ad-GFP control. A similar lack of effects was observed with HPV-negative cervical carcinoma cells, C33A. In contrast, Ad-Notch1 expression caused strong inhibition of DNA synthesis in all tested HPV-positive cervical cancer cells (HeLa, C4-I, C4-II, SiHa, Caski), except HT-3 cells, which, as mentioned before, contain mutated p53 and p105-Rb genes (Naeger et al. 1999).
Figure 5.
Specific growth inhibitory effects of activated Notch1 in cervical carcinoma cell lines. (A) Epidermoid carcinoma (A-431), breast (MDA-MB-453), colon (HCT116), and indicated cervical carcinoma cells were infected with a recombinant adenovirus expressing activated Notch1 (Ad-Notch1) or green fluorescence protein (Ad-GFP). (Upper panel) Total cell extracts from the indicated cell lines infected with the Ad-Notch1 virus were examined for levels of activated Notch1 expression by immunoblotting with the corresponding antibody. (Lower panel) Cellular DNA synthesis was determined by 3H-thymidine incorporation assay 36 h after infection. Cells were tested in triplicated wells, and the standard deviation is indicated. The slight differences in sensitivity of the various HPV-positive carcinoma cell lines to Ad-Notch1 growth inhibition are likely owing to the different growth patterns of these various cells. In fact, some of these cell lines tend to grow in tightly packed clusters, with cells at the center of these clusters being less susceptible to adenoviral infection (as evaluated by expression of the GFP marker, which is also transduced by the Ad-Notch1 virus). (B) HeLa cells were transfected with a plasmid expression vector for activated Notch1 (Notch1ICD) or empty vector control (Ctr) together with trace amounts of a GFP expression vector for identification of transfected cells. Cells were labeled with BrdU for 3 h prior to termination of the experiment (36 h after transfection). The BrdU-labeling index of GFP-positive transfected cells was determined by counting in each case a minimum of 120 cells from six independent fields. Values are expressed as percentages relative to the control. Similar results were obtained in two other independent experiments. (C) HeLa cells were transfected with a plasmid expression vector for activated Notch1 (Notch1ICD) or empty vector control (Ctr) together with trace amounts of an expression vector for G418 resistance. The same number of transfected cells was plated and cultured in triplicate dishes in the presence of antibiotic selection for 2 wk. The number of macroscopically visible colonies formed by cells transfected with the control versus activated Notch1 expression vector was 311 (±20) and 68 (±8), respectively. Similar differences were observed in two other independent experiments. (D) HeLa cells at 24 h after transfection with the activated Notch1 vector (Notch1ICD), and cells from six independent colonies (1–6) that emerged from cultures transfected with the same vector after 2 wk of G418 selection (as in C), were analyzed for levels of Notch1 expression by immunoblotting with the corresponding specific antibodies. (Ctr) Cells from a colony of HeLa cells transfected with empty vector control. The immunoblot was reprobed with anti-cdk2 antibody for an equal loading control.
To confirm the above results by an adenovirus-independent approach, HeLa cells were transfected with a plasmid expression vector for activated Notch1 or empty vector control, plus a GFP expression vector for identification of the transfected cell population. Even in this case, DNA synthesis, as determined by a BrdU-labeling index of transfected cells, was strongly suppressed by activated Notch1 expression (Fig. 5B). In a long-term growth colony assay, cotransfection of HeLa cells with a plasmid for G418 resistance plus the activated Notch1 vector caused a greater-than-fivefold reduction in colony formation relative to cotransfection with the empty vector control (Fig. 5C). Whereas expression of activated Notch1 could be readily detected in HeLa cells at 24 h after transfection, no expression was found in six independent colonies that emerged from cultures transfected with the same vector after prolonged selection (Fig. 5D). This indicates that there is strong selection pressure for loss of Notch1 expression, even if it may still be possible to recover cells with activated Notch1 expression that harbor secondary mutations rendering them resistant to the Notch1 growth inhibitory effects.
Down-modulation of HPV E6/7 expression by activated Notch1 expression
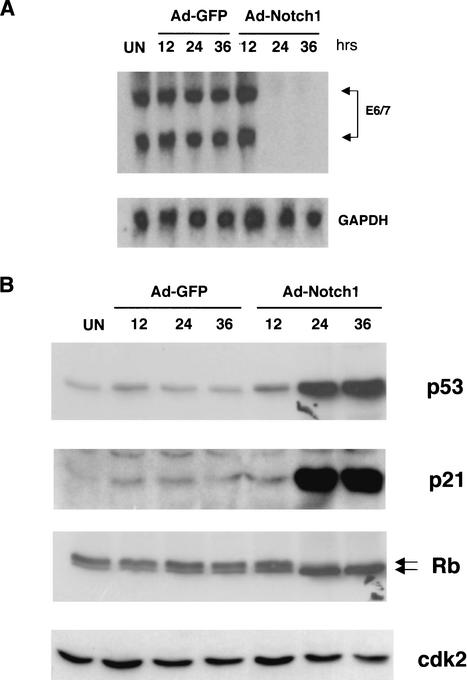
As growth of HPV-transformed cervical cancer cells is dependent on sustained E6 and E7 expression (Scheffner et al. 1994; zur Hausen 2000), we tested whether the specific growth inhibitory effects of activated Notch1 on these cells could be explained by down-modulation of the E6 and E7 genes. Northern blot analysis of HeLa cells at various times after adenovirus infection revealed a drastic block of E6 and E7 mRNA expression as a consequence of activated Notch1 expression (Fig. 6A).
Figure 6.
Suppression of E6/E7 mRNA expression by activated Notch1 and associated downstream effects. (A) HeLa cells were either uninfected (UN) or infected with the Ad-Notch1 and Ad-GFP virus for the indicated times (in hours). Levels of HPV18 E6/E7 mRNA expression were determined by Northern blot analysis with corresponding specific probes. Densitomeric scanning of the autoradiographs indicated that activated Notch1 expression caused a 95-fold reduction of E6/E7 mRNA levels by 24 h of infection (i.e., by the time the activated Notch1 protein is expressed). (B) HeLa cells were infected as in A, followed by immunoblot analysis of total cell extracts with antibodies against the p53, p21, and p105-Rb proteins as indicated. Immunoblotting with antibodies against Cdk2 was used for an equal loading control.
The p53 and p105-Rb tumor suppressor pathways are intact in HeLa cells, and repression of E6 and E7 expression results in activation of these pathways with consequent growth suppression (Goodwin and DiMaio 2000; Wells et al. 2000). In parallel with the block of E6/E7 expression, expression of activated Notch1 in HeLa cells caused a sustained increase of p53 protein levels together with induction of the p53-responsive p21WAF1/Cip1 gene (Fig. 6B). Concomitantly, p105-Rb was mostly converted to its hypophosphorylated form, as was reported previously for HeLa cells growth-arrested by viral E2 expression (Goodwin and DiMaio 2000).
Selective suppression of HPV-driven transcription by the Notch1 but not Notch2 protein, independently of RBP-Jκ function
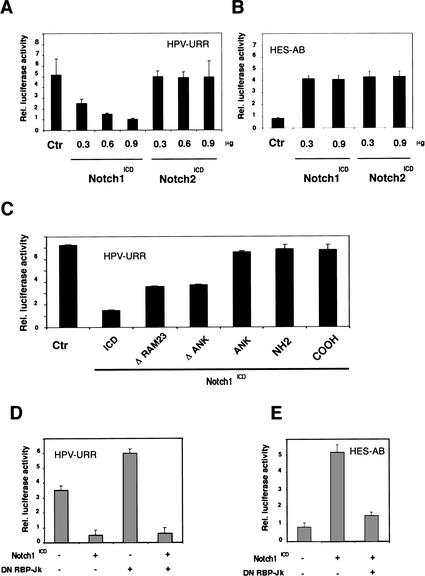
Transcription of the E6 and E7 genes is driven by the viral URR promoter, which is maintained intact and active in HPV-transformed cancer cells (zur Hausen 2000). To assess whether the block of E6/E7 expression by activated Notch1 occurs at the level of URR promoter activity, HeLa cells were transfected with a luciferase reporter plasmid for the URR promoter (Goodwin et al. 1998) plus or minus an expression vector for activated Notch1 in various amounts. As shown in Figure 7A, URR promoter activity was drastically reduced by cotransfection of the activated Notch1 expression vector in a dose-dependent manner.
Figure 7.
Transcriptional repression of HPV-URR promoter activity by activated Notch1. (A,B) HeLa cells were transfected with either the HPV-URR (A) or HES-AB (B) luciferase reporter plasmids plus/minus expression vectors for the activated cytoplasmic forms of Notch1 and Notch2 in increasing amounts as indicated. Luciferase activity was determined 48 h after transfection and normalized with a Renella reporter internal control. Values are expressed as arbitrary units. All conditions were tested in triplicate samples, and the standard deviation is indicated. (C) HeLa cells were transfected with the HPV-URR reporter plus/minus expression vectors for cytoplasmic activated Notch1 in an intact form (ICD) or its separate (NH2) N- (amino acids 1752–2123) and (COOH) C-terminal (amino acids 2124–2555) regions, or with internal deletions of the (ΔRAM23) RAM23 (amino acids 1751–1851) or (ΔANK) ANK (amino acids 1851–2096) domains, or (ANK) a vector expressing the ANK domain of Notch1 (amino acids 1852–2196) devoid of any other domains. Luciferase activity was determined 48 h after transfection as in A. (D) HeLa cells were transfected with either the HPV-URR (left panel) or HES-AB (right panel) luciferase reporter plasmids plus/minus the expression vector for activated Notch1 (0.90 μg) alone or together with an expression vector for dominant negative RBP-Jκ (0.90 μg) as indicated. Luciferase activity was determined 48 h after transfection as in A.
The Notch1 protein shares a high degree of homology with Notch2 (Artavanis-Tsakonas et al. 1999; Mumm and Kopan 2000). The selective down-modulation of the Notch1 protein in cervical cancer cells with normal Notch2 levels (Fig. 1) suggested that in the context of HPV-driven transcription and transformation, the two proteins are not functionally equivalent. To investigate this possibility, HeLa cells were transfected in parallel with expression vectors for the activated cytoplasmic forms of Notch1 and Notch2 (Capobianco et al. 1997). Expression of the two proteins in this equivalent activated form induced activity of the Notch/RBP-Jκ-responsive promoter HES-AB to a similar extent (Fig. 7B). In contrast, expression of activated Notch2 failed to cause suppression of HPV-URR promoter activity at any tested concentrations, although this was observed readily after expression of activated Notch1 (Fig. 7A).
The Notch1 cytoplasmic region is composed of several functional domains involved in distinct protein–protein interactions, including the Ram23 region involved in RBP-Jκ binding and activation and the ankyrin-repeat domain (ANK), which provides a second site for RBP-Jκ binding and activation, and also interacts with a number of other cell signaling molecules (Artavanis-Tsakonas et al. 1999; Mumm and Kopan 2000). Mutants lacking the Ram23 or ANK domains were found to suppress URR promoter activity, although to a lower extent than intact the Notch1 cytoplasmatic region. Expression of the Notch1 ANK domain or of the separate N- and C-terminal regions exerted no such effect (Fig. 7C).
As mentioned, the best characterized mediator of Notch activation is the DNA-binding protein RBP-Jκ (CSL), which is converted from a repressor into an activator of transcription by Notch (Artavanis-Tsakonas et al. 1999; Mumm and Kopan 2000). Expression of a dominant-negative mutant of RBP-Jκ (R218H), which suppresses activation of endogenous RBP-Jκ (Kurooka et al. 1998), had no effect on the ability of activated Notch1 to suppress HPV-URR promoter activity, but under the same conditions it blocked induction of a classical Notch/RBP-Jκ-responsive promoter (HES-AB; Jarriault et al. 1995; Fig. 7D). Thus, HPV-URR activity is specifically suppressed by the activated Notch1, but not the Notch2, protein, and is independent of RBP-Jκ function.
Suppression of URR-driven transcription by activated Notch1 through differential modulation of specific AP-1 family members
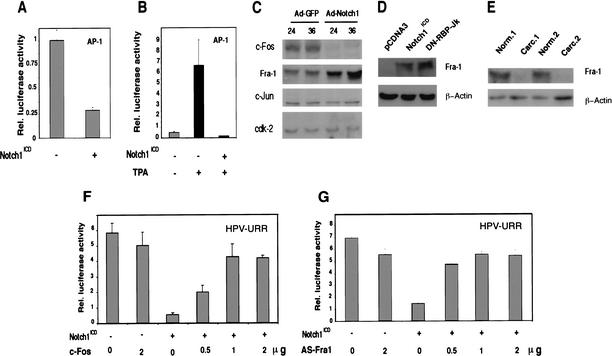
The AP-1 complex plays a key role in initiating and maintaining transcription from the URR promoter (Thierry et al. 1992). Transient transfection of HeLa cells with an AP-1 reporter plasmid plus/minus a vector for activated Notch1 showed that increased Notch1 activity represses AP-1 activity both under basal conditions and after induction by the phorbol ester TPA (Fig. 8A,B). AP-1 is a heterodimeric DNA-binding complex formed by proteins of the c-Jun and c-Fos families (Angel and Karin 1991). Despite their high degree of sequence homology, individual members of these families differ in their ability to activate target genes and can also function in an inhibitory fashion. In particular, whereas c-Fos-containing heterodimers lead to AP-1 active complexes, the c-Fos-related Fra-1 protein exerts, in specific contexts, a suppressive effect (Yoshioka et al. 1995). Immunoblotting with antibodies against various AP-1 family members revealed that in HeLa cells infected with the Ad-Notch1 virus, levels of c-Jun were unaffected, expression of c-Fos was significantly suppressed, and that of Fra-1 was increased (Fig. 8C). To assess whether induction of Fra-1 expression by activated Notch1 is RBP-Jκ-dependent, HeLa cells were transfected with a plasmid expression vector for activated Notch1 plus/minus a vector for an RBP-Jκ dominant-negative mutant. As shown in Figure 8D, activated Notch1 induced Fra-1 expression in both situations. On the basis of the above results, in cervical tumors with decreased Notch1 expression, Fra-1 protein levels should also be down-modulated. Immunoblotting of two independent tumor extracts in parallel with matched surrounding normal regions (the same as analyzed for levels of Notch1 expression in Fig. 2B) showed that this is, indeed, the case (Fig. 8E).
Figure 8.
Suppression of HPV-URR promoter activity by activated Notch1 through differential modulation of specific AP-1 components. (A,B) HeLa cells were transfected with an AP-1 reporter plasmid plus/minus an expression vector for activated Notch1 or empty vector control. Promoter activity was measured under (A) basal conditions and (B) at 12 h of AP-1 activation by phorbol ester TPA treatment (100 ng/mL). (C) HeLa cells were infected with the Ad-GFP or Ad-Notch1 viruses for the indicated times (in hours), and total cell extracts were analyzed by immunoblotting with antibodies against various AP-1 components as indicated. Immunoblotting with antibodies against Cdk2 was used as an equal loading control. Densitometric scanning of the autoradiographs indicated that, relative to the control, expression of Fra-1 was increased 3.2- and 8.5-fold at 24 and 36 h, respectively, after infection with the Ad-Notch1 virus. Conversely, expression of c-Fos was decreased >10-fold at 24 h and 36 h after Ad-Notch1 infection. (D) HeLa cells were transfected with either the pCDNA3 or Notch1ICD plasmids plus/minus the expression vector for dominant-negative RBP-Jκ as indicated. Transient transfection efficiency, as assessed by expression of a GFP expression vector added in trace amounts, was >80%. At 48 h after transfection, total cell extracts were analyzed by immunoblotting with antibodies against Fra-1 or β-actin as indicated. (E) Total extracts from surgically excised cervical carcinomas and corresponding surrounding normal regions (examined for levels of Notch1 and Notch2 expression in Fig. 2B) were analyzed by immunoblotting with antibodies against Fra-1 or β-actin as indicated. (F,G) HeLa cells were transfected with the HPV-URR reporter plus/minus expression vectors for activated Notch1 and c-Fos (F) or anti-sense Fra-1 cDNA (G) in various amounts (in micrograms) as indicated. Luciferase activity was determined at 48 h after infection as in Fig. 7.
Finally, to assess whether these changes in AP-1 components can account for suppression of URR promoter activity, HeLa cells were transfected with the URR reporter plasmid plus/minus vectors for activated Notch1, c-Fos, or anti-sense Fra-1 cDNA. As shown in Figure 8F, suppression of URR promoter activity by activated Notch1 was reversed by increased c-Fos expression in a dose-dependent manner. Similar counteracting effects were observed after expression of the anti-sense Fra1 cDNA (Fig. 8G).
Discussion
Although the link between HPV infection and cervical carcinogenesis is well established, additional changes in the host cell and/or surrounding environment are a prerequisite for tumor development (zur Hausen 2000). In particular, studies of keratinocytes at various stages of HPV-induced transformation indicate that sustained and deregulated expression of the HPV E6 and E7 oncogenes is required for malignant conversion, rendering cells resistant to intercellular forms of negative growth regulation (zur Hausen 2000). Activation of the Notch signaling pathway is an important form of intercellular communication that serves to coordinate differentiation of keratinocytes within overlying layers of the stratified epithelium (Rangarajan et al. 2001b). Cervical cancer represents a major type of keratinocyte-derived tumor. Our findings indicate that Notch1 exerts specific counteracting effects against HPV-induced transformation and E6/E7 expression, and down-modulation of Notch1 expression is likely to play an important role in the later stages of HPV-induced carcinogenesis.
Previous immunohistochemical data have indicated that Notch1 expression is elevated in squamous metaplasia of the columnar epithelium, and in early HPV-induced lesions (CINI-III) and well-differentiated superficial carcinomas of the cervix (Zagouras et al. 1995; Daniel et al. 1997; Gray et al. 1999). We have shown here that in invasive cervical cancers, Notch1 expression is substantially reduced. This dual pattern of Notch1 expression suggests that this protein may play a permissive and/or tumor-promoting function in the early stages of cervical carcinogenesis, and a suppressive function in the later stages. Consistent with this possibility, in HaCaT cells, an established keratinocyte cell line with mutated p53, activated Notch1 synergizes with exogenously expressed E6/E7 by protecting cells against suspension-induced apoptosis (Rangarajan et al. 2001a). Conversely, we have shown here that activated Notch1 causes strong growth inhibition of a panel of malignant HPV-positive cervical carcinoma cells, all characterized by very low expression of the endogenous Notch1 gene.
HeLa cells provide a classical model system for mechanistic studies of HPV-induced malignant transformation in cells with otherwise intact p53 and p105-Rb tumor suppressor pathways (Goodwin et al. 2000; Wells et al. 2000; and references therein). Unlike in HeLa, activated Notch1 caused no growth inhibition of HPV-negative cervical carcinoma cells (C33A) or HPV-positive cells (HT-3) with secondary mutations in the p53 and p105-Rb tumor suppressor genes, suggesting that Notch1-induced growth arrest depends on restoration of normal tumor suppressor pathways abrogated by E6/E7 expression. We have found that RBP-Jκ-dependent transcription, which is induced by both activated Notch1 and Notch2 in HeLa cells, is not involved in suppression of HPV-driven transcription (nor in Notch1-induced growth suppression; data not shown). Rather, E6/E7 expression is suppressed specifically by activated Notch1 through changes in expression of individual components of the AP-1 transcription complex. The importance of AP-1 as a positive regulator of HPV-driven transcription has been documented in a number of previous studies (Thierry et al. 1992; Soto et al. 1999 and references therein). Our work provides a link between Notch signaling and negative control of AP-1 activity, achieved through changes in the intracellular balance of individual components. In particular, activated Notch1 was found to suppress expression of the c-Fos protein, while inducing that of another Fos family member, Fra-1. The latter protein has the same DNA-binding specificity as c-Fos but, unlike c-Fos, lacks a transcription activating domain and can thus function as a suppressor rather than an inducer of AP-1-dependent transcription (Yoshioka et al. 1995). Fra-1 is expressed in vivo in the suprabasal layers of the epidermis (Welter and Eckert 1995) and has been connected with positive control of keratinocyte differentiation marker expression, specifically involucrin (Welter et al. 1995). We have shown previously that increased Notch signaling in primary keratinocytes induces involucrin expression through an RBP-Jκ-independent pathway (Rangarajan et al. 2001b). An attractive possibility is that the role of Notch in keratinocyte differentiation marker expression, as well as in negative regulation of HPV-dependent transcription, are both connected, at least in part, with increased Fra-1 expression. Biologically, it is worth stressing the intimate connection between the HPV life cycle and keratinocyte differentiation control (McMurray et al. 2001). Although replication and transcription of the virus are limited to the upper differentiated layers of the stratified epithelium formed by cervical keratinocytes, the fine balance between replication of the viral genome and production of viral particles is likely to require not only positive but also negative regulatory mechanisms (McMurray et al. 2001). Elevated Notch1 activity, coupled with increased Fra-1 expression, could provide one such mechanism that needs to be inactivated for sustained expression of viral transforming genes in carcinogenesis. Consistent with this view are our present findings as well as the fact that the chromosomal locus containing the fra-1 gene is structurally deleted or rearranged in a subset of cervical carcinoma cells (Jesudasan et al. 1995). In nonmalignant HeLa hybrid cells, exposure to TNF-α causes a suppression of HPV transcription through a similar mechanism described here, involving up-regulation of Fra-1 expression, whereas the malignant parental cells are totally resistant to the TNF-α effect (Soto et al. 1999). As HeLa cells are still capable of increased Fra-1 expression in response to activated Notch1, their failure to respond to TNF-α could be linked to down-modulation of endogenous Notch1 signaling and/or blocked activation. In the panel of cervical carcinoma cells that we have examined, expression of the Notch1 gene was markedly reduced at the level of transcription, but expression of Notch2 was unaffected. Although the regulatory regions of the two genes are still poorly characterized, an elucidation of the specific down-modulation of Notch1 in cervical carcinoma cells is likely to generate further insights into how cervical tumors originate.
Materials and methods
Cells and viruses
All cells were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum. Transient transfection/promoter activity assays were performed using a Dual-Luciferase/Renella Reporter Assay System (Promega). All conditions were tested in triplicate samples, and experiments were repeated at least three times. For BrdU-labeling assays, cells were incubated for 3 h with BrdU (cell proliferation labeling reagent; Amersham-Pharmacia Biotech) at 48 h after transfection. For long-term colony growth assays, cells were transfected with 5 μg of Notch1ICD plasmid or empty vector control and 1 μg of a G418 resistance vector (pCDNA3; Invitrogen). At 24 h after transfection, cells were trypsinized and replated in the same number (1 × 105 per 60-mm dish) in medium containing 900 μg/mL G418 (Gemini Bio Products, Inc.) for 2 wk.
Recombinant adenoviruses expressing the constitutively active form of human Notch1 and the green fluorescent protein (GFP), and conditions for viral amplification, purification, and infection of cells were previously described (Rangarajan et al. 2001b). Viruses were used at a multiplicity of infection of 100.
Plasmids
Expression vectors for activated Notch1 and Notch2 (Capobianco et al. 1997), activated Notch1 mutants (Rangarajan et al. 2001b), dominant-negative RBP-Jκ (Kato et al. 1997), c-Fos, and Fra-1 (Cohen and Curran 1988) were described previously. The expression vector for anti-sense Fra-1 was constructed by inserting the corresponding cDNA in an anti-sense orientation in pCDNA3. Expression vectors for cytoplasmic activated Notch1 in its N-terminal (amino acids 1752–2123) or C-terminal (amino acids 2124–2555) regions were obtained by inserting the corresponding cDNAs into pCDNA3 plasmid. The HPV E6 and E7 probes for Northern analysis and the luciferase reporter plasmid for HPV18-URR (pGL3-HPV18LCR) were kindly provided by D. DiMaio (Genetics Department, Yale University, New Haven, CT; Goodwin et al. 1998), and the luciferase reporter plasmid for AP-1 was from Stratagene.
Antibodies
The anti-Notch-1 Tan-20 and anti-Notch-2 C651.6DBHN antibodies were from the Developmental Studies Hybridoma Bank, University of Iowa. The anti-Notch1 (C-20) and anti-Notch3 (M-20) antibodies and antibodies against p21, p53, p105-Rb, CDK2, c-Fos, Fra-1, and c-Jun were from Santa Cruz Biotechnology. HRP-conjugated secondaries were from Amersham. Immunoblots were developed with ECL (NEN).
RNA analysis
Same amounts of total cellular RNAs (1 μg) were converted into cDNAs by reverse transcription (Clontech Advantage RT-for-PCR). Real-time PCR was performed using the Icycler IQ Real-Time detection System (Bio-Rad) according to the manufacturer's recommendations. Amplification of the β-actin cDNA was used as the endogenous normalization standard. The 50-μL amplification reaction mixture contained 23 μL of cDNA (1:200 dilution), 25 μL of 2× SYBR Green PCR master mix (Applied Biosystems), and 1 μL each (200 nM) of β-actin or Notch primers. Each sample was amplified in triplicate. A two-step cycling protocol with combined primer annealing and elongation at 55°C was used. The following forward and reverse primers were used: β-actin, 5′-TCACCCACACTGTGCCCATCTACGA-3′ and 5′-CAGCGGAACCGCTCATTGCCAATGG-3′; Notch1, 5′-GGCCACCTGGGCCGGAGCTTC-3′ and 5′-GCGATCTG GGACTGCATGCTG-3′; Notch2, 5′-GGCCCCCTGCCCACC ATGTAC-3′ and 5′-CCCGCTGACCTCCTCCAGC-3′. The β-actin, Notch1, and Notch2 PCR products were 295, 365, and 343 bp, respectively.
Laser capture microscopy
All frozen tissues used for this study were obtained from The Cooperative Human Tissue Network (Gynecology and Oncology Group, Tissue Bank, Children's Hospital of Columbus, OH). The frozen tissue blocks were sectioned at 8 μm in a cryostat, mounted on uncoated glass slides, and immediately stored at −80°C. The frozen sections were immediately fixed in 70% ethanol for 30 sec, stained with H&E, followed by 5-sec dehydration steps in 70%, 95%, and 100% ethanol and a final 5-min dehydration step in xylene. Once air-dried, the sections were laser-microdissected with a PixCell I and II LCM system from Arcturus Engineering, using a standard protocol (Emmert-Buck et al. 1996). Each sample was estimated to be >98% homogeneous as determined by microscopic visualization of the captured cells. Total RNA from each population of laser captured cells was extracted by means of a microisolation protocol and converted to double-stranded cDNA as described (Sgroi et al. 1999). The produced cDNAs were quantified with PicoGreen (Molecular Probes), and identical amounts (2.5 ng) were analyzed by real-time PCR using the same set of oligonucleotide primers indicated above for analysis of RNAs derived from the cultured cells.
Acknowledgments
We are grateful to D. DiMaio, P. Howley, and T. Curran for their gifts of reporter and expression plasmids, J. Rheinwald and M. Stanley for their gift of primary cervical keratinocytes and W12 cells, respectively, and C. Brisken for critical reading of the manuscript. We are also very thankful to P. Zagouras, M.L. Carcangiu, S. Artavanis-Tsakonas, and A. Rangarajan for constructive discussions and for sharing some of their unpublished results. We thank J. Gaudet and E. Chatfield for their skillful execution of laser capture microscopy and tissue mRNA purification. This work was supported by NIH Grants AR39190, CA16038, and CA73796 to G.P.D., and, in part, by the Cutaneous Biology Research Center through the Massachusetts General Hospital/Shiseido Co. Ltd. Agreement.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL paolo.dotto@cbrc2.mgh.harvard.edu; FAX (617) 724-9572.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.988902.
References
- Angel P, Karin M. The role of Jun, Fos and the AP-1 complex in cell-proliferation and transformation. Biochim Biophys Acta. 1991;1072:129–157. doi: 10.1016/0304-419x(91)90011-9. [DOI] [PubMed] [Google Scholar]
- Artavanis-Tsakonas S, Rand MD, Lake RJ. Notch signaling: Cell fate control and signal integration in development. Science. 1999;284:770–776. doi: 10.1126/science.284.5415.770. [DOI] [PubMed] [Google Scholar]
- Capobianco AJ, Zagouras P, Blaumueller CM, Artavanis-Tsakonas S, Bishop JM. Neoplastic transformation by truncated alleles of human NOTCH1/TAN1 and NOTCH2. Mol Cell Biol. 1997;17:6265–6273. doi: 10.1128/mcb.17.11.6265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen DR, Curran T. fra-1: A serum-inducible, cellular immediate-early gene that encodes a fos-related antigen. Mol Cell Biol. 1988;8:2063–2069. doi: 10.1128/mcb.8.5.2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel B, Rangarajan A, Mukherjee G, Vallikad E, Krishna S. The link between integration and expression of human papillomavirus type 16 genomes and cellular changes in the evolution of cervical intraepithelial neoplastic lesions. J Gen Virol. 1997;78:1095–1101. doi: 10.1099/0022-1317-78-5-1095. [DOI] [PubMed] [Google Scholar]
- Dyson N, Howley PM, Munger K, Harlow E. The human papilloma virus-16 E7 oncoprotein is able to bind to the retinoblastoma gene product. Science. 1989;243:934–937. doi: 10.1126/science.2537532. [DOI] [PubMed] [Google Scholar]
- Dyson N, Guida P, Munger K, Harlow E. Homologous sequences in adenovirus E1A and human papillomavirus E7 proteins mediate interaction with the same set of cellular proteins. J Virol. 1992;66:6893–6902. doi: 10.1128/jvi.66.12.6893-6902.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellisen LW, Bird J, West DC, Soreng AL, Reynolds TC, Smith SD, Sklar J. TAN-1, the human homolog of the Drosophila notch gene, is broken by chromosomal translocations in T lymphoblastic neoplasms. Cell. 1991;66:649–661. doi: 10.1016/0092-8674(91)90111-b. [DOI] [PubMed] [Google Scholar]
- Emmert-Buck MR, Bonner RF, Smith PD, Chuaqui RF, Zhuang Z, Goldstein SR, Weiss RA, Liotta LA. Laser capture microdissection. Science. 1996;274:998–1001. doi: 10.1126/science.274.5289.998. [DOI] [PubMed] [Google Scholar]
- Funk JO, Waga S, Harry JB, Espling E, Stillman B, Galloway DA. Inhibition of CDK activity and PCNA-dependent DNA replication by p21 is blocked by interaction with the HPV-16 E7 oncoprotein. Genes & Dev. 1997;11:2090–2100. doi: 10.1101/gad.11.16.2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin EC, DiMaio D. Repression of human papillomavirus oncogenes in HeLa cervical carcinoma cells causes the orderly reactivation of dormant tumor suppressor pathways. Proc Natl Acad Sci. 2000;97:12513–12518. doi: 10.1073/pnas.97.23.12513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin EC, Naeger LK, Breiding DE, Androphy EJ, DiMaio D. Transactivation-competent bovine papillomavirus E2 protein is specifically required for efficient repression of human papillomavirus oncogene expression and for acute growth inhibition of cervical carcinoma cell lines. J Virol. 1998;72:3925–3934. doi: 10.1128/jvi.72.5.3925-3934.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin EC, Yang E, Lee CJ, Lee HW, DiMaio D, Hwang ES. Rapid induction of senescence in human cervical carcinoma cells. Proc Natl Acad Sci. 2000;97:10978–10983. doi: 10.1073/pnas.97.20.10978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray GE, Mann RS, Mitsiadis E, Henrique D, Carcangiu ML, Banks A, Leiman J, Ward D, Ish-Horowitz D, Artavanis-Tsakonas S. Human ligands of the Notch receptor. Am J Pathol. 1999;154:785–794. doi: 10.1016/S0002-9440(10)65325-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang MS, Zlobin A, Kast WM, Miele L. Notch signaling as a target in multimodality cancer therapy. Curr Opin Mol Ther. 2000;2:55–65. [PubMed] [Google Scholar]
- Jarriault S, Brou C, Logeat F, Schroeter EH, Kopan R, Israel A. Signalling downstream of activated mammalian Notch. Nature. 1995;377:355–358. doi: 10.1038/377355a0. [DOI] [PubMed] [Google Scholar]
- Jesudasan RA, Rahman RA, Chandrashekharappa S, Evans GA, Srivatsan ES. Deletion and translocation of chromosome 11q13 sequences in cervical carcinoma cell lines. Am J Hum Genet. 1995;56:705–715. [PMC free article] [PubMed] [Google Scholar]
- Jones DL, Alani RM, Munger K. The human papillomavirus E7 oncoprotein can uncouple cellular differentiation and proliferation in human keratinocytes by abrogating p21Cip1-mediated inhibition of cdk2. Genes & Dev. 1997;11:2101–2111. doi: 10.1101/gad.11.16.2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato H, Taniguchi Y, Kurooka H, Minoguchi S, Sakai T, Nomura-Okazaki S, Tamura K, Honjo T. Involvement of RBP-J in biological functions of mouse Notch1 and its derivatives. Development. 1997;124:4133–4141. doi: 10.1242/dev.124.20.4133. [DOI] [PubMed] [Google Scholar]
- Klingelhutz AJ, Foster SA, McDougall JK. Telomerase activation by the E6 gene product of human papillomavirus type 16. Nature. 1996;380:79–82. doi: 10.1038/380079a0. [DOI] [PubMed] [Google Scholar]
- Kurooka H, Kuroda K, Honjo T. Roles of the ankyrin repeats and C-terminal region of the mouse notch1 intracellular region. Nucleic Acids Res. 1998;26:5448–5455. doi: 10.1093/nar/26.23.5448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowell S, Jones P, Le Roux I, Dunne J, Watt FM. Stimulation of human epidermal differentiation by delta-notch signalling at the boundaries of stem-cell clusters. Curr Biol. 2000;10:491–500. doi: 10.1016/s0960-9822(00)00451-6. [DOI] [PubMed] [Google Scholar]
- McMurray HR, Nguyen D, Westbrook TF, McAnce DJ. Biology of human papillomaviruses. Intl J Exp Pathol. 2001;82:15–33. doi: 10.1046/j.1365-2613.2001.00177.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miele L, Osborne B. Arbiter of differentiation and death: Notch signaling meets apoptosis. J Cell Physiol. 1999;181:393–409. doi: 10.1002/(SICI)1097-4652(199912)181:3<393::AID-JCP3>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- Morrison SJ, Perez SE, Qiao Z, Verdi JM, Hicks C, Weinmaster G, Anderson DJ. Transient Notch activation initiates an irreversible switch from neurogenesis to gliogenesis by neural crest stem cells. Cell. 2000;101:499–510. doi: 10.1016/s0092-8674(00)80860-0. [DOI] [PubMed] [Google Scholar]
- Mumm JS, Kopan R. Notch signaling: From the outside in. Dev Biol. 2000;228:151–165. doi: 10.1006/dbio.2000.9960. [DOI] [PubMed] [Google Scholar]
- Naeger LK, Goodwin EC, Hwang ES, DeFilippis RA, Zhang H, DiMaio D. Bovine papillomavirus E2 protein activates a complex growth-inhibitory program in p53-negative HT-3 cervical carcinoma cells that includes repression of cyclin A and cdc25A phosphatase genes and accumulation of hypophosphorylated retinoblastoma protein. Cell Growth Differ. 1999;10:413–422. [PubMed] [Google Scholar]
- Rangarajan A, Syal R, Selvarajah S, Chakrabarti O, Sarin A, Krishna S. Activated Notch1 signaling cooperates with papillomavirus oncogenes in transformation and generates resistance to apoptosis on matrix withdrawal through PKB/Akt. Virology. 2001a;286:23–30. doi: 10.1006/viro.2001.0867. [DOI] [PubMed] [Google Scholar]
- Rangarajan A, Talora C, Okuyama R, Nicolas M, Mammucari C, Oh H, Aster JC, Krishna S, Metzger D, Chambon P, et al. Notch signaling is a direct determinant of keratinocyte growth arrest and entry into differentiation. EMBO J. 2001b;20:3427–3436. doi: 10.1093/emboj/20.13.3427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheffner M, Werness BA, Huibregtse JM, Levine AJ, Howley PM. The E6 oncoprotein encoded by human papillomavirus types 16 and 18 promotes the degradation of p53. Cell. 1990;63:1129–1136. doi: 10.1016/0092-8674(90)90409-8. [DOI] [PubMed] [Google Scholar]
- Scheffner M, Romanczuk H, Munger K, Huibregtse JM, Mietz JA, Howley PM. Functions of human papillomavirus proteins. Curr Top Microbiol Immunol. 1994;186:83–99. doi: 10.1007/978-3-642-78487-3_5. [DOI] [PubMed] [Google Scholar]
- Schroeder T, Just U. Notch signalling via RBP-J promotes myeloid differentiation. EMBO J. 2000;19:2558–2568. doi: 10.1093/emboj/19.11.2558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sgroi DC, Teng S, Robinson G, LeVangie R, Hudson JR, Jr, Elkahloun AG. In vivo gene expression profile analysis of human breast cancer progression. Cancer Res. 1999;59:5656–5661. [PubMed] [Google Scholar]
- Shou J, Ross S, Koeppen H, de Sauvage FJ, Gao WQ. Dynamics of notch expression during murine prostate development and tumorigenesis. Cancer Res. 2001;61:7291–7297. [PubMed] [Google Scholar]
- Soto U, Das BC, Lengert M, Finzer P, zur Hausen H, Rosl F. Conversion of HPV 18 positive non-tumorigenic HeLa-fibroblast hybrids to invasive growth involves loss of TNF-α mediated repression of viral transcription and modification of the AP-1 transcription complex. Oncogene. 1999;18:3187–3198. doi: 10.1038/sj.onc.1202765. [DOI] [PubMed] [Google Scholar]
- Stanley MA, Browne HM, Appleby M, Minson AC. Properties of a non-tumorigenic human cervical keratinocyte cell line. Intl J Cancer. 1989;43:672–676. doi: 10.1002/ijc.2910430422. [DOI] [PubMed] [Google Scholar]
- Thierry F, Spyrou G, Yaniv M, Howley P. Two AP1 sites binding JunB are essential for human papillomavirus type 18 transcription in keratinocytes. J Virol. 1992;66:3740–3748. doi: 10.1128/jvi.66.6.3740-3748.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uyttendaele H, Marazzi G, Wu G, Yan Q, Sassoon D, Kitajewski J. Notch4/int-3, a mammary proto-oncogene, is an endothelial cell-specific mammalian Notch gene. Development. 1996;122:2251–2259. doi: 10.1242/dev.122.7.2251. [DOI] [PubMed] [Google Scholar]
- Wells SI, Francis DA, Karpova AY, Dowhanick JJ, Benson JD, Howley PM. Papillomavirus E2 induces senescence in HPV-positive cells via pRB- and p21(CIP)-dependent pathways. EMBO J. 2000;19:5762–5771. doi: 10.1093/emboj/19.21.5762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welter JF, Eckert RL. Differential expression of the fos and jun family members c-fos, fosB, Fra-1, Fra-2, c-jun, junB and junD during human epidermal keratinocyte differentiation. Oncogene. 1995;11:2681–2687. [PubMed] [Google Scholar]
- Welter JF, Crish JF, Agarwal C, Eckert RL. Fos-related antigen (Fra-1), junB, and junD activate human involucrin promoter transcription by binding to proximal and distal AP1 sites to mediate phorbol ester effects on promoter activity. J Biol Chem. 1995;270:12614–12622. doi: 10.1074/jbc.270.21.12614. [DOI] [PubMed] [Google Scholar]
- Yoshioka K, Deng T, Cavigelli M, Karin M. Antitumor promotion by phenolic antioxidants: Inhibition of AP-1 activity through induction of Fra expression. Proc Natl Acad Sci. 1995;92:4972–4976. doi: 10.1073/pnas.92.11.4972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zagouras P, Stifani S, Blaumueller CM, Carcangiu ML, Artavanis-Tsakonas S. Alterations in Notch signaling in neoplastic lesions of the human cervix. Proc Natl Acad Sci. 1995;92:6414–6418. doi: 10.1073/pnas.92.14.6414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- zur Hausen H. Papillomaviruses causing cancer: Evasion from host-cell control in early events in carcinogenesis. J Natl Cancer Inst. 2000;92:690–698. doi: 10.1093/jnci/92.9.690. [DOI] [PubMed] [Google Scholar]